Abstract
The CCR4–NOT complex is the major enzyme catalyzing mRNA deadenylation in Saccharomyces cerevisiae. We have identified homologs for almost all subunits of this complex in the Drosophila genome. Biochemical fractionation showed that the two likely catalytic subunits, CCR4 and CAF1, were associated with each other and with a poly(A)-specific 3′ exonuclease activity. In Drosophila, the CCR4 and CAF1 proteins were ubiquitously expressed and present in cytoplasmic foci. Individual knock-down of several potential subunits of the Drosophila CCR4–NOT complex by RNAi in tissue culture cells led to a lengthening of bulk mRNA poly(A) tails. Knock-down of two individual subunits also interfered with the rapid deadenylation of Hsp70 mRNA during recovery from heat shock. Similarly, ccr4 mutant flies had elongated bulk poly(A) and a defect in Hsp70 mRNA deadenylation. A minor increase in bulk poly(A) tail length was also observed in Rga mutant flies, which are affected in the NOT2 subunit. The data show that the CCR4–NOT complex is conserved in Drosophila melanogaster and plays a role in general and regulated mRNA deadenylation.
Keywords: deadenylation, Drosophila, mRNA turnover, poly(A) tails, post-transcriptional regulation
Introduction
A characteristic feature of mRNA is its rapid turnover, permitting protein production to be continuously adjusted to the physiological requirements. Each mRNA has a characteristic half-life; within a given cell, the half-lives of different mRNAs differ by more than a factor of 10. A short half-life leads to a rapid adjustment of the steady-state level of an mRNA upon either an up- or downregulation of the rate of transcription (Ross, 1995). The half-lives of some mRNAs can also be regulated. For example, pronounced instability of the mRNA encoding the Hsp70 heat shock protein contributes, in addition to transcriptional repression, to its very low steady-state level at normal temperatures, and stabilization of the message plays a role in Hsp70 induction upon heat shock (Petersen and Lindquist, 1988, 1989; Dellavalle et al, 1994; Laroia et al, 1999).
Two general pathways of mRNA decay have been characterized in yeast (Parker and Song, 2004). Exonucleolytic removal of the poly(A) tail (deadenylation) is the first step in both, and this can also be the rate-limiting and regulated step. In the major decay pathway, the second step is the hydrolytic cleavage of the m7GpppN cap, which occurs only after the poly(A) tail has been shortened to about 10 nt. The RNA is then degraded from its 5′ end. In the minor pathway, the step following deadenylation consists of 3′ exonucleolytic degradation of the mRNA body. In yeast, two different enzyme complexes are involved in deadenylation. The more important one contains Ccr4p as the major catalytic subunit (Tucker et al, 2001, 2002; Chen et al, 2002). A second subunit, Pop2p/Caf1p, is a member of the RNase D family of 3′ exonucleases and also has 3′ exonuclease activity (Daugeron et al, 2001; Thore et al, 2003), but under normal conditions the activity of Ccr4p dominates (Chen et al, 2002; Tucker et al, 2002) (C Temme and E Wahle, unpublished data). Additional polypeptides in the complex are Not1–5p, Caf40p and Caf130p (Chen et al, 2001). ccr4Δ and pop2Δ/caf1Δ mutants grow slowly and display a reduction in the rate of mRNA deadenylation (Tucker et al, 2001). not2Δ and not5Δ also affect mRNA deadenylation, albeit more weakly than ccr4 and pop2/caf1 mutations (Tucker et al, 2002). The CCR4–NOT complex is also believed to play a role in transcriptional regulation (Collart, 2003; Denis and Chen, 2003). The second deadenylase of Saccharomyces cerevisiae is composed of the Pan2 and Pan3 proteins (Boeck et al, 1996; Brown et al, 1996). Deletion mutants show an increase in the steady-state length of poly(A) tails, and the Pan2p/Pan3p complex catalyzes the residual deadenylation observed in ccr4 or pop2/caf1 mutants (Tucker et al, 2001). The Pan2p/Pan3p complex appears to be involved in an initial shortening of overly long poly(A) tails to a length of 50–90 nt (Brown and Sachs, 1998).
In metazoans, like in yeast, deadenylation is the first and often rate-limiting step of mRNA decay (Chen and Shyu, 1995). For example, the unstable Hsp70 mRNA mentioned above is rapidly deadenylated at normal growth temperature; retarded deadenylation during heat shock is at least partially responsible for stabilization of the message (Dellavalle et al, 1994). Subsequent decay steps proceed rapidly and without detectable intermediates in vivo. However, it is generally accepted that both pathways of mRNA decay described above also exist in animal cells (Couttet et al, 1997; Decker and Parker, 2002; van Hoof and Parker, 2002). The situation is more complex with respect to deadenylating enzymes. The CCR4–NOT complex is conserved in man and other species, but the human genome contains two homologs of yeast POP2/CAF1 (Albert et al, 2000), and metazoans have four different homologs of the yeast CCR4 gene (Dupressoir et al, 2001). Two of these, Xenopus Nocturnin and human CCR4, have been demonstrated to have poly(A)-degrading activity in vitro (Chen et al, 2002; Baggs and Green, 2003). Thus, there may be different versions of the CCR4–NOT complex in higher eukaryotes. In human cells, the Pan2/Pan3 complex has also been characterized (Uchida et al, 2004). Finally, a third enzyme, the poly(A)-specific ribonuclease (PARN; initially called DAN), has been identified in vertebrates and is thought to be responsible for the so-called default deadenylation during Xenopus oocyte maturation, a developmentally controlled general poly(A) tail degradation serving in translational silencing of mRNAs (Körner et al, 1998; Wickens et al, 2000).
Here, we report on the conservation of the CCR4–NOT complex in Drosophila melanogaster and its role both in bulk mRNA deadenylation and in regulated deadenylation of Hsp70 mRNA in vivo.
Results
Drosophila homologs of known poly(A)-degrading enzymes
As reported before (Dupressoir et al, 2001), the genome of D. melanogaster contains a close homolog of the yeast CCR4 gene. The predicted protein contains the exonuclease domain as well as the leucine-rich repeat, which is required for the interaction of the CCR4 protein with other polypeptides and distinguishes what is considered the ‘true' CCR4 homolog from other CCR4-like genes (Draper et al, 1995; Dupressoir et al, 2001; Clark et al, 2004) (Figure 1). The fly genome also contains a gene homologous to yeast POP2/CAF1 (Daugeron et al, 2001) (Figure 1). All three exonuclease motifs characterizing the RNaseD family of 3′ exonucleases are well conserved in Drosophila CAF1. In addition, single fly genes homologous to yeast NOT1, NOT2 and NOT4, respectively, were identified (Figure 1). A single Drosophila gene homologous to both yeast NOT3 and yeast NOT5 was found; the two yeast genes also have sequence similarity to each other (Oberholzer and Collart, 1998). A homolog of the yeast CAF40 gene was also detected, but no gene with similarity to CAF130 was detected. Whereas genes corresponding to the yeast Pan2p/Pan3p deadenylase are present in the fly genome (CG8232 and CG11486), no gene with convincing similarity to mammalian PARN could be found.
Figure 1.
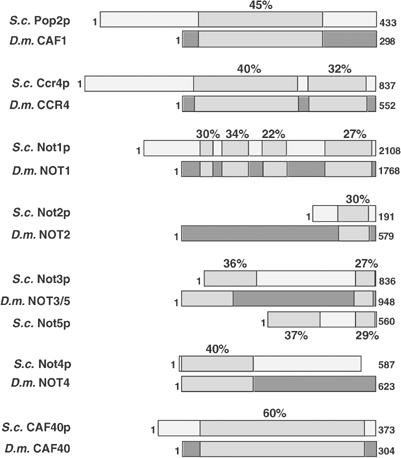
Sequence alignments of the core components from the CCR4–NOT complex from S. cerevisiae (S.c.) and D. melanogaster (D.m.). Areas of high sequence identity are hatched with the amount of identity indicated above. The length of the individual proteins is given on the right. Gene references in FlyBase are as follows: CCR4, CG31137; CAF1, CG5684; NOT1, CG1884; NOT2, CG2138; NOT3/5, CG8426; NOT4, CG31716; CAF40, CG14213.
The Drosophila CAF1 and CCR4 proteins are associated with each other and with a poly(A)-specific 3′ exonuclease activity
Cytoplasmic extracts of Drosophila S2 cells were fractionated by DEAE column chromatography. Fractions were tested for poly(A)-degrading activity by an assay that measures the release of acid-soluble material from homogeneously labeled poly(A). Under the conditions used here, a single activity peak was obtained (Figure 2A). In addition, activity was detected in the flow-through; this was not further investigated. Western blotting with antibodies raised against recombinant Drosophila CCR4 and CAF1 proteins detected proteins of the anticipated molecular masses (CAF1, 34 kDa; CCR4, 63 kDa), which were co-eluted from the column (Figure 2B). Although both proteins were present in the column fractions containing the poly(A)-degrading activity, their peak (frs. 9–11) was displaced from the activity peak (fr. 7) toward higher salt concentrations. The poly(A)-degrading activity overlapped with an inhibitory activity, which was eluted from the DEAE column at slightly higher salt concentration (Supplementary Figure 1). Thus the ‘true' peak of the poly(A)-degrading nuclease is likely to be in a later fraction and may well correspond to the CCR4/CAF1 peak. As shown by Western blotting, the inhibitory fractions contained the cytoplasmic poly(A)-binding protein, a known inhibitor of yeast Ccr4p in vitro (Tucker et al, 2002), but preliminary evidence suggests that this protein is not primarily responsible for the inhibition (S Meyer, C Temme and E Wahle, unpublished data). In other DEAE columns, run under different conditions, the poly(A)-degrading activity was separated into two peaks, one of which did not comigrate with CCR4 and CAF1, and this second activity may also have contributed to the displacement of the activity peak from the CCR4/CAF1 peak. As a further test of the association of CCR4, CAF1 and the poly(A)-degrading activity, an aliquot from the activity peak of the DEAE column (fr. 7) was analyzed by gel filtration chromatography. A single activity peak was found, which coincided with both CCR4 and CAF1 (Figure 3). Both proteins were eluted from the column later than predicted even from their individual molecular masses, suggesting that the separation was affected by their interaction with the gel matrix. In contrast, when a sample of crude cytoplasmic S2 extract was separated by gel filtration on a larger scale, the bulk of CCR4 and CAF1 co-eluted ahead of the largest marker protein (ferritin, 440 kDa) (data not shown). We have also observed co-elution of both CCR4 and CAF1 with poly(A)-degrading activity in a cation exchange column (data not shown).
Figure 2.
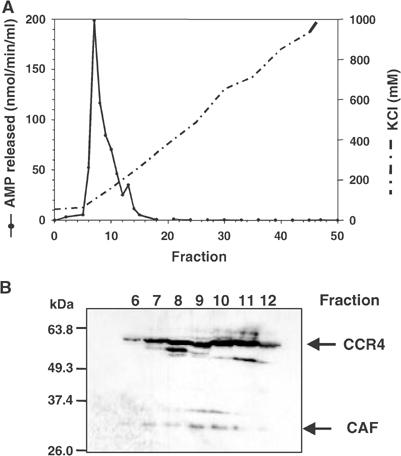
Detection of poly(A)-degrading activity in extracts from S2 cells. (A) S2 cytoplasmic extract was fractionated over a DEAE column and the poly(A)-degrading activity of each fraction was measured in a standard poly(A) degradation assay. (B) Western blot of DEAE column fractions. A 15 μl portion of each fraction was separated on a 10% SDS–PAGE, blotted onto a nitrocellulose membrane and probed first with antiserum against CCR4 and then with affinity-purified CAF1 antibodies.
Figure 3.
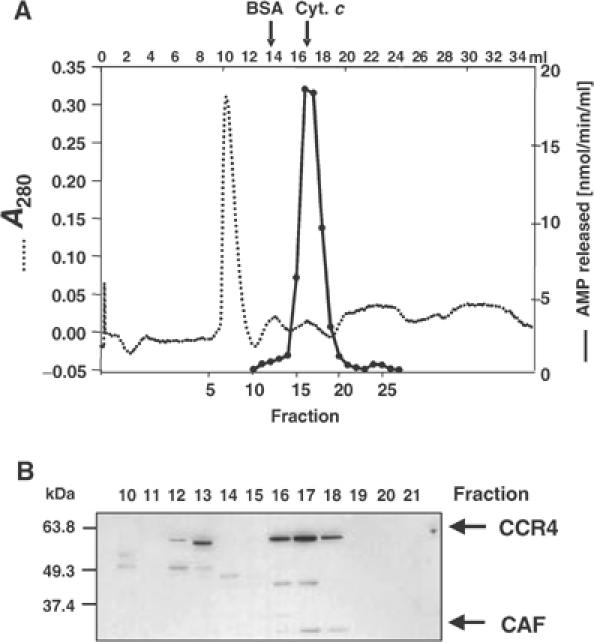
Analysis of poly(A)-degrading activity by gel filtration. (A) A 200 μl portion of fraction 7 of the DEAE column (Figure 2A) was loaded on a Superdex 200 HS gel filtration column. After 5 ml, fractions of 0.7 ml were collected. Activity was measured in a standard poly(A) degradation assay. The elution volumes of BSA and cytochrome c are indicated on top. (B) Western blot against CAF1 and CCR4. Protein from 300 μl of each fraction was precipitated with TCA, separated on a 10% SDS–PAGE, blotted onto a nitrocellulose membrane and probed with CCR4 and CAF1 antiserum.
Antibodies against Drosophila CCR4 and CAF1 were used to deplete the poly(A)-degrading activity from the DEAE peak fraction. Anti-CAF1 antibodies reduced the activity 25-fold in contrast to control antibodies, which reduced the activity only two-fold, presumably due to nonspecific adsorption or inactivation. Accordingly, nine times more poly(A)-degrading activity was detected in the immunoprecipitate of CAF1 than in the control precipitate (Supplementary Figure 2 and data not shown). A similar depletion of activity was obtained with anti-CCR4 serum (Supplementary Figure 2). In summary, the CCR4 and CAF1 proteins of Drosophila, like those of yeast, are associated with each other and with poly(A)-degrading activity and are likely to be in a complex with other polypeptides, conceivably the Drosophila NOT proteins.
In the DEAE column fractions, accumulation of acid-soluble product was linear with time, consistent with an exonuclease activity. The product was 5′ AMP (data not shown). A fraction from the activity peak of a DEAE column was also assayed for the degradation of a capped, polyadenylated RNA carrying a 32P-label in the cap. Products were analyzed by denaturing gel electrophoresis (Figure 4). Time-dependent gradual shortening of this 5′-labeled RNA demonstrated a 3′ exonucleolytic mode of degradation. Accordingly, gradual shortening was not observed when the RNA was modified at the 3′ end with poly(A) polymerase and 3′ dATP (data not shown). Transient accumulation of completely deadenylated RNA suggests that the 3′ exonuclease is poly(A) specific, although a protection of the RNA ‘body' by RNA-binding proteins cannot be excluded in these crude fractions. Deadenylated RNA did not accumulate when the 3′ end was modified. Although additional nuclease activities were clearly present in the column fraction, the polyadenylated RNA was stabilized by immunodepletion with anti-CCR4 or anti-CAF1 (data not shown).
Figure 4.
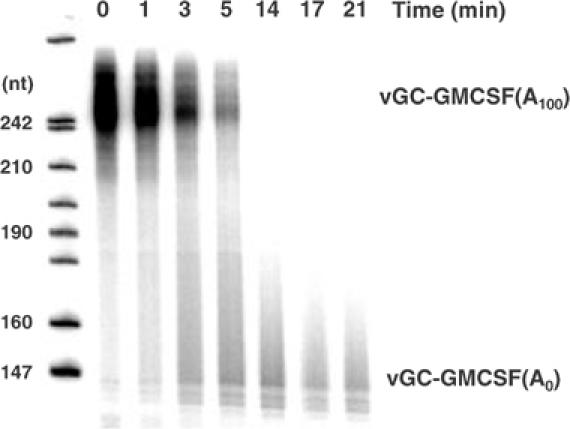
Deadenylation of a capped and polyadenylated RNA. A 1 μl portion of a fraction from a DEAE column similar to the one in Figure 2 was mixed on ice with 0.63 pmol of cap-labeled polyadenylated vGC-GMCSF RNA in a 20 μl standard reaction. A 2.5 μl volume of the reaction was stopped immediately after addition of the protein (t=0). The rest of the mixture was transferred to 30°C. Aliquots were withdrawn and stopped at the time points indicated. The RNA was analyzed on a 5% polyacrylamide–urea gel.
CCR4 and CAF1 concentrate in cytoplasmic foci
The expression pattern and subcellular distribution of CCR4 and CAF1 proteins were analyzed in Drosophila. CCR4 and CAF1 were present at all developmental stages from embryos to adults, with a peak of expression at zygotic transcription (2- to 3-h-old embryos) for CCR4 and a low level of expression in adults for CAF1 (Figure 5A and B). CCR4 protein was not detected in two ccr4 mutant backgrounds (see below) (Figure 5A). Distribution of CCR4 and CAF1 was analyzed by immunostaining of ovaries (Figure 5C–P). CCR4 was mostly cytoplasmic (Figure 5C and E–G), as expected for an enzyme involved in mRNA deadenylation. Strikingly, the protein distribution was not homogeneous; in addition to a diffuse pattern in the cytoplasm, CCR4 concentrated in discrete foci (Figure 5E–G). Similarly, CAF1 was abundant in the cytoplasm with a punctate distribution, including foci similar in size to CCR4 foci (Figure 5J and L–N). As expected, anti-CCR4 staining was reduced to background level in the ccr4 mutant (Figure 5H and I). Surprisingly, the CAF1 level was also dramatically reduced in this mutant (Figure 5O and P), suggesting that CAF1 stability may require CCR4.
Figure 5.
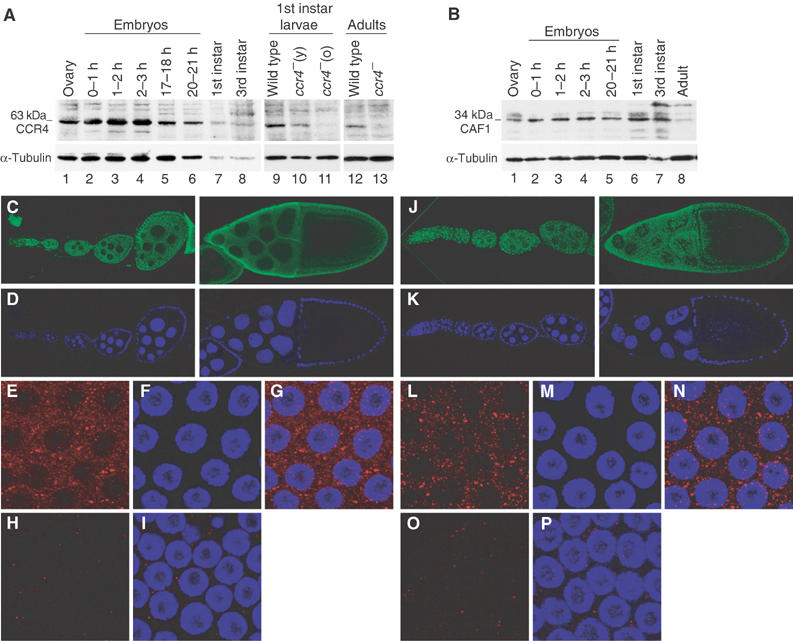
Expression and subcellular distribution of CCR4 and CAF1 in Drosophila. (A, B) Western blots probed with anti-CCR4 (A) and affinity-purified anti-CAF1 (B) showing the presence of CCR4 and CAF1 in different developmental stages, and the lack of CCR4 in ccr4 mutants (A, right panels). Protein extracts loaded were from 0.75 ovary, 20 embryos, 20 first instar larvae, 0.75 third instar larva and 0.3 adult male. Shown are wild-type extracts (A, lanes 1–9, 12 and B, lanes 1–8), extracts from ccr4KG877/ccr4KG877 first instar larvae, taken 5–15 h after eclosion (A, lane 10, y: young) or about 30 h after eclosion (A, lane 11, o: old), and extract from ccr4KG877/Df(3R)crb-F89-4 adult males (A, lane 13). The blots were then probed with anti-α-tubulin as a loading control. (C–P) Immunodetection of CCR4 and CAF1 in ovaries. (C, D, J, K) Confocal images showing that CCR4 and CAF1 are present at all stages of oogenesis and in the different cell types. Earliest stages of oogenesis (left panels), stage 10 (right panels), anti-CCR4 (C), anti-CAF1 (J) and DAPI (D, K). (E–I, L–P) Enlargement of stage-10 egg chamber follicle cells showing accumulation of CCR4 (E–G) and CAF1 (L–N) in cytoplasmic foci in the wild type and a strong decrease in CCR4 (H, I) and CAF1 (O, P) levels in ccr4KG877/Df(3R)crb-F89-4 mutant females. Anti-CCR4 in red (E, G, H, I), Anti-CAF1 in red (L, N, O, P), DAPI in blue (F, G, I, M, N, P), merge anti-CCR4/DAPI (G, I) and anti-CAF1/DAPI (N, P).
CAF1, CCR4 and NOT proteins are involved in bulk mRNA deadenylation
RNA interference (RNAi) was used to analyze the physiological role of the Drosophila CCR4–NOT complex. S2 cells were treated with double-stranded RNAs encoding selected subunits of the complex and analyzed after 5 days. Western blots showed a significant reduction of CAF1 protein in cells treated with double-stranded caf1 RNA, but not in cells treated with ccr4 RNA, Rga RNA (encoding the NOT2 protein; see below) or control RNA. In contrast, a reduction of CCR4 protein was seen not only in cells treated with ccr4 RNA but also in cells treated with caf1 RNA (Figure 6A). Yeast Ccr4p is thought to be attached to the rest of the complex via Pop2p/Caf1p (Bai et al, 1999). Upon depletion of CAF1, CCR4 may thus be lost from the complex and degraded. Antibodies to NOT2 were not available. The cell number in the culture treated with caf1 RNA was about 25% lower than in the control, but cells appeared healthy when examined under the microscope. Treatment with ccr4 or Rga RNA had no detectable effect on cell growth. The poly(A) tail length distribution of total cellular RNA was analyzed by 3′-end labeling, digestion with RNases A and T1, which leaves only poly(A) intact, and denaturing gel electrophoresis. Depletion of CCR4 protein had little if any effect on the steady-state poly(A) tail length of bulk RNA (Figure 6B). In contrast, depletion of CAF1 led to a strong increase in the average poly(A) length: The amount of poly(A) longer than 70 nt was increased, and the amount of poly(A) shorter than 40 nt was decreased. A similar but less pronounced effect was seen upon knock-down of NOT2 (Figure 6B). This suggests that Drosophila CAF1 and NOT2 proteins are required for poly(A) tail shortening in vivo.
Figure 6.
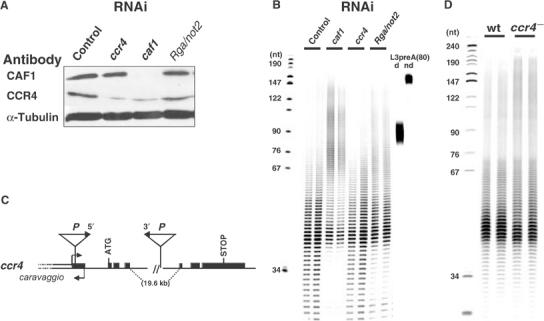
The CCR4–NOT complex plays a role in general deadenylation. (A) After 5 days of incubation with either luciferase (control), caf1, ccr4 or Rga/not2 dsRNA, S2 cells (one well for each dsRNA) were resuspended in 150 μl lysis buffer and lysed by two cycles of freezing and thawing. A 15 μl portion of the resulting extracts was separated on a 10% SDS–PAGE and standard Western blots were performed against CAF1, CCR4 and α-tubulin as a loading control. (B) Analysis of the length of bulk poly(A) in S2 cells after 5 days of incubation with dsRNA as in (A). 3′-labeled and digested RNAs were separated on a 10% polyacrylamide–urea gel. As a control, in vitro-synthesized L3preA(80) RNA was treated like the experimental RNA samples (d, digested; nd, nondigested). Lengths of DNA markers (in nt) are indicated on the left. (C) Schematic representation of the ccr4 locus and ccr4KG877 allele. Solid boxes are ccr4 exons, thin lines are introns, the white box to the left indicates another transcription unit with the transcription start site (lower horizontal arrow) and the upper horizontal arrow shows the ccr4 transcription start site. The ccr4 coding sequence is indicated by the start (ATG) and stop codons. The two P-elements inserted in the ccr4KG877 mutant are indicated (not drawn to scale). Both P-elements are incomplete, the element inserted in the 5′ UTR lacking its 3′ end and the element inserted in intron 4 lacking its 5′ end. The arrowheads indicate the repeat sequence at both the ends of the P-element. The locus was drawn according to information from FlyBase and BDGP; in addition, the P-element deletions were verified by PCR analysis and the P-element insertion sites by sequencing. (D) Analysis of the length of bulk poly(A) from wild-type (wt) and ccr4KG877/Df(3R)crb-F89-4 (ccr4−) flies. Total RNA from two independent RNA preparations from wild-type (wt) or ccr4 mutant adult males, respectively, was 3′ labeled, digested and separated on a 10% polyacrylamide–urea gel.
Treatment of S2 cells with double-stranded RNA encoding either NOT1 or NOT3/5 also led to a decrease in short poly(A) and an increase of poly(A) between 70 and 120 nt (Supplementary Figure 3). Although depletion of the proteins could not be assayed due to the lack of suitable antibodies, and the effects were again less pronounced than those observed upon a knock-down of CAF1, the results suggest that these two NOT proteins are also involved in bulk deadenylation. Preliminary experiments showed a similar result for NOT4 but not for CAF40 (data not shown). In summary, it is presumably the entire CCR4/CAF/NOT complex that is catalyzing poly(A) tail degradation.
We sought to confirm the results of RNAi experiments by using Drosophila mutants affecting the CCR4–NOT complex. Drosophila NOT2 is encoded by the Regena (Rga) gene. The Rgal(3)03834 mutant, a P-element insertion, is thought to be a null allele (Frolov et al, 1998). Rgal(3)03834/Rgal(3)03834 individuals died as second instar larvae, although rare adult escapers survived. In agreement with a role of Drosophila NOT2 in deadenylation, there was a slight but reproducible lengthening of the steady-state poly(A) tails in RNA from Rgal(3)03834 mutant larvae, with more poly(A) tails between 55 and 80 nt compared to wild type, although the longest poly(A) tails reached the same length in mutant and in wild-type larvae (160 nt) (Supplementary Figure 4).
A ccr4 mutant, ccr4KG877, generated by the Berkeley Drosophila Genome Project (BDGP) contains two P-elements inserted in the ccr4 gene, one in the 5′ UTR and one in the large fourth intron (Figure 6C). ccr4KG877/ccr4KG877 individuals died as first instar larvae. However, this lethality was due to an associated mutation and did not result from a lack of CCR4 protein, as ccr4KG877/Df(3R)crb-F89-4 (a large deletion including the ccr4 locus) flies were viable. Western blot analysis indicated that ccr4KG877 is a strong allele, as the CCR4 protein was not detected in ccr4KG877/ccr4KG877 larvae and in ccr4KG877/Df(3R)crb-F89-4 adult males (Figure 5A). A significant lengthening of poly(A) tails was observed in ccr4KG877/Df(3R)crb-F89-4 adult males lacking the CCR4 protein: the amount of poly(A) tails around 50 nt was slightly decreased, whereas the amount of poly(A) tails above 100 nt was increased compared to wild type. In addition, the longest poly(A) tails reached 200 nt in the mutant adults, 40 nt longer than in wild-type adults (Figure 6D).
These results confirm a role of Drosophila NOT2 and CCR4 proteins in mRNA deadenylation in vivo and show, in contrast to RNAi experiments, that the lack of CCR4 protein leads to a stronger poly(A) tail lengthening than the lack of NOT2. This suggests a more direct role of CCR4 in deadenylation, in agreement with its expected catalytic activity.
The Drosophila CAF1 and CCR4 proteins are required for regulated deadenylation of the Hsp70 mRNA
The Hsp70 mRNA of Drosophila has been identified as a very unstable RNA whose degradation is initiated by rapid deadenylation (see Introduction). Transcription of the Hsp70 gene is induced by heat shock and ceases when cells are returned to their normal growth temperature. Thus, a transcriptional pulse-chase experiment to assess the time course of deadenylation is possible without artificial inhibition of transcription. After 5 days of treatment with double-stranded caf1, ccr4, Rga or control RNA, S2 cells were exposed to a temperature of 35.6°C for 30 min and then allowed to recover at their normal growth temperature of 25°C. RNA was prepared at different times, and the poly(A) tail length of the Hsp70 mRNA was measured by a ligation-mediated RT–PCR assay, the PAT assay (Sallés and Strickland, 1999). An in vitro transcript carrying a poly(A) tail of about 45 nt (L3preA45) served as an internal control. As expected, no Hsp70 PCR product was obtained with RNA isolated from cells before heat shock (Figure 7A). RNA isolated from heat-shocked cells resulted in a PCR product with a size range corresponding to poly(A) tails of up to more than 130 nt. Upon recovery from heat shock, the poly(A) tails in control cells were rapidly shortened, and RNAs with very short oligo(A) tails accumulated transiently (Figure 7A). At later time points, the amount of PCR product was strongly reduced. As the RT–PCR assay requires a small number of A residues at the RNA's 3′ end for primer binding, the absence of product may reflect either the accumulation of completely deadenylated Hsp70 mRNA or, more likely, its degradation. In the cells treated with double-stranded caf1 or Rga RNA, the rate of deadenylation was strongly reduced. Poly(A) tails of well over 60 nt in length persisted for at least 3 h after heat shock. Also, there was no accumulation of RNA with very short oligo(A) tails, and significantly more polyadenylated Hsp70 mRNA was left at the end of the time course compared to the RNA from control cells. In contrast, knock-down of CCR4 had little effect. A control experiment using actinomycin D showed that the time course of deadenylation was not influenced by continuing Hsp70 mRNA synthesis and purely reflected the fate of mRNA synthesized during the heat shock. We conclude that Hsp70 mRNA deadenylation during recovery from heat shock is catalyzed by the CCR4–NOT complex.
Figure 7.
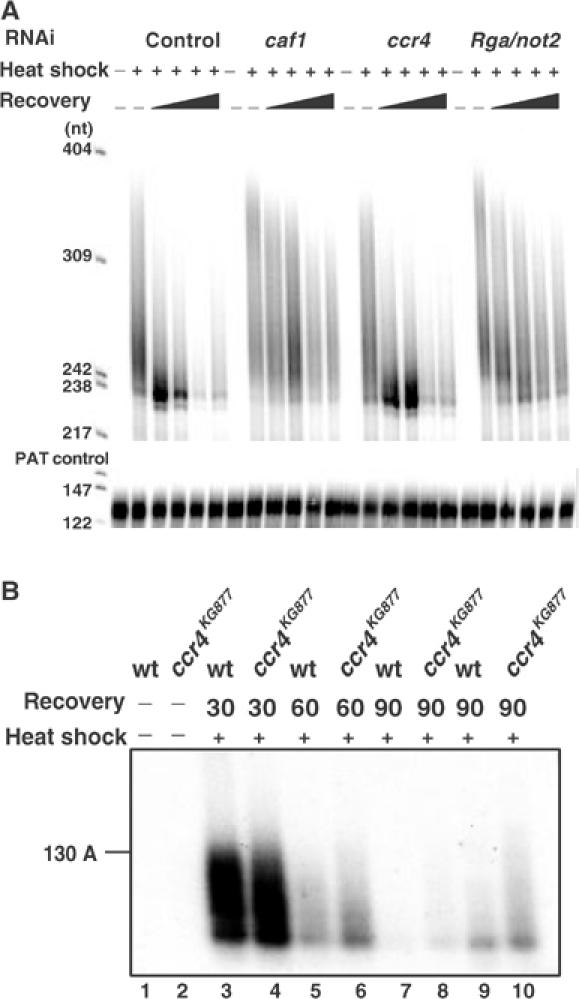
Determination of the poly(A) tail length of the Hsp70 transcript during recovery from heat shock. (A) S2 cells treated with dsRNA against luciferase (control), caf1, ccr4 or Rga/not2, respectively, were harvested before heat shock or at 0, 30, 60, 120 and 180 min of recovery from a 30 min heat shock at 35.6°C. Total RNA was isolated and mixed with in vitro-synthesized L3pre(A45) RNA as a control. cDNA was generated, and either the Hsp70 cDNA or L3pre cDNA including the poly(A) tails was PCR-amplified with radioactively labeled primers. The products were separated on a 5% polyacrylamide–urea gel. (B) PAT assays measuring poly(A) tail lengths of Hsp70 mRNA in larvae. RNA was from wild-type (wt) (lanes 1, 3, 5, 7 and 9) or ccr4KG877/ccr4KG877 (lanes 2, 4, 6, 8 and 10) first instar larvae, without heat shock (lanes 1 and 2), or with a 30 min heat shock at 36.5°C, followed by 30 min (lanes 3 and 4), 60 min (lanes 5 and 6) or 90 min of recovery (lanes 7–10) at 25°C. In lanes 9 and 10, three times more PCR product was loaded than in the other lanes to visualize the low amount of Hsp70 mRNA remaining.
These results were confirmed in vivo with the help of the ccr4KG877 mutant. Wild-type and ccr4KG877 homozygous mutant first instar larvae were heat shocked at 36.5°C for 30 min and allowed to recover at 25°C. In wild-type larvae, Hsp70 mRNA deadenylation and degradation were intermediate after 1 h recovery and almost complete after 1.5 h (Figure 7B, lanes 3, 5, 7 and 9). In ccr4KG877 mutant larvae, deadenylation and degradation of Hsp70 mRNA were delayed. Both after 1 and 1.5 h of recovery, longer Hsp70 poly(A) tails and more polyadenylated Hsp70 mRNA were present in mutant larvae compared to wild type (compare lanes 5 and 6, and lanes 9 and 10). This corroborates the results of the RNAi experiments and indicates a role of the CCR4–NOT complex in regulated deadenylation of Hsp70 mRNA.
Discussion
The CCR4–NOT complex, identified as the major cytoplasmic mRNA deadenylase in S. cerevisiae, is conserved in D. melanogaster. Homologs for most subunits of the yeast complex were found in the Drosophila genome, as recently also reported by others (Denis and Chen, 2003). The Caf130 protein of yeast does not have any obvious counterpart in the fly, and such a protein also does not appear to be encoded in the human or other completely sequenced genomes (Denis and Chen, 2003). The Not3 and Not5 proteins of yeast have sequence similarity to each other and are represented by a single protein in Drosophila as is the case in humans (Albert et al, 2000). Most importantly, the two catalytic subunits, CCR4 and CAF1, are conserved in Drosophila. In several different chromatographic separations, these two proteins were found to be associated with each other. However, we have sometimes seen a partial displacement of the CAF1 peak with respect to the CCR4 peak, suggesting that the association may not be extremely tight. The nuclease activity associated with the two proteins was classified as a 3′ exonuclease based on (i) the linear accumulation of acid-soluble product, (ii) gradual shortening of a 5′-labeled RNA and (iii) protection of the RNA by modification of the 3′ end. The nuclease is likely to be poly(A) specific since degradation of a polyadenylated RNA resulted in the accumulation of a deadenylated product. A more thorough characterization will be meaningful only after further purification of the enzyme.
The composition of the CCR4–NOT complex is not entirely clear. A complex containing Ccr4p, Caf1p, Not1–5p, Caf40p and Caf130p has been purified from yeast (Chen et al, 2001). In contrast, TAP-tag purifications consistently revealed co-purification of Ccr4p, Not1p, Caf1p, Caf40p and Caf130p, but there was little evidence for an association of these proteins with Not2–5p (Gavin, 2002). Similarly, purification of TAP-tagged human NOT2 resulted in co-purification of NOT1, both human homologs of POP2/CAF1, CAF40 and CCR4, but not of NOT3 and NOT4 (Gavin, 2002). The possibility has been discussed that the Not2–5 proteins may play distinct roles as opposed to Caf1p and Ccr4p (Collart, 2003; Denis and Chen, 2003). However, in S. cerevisiae, not2Δ and not5Δ mutations cause a significant decrease in the rate of poly(A) tail shortening, and not3Δ and not4Δ mutants also have slight defects (Tucker et al, 2002). These observations are consistent with an involvement of the entire complex in mRNA deadenylation. Our data also support this idea: All subunits tested except CAF40 were found to be involved in deadenylation either by the analysis of mutant flies or in RNAi knock-down experiments. While knock-down of CAF40 had no effect on steady-state poly(A) length, this result does not rule out an involvement of the protein in deadenylation, since the success of RNAi could not be checked due to a lack of suitable antibodies.
Both ccr4Δ and pop2/caf1Δ mutants of S. cerevisiae have pronounced growth defects, at least in some genetic backgrounds, but they are viable. In contrast, NOT1 is an essential gene, and some combinations of mutations in subunits of the CCR4–NOT complex are synthetically lethal (Collart, 2003). The ccr4 allele of Drosophila described here is also viable. Although the level of the CCR4 protein is strongly reduced, the mutation is not detrimental to Drosophila development and the defect in deadenylation is relatively weak. An RNAi knock-down of CCR4 in S2 cells had essentially no detectable consequences for steady-state poly(A) tail length or the rate of Hsp70 deadenylation. While an incomplete depletion of the protein may contribute to the weakness of the knock-down phenotype, a more fundamental explanation for the relatively weak effect of CCR4 mutation or depletion may be the existence of other CCR4-like proteins. At least one of them, Nocturnin, is expected to contribute to mRNA deadenylation (Baggs and Green, 2003), but the other two CCR4 paralogs might have a similar role. Interestingly, this functional redundancy does not occur in the female germ line, as decreasing the level of CCR4 leads to female sterility (S Zaessinger and M Simonelig, unpublished data). Our biochemical fractionation experiments have also provided evidence for at least one additional poly(A)-degrading enzyme in S2 cells: Under slightly different conditions, we have separated a distinct poly(A)-specific 3′ exonuclease from the one associated with CCR4 and CAF1 (C Temme and E Wahle, unpublished data). This nuclease and the one present in the flow-through of the DEAE column remain to be identified.
Immunostaining detected the CCR4 and CAF1 proteins in distinct cytoplasmic foci. Similar foci containing the mRNA decapping and degradation enzymes have been described in mammalian cells and in yeast and are thought to be the sites of mRNA decapping and decay (Ingelfinger et al, 2002; van Dijk et al, 2002; Eystathioy et al, 2003; Sheth and Parker, 2003; Cougot et al, 2004). The mammalian foci also contain the CCR4 protein (Cougot et al, 2004), whereas an association of yeast Ccr4p with cytoplasmic foci is uncertain (Sheth and Parker, 2003). The Drosophila CCR4 and CAF1 foci probably correspond to these cytoplasmic sites of mRNA decay found in other species. Decapping and degradation of the mRNA body are rapid events following complete deadenylation, and the mRNA's association with the proteins required for decapping appears to be accompanied (or even caused) by a loss of translation factors like eIF4E, eIF4G and Pab1p (Tharun and Parker, 2001). Thus, it is not hard to imagine that this mRNP rearrangement also involves the association with a particular cytoplasmic structure. Deadenylation, in contrast, is a continuous process during the mRNA's cytoplasmic lifetime. Thus, deadenylation presumably occurs while the RNA is being translated and would not be expected to be localized. In agreement with this, a substantial proportion of CCR4 seems to be widespread in the cytoplasm. Further experiments will be necessary to examine the functional significance of the presence of CCR4 and CAF1 in foci.
Our data show that the CCR4–NOT complex of Drosophila is involved both in basal and regulated deadenylation. Deadenylation of the Hsp70 mRNA is regulated in two ways: First, any Hsp70 RNA made at normal growth temperature is deadenylated and degraded very rapidly (Petersen and Lindquist, 1988, 1989; Dellavalle et al, 1994). Although sequence dependence of deadenylation has not been directly examined, rapid decay of the Hsp70 message depends on the 3′ UTR (Petersen and Lindquist, 1989). Presumably, 3′ UTR sequences activate the CCR4–NOT complex directly or indirectly via bound proteins. Second, heat shock leads to a stabilization of the message, apparently partly due to a reduced rate of deadenylation (Petersen and Lindquist, 1988; Dellavalle et al, 1994). As other rapidly deadenylated, unstable mRNAs are also stabilized during heat shock (Andrews et al, 1987; Sadis et al, 1988; Laroia et al, 1999), it is possible that the deadenylating enzyme itself is a target of regulation rather than mRNA-specific factors. In agreement with this idea, growth conditions regulate a number of phosphorylation events in the yeast CCR4–NOT complex, some of proven physiological significance (Liu et al, 1997; Moriya et al, 2001; Lenssen et al, 2002). The double-stranded RNA-dependent protein kinase has been implicated in heat-shock regulation of mRNA stability in mammalian cells (Zhao et al, 2002), but the relevant target is unknown. The ubiquitin system has also been implicated in the regulation of mRNA stability during heat shock (Laroia et al, 1999). A further biochemical characterization of the CCR4–NOT complex will be essential in order to find out if and how this enzyme is regulated by heat shock.
Materials and methods
Drosophila stocks
The w1118 stock was used as a control. Rga or ccr4 homozygous mutant larvae were selected using the balancer chromosome TM3, Serrate-pAct-GFP. For bulk poly(A) measurements, RNA from 50 w1118 or Rgal(3)03834 homozygous second instar larvae and from 15 w1118 or ccr4KG877/Df(3R)crb-F89-4 adult males was prepared. For PAT assays, RNA was prepared from 15 w1118 or ccr4KG877 homozygous first instar larvae raised at 25°C and collected without a heat shock, or after a 30 min heat shock at 36.5°C followed by 30, 60 or 90 min of recovery at 25°C.
Cell culture, extract preparation and fractionation
Drosophila S2 cells (Invitrogen) were cultured as semi-adherent cells at 25°C in SF-900 serum-free medium (Invitrogen) with antibiotics and antimycotics. Suspension cultures were grown in the same medium containing 0.05% Pluronic (Invitrogen) in Erlenmayer flasks on an orbital shaker at 135 rpm. For preparation of cytoplasmic extract, 600 ml of S2 suspension culture (2 × 107 cells/ml) was harvested and washed with PBS. The cell pellet was resuspended in 35 ml of ice-cold hypotonic buffer (10 mM Hepes (pH 7.9), 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.2 mM PMSF, 2 μg/ml leupeptin, 1 μg/ml pepstatin) and kept on ice for 10 min. The cells were lysed by 20 strokes in a dounce homogenizer and adjusted to 10% glycerol and 0.02% NP-40. After centrifugation at 20 000 g at 4°C for 1 h, the supernatant was frozen in liquid nitrogen and stored at −70°C.
Fractionations were carried out at 4–8°C. Cytoplasmic extract (400 mg protein) was adjusted to 50 mM Tris–HCl (pH 7.5) and loaded on a 40 ml DEAE–Sepharose column (Pharmacia) equilibrated in 50 mM Tris–HCl (pH 7.5), 10% glycerol, 20 mM KCl, 0.02% NP-40, 0.5 mM DTT, 0.2 mM PMSF, 2 μg/ml leupeptin and 1 μg/ml pepstatin. The column was washed with four column volumes of buffer and developed with a 10 column volume gradient from 50 mM–1 M KCl. A 200 μl portion of fraction 7 of the DEAE column was applied to a prepacked Superdex 200 HS FPLC column (Pharmacia) in DEAE buffer containing 150 mM KCl at a flow rate of 0.25 ml/min. In a separate run, BSA (66 kDa) and cytochrome c (12.5 kDa) were analyzed as markers.
Activity assays
Homogeneously labeled poly(A) was made by extension of an oligo(A) primer by poly(A) polymerase with [α-32P]ATP (Körner and Wahle, 1997). The enzyme used was a recombinant C-terminal deletion variant of bovine poly(A) polymerase, 1–513 (Martin and Keller, 1996). Poly(A) degradation and deadenylation assays were performed as described (Körner and Wahle, 1997). The reaction buffer was 20 mM Hepes (pH 7.05), 150 mM KCl, 1 mM Mg2+ acetate, 1 mM DTT, 10% glycerol, 0.02% NP-40 and 0.2 mg/ml methylated BSA. vGC-GMCSF RNA (240 nt) contains the 5′ UTR of the human globin gene, a modified 3′ UTR of the human globin gene containing a mutant ARE and a poly(A) tail of 100 nt. The plasmid encoding this RNA was derived from the one described by Voeltz et al (2001) by deletion of the coding sequence. RNAs were either capped co-transcriptionally or cap-labeled as described (van Dijk et al, 2002) with the help of recombinant capping enzyme and [α-32P]GTP. Capping efficiency was 12%.
RNAi experiments
RNAi experiments were performed as described ( www.dixonlab.biochem.med.umich.edu). PCR products bearing the T7 promoter sequence on the 5′ ends of both strands were generated using a reverse transcription reaction from total S2 RNA as a template. Primer sequences are listed in Supplementary material. Double-stranded RNA was produced with the T7-MEGAscript-Kit (Ambion). Cells were harvested after 5 days of RNA treatment.
Antibodies and immunological procedures
The Drosophila caf1 coding sequence was amplified from a cDNA library (primer 1: 5′ATGAAATGGACAATGCCCTCG3′; primer 2: 5′TCATGAAGCGCTGTTCGTCTCA3′) and cloned into the EcoRV site of a pRSET expression vector (Invitrogen) in-frame with an N-terminal His6 tag. A 3′-terminal fragment of the Drosophila ccr4 coding sequence was amplified and cloned in the same way (primer 1: 5′ATGCTGGACAACTTGTCATTTA3′; primer 2: 5′CTACCGGCGATTGATCAGCC3′). Both proteins were expressed in Escherichia coli BL21 Codon+cells and purified. Polyclonal rabbit antibodies were produced by Eurogentec (Belgium). Antibodies against Drosophila CAF1 were affinity-purified on 100 μg of recombinant CAF1 separated by SDS–PAGE and blotted onto a nitrocellulose membrane. Rabbit antibodies against Drosophila PABPC were a gift of Nahum Sonenberg (Roy et al, 2004).
Western blots were blocked in 50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.2% Tween-20 and 2.5% BSA. The same buffer was also used for incubation with antisera and washing. As loading control, α-tubulin antibody (Sigma) was used at a 1:500 or 1:1000 dilution. Antibodies were detected by peroxidase-conjugated swine anti-rabbit immunoglobulins (DAKO, Glostrup, Denmark) and chemiluminescence staining (SuperSignal Kit, Pierce). Immunostaining of Drosophila tissues and Western blots with Drosophila extracts were performed as described (Benoit et al, 1999). Anti-CCR4 dilutions were 1:300 for immunostaining and 1:1000 for Westerns blots.
RNA analysis
Total RNA was isolated from S2 cells or from various Drosophila stages by the TRIZOL method (Invitrogen). For determination of bulk poly(A) length, 1–1.5 μg total RNA was incubated in a 20 μl reaction volume with 10 μCi [α-32P]3′-dATP and 100 U bovine poly(A) polymerase 1–513 (Martin and Keller, 1996) in 25 mM Tris–HCl (pH 8.3), 40 mM KCl, 10% glycerol, 0.05 mM EDTA, 0.5 mM MnCl2, 0.2 mg/ml BSA, 0.02% NP-40 and 0.5 mM DTT for 1 h at 37°C. RNA was purified by phenol/chloroform extraction and ethanol precipitation and dissolved in 50 μl 20 mM Tris–HCl (pH 8). Labeled RNA (100 000–500 000 cpm) was digested in a 20 μl reaction with 25 U RNase T1 (Sigma) and 5 ng RNase A (Roth) in 50 mM Tris–HCl (pH 7.5) and 100 mM KCl together with 20 μg yeast RNA (Merck) for 30 min at 30°C. The reaction was stopped by addition of the same volume of 200 mM Tris–HCl (pH 7.9), 300 mM NaCl, 25 mM EDTA, 2% SDS containing 20 μg proteinase K, 1 μg rRNA and 1 μg glycogen. As a control for the specificity of the RNase digestion, in vitro-synthesized L3pre RNA (65 nt) bearing a DNA-encoded poly(A) tail of 80 nt (L3preA80; from U Kühn) was treated in the same way. After a 30 min incubation at 37°C, the remaining RNA was ethanol precipitated and separated on a 10% polyacrylamide–urea gel. Gels were analyzed on a PhosphorImager and data were analyzed with the help of the ImageQuant program.
Poly(A) tails of the Hsp70 mRNA were analyzed by the PAT assay (Sallés and Strickland, 1999). A 3 μg portion of total RNA from S2 cells was mixed with 2 ng of in vitro-synthesized L3preA45-RNA (Kerwitz et al, 2003) as an internal control. The oligo(dT) anchor primer was 5′GCGAGCTCCGCGGCCGCGTTTTTTTTTTTT3′ and the Hsp70-specific primer was 5′GTCGACTAAGGCCAAAGA3′, which could prime both on the Hsp70 Aa and the Hsp70 Ab mRNA. The predicted minimal length of the product derived from almost completely deadenylated Hsp70 RNA is 236 bp under the assumption that the anchor primer primes on the very first A of the remaining oligo(A) tail. The L3pre-specific primer was 5′GAATACAAGCTTGGGCTGCAGGT3′. The predicted product size from L3preA45 is approximately 130 bp. The primers were radioactively labeled at their 5′ ends. PCR cycles were as follows: 3 min 94°C followed by 35 cycles of 30 s, 94°C; 45 s, 58°C; 2 min, 72°C. PCR products were purified by phenol/chloroform extraction and ethanol precipitation, separated on a 10% polyacrylamide–urea gel and detected on a PhosphorImager. For PAT assays from Drosophila larval RNA, the L3preA45-RNA was omitted, and the Hsp70 primer was 5′TTTGTTCATCAATGGGTTAT3′. The predicted minimal product size is 210 bp. Nonradioactive PCR products were run on a 2% agarose gel and detected by Southern blotting with an Hsp70 3′ UTR radioactive probe.
Supplementary Material
Supplement
Acknowledgments
We thank Kleomenis Dardoussis, Uwe Kühn, Nahum Sonenberg, Joan Steitz and Ghia Voeltz for reagents, the Bloomington stock center for Drosophila stocks and Rebekka Weißbach for help with some experiments. This work was supported by grants from the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie (to EW) and the Centre National de la Recherche Scientifique (UPR 1142) and l'Association Française contre les Myopathies (to MS). SZ held an award from the Ministère de l'Enseignement Supérieur et de la Recherche.
References
- Albert TK, Lemaire M, van Berkum NL, Gentz R, Collart M, Timmers HTM (2000) Isolation and characterization of human orthologs of yeast CCR4–NOT complex subunits. Nucleic Acids Res 28: 809–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews GK, Harding MA, Calvet JP, Adamson ED (1987) The heat shock response in HeLa cells is accompanied by elevated expression of the c-fos proto-oncogene. Mol Cell Biol 7: 3452–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baggs JE, Green CB (2003) Nocturnin, a deadenylase in Xenopus laevis retina: a mechanism for posttranscriptional control of circadian-related mRNA. Curr Biol 13: 189–198 [DOI] [PubMed] [Google Scholar]
- Bai Y, Salvadore C, Chiang Y-C, Collart M, Liu H-Y, Denis CL (1999) The CCR4 and CAF1 proteins of the CCR4–NOT complex are physically and functionally separated from NOT2, NOT4 and NOT5. Mol Cell Biol 19: 6642–6651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit B, Nemeth A, Aulner N, Kühn U, Simonelig M, Wahle E, Bourbon H-M (1999) The Drosophila poly(A)-binding protein II is ubiquitous throughout Drosophila development and has the same function in mRNA polyadenylation as its bovine homolog in vitro. Nucleic Acids Res 27: 3771–3778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeck R, Tarun S, Rieger M, Deardorff JA, Müller-Auer S, Sachs AB (1996) The yeast Pan2 protein is required for poly(A)-binding protein-stimulated poly(A)-nuclease activity. J Biol Chem 271: 432–438 [DOI] [PubMed] [Google Scholar]
- Brown CE, Sachs AB (1998) Poly(A) tail length control in Saccharomyces cerevisiae occurs by message-specific deadenylation. Mol Cell Biol 18: 6548–6559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CE, Tarun SZ, Boeck R, Sachs AB (1996) PAN3 encodes a subunit of the Pab1p-dependent poly(A) nuclease in Saccharomyces cerevisiae. Mol Cell Biol 16: 5744–5753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-Y, Shyu A-B (1995) AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci 20: 465–470 [DOI] [PubMed] [Google Scholar]
- Chen J, Chiang Y, Denis CL (2002) CCR4, a 3′–5′ poly(A) RNA and ssDNA exonuclease, is the catalytic component of the cytoplasmic deadenylase. EMBO J 21: 1414–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Rappsilber J, Chiang Y-C, Russell P, Mann M, Denis CL (2001) Purification and characterization of the 1.0 MDa CCR4–NOT complex identifies two novel components of the complex. J Mol Biol 314: 683–694 [DOI] [PubMed] [Google Scholar]
- Clark LB, Viswanathan P, Quigley G, Chiang Y-C, McMahon JS, Yao G, Chen J, Nelsbach A, Denis CL (2004) Systematic mutagenesis of the leucine-rich repeat (LRR) domain of CCR4 reveals specific sites for binding to CAF1 and a separate critical role for the LRR in CCR4 deadenylase activity. J Biol Chem 279: 13516–13623 [DOI] [PubMed] [Google Scholar]
- Collart M (2003) Global control of gene expression in yeast by the Ccr4–Not complex. Gene 313: 1–16 [DOI] [PubMed] [Google Scholar]
- Cougot N, Babajko S, Seraphin B (2004) Cytoplasmic foci are sites of mRNA decay in human cells. J Cell Biol 165: 31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couttet P, Fromont-Racine M, Steel D, Pictet R, Grange T (1997) Messenger RNA deadenylation precedes decapping in mammalian cells. Proc Nat Acad Sci USA 94: 5628–5633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugeron M-C, Mauxion F, Seraphin B (2001) The yeast POP2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res 29: 2448–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker CJ, Parker R (2002) mRNA decay enzymes: decappers conserved between yeast and mammals. Proc Natl Acad Sci USA 99: 12512–12514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellavalle RP, Petersen R, Lindquist S (1994) Preferential deadenylation of Hsp70 mRNA plays a key role in regulating Hsp70 expression in Drosophila melanogaster. Mol Cell Biol 14: 3646–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis CL, Chen J (2003) The CCR4–NOT complex plays diverse roles in mRNA metabolism. Prog Nucleic Acids Res Mol Biol 73: 221–250 [DOI] [PubMed] [Google Scholar]
- Draper MP, Salvadore C, Denis CL (1995) Identification of a mouse protein whose homolog in Saccharomyces cerevisiae is a component of the CCR4 transcriptional regulatory complex. Mol Cell Biol 15: 3487–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupressoir A, Morel A-P, Barbot W, Loireau M-P, Corbo L, Heidmann T (2001) Identification of four families of yCCR4- and Mg-dependent endonuclease-related proteins in higher eukaryotes, and characterization of orthologs of yCCR4 with a conserved leucine-rich repeat essential for hCAF1/hPOP2 binding. BMC Genomics 2: 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eystathioy T, Jakymiw A, Chan EKL, Seraphin B, Cougot N, Fritzler MJ (2003) The GW182 protein colocalizes with mRNA degradation associated proteins hDcp1 and hLSm4 in cytoplasmic GW bodies. RNA 9: 1171–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frolov MV, Benevolenskaya EV, Birchler JA (1998) Regena (Rga), a Drosophila homolog of the global negative transcriptional regulator CDC36 (NOT2) from yeast, modifies gene expression and suppresses position effect variegation. Genetics 148: 317–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin ACea (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Ingelfinger D, Arndt-Jovin DJ, Lührmann R, Achsel T (2002) The human LSm1–7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrn1 in distinct cytoplasmic foci. RNA 8: 1489–1501 [PMC free article] [PubMed] [Google Scholar]
- Kerwitz Y, Kühn U, Lilie H, Knoth A, Scheuermann T, Friedrich H, Schwarz E, Wahle E (2003) Stimulation of poly(A) polymerase through a direct interaction with the nuclear poly(A) binding protein allosterically regulated by RNA. EMBO J 22: 3705–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Körner C, Wahle E (1997) Poly(A) tail shortening by a mammalian poly(A)-specific 3′-exoribonuclease. J Biol Chem 272: 10448–10456 [DOI] [PubMed] [Google Scholar]
- Körner CG, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E (1998) The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J 17: 5427–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroia G, Cuesta R, Brewer G, Schneider RJ (1999) Control of mRNA decay by heat shock–ubiquitin–proteasome pathway. Science 284: 499–502 [DOI] [PubMed] [Google Scholar]
- Lenssen E, Oberholzer U, Labarre J, De Virgilio C, Collart M (2002) Saccharomyces cerevisiae Ccr4–Not complex contributes to the control of Msn2p-dependent transcription by the Ras/cAMP pathway. Mol Microbiol 43: 1023–1037 [DOI] [PubMed] [Google Scholar]
- Liu H-Y, Toyn JH, Chiang Y-C, Draper MP, Johnston LH, Denis CL (1997) DBF2, a cell cycle-regulated protein kinase, is physically and functionally associated with the CCR4 transcriptional regulatory complex. EMBO J 16: 5289–5298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Keller W (1996) Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and a catalytic domain, homologous to the family X polymerases, and to other nucleotidyl transferases. EMBO J 15: 2593–2603 [PMC free article] [PubMed] [Google Scholar]
- Moriya H, Shimizu-Yoshida Y, Omori A, Iwashita S, Katoh M, Sakai A (2001) Yak1p, a DYRK family kinase, translocates to the nucleus and phosphorylates yeast Pop2p in response to a glucose signal. Genes Dev 15: 1217–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer U, Collart M (1998) Characterization of NOT5 that encodes a new component of the Not protein complex. Gene 207: 61–69 [DOI] [PubMed] [Google Scholar]
- Parker R, Song H (2004) The enzymes and control of eukaryotic mRNA turnover. Nat Struct Mol Biol 11: 121–127 [DOI] [PubMed] [Google Scholar]
- Petersen R, Lindquist S (1988) The Drosophila hsp70 message is rapidly degraded at normal temperature and stabilized by heat shock. Gene 72: 161–168 [DOI] [PubMed] [Google Scholar]
- Petersen RB, Lindquist S (1989) Regulation of HSP70 synthesis by messenger RNA degradation. Cell Regul 1: 135–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J (1995) mRNA stability in mammalian cells. Microbiol Rev 59: 423–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy G, Miron M, Khaleghpour K, Lasko P, Sonenberg N (2004) The Drosophila poly(A) binding protein-interacting protein, dPaip2, is a novel effector of cell growth. Mol Cell Biol 24: 1143–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadis S, Hickey E, Weber LA (1988) Effect of heat shock on RNA metabolism in HeLa cells. J Cell Physiol 135: 377–386 [DOI] [PubMed] [Google Scholar]
- Sallés FJ, Strickland S (1999) Analysis of poly(A) tail lengths by PCR: the PAT assay. In Methods in Molecular Biology, Haynes S (ed) Vol. 118, pp 441–448. Totowa, NJ: Humana Press [DOI] [PubMed] [Google Scholar]
- Sheth U, Parker R (2003) Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science 300: 805–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharun S, Parker R (2001) Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p–7p complex on deadenylated yeast mRNAs. Mol Cell 8: 1075–1083 [DOI] [PubMed] [Google Scholar]
- Thore S, Mauxion F, Seraphin B, Suck D (2003) X-ray structure and activity of the yeast Pop2 protein: a nuclease subunit of the mRNA deadenylase complex. EMBO Rep 4: 1150–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Staples RR, Valencia-Sanchez MA, Muhlrad D, Parker R (2002) Ccr4p is the catalytic subunit of a Ccr4p/Pop2/Notp mRNA deadenylase complex in Saccharomyces cerevisiae. EMBO J 21: 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R (2001) The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell 104: 377–386 [DOI] [PubMed] [Google Scholar]
- Uchida N, Hoshino S, Katada T (2004) Identification of a human cytoplasmic poly(A) nuclease complex stimulated by poly(A)-binding protein. J Biol Chem 279: 1383–1391 [DOI] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Séraphin B (2002) Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 21: 6915–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoof A, Parker R (2002) Messenger RNA degradation: beginning at the end. Curr Biol 12: R285–R287 [DOI] [PubMed] [Google Scholar]
- Voeltz GK, Ongkasuwan J, Standart N, Steitz JA (2001) A novel embryonic poly(A) binding protein, ePAB, regulates mRNA deadenylation in Xenopus egg extracts. Genes Dev 15: 774–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickens M, Goodwin EB, Kimble J, Strickland S, Hentze M (2000) Translational control of developmental decisions. In Translational Control of Gene Expression, Sonenberg N, Hershey JWB, Mathews MB (eds), pp 295–370. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Zhao M, Tang D, Lechpammer S, Hoffman A, Ascea A, Stevenson MA, Calderwood SK (2002) Double-stranded RNA-dependent protein kinase (pkr) is essential for thermotolerance, accumulation of HSP70, and stabilization of ARE-containing HSP70 mRNA during stress. J Biol Chem 277: 44539–44547 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement


