Abstract
Synaptic specificity is a defining property of neural networks. In the cerebellum, synapses between parallel fiber neurons and Purkinje cells are specified by the simultaneous interactions of secreted protein Cerebellin with presynaptic Neurexin and postsynaptic delta-type Glutamate receptors (GluD). Here, we determined the crystal structures of the trimeric C1q-like domain of rat Cerebellin-1, and the first complete ectodomain of a GluD, rat GluD2. Cerebellin binds to the LNS6 domain of α- and β-Neurexin-1 through a high-affinity interaction that involves its highly flexible N-terminal domain. In contrast, we show that the interaction of Cerebellin with isolated GluD2 ectodomain is low affinity, which is not simply an outcome of lost avidity when compared to binding with a tetrameric full-length receptor. Rather, high-affinity capture of Cerebellin by post-synaptic terminals is likely controlled by long-distance regulation within this trans-synaptic complex. Altogether, our results suggest unusual conformational flexibility within all components of the complex.
Keywords: Synapse, Neurexin, Glutamate Receptor Delta-2, Cerebellin, Crystal structure, Negative-stain single-particle electron microscopy, Structural flexibility
eTOC Blurb
Cerebellin, Neurexin and Glutamate receptor delta-2 create a trans-synaptic protein complex that organizes synapses. Using crystallography, single-particle electron microscopy and affinity measurements, Cheng et al. demonstrate large conformational plasticity and flexibility within the complex, and provide insights into high- and low-affinity components of the system.
Introduction
Neurexins (Nrxn) are cell adhesion molecules that are heavily expressed in the brain, and especially in the presynaptic terminals in neurons (Ullrich et al., 1995; Ushkaryov et al., 1992). They have been implicated in synapse specification, maturation and function (Graf et al., 2004; Ko et al., 2009; Scheiffele et al., 2000), and are known to create trans-synaptic complexes through interactions with several post-synaptic proteins (reviewed in Reissner et al., 2013). Rare mutations and copy number variations of Neurexins and of proteins known to associate with Neurexins have been identified in multiple neurodevelopmental disorders, especially autism (Südhof, 2008). Mammalian Neurexins are known to be expressed from two alternate promoters, resulting in the longer α and shorter β forms, and also go through alternative splicing at five or six sites, resulting in >1,000 unique forms for a given Neurexin gene (Treutlein et al., 2014; Ullrich et al., 1995). This large potential of diversity in Neurexin sequences is neither accidental nor unutilized: a large range of variants are actually transcribed (Treutlein et al., 2014), and different neuron subtypes have been shown to express defined repertoires of Neurexin variants (Fuccillo et al., 2015; Schreiner et al., 2014). Furthermore, interactions of several cell surface and extracellular proteins with Neurexin have been demonstrated to be dependent on specific splice variation (Reissner et al., 2013). Therefore, Neurexins provide support to the paradigm that molecular cues (i.e. molecular identity tags) presented on neurons dictate neuronal connectivity through specific molecular interactions (Shen and Scheiffele, 2010; Zipursky and Sanes, 2010). Accordingly, Neurexins have been proposed to create a “synaptic adhesion code” (Fuccillo et al., 2015).
Contributing to this connection between neuronal connectivity and molecular diversity in the brain is the recent discovery of secreted proteins called Cerebellins, and their simultaneous interactions with Neurexin and the delta-type Glutamate receptors (GluD) on presynaptic and postsynaptic terminals, respectively (Matsuda et al., 2010; Uemura et al., 2010). Cerebellin-1 (Cbln1) is secreted from presynaptic terminals in granule cells within parallel fibers in the cerebellum (Hirai et al., 2005). Cbln1 has been shown to interact with β-Neurexin carrying the fourth splice site (+SS4) expressed on presynaptic membranes of parallel fibers (PF), and with post-synaptic delta-type Glutamate receptors expressed on Purkinje cells (PC) (Uemura et al., 2010). The trans-synaptic complex made up of Nrxn, Cbln, and GluD allows for specificity to be established for PF-PC synapses, while the complex initiates signaling for synaptic differentiation presumably on both terminals. As a tell-tale sign of the centrality of this complex, artificial synapses can be formed between HEK293 cells and neurons if GluD and Nrxn are presented by the two cells, respectively, and Cblns are provided in growth media (Elegheert et al., 2016; Matsuda and Yuzaki, 2011). In the best studied case of a trans-synaptic complex involving Nrxn, a constitutively dimeric post-synaptic protein, Neuroligin, is known to bind either α- or β-Neurexin lacking the SS4 in a 2:2 stoichiometric complex (Araç et al., 2007; Chen et al., 2008; Comoletti et al., 2006; Fabrichny et al., 2007; Ichtchenko et al., 1995), and with comparable results in artificial synapse formation assays. It remains to be seen whether the complexes of Nrxn with Cbln/GluD and with Neuroligin share similar architectural, structural and functional properties, or if Nrxn has evolved multiple structural and signaling modes of action depending on its splice variation and binding partners.
The “molecular glue” between pre- and post-synaptic receptors at PF-PC synapses, Cerebellin-1 was initially identified as the precursor protein from which a ubiquitous 16-amino acid neuropeptide is generated, although the physiological function of this peptide is not clear (Mugnaini et al., 1988; Slemmon et al., 1984; Urade et al., 1991). All four Cerebellins contain a C1q and Tumor Necrosis Factor-α-like domain (henceforth called the C1q domain), preceded by a cysteine-rich N-terminal domain (CRN). The cysteine-rich N-terminal domain of Cerebellins has been shown to be necessary for the creation of hexamers from the C1q trimers (Bao et al., 2005).
On the other hand, the post-synaptic delta-type Glutamate receptors display a complicated domain and organizational structure. GluD extracellular N-terminal domains (ATD) have been implicated in Cbln binding (Matsuda et al., 2010; Uemura et al., 2010). ATD is followed by a ligand-binding domain (LBD), which is a bipartite domain divided in the middle by two transmembrane (TM) helices, and is followed by the third helix of the TM domain and a C-terminal cytoplasmic domain. GluDs are a class of ionotropic Glutamate receptors (iGluR), which are tetrameric. iGluRs are known for their ability to bind neurotransmitters, metal ions, many agonists, antagonists and allosteric modulators, which regulate their conformational states and ion channel activities (Karakas et al., 2015).
The key questions yet to be answered are how trans-synaptic Nrxn-Cbln-GluD complexes can form, how they are organized, and how they signal. The answer to these will depend on the molecular architecture and biochemical properties of individual components and the complexes. Here, we report our structural and biochemical insights into these trans-synaptic complexes. We determined the crystal structure of the trimeric Cbln1 and demonstrated that Cbln1 complexes can be formed with α- and β-Neurexin in a splicing-dependent fashion. Using single-particle electron microscopy, we reveal the overall architecture of Cbln complexed with α-Neurexin and with β-Neurexin, and show that the interaction depends on the Cysteine-rich N-terminal domain of Cbln1, creating a flexible complex where Neurexin is able to sample large volumes within the synaptic cleft. On the post-synaptic end of this complex, we present the first full-length dimeric ectodomain of a GluD, which shows a previously unobserved conformation of iGluR ectodomains, where the ligand-binding domains are “swung-out” in a fashion alike the desensitized state of iGluRs. Combined with the recently published structure of the GluD2-Cbln complex by Elegheert et al. (2016), our results open the way for further studies on the regulation and signaling through this trans-synaptic complex.
Results
Cerebellin-1 binds with α and β-Neurexin in a splice variant-dependent fashion
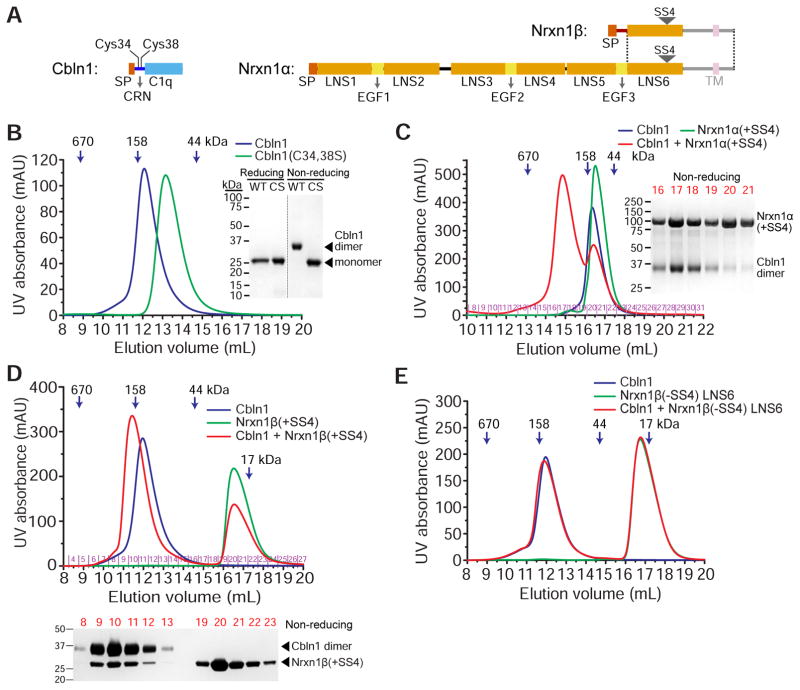
To study the biochemical properties of Cerebellin-Neurexin complexes, we first expressed and purified full-length rat Cerebellin-1 and the structured ectodomain portions of rat Neurexin-1α (Domains LNS1 or LNS2 to LNS6) and Neurexin-1β (N-terminal β-unique region plus LNS6, or LNS6 only), with and without the fourth splice site (+/−SS4) (Figure 1A). Cerebellin is expected to form disulfide-mediated dimers of the C1q-like trimers, resulting in a hexameric structure (Bao et al., 2005). We observe the dimerization due to disulfide bonds of Cbln1 when purified samples are run over polyacrylamide gels without reducing agents (Figure 1B). Furthermore, while wild-type Cbln1 elutes at hexamer-like retention volumes on size-exclusion columns, the C34,38S double mutant runs as a trimer (Figure 1B). As expected, Neurexins elute from size exclusion columns at volumes compatible with monomers (Figures 1C–1E). We then tested whether high-affinity complexes can be observed in size-exclusion chromatography runs. Both α- and β-Neurexins containing the SS4 (+SS4) co-elute with Cbln1 hexamers (Figures 1C and 1D). Specifically, the Nrxn1 LNS6 domain +SS4 is sufficient for Cbln1 binding (Figure S1A). As expected, Nrxn1β without SS4 (−SS4) does not co-elute with Cbln1, demonstrating that SS4 is necessary for the recognition of Nrxn by Cbln (Figure 1E).
Figure 1. Rat Cbln1 hexamers with intact CRN domains bind rat α- or β-Nrxn1 including SS4.
A. Domain structure of Cbln1, Nrxn1α and Nrx1β, drawn to scale. The shaded regions, the transmembrane (TM) helix and the unstructured juxtamembrane domains (intracellular and extracellular), are excluded from our constructs. The dotted lines mark the boundaries of the region expressed by exons shared by α and β-Neurexins. SP: Signal peptide.
B. Cbln1 (blue curve) runs as hexamers on a Superdex 200 size-exclusion column when its cysteines in the CRN domain are intact. C34,38S (CS) double mutation causes it to run as trimers (green curve). Wild-type (WT) Cbln1 runs as a dimer on non-reducing denaturing gels, while CS runs as a monomer.
C. Cbln1 binds Nrxn1α(+SS4) domains LNS2 to LNS6, as observed on a Superose 6 size-exclusion column. Size-exclusion fractions for the Cbln1+Nrxn1α(+SS4) sample (red curve) are run on a non-reducing gel.
D. Cbln1 binds Nrxn1β(+SS4) as observed on a Superdex 200 size-exclusion column. Nrxn1β construct contains residues S48 to P292, including the β form-unique N-terminal region and the LNS6 domain. Size-exclusion fractions for the Cbln1+Nrxn1β(+SS4) sample (red curve) are run on a non-reducing gel. For Cbln1 binding to an LNS6-only Nrxn1 construct, see Figure S1A.
E. Cbln1 does not bind Nrxn1β-LNS6 without SS4 (−SS4).
See also Figure S1.
We also tested whether the hexameric state or the cysteines within the CRN domain are necessary for Neurexin binding. Similar to the observations of Elegheert et al. (2016) and Uemura et al. (2010) for human and mouse Cblns, the trimeric Cys-to-Ser mutant no longer binds Neurexin (Figure S1B). This could be either due to the necessity for the short Cysteine-containing N-terminal domain to create a highly-avid hexameric state, or a result of the Cys-to-Ser mutations abolishing the Neurexin-binding site on Cerebellin-1. Overall, these results are in agreement with other studies investigating Cerebellin-Neurexin complexes.
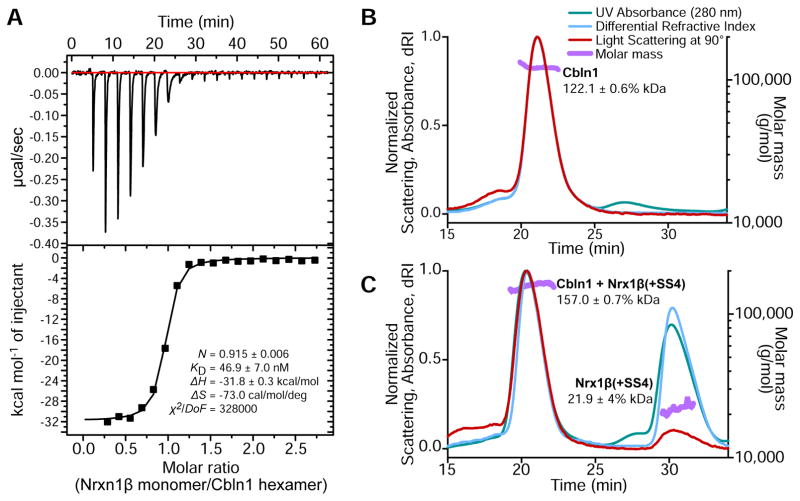
Cerebellin–Neurexin complex has 6:1 stoichiometry
Our size-exclusion chromatography results (Figures 1C and 1D) suggested strong binding between Cerebellins and Neurexins. To quantify the affinity of this interaction, we performed isothermal titration calorimetry experiments for Cbln1 against β-Neurexin-1. As predicted the binding was strong with a dissociation constant of 47 nM (Figure 2A). Interestingly, the observed stoichiometry in this experiment was 6:1, or one Cbln1 hexamer for one Neurexin-1β. Our results disagree with the published calorimetry experiments by Lee et al. (2012), who proposed a 6:2 stoichiometric model, but agree with the recent work by Elegheert, et al. (2016). To further investigate this issue, we measured the molecular size of the Cbln1-Nrxn1β complex using multi-angle light scattering (MALS). The Cbln1 hexamers were measured to be 122 kDa, closely matching the expected hexameric size of 126.6 kDa (Figure 2B). The Cbln1-Nrxn1β complex molecular mass was 157 kDa, matching a putative 6:1 complex with an expected size of 159.2 kDa (Figure 2C).
Figure 2. Rat Cbln1 binds Nrxn1β with high affinity and a stoichiometry of 1 hexamer to 1.
A. Isothermal titration calorimetry experiments for Cbln1. Molar ratio represents the ratio of Neurexin monomers to Cerebellin hexamers.
B–C. MALS analysis confirms molar mass for Cbln1 and Cbln1-Nrxn1β complex to match one hexamer (B), and one hexamer + one monomer (C), respectively.
Cerebellin-1 C1q domain adopts the canonical C1q-like fold
Next, we set out to determine the structure of a Nrxn-Cbln complex to reveal the molecular determinants of specificity and the high-resolution architecture of this complex. All our trials have led to Cbln1-only crystals. These crystals appeared as triangular pyramids connected at the tips, often fusing into triangular prism-like structures. We were able to collect a dataset from a crystal grown in a Formate condition that displayed no signs of a multi-crystalline nature or twinning based on its diffraction pattern and diffraction intensity statistics. This crystal yielded a 1.8 Å-resolution dataset in the hexagonal P6 space group. We determined the structure of Cbln1 using the monomer for the C1q-like structure of human Caprin-2 (PDB: 4OUM) (Miao et al., 2014) as a molecular replacement model, which has 39% sequence identity to the C1q-like domain of Cbln1. The crystallographic data and refinement statistics are in Table 1.
Table 1. Crystallographic data and refinement statistics for the Cbln1 structure.
The values in parentheses are for the highest-resolution bin.
| Data Collection | Cbln1 |
|---|---|
| Space Group | P 6 |
| Cell Dimensions | |
| a, b, c (Å) | 82.74, 82.74, 50.37 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 50-1.80 (1.84-1.80) |
| Rsym (%) | 4.4 (76.4) |
| CC1/2 | 1.00 (0.69) |
| <I>/<σ(I)> | 22.50 (1.74) |
| Completeness (%) | 98.2 (83.8) |
| Redundancy | 6.5 (3.2) |
| Refinement | |
| Resolution (Å) | 50-1.80 (1.84-1.80) |
| Reflections | 18144 (1162) |
| Rcryst (%) | 16.01 (33.15) |
| Rfree (%) | 18.88 (33.43) |
| Number of atoms | |
| Protein | 1119 |
| Ligand | 38 |
| Water | 81 |
| Average B-factors (A2) | |
| All | 41.0 |
| Protein | 39.0 |
| Ligand | 86.8 |
| Solvent | 47.1 |
| R.m.s. deviations from ideality | |
| Bond Lengths (Å) | 0.005 |
| Bond Angles (°) | 0.740 |
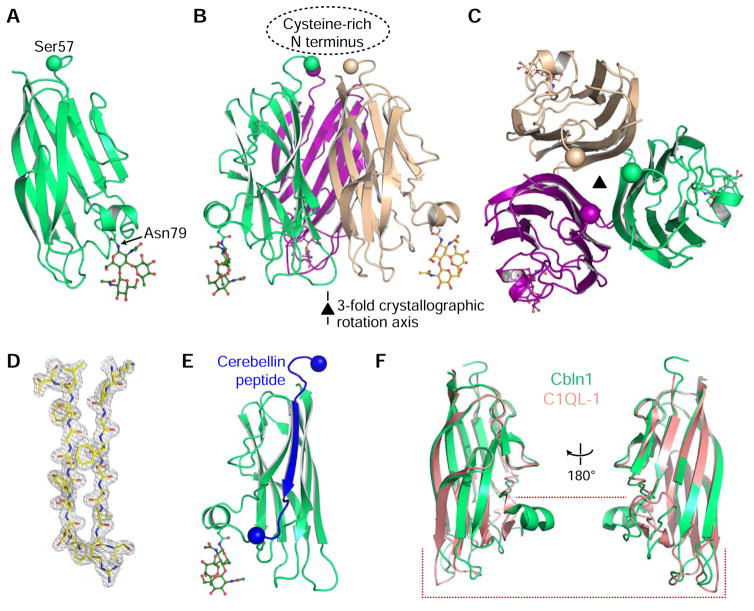
The fully refined crystal structure shows the canonical trimeric structure, which can be generated from the monomeric asymmetric unit using the three-fold symmetry of the crystal lattice (Figures 3A–D). We observed no sign of the Cysteine-rich N-terminal domain beyond residue Ser-57 or Gly-58 in electron density maps (Figure 3C). Furthermore, the crystallographic model does not leave enough space for the 35-aa N-terminal domain due to packing within the crystal. These observations strongly suggest that the protein degraded and shed its N terminus during the crystallization experiment. Ser-57 and Gly-58 are the first residues of the 16- and 15-amino acid neuropeptides cerebellin and des-Ser1-cerebellin (Slemmon et al., 1984), confirming the in vivo observation that Cerebellin is naturally prone to be processed at these residues. The cerebellin peptides end at residue His-73, which is placed within the next loop and solvent-accesible region in Cbln1; i.e. cerebellin peptide comprises the first beta-strand of the C1q domain (Figure 3E, colored bright blue).
Figure 3. Crystal structure of Cbln1.
A. Cartoon model of the Cerebellin-1 C1q monomer. N-linked glycan attached to Asn79 side chain is shown in stick representation. The Cα atom of the last residue visible in the electron density is shown as a ball.
B. Cbln1 trimer can be formed by applying the crystallographic three-fold symmetry operation. The location of the missing Cysteine-rich N-terminal (CRN) domain is highlighted.
C. Looking at the Cbln1 trimer along the symmetry axis from the top, where CRN would be positioned.
D. Representative 2mFo-DFc electron density electron density.
E. Source of the Cerebellin peptide within the Cbln1 protein. Balls highlight Cα atoms of the first and last residues of the peptide.
F. Comparison of Cbln1 C1q domain to the C1q domain of C1QL-1 (PDB: 4D7Y), where most differences are towards the “bottom half” of the domain. For comparisons with other C1q domains, see Figure S2.
The C1q domain of Cbln1 preserves the secondary structure topology of the most similar C1q domains found in Caprin-2 and C1QL proteins 1 to 3, while having unique features (Kakegawa et al., 2015; Miao et al., 2014; Ressl et al., 2015). The “top half” of the C1q domain in proximity to the CRN can be closely superimposed with the C1q crystal structures of Caprin-2 and C1QL proteins, while the bottom half of Cbln1 C1q domain shows a large divergence from its homologs (Figures 3F, S2A–C). This part of the C1q domain has been shown to bind Calcium ions in Caprin-2 and C1QLs, while there is no evidence that Cerebellins bind Calcium. More importantly, this deviation at the bottom half from other C1q domain proteins may be the result of specialization to protein binding in Cblns, as Elegheert et al. has recently demonstrated this part of Cbln1 to be the binding site for GluD2.
Architecture of the Cerebellin – Neurexin complex
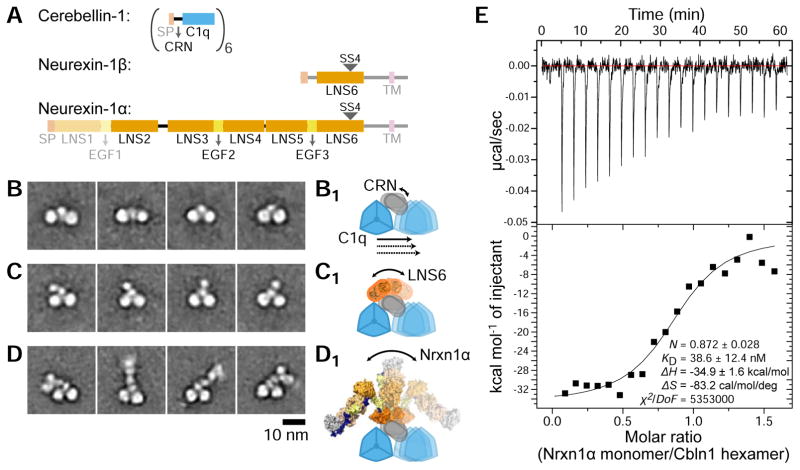
As the stoichiometry of the Cbln-Nrxn complex is established at 6:1, we could not explain how a single copy of the monomeric Neurexin could interact with the two-fold and three-fold symmetric Cerebellin hexamers. To solve this conundrum and to understand how Cerebellin can serve as the “molecular glue” for pre- and post-synaptic receptors, we employed single-particle negative-stain electron microscopy (EM) analysis to visualize Cerebellin and Cerebellin-Neurexin complexes. The EM averages show that Cbln1 is made by two three-fold-symmetric C1q-like domains connected by the hexameric 35-aa CRN domain (Figures 4B, 4B1). The distance between the centers of the C1q domains varies significantly among class averages, strongly indicating that the CRN domains serve as a flexible hinge between the two C1q trimers. In EM images for Cbln1-Nrxn1β, we observed only one copy of the Nrxn1 LNS6 domain, a proxy for the short β isoform, interacting with the hexameric CRN region (Figure 4C). This is in full agreement with the 6:1 stoichiometry we have measured for Cbln1:Nrxn1β (Figures 2A and 2C). We also observe β-Nrxn to assume variable positioning with respect to the two Cerebellin trimers, indicating flexibility of the CRN in relation to the C1q domains (Figure 4C1).
Figure 4. Cerebellin binds both α- and β-Neurexin with its flexible CRN domain.
A. Domain compositions of Cbln1, Nrxn1α and 1β. The shaded domains were not included in expressed constructs used for electron microscopy.
B–D. Representative EM 2D class averages of Cbln1 (B), Cbln1+Nrxn1β (C) and Cbln1+Nrxn1α (D). See also Figure S3.
B1–D1. Schematized representations of movements observed in class averages shown in (B) to (D).
E. One of the three isothermal titration calorimetry experiments for Cbln1 and Nrxn1α binding. The results are fit to a “one set of sites” binding model.
The majority of Neurexin-1 expressed in the brain is in the α form (Ullrich et al., 1995), and the α form is strongly expressed in the cerebellum (Fuccillo et al., 2015). To see if Nrxn1α could interact with Cbln1 in a manner similar to Nrxn1β, we applied single-particle negative-stain EM to examine the complex of Cbln1 bound to Nrxn1α domains LNS2 to LNS6, the region of Nrxn1α previously determined to be relatively rigid (Chen et al., 2011; Comoletti et al., 2010; Miller et al., 2011). Similarly, we could observe Nrxn1 LNS6 domain interacting with Cbln1 CRN, and the N-terminal domains of Nrxn1α positioned away from the two C1q domain trimers, avoiding steric clashes with Cbln1 (Figures 4D, 4D1). Based on these results, we expect that +SS4 variants of Nrxn1α should be able to function like Nrxn1β, and the affinity of Cbln1 for Nrxn1α and 1β should be similar, despite a report showing differences in affinity between α and β forms of Neurexins (Joo et al., 2011). Using isothermal titration calorimetry, we measured the affinity between Cbln1 and Nrxn1α LNS2-to-LNS6 to be 35 nM ± 8 nM (standard deviation over three measurements) with a hexamer to monomer stoichiometry of 0.912 ± 0.045 (Figure 4E), similar to what we observed for Nrxn1β (Figure 2A). Besides confirming the high flexibility of the CRN with respect to the C1q domains of Cerebellin, the EM averages also reveal additional variability in the disposition of the N-terminal LNS domains (LNS2–LNS3). The observed flexibility of Cbln1 CRN and Nrxn1α LNS domains suggests that the neurexin arm can probe three-dimensional space with a sweep of ~110° or greater (Figure 4D1).
GluD2 extracellular domain dimers or tetramers do not bind Cerebellin with high affinity
To bridge the synaptic cleft, Cerebellins interact with delta-type ionotropic Glutamate receptors expressed on postsynaptic membranes. GluDs, similar to other tetrameric ionotropic glutamate receptors, are ion channels that contain a three-helix membrane domain near its C-terminal end. GluDs are the least characterized family of iGluRs, especially since their channel activity cannot be recapitulated in the absence of an activating ligand, which is yet to be identified (Naur et al., 2007). Unlike the other iGluR families, no structure of a GluD beyond isolated single domains has been determined (Elegheert et al., 2016; Naur et al., 2007).
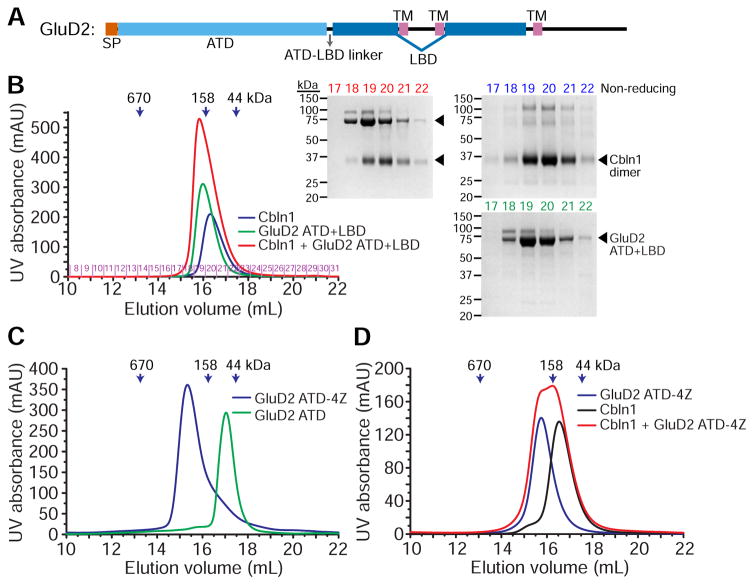
GluD ectodomains are made up of an N-terminal domain (ATD), which Cerebellins are known to bind (Matsuda et al., 2010; Uemura et al., 2010), followed by a ligand-binding domain (LBD), which binds D-Serine and Glycine, but not Glutamate (Naur et al., 2007) (Figure 5A). The LBD is split in the middle by two of the transmembrane helices. To study the binding of Cerebellin to GluD receptors, we created soluble expression constructs of the ectodomain, where we bypassed the first two membrane helices by replacing them with a Gly-Thr linker based on strategies established for iGluRs (Armstrong and Gouaux, 2000; Naur et al., 2007).
Figure 5. GluD2-Cbln affinity is weak regardless of oligomeric state.
A. Domain structure of GluD2. All domain and structural elements are drawn to scale.
B. Cbln1 bind does not bind GluD2 ectodomain (ATD+LBD) with high affinity, as observed by lack of co-elution on size-exclusion columns. Colors of chromatograms match colors of fractions as they are labeled on non-reducing gels.
C. GluD2 ATD domain can be tetramerized using a helical coiled coil zipper (tetrameric zipper, or “4Z”).
D. Cbln1 bind does not bind tetrameric GluD2 ATD-4Z with high affinity, as observed by lack of co-elution on size-exclusion columns, or by lack of heat release or uptake during ITC experiments in the presence or absence of calcium (Figure S4).
To characterize a Cbln-GluD complex, we have expressed and purified constructs of GluD2 containing the ATD and both the ATD and LBD, but did not observe complexes of sufficiently high affinity to co-elute via size exclusion (Figure 5B). Similarly, we could not observe particles representing the GluD2-Cbln1 complex in EM images (data not shown). Interestingly, both ATD and ATD+LBD ran as dimers on size-exclusion columns, unlike the tetramers known for full-length receptors. As recently proposed by Elegheert et al., we suspected that tetrameric nature of the full-length receptors could provide the necessary avidity to create high-affinity GluD-Cbln complexes. Therefore, we created GluD2 ATD tetramers (GluD2 ATD-4Z) using an engineered tetrameric coiled-coil zipper based on GCN4 (Harbury et al., 1993) (Figure 5C). However, GluD2 ATD tetramers could still not form high-affinity Cbln complexes (Figures 5D and S4). This is despite the reported affinities between GluD2 and Cbln1, which range between 16.5 to 167 nM (Elegheert et al., 2016; Matsuda et al., 2010; Uemura et al., 2010), but in agreement with the observation where a stable complex of Cbln1 with detergent-solubilized, full-length tetramers of GluD2 did not form during size-exclusion runs (Elegheert et al., 2016, Figure S5B therein), and a stable complex could only be crystallized after fusing Cbln1 and GluD2 ATD into a single-chain construct. While tetrameric avidity could still be a factor in high-affinity Cerebellin binding, we suggest that long-distance allosteric effects and conformational changes emanating from the membrane domain, the ligand-binding domain, or Cbln-Nrxn-mediated activation or clustering are also likely effectors of higher-affinity Cerebellin binding to GluD receptors.
GluD2 ectodomain architecture
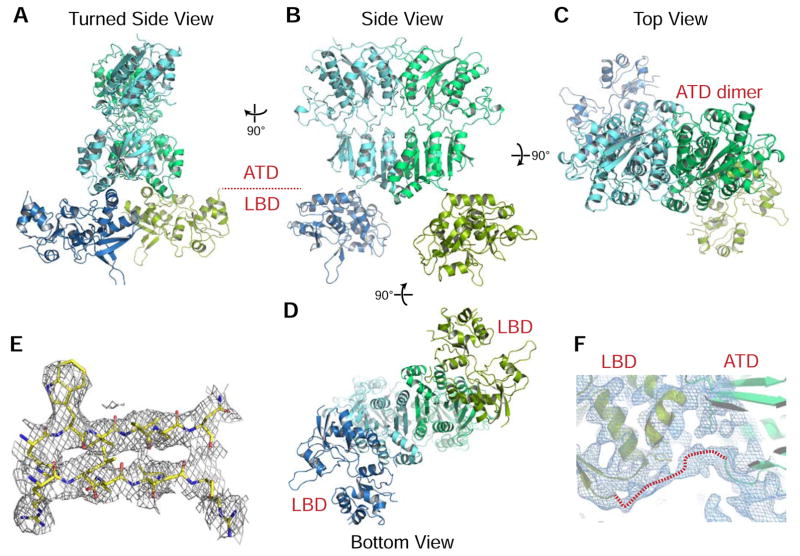
To further characterize this understudied class of ligand-gated ion channels, we next set out to determine the first complete ectodomain structure of a delta-type Glutamate receptor. Here, we present the crystal structure of the GluD2 ectodomain in a dimeric state (Figure 6A–D, Table 2). Despite the limited resolution, electron density shows side chain positions in a majority of residues (Figure 6E), and also includes features that are not part of the model built. The high quality of the refined model and the detail in electron density maps is likely due to the use of high-resolution models as starting points of refinement (Brunger et al., 2009). The most remarkable feature present in electron density despite being absent in the modeled coordinates is the 10-residue linker between the ATD and LBD (Figure 6F). The ordered nature of this linker implies that the orientation of the LBD with regards to the ATD is unlikely a random pose or a crystal artifact, and represents a putative conformational state for GluD receptors.
Figure 6. Structure of the symmetric dimers of rat GluD2 ectodomain.
A–D. GluD2 ectodomain dimers in four different views.
E. Representative 2mFo-DFc electron density from the beta-strands of the C-terminal lobe of the ATD domain at 1.0 σ.
F. NCS-averaged 2mFo-DFc electron density for the unmodeled ATD-LTD linker, which packs both on to the ATD and LBD.
Table 2. Crystallographic data and refinement statistics for the GluD2 ectodomain structure.
The values in parentheses are for the highest-resolution bin.
| Data Collection | GluD2 ATD+LBD |
|---|---|
| Space Group | P 3221 |
| Cell Dimensions | |
| a, b, c (Å) | 179.172, 179.172, 214.390 |
| α, β, γ (°) | 90, 90, 120 |
| Resolution (Å) | 50-4.15 (4.22-4.15) |
| Rsym (%) | 15.7 (79.5) |
| CC1/2 | 0.99 (0.46) |
| <I>/<σ(I)> | 8.9 (1.6) |
| Completeness (%) | 100.0 (100.0) |
| Redundancy | 4.2 (3.9) |
| Refinement | |
| Resolution (Å) | 50-4.15 (4.26-4.15) |
| Reflections | 30,109 |
| Twin law and fraction | -h, -k, l |
| Twin fraction | 0.49 |
| Rcryst (%) | 21.12 (21.36) |
| Rfree (%) | 26.04 (27.17) |
| Number of atoms | |
| Protein | 15,408 |
| Ligand | 0 |
| Water | 0 |
| Average B-factors (A2) | |
| All | 59.8 |
| Protein | 59.8 |
| R.m.s. deviations from ideality | |
| Bond Lengths (Å) | 0.003 |
| Bond Angles (°) | 0.613 |
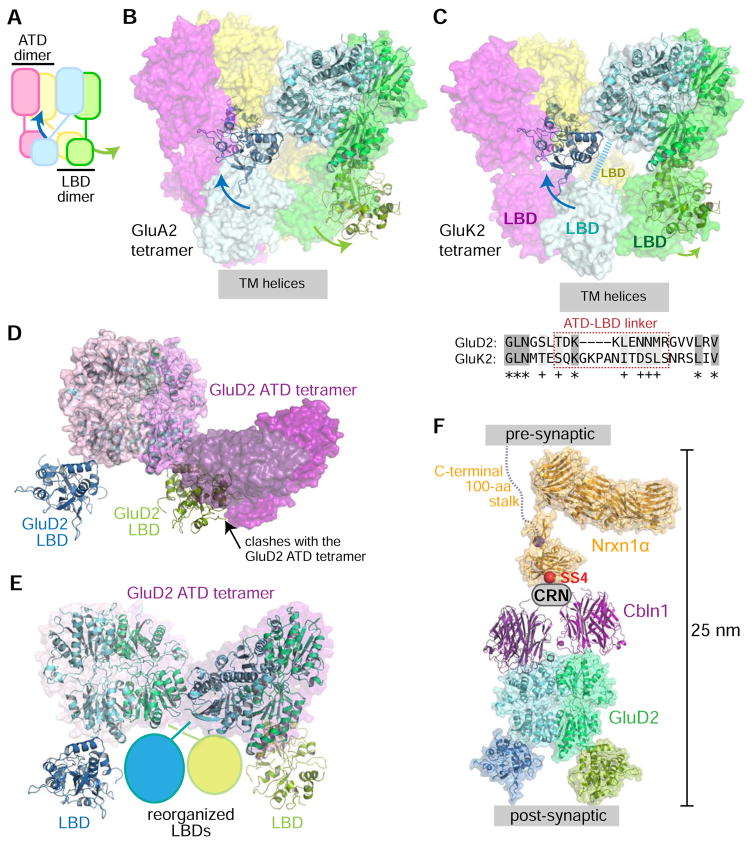
iGluR tetramers are dimers of dimers, where ATD dimers and LBD dimers are formed using different pairs of subunits (Figure 7A), leading to an overall tetrameric state, which is reinforced by the tetrameric channel domain. The organization of domains and local interactions between the domains within iGluR tetramers are correlated with their functional states (reviewed in Karakas et al., 2015). While the well-established ATD dimer is observed in our structure, the LBD dimer (Naur et al., 2007) no longer forms, resulting in ATD-mediated dimers only. This holds for all three independent copies of the GluD2, and the two independent GluD2 dimers within the crystallographic asymmetric unit (one dimer is created through crystallographic two-fold symmetry). Although isolated LBD domains and LBDs within full-length receptors have been observed almost exclusively to form dimers in crystal structures, there are exceptions where the LBD dimers break, especially in the desensitized state of the receptor (Dürr et al., 2014; Meyerson et al., 2014), and in the case of the D-Ser-bound LBD of GluD2 (PDB: 2V3U) (Naur et al., 2007).
Figure 7. GluD2 dimers are in a conformation not observed before in iGluR proteins.
A. Schematic representation of iGluR tetramers, with the swing-out motion in the GluD2 ectodomain structure mapped on top. The swinging-out of LBDs in the GluD2 ectodomain dimer structure are shown with arrows.
B. GluD2 dimer superposed on GluA2 antagonist bound structure (PDB: 3KG2).
C. GluD2 dimer superposed on the desensitized state structure of GluK2 (PDB: 4UQQ). The shorter ATD-LBD linker in GluD2 compared to GluK2 does not allow for GluDs to adopt a similar desensitized state structure. The ATD-LBD linker in the cyan subunit of GluK2 was not resolved in the structure, and is depicted as a thick dashed line.
D. GluD2 ectodomain structure is not compatible with tetramerization as observed for GluD2 ATD and the GluD2 ATD + Cbln complex (Elegheert et al.). GluD2 ATD tetramer (PDB: 5KCA) is shown in surface representation, with each monomer as a different hue of pink to purple, while the GluD2 ectodomain dimer is drawn in cartoon representation. The green-colored LBD severely clashes with an ATD (dark purple) from the tetrameric structure.
E. GluD2 symmetric dimers can be made to form tetramers if one LBD from each dimer moves out of the way.
F. The Nrxn-Cbln-GluD complex (modeled) fits within the synaptic cleft.
Discussion
Ionotropic glutamate receptors (iGluRs) have recently been established as receptors for protein ligands involved in synaptic development (Matsuda et al., 2016; Uemura et al., 2010). iGluR interactions with soluble C1q-domain proteins provide a pathway for trans-synaptic interactions that are crucial for synapse formation and bi-directional synaptic differentiation. iGluR/C1q complexes establish trans-synaptic bridges using Neurexins on the pre-synaptic side, a protein family well-established as synaptic development molecules.
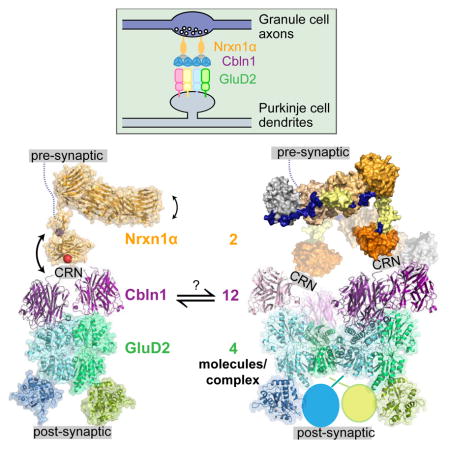
Assembly with Cbln and GluD may dimerize Neurexin
The work we present here, alongside other studies, have established that one Cerebellin hexamer can (1) recruit one α- or β-Neurexin on presynaptic terminals with high affinity, and (2) recruit one GluD dimer on the post-synaptic side, where the affinity may be lower or may need to be regulated by other factors (Elegheert et al., 2016; Lee et al., 2012). Since full-length GluDs are tetramers, this results in a 2 Nrxn:12 Cbln:4 GluD complex, where the pre- and post-synaptic signaling molecules, Nrxn and GluD, may be in their signaling-competent oligomeric states.
The Nrxn(−SS4)-Neuroligin trans-synaptic signaling axis, which can similarly initiate synapse formation in culture and is necessary for synaptic maturation in vivo, serves as a useful analogy to the Nrxn(+SS4)-Cbln-GluD system. In this complex, Neurexin is dimerized by binding the constitutively dimeric Neuroligin, which then leads to differentiation on the presynaptic end. The same dimerization effect is achieved by GluD with Cbln as an intermediary. Similarly, interactions of Neurexin with other cell surface receptors and intracellular scaffolding proteins that can dimerize itself may reinforce the tetrameric, functional form of GluD.
Oligomeric and functional states of GluD
Our structure of the GluD2 extracellular domain shows a novel tetramer-incompatible conformation (Figure 7B and 7C, showing clashes with dimer of dimers for GluA2 and GluK2, respectively). In the GluD2 ectodomain structure, the highly regulated ligand-binding domain is in a “swung-out” monomeric state, where neighboring LBDs do not interact and the ectodomain is dimerized only via the interactions of the ATD. iGluRs have on occasion been observed to have monomeric LBDs, which are attributed to the desensitized state of the channel.
While the GluD2 ectodomain structure likely represents a desensitized state, the ATD-LBD architecture within this structure still does not resemble the known GluK2 (Figure 7C) and GluA2 structures captured in the desensitized state (Dürr et al., 2014; Meyerson et al., 2014). One contributing factor is the linker between the ATD and LBD domains, as a result of which the GluK2 desensitized conformation in Figure 7C may not be accessible to GluDs due to a shorter ATD-LBD linker. Another possible factor is that our structure is in an apo state, and it is conceivable that ligands may cause GluD2 to switch to previously known conformational and functional states. Contrary to this idea, however, the ligand-bound form of the isolated GluD2 LBD crystallizes in the rare monomeric form, while the apo LBD is observed as a dimer (Naur et al., 2007). Therefore, it remains to be determined whether GluDs are a special case among iGluRs, where LBD dimers are uniquely prone to break.
Finally, we imagine that the ATD-LBD orientation within dimers observed in the GluD2 ectodomain structure may break its symmetry in the context of the full-length membrane protein. This would result in one subunit preserving the ATD-LBD contacts, and the other swinging in, allowing for LBD dimers and GluD tetramers to form (Figures 7D and 7E), as has been observed in one of the two GluD ATD-only structures (Elegheert et al., 2016). Such a rearrangement would allow for the sampling of the myriad conformations seen in the numerous iGluR structures published to date. With the structural insights gained here, the next step in understanding the GluD conformation-function relationship is to create mutants locked in the “swung-out” state and test functional consequences.
Is the tetrameric state, i.e. avidity, necessary for high-affinity Cerebellin-GluD interactions?
A mechanistic conundrum on soluble GluD-Cbln interactions is the apparent weak affinity between the two proteins. While mid-nM affinities have been reported using surface plasmon resonance (Elegheert et al., 2016; Lee et al., 2012; Matsuda et al., 2010; Uemura et al., 2010), the lack of a stable complex over size-exclusion columns is puzzling. Elegheert et al. has proposed avidity as the major source of higher apparent affinity in the context of the full-length tetrameric receptors, which is likely part of the answer. However, we still could not observe detectable affinities into the micromolar range when GluD2 ATD is tetramerized (Figures 5D and S4). Therefore, we propose that long-distance conformational effects originating from the LBD or the membrane (channel) domain, possibly controlled by small-molecule ligand binding, may play a role in generating high-affinity GluD receptors able to capture the Nrxn-Cbln complex. Another possibility is that Cbln-GluD affinities may be physiologically weak. Once Cbln is bound to Nrxn, it changes from a soluble, secreted ligand to a membrane-bound and highly clustered cell adhesion molecule (CAM) on the presynaptic side. CAMs can function at lower monomeric affinities, and therefore there may no longer be a need for a high-affinity capture of Cbln by the post-synaptic GluD receptors.
Exceptional flexibility in Nrxn-containing trans-synaptic complexes
Despite many advances since the discovery of the Nrxn-Cbln-GluD complex in 2010, signals generated by Nrxn and GluD resulting in synaptic differentiation are still not clear. One theory that has found support in certain classes of synaptic and neural adhesion molecules such as immunoglobulin superfamily proteins (e.g., SYGs) and cadherins (Nagar et al., 1996; Özkan et al., 2014) is receptor rigidity as a crucial component of receptor signaling. However, in both the Nrxn-Neuroligin and Nrxn-Cbln-GluD trans-synaptic complexes, results point towards very pronounced mobility instead, resulting from flexibility in juxtamembrane regions of Nrxns and Neuroligins, flexibility of CRN within Cerebellins, and the conformational plasticity of GluD receptors. While this flexibility likely has structural and functional consequences, it is not yet clear how. One obvious and critical outcome of the flexibility is the ability of the narrow synaptic cleft to accommodate these complexes. The flexibility at the CRN domain of Cblns, the L-shape of the α-Neurexins, and further flexibility within α-Neurexins allow for this complex to fit within the 20–25 nm-wide synaptic cleft (Figure 7F), and sample the extracellular space for other protein ligands of Neurexins (such as GABAAα receptors, Neurexophilins, Dystroglycan, and others).
The work we have detailed here and the recent study by Elegheert et al. (2016) lay the structural framework, namely the molecular determinants of interactions and conformational flexibility within the complex, and poise the field for future advances. Prospective studies will have to explain the functional relevance of conformational plasticity within the complex including the membrane domain of GluDs, as well as how the complex is regulated by small molecules at the synaptic cleft, and how the formation of the complex leads to bidirectional signaling. We expect such studies to also explain the apparent low affinity of Cblns for GluDs, and possibly elucidate the elusive channel activity of GluDs.
Experimental Procedures
Expression and purification of Cbln1, β- and α-Neurexin-1 and GluD2
Rat Cbln1, Nrxn1α, Nrxn1β and GluD2 constructs with C-terminal hexahistidine tags were expressed in lepidopteran High Five cells using baculoviruses. All proteins were secreted into expression media (Insect-XPRESS, Lonza) using the native signal peptide for Cbln1 or baculoviral gp64 leader sequence for others. Proteins were purified with immobilized metal-affinity chromatography (Ni2+-NTA Agarose resin, QIAGEN), followed by size-exclusion chromatography (Superdex 200 Increase and/or Superose 6 Increase 10/300 columns, GE Healthcare) in 10 mM HEPES, pH 7.2, 150 mM NaCl.
Isothermal titration calorimetry
Calorimetry experiments were performed using a MicroCal iTC200 (Malvern Instruments) at 25°C. Nrxn1α binding to Cbln1 was measured three times due to lower levels of signal/noise in measurements, caused by lower protein concentrations used during the experiment, since higher concentrations resulted in protein precipitation and large spikes in the heat curves. The number reported in the text is the average and sample standard deviation of these three measurements.
Multi-angle light scattering for β-Neurexin + Cerebellin
Molecular sizes for Cbln and Cbln-Nrxn were determined using a setup combining size-exclusion chromatography (Superdex 200 Increase 10/300 column), a multi-angle light scattering instrument (DAWN Heleos, Wyatt), and a refractometer (Optilab rEX, Wyatt Technology), run by an ÄKTA FPLC (GE Healthcare). Refractive index measurements were used for concentration measurements. dn/dc values of 0.180 and 0.181, corrected for the presence of N-linked glycan groups, were used for Cbln1 and Cbln1-Nrxn1 mass measurements, respectively.
X-ray crystallography of Cerebellin-1 and GluD2
Cbln1 crystals can be grown in high-molecular weight polyethylene glycol solutions or in carboxylic organic salts such as tartrate and formate. Best crystals were grown from Cbln1 treated with Carboxypeptidase A and B to remove the hexahistidine tag in a solution of 0.1 M Tris, pH 7.5, 3 M Sodium formate at 21°C. Diffraction data was reduced with XDS (Kabsch, 2010). GluD2 ectodomain crystals were grown in 0.1 M Sodium cacodylate, pH 6.6, 1.3 M Ammonium dihydrogen phosphate at 21°C, and diffraction data was processed with HKL-2000 using automatic corrections (Otwinowski and Minor, 1997). The correct space group for GluD2 crystals (P3221) could only be determined by a systematic molecular replacement search in all possible crystallographic subgroups of the apparent space group (P62/422) due to twinning. Only monomers of the ATD and LBD domains were used for molecular replacement to avoid biases towards any dimeric conformation. Both structures were solved with molecular replacement using PHASER, followed by model refinement with phenix.refine and model building in Coot (Afonine et al., 2012; Emsley et al., 2010; McCoy et al., 2007). Model validation was performed using Molprobity tools within the PHENIX suite (Adams et al., 2010; Chen et al., 2010). All structural figures were drawn in PyMOL (Schrödinger, LLC). The atomic coordinates and structure factors have been deposited in the Protein Data Bank (http://wwpdb.org/) with the PDB codes 5KWR and 5L2E.
Negative-stain electron microscopy of Cerebellin-1 and Neurexin
All protein samples were prepared for negative stain EM as described previously (Peisley and Skiniotis, 2015). Images were recorded at room temperature with a Tecnai T12 transmission electron microscope operated at 120 kV on a Gatan US4000 CCD camera at a magnification of ×71,138 and a defocus value of ~1.5 μm. 3539, 5453 and 9200 particle projections for Cbln1, Cbln1+Nrxn1β and Cbln1+Nrxn1α, respectively, were subjected to two-dimensional reference-free alignment and classification using ISAC (Yang et al., 2012).
Supplementary Material
Highlights.
Secreted Cerebellin and presynaptic Neurexin form highly flexible complex.
Cerebellin-Neurexin complex is high affinity with a 6:1 stoichiometry.
Postsynaptic Glutamate receptor-delta2 (GluD2) binds Cerebellin with low affinity.
GluD2 ectodomain is dimeric and can adopt a novel, desensitized conformational state.
Acknowledgments
We thank Drs. Michael Birnbaum, Demet Arac and Eduardo Perozo for reading and providing feedback on the manuscript. We also thank Dr. Tian Li for help with MALS equipment, and Jeffrey Tarrasch and Agnieszka Olechwier for technical assistance. We are grateful to Dr. Michael Hollmann at Ruhr-Universität Bochum, Germany for the cDNAs of GluD1 and GluD2. We acknowledge Dr. Elena Solomaha and the University of Chicago BioPhysics Core Facilities for training with and access to ITC. This work was supported in part by National Institutes of Health Grants R01 NS097161 (to E. Ö.) and R01 DK090165 (to G. S.), Klingenstein-Simons Fellowship Award in the Neurosciences (to E. Ö.), and a Core Facility Grant from the University of Chicago Institute of Translational Medicine (to E. Ö.), which was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health through Grant Number UL1 TR000430. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. We thank GM/CA@APS, which has been funded in whole or in part with Federal funds from the National Cancer Institute (ACB-12002) and the National Institute of General Medical Sciences (AGM-12006).
Footnotes
Author Contributions
S. C., G. S. and E. Ö. conceived and coordinated the study. S. C. and E. Ö. wrote the paper with contributions from all authors. S. C. expressed, purified and crystallized all proteins, and S. C. and E. Ö. solved crystal structures and performed light scattering experiments. A. B. S. and G. S. performed electron microscopy experiments and analyzed data. J. W. and S. C. performed calorimetry experiments. All authors reviewed the results and approved the final version of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix. refine Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araç D, Boucard AA, Özkan E, Strop P, Newell E, Südhof TC, Brunger AT. Structures of neuroligin-1 and the neuroligin-1/neurexin-1 beta complex reveal specific protein-protein and protein-Ca2+ interactions. Neuron. 2007;56:992–1003. doi: 10.1016/j.neuron.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- Bao D, Pang Z, Morgan JI. The structure and proteolytic processing of Cbln1 complexes. J Neurochem. 2005;95:618–629. doi: 10.1111/j.1471-4159.2005.03385.x. [DOI] [PubMed] [Google Scholar]
- Brunger AT, DeLaBarre B, Davies JM, Weis WI. X-ray structure determination at low resolution. Acta Crystallogr D Biol Crystallogr. 2009;65:128–133. doi: 10.1107/S0907444908043795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Venugopal V, Murray B, Rudenko G. The structure of neurexin 1α reveals features promoting a role as synaptic organizer. Structure. 2011;19:779–789. doi: 10.1016/j.str.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Liu H, Shim AHR, Focia PJ, He X. Structural basis for synaptic adhesion mediated by neuroligin-neurexin interactions. Nat Struct Mol Biol. 2008;15:50–56. doi: 10.1038/nsmb1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comoletti D, Flynn RE, Boucard AA, Demeler B, Schirf V, Shi J, Jennings LL, Newlin HR, Südhof TC, Taylor P. Gene selection, alternative splicing, and post-translational processing regulate neuroligin selectivity for beta-neurexins. Biochemistry. 2006;45:12816–12827. doi: 10.1021/bi0614131. [DOI] [PubMed] [Google Scholar]
- Comoletti D, Miller MT, Jeffries CM, Wilson J, Demeler B, Taylor P, Trewhella J, Nakagawa T. The macromolecular architecture of extracellular domain of alphaNRXN1: domain organization, flexibility, and insights into trans-synaptic disposition. Structure. 2010;18:1044–1053. doi: 10.1016/j.str.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr KL, Chen L, Stein RA, De Zorzi R, Folea IM, Walz T, Mchaourab HS, Gouaux E. Structure and dynamics of AMPA receptor GluA2 in resting, pre-open, and desensitized states. Cell. 2014;158:778–792. doi: 10.1016/j.cell.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elegheert J, Kakegawa W, Clay JE, Shanks NF, Behiels E, Matsuda K, Kohda K, Miura E, Rossmann M, Mitakidis N, Motohashi J, Chang VT, Siebold C, Greger IH, Nakagawa T, Yuzaki M, Aricescu AR. Structural basis for integration of GluD receptors within synaptic organizer complexes. Science. 2016;353:295–299. doi: 10.1126/science.aae0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrichny IP, Leone P, Sulzenbacher G, Comoletti D, Miller MT, Taylor P, Bourne Y, Marchot P. Structural analysis of the synaptic protein neuroligin and its beta-neurexin complex: determinants for folding and cell adhesion. Neuron. 2007;56:979–991. doi: 10.1016/j.neuron.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuccillo MV, Földy C, Gökce Ö, Rothwell PE, Sun GL, Malenka RC, Südhof TC. Single-Cell mRNA Profiling Reveals Cell-Type-Specific Expression of Neurexin Isoforms. Neuron. 2015;87:326–340. doi: 10.1016/j.neuron.2015.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell. 2004;119:1013–1026. doi: 10.1016/j.cell.2004.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbury PB, Zhang T, Kim PS, Alber T. A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants. Science. 1993;262:1401–1407. doi: 10.1126/science.8248779. [DOI] [PubMed] [Google Scholar]
- Hirai H, Pang Z, Bao D, Miyazaki T, Li L, Miura E, Parris J, Rong Y, Watanabe M, Yuzaki M, Morgan JI. Cbln1 is essential for synaptic integrity and plasticity in the cerebellum. Nat Neurosci. 2005;8:1534–1541. doi: 10.1038/nn1576. [DOI] [PubMed] [Google Scholar]
- Ichtchenko K, Hata Y, Nguyen T, Ullrich B, Missler M, Moomaw C, Südhof TC. Neuroligin 1: a splice site-specific ligand for beta-neurexins. Cell. 1995;81:435–443. doi: 10.1016/0092-8674(95)90396-8. [DOI] [PubMed] [Google Scholar]
- Joo JY, Lee SJ, Uemura T, Yoshida T, Yasumura M, Watanabe M, Mishina M. Differential interactions of cerebellin precursor protein (Cbln) subtypes and neurexin variants for synapse formation of cortical neurons. Biochem Biophys Res Commun. 2011;406:627–632. doi: 10.1016/j.bbrc.2011.02.108. [DOI] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakegawa W, Mitakidis N, Miura E, Abe M, Matsuda K, Takeo YH, Kohda K, Motohashi J, Takahashi A, Nagao S, Muramatsu S, Watanabe M, Sakimura K, Aricescu AR, Yuzaki M. Anterograde C1ql1 signaling is required in order to determine and maintain a single-winner climbing fiber in the mouse cerebellum. Neuron. 2015;85:316–329. doi: 10.1016/j.neuron.2014.12.020. [DOI] [PubMed] [Google Scholar]
- Karakas E, Regan MC, Furukawa H. Emerging structural insights into the function of ionotropic glutamate receptors. Trends Biochem Sci. 2015;40:328–337. doi: 10.1016/j.tibs.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko J, Zhang C, Arac D, Boucard AA, Brunger AT, Südhof TC. Neuroligin-1 performs neurexin-dependent and neurexin-independent functions in synapse validation. EMBO J. 2009;28:3244–3255. doi: 10.1038/emboj.2009.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Uemura T, Yoshida T, Mishina M. GluRδ2 assembles four neurexins into trans-synaptic triad to trigger synapse formation. J Neurosci. 2012;32:4688–4701. doi: 10.1523/JNEUROSCI.5584-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Budisantoso T, Mitakidis N, Sugaya Y, Miura E, Kakegawa W, Yamasaki M, Konno K, Uchigashima M, Abe M, Watanabe I, Kano M, Watanabe M, Sakimura K, Aricescu AR, Yuzaki M. Transsynaptic Modulation of Kainate Receptor Functions by C1q-like Proteins. Neuron. 2016;90:752–767. doi: 10.1016/j.neuron.2016.04.001. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Miura E, Miyazaki T, Kakegawa W, Emi K, Narumi S, Fukazawa Y, Ito-Ishida A, Kondo T, Shigemoto R, Watanabe M, Yuzaki M. Cbln1 is a ligand for an orphan glutamate receptor delta2, a bidirectional synapse organizer. Science. 2010;328:363–368. doi: 10.1126/science.1185152. [DOI] [PubMed] [Google Scholar]
- Matsuda K, Yuzaki M. Cbln family proteins promote synapse formation by regulating distinct neurexin signaling pathways in various brain regions. Eur J Neurosci. 2011;33:1447–1461. doi: 10.1111/j.1460-9568.2011.07638.x. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S. Structural mechanism of glutamate receptor activation and desensitization. Nature. 2014;514:328–334. doi: 10.1038/nature13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H, Jia Y, Xie S, Wang X, Zhao J, Chu Y, Zhou Z, Shi Z, Song X, Li L. Structural insights into the C1q domain of Caprin-2 in canonical Wnt signaling. J Biol Chem. 2014;289:34104–34113. doi: 10.1074/jbc.M114.591636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MT, Mileni M, Comoletti D, Stevens RC, Harel M, Taylor P. The crystal structure of the α-neurexin-1 extracellular region reveals a hinge point for mediating synaptic adhesion and function. Structure. 2011;19:767–778. doi: 10.1016/j.str.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugnaini E, Dahl AL, Morgan JI. Cerebellin is a postsynaptic neuropeptide. Synapse. 1988;2:125–138. doi: 10.1002/syn.890020204. [DOI] [PubMed] [Google Scholar]
- Nagar B, Overduin M, Ikura M, Rini JM. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 1996;380:360–364. doi: 10.1038/380360a0. [DOI] [PubMed] [Google Scholar]
- Naur P, Hansen KB, Kristensen AS, Dravid SM, Pickering DS, Olsen L, Vestergaard B, Egebjerg J, Gajhede M, Traynelis SF, Kastrup JS. Ionotropic glutamate-like receptor delta2 binds D-serine and glycine. Proc Natl Acad Sci USA. 2007;104:14116–14121. doi: 10.1073/pnas.0703718104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. [20] Processing of X-ray diffraction data collected in oscillation mode. In: Carter JCW, Sweet RM, editors. Macromolecular Crystallography, Part A, Methods in Enzymology. Academic Press; New York: 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Özkan E, Chia PH, Wang RR, Goriatcheva N, Borek D, Otwinowski Z, Walz T, Shen K, Garcia KC. Extracellular Architecture of the SYG-1/SYG-2 Adhesion Complex Instructs Synaptogenesis. Cell. 2014;156:482–494. doi: 10.1016/j.cell.2014.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peisley A, Skiniotis G. 2D Projection Analysis of GPCR Complexes by Negative Stain Electron Microscopy. Methods Mol Biol. 2015;1335:29–38. doi: 10.1007/978-1-4939-2914-6_3. [DOI] [PubMed] [Google Scholar]
- Reissner C, Runkel F, Missler M. Neurexins. Genome Biol. 2013;14:213. doi: 10.1186/gb-2013-14-9-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressl S, Vu BK, Vivona S, Martinelli DC, Südhof TC, Brunger AT. Structures of C1q-like Proteins Reveal Unique Features among the C1q/TNF Superfamily. Structure. 2015 doi: 10.1016/j.str.2015.01.019. [DOI] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- Schreiner D, Nguyen TM, Russo G, Heber S, Patrignani A, Ahrné E, Scheiffele P. Targeted combinatorial alternative splicing generates brain region-specific repertoires of neurexins. Neuron. 2014;84:386–398. doi: 10.1016/j.neuron.2014.09.011. [DOI] [PubMed] [Google Scholar]
- Shen K, Scheiffele P. Genetics and cell biology of building specific synaptic connectivity. Annu Rev Neurosci. 2010;33:473–507. doi: 10.1146/annurev.neuro.051508.135302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slemmon JR, Blacher R, Danho W, Hempstead JL, Morgan JI. Isolation and sequencing of two cerebellum-specific peptides. Proc Natl Acad Sci USA. 1984;81:6866–6870. doi: 10.1073/pnas.81.21.6866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455:903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein B, Gokce O, Quake SR, Südhof TC. Cartography of neurexin alternative splicing mapped by single-molecule long-read mRNA sequencing. Proc Natl Acad Sci USA. 2014;111:E1291–1299. doi: 10.1073/pnas.1403244111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura T, Lee SJ, Yasumura M, Takeuchi T, Yoshida T, Ra M, Taguchi R, Sakimura K, Mishina M. Trans-synaptic interaction of GluRdelta2 and Neurexin through Cbln1 mediates synapse formation in the cerebellum. Cell. 2010;141:1068–1079. doi: 10.1016/j.cell.2010.04.035. [DOI] [PubMed] [Google Scholar]
- Ullrich B, Ushkaryov YA, Südhof TC. Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14:497–507. doi: 10.1016/0896-6273(95)90306-2. [DOI] [PubMed] [Google Scholar]
- Urade Y, Oberdick J, Molinar-Rode R, Morgan JI. Precerebellin is a cerebellum-specific protein with similarity to the globular domain of complement C1q B chain. Proc Natl Acad Sci USA. 1991;88:1069–1073. doi: 10.1073/pnas.88.3.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushkaryov YA, Petrenko AG, Geppert M, Südhof TC. Neurexins: synaptic cell surface proteins related to the alpha-latrotoxin receptor and laminin. Science. 1992;257:50–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- Yang Z, Fang J, Chittuluru J, Asturias FJ, Penczek PA. Iterative stable alignment and clustering of 2D transmission electron microscope images. Structure. 2012;20:237–247. doi: 10.1016/j.str.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipursky SL, Sanes JR. Chemoaffinity revisited: dscams, protocadherins, and neural circuit assembly. Cell. 2010;143:343–353. doi: 10.1016/j.cell.2010.10.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.