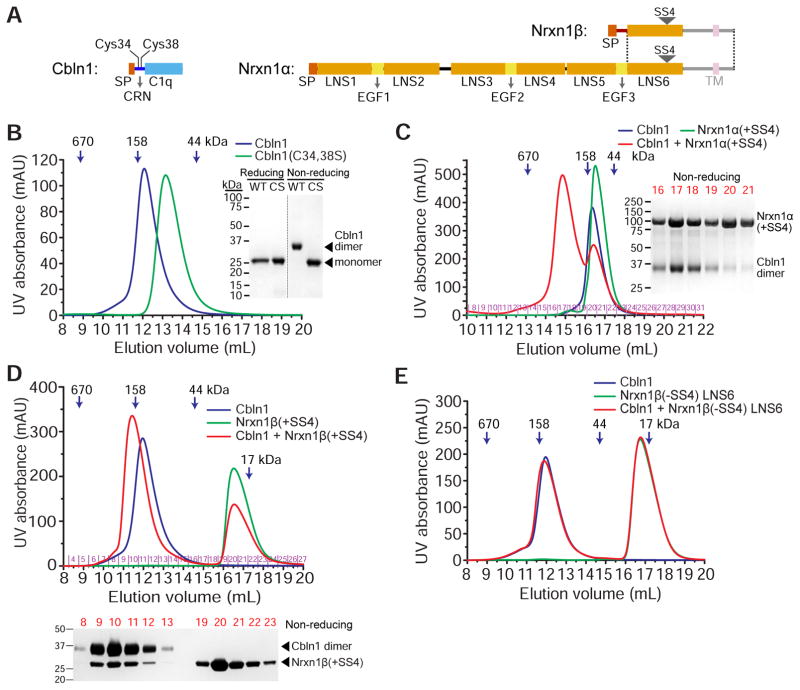
Figure 1. Rat Cbln1 hexamers with intact CRN domains bind rat α- or β-Nrxn1 including SS4.
A. Domain structure of Cbln1, Nrxn1α and Nrx1β, drawn to scale. The shaded regions, the transmembrane (TM) helix and the unstructured juxtamembrane domains (intracellular and extracellular), are excluded from our constructs. The dotted lines mark the boundaries of the region expressed by exons shared by α and β-Neurexins. SP: Signal peptide.
B. Cbln1 (blue curve) runs as hexamers on a Superdex 200 size-exclusion column when its cysteines in the CRN domain are intact. C34,38S (CS) double mutation causes it to run as trimers (green curve). Wild-type (WT) Cbln1 runs as a dimer on non-reducing denaturing gels, while CS runs as a monomer.
C. Cbln1 binds Nrxn1α(+SS4) domains LNS2 to LNS6, as observed on a Superose 6 size-exclusion column. Size-exclusion fractions for the Cbln1+Nrxn1α(+SS4) sample (red curve) are run on a non-reducing gel.
D. Cbln1 binds Nrxn1β(+SS4) as observed on a Superdex 200 size-exclusion column. Nrxn1β construct contains residues S48 to P292, including the β form-unique N-terminal region and the LNS6 domain. Size-exclusion fractions for the Cbln1+Nrxn1β(+SS4) sample (red curve) are run on a non-reducing gel. For Cbln1 binding to an LNS6-only Nrxn1 construct, see Figure S1A.
E. Cbln1 does not bind Nrxn1β-LNS6 without SS4 (−SS4).
See also Figure S1.