Figure 7. GluD2 dimers are in a conformation not observed before in iGluR proteins.
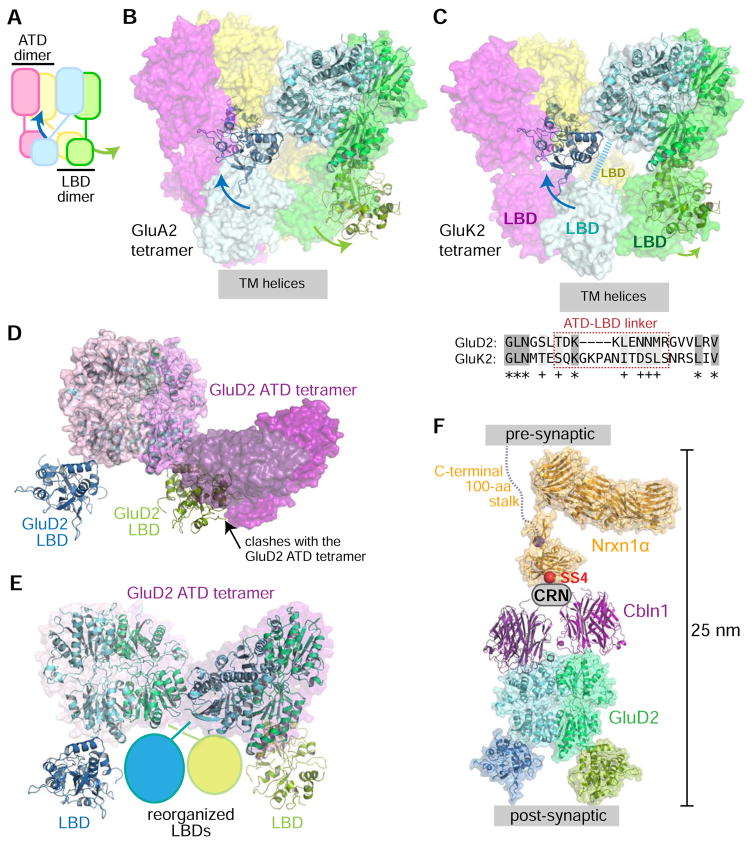
A. Schematic representation of iGluR tetramers, with the swing-out motion in the GluD2 ectodomain structure mapped on top. The swinging-out of LBDs in the GluD2 ectodomain dimer structure are shown with arrows.
B. GluD2 dimer superposed on GluA2 antagonist bound structure (PDB: 3KG2).
C. GluD2 dimer superposed on the desensitized state structure of GluK2 (PDB: 4UQQ). The shorter ATD-LBD linker in GluD2 compared to GluK2 does not allow for GluDs to adopt a similar desensitized state structure. The ATD-LBD linker in the cyan subunit of GluK2 was not resolved in the structure, and is depicted as a thick dashed line.
D. GluD2 ectodomain structure is not compatible with tetramerization as observed for GluD2 ATD and the GluD2 ATD + Cbln complex (Elegheert et al.). GluD2 ATD tetramer (PDB: 5KCA) is shown in surface representation, with each monomer as a different hue of pink to purple, while the GluD2 ectodomain dimer is drawn in cartoon representation. The green-colored LBD severely clashes with an ATD (dark purple) from the tetrameric structure.
E. GluD2 symmetric dimers can be made to form tetramers if one LBD from each dimer moves out of the way.
F. The Nrxn-Cbln-GluD complex (modeled) fits within the synaptic cleft.