Abstract
The current clinical practice is based on the assumption of strong correlation between diffuse glutamine synthetase expression and β-catenin activation in hepatocellular adenoma and hepatocellular carcinoma. This high correlation is based on limited data, and may represent an oversimplification as glutamine synthetase staining patterns show wide variability in clinical practice. Standardized criteria for interpreting diverse glutamine synthetase patterns, and the association between each pattern and β-catenin mutations is not clearly established. This study examines the correlation between glutamine synthetase staining patterns and β-catenin mutations in 15 typical hepatocellular adenomas, 5 atypical hepatocellular neoplasms and 60 hepatocellular carcinomas. Glutamine synthetase staining was classified into one of three patterns: (a) diffuse homogeneous: moderate to strong cytoplasmic staining in more than 90% of lesional cells, without a map-like pattern, (b) diffuse heterogeneous: moderate to strong staining in 50–90% of lesional cells, without a map-like pattern, and (c) patchy: moderate to strong staining in <50% of lesional cells (often perivascular), or weak staining irrespective of extent, and all other staining patterns (including negative cases). Sanger sequencing of CTNNB1 exon 3 was performed in all cases. Of hepatocellular tumors with diffuse glutamine synthetase staining (homogeneous or heterogeneous), an exon 3 β-catenin mutation was detected in 33% (2/6) of typical hepatocellular adenoma, 75% (3/4) of atypical hepatocellular neoplasm and 17% (8/47) of hepatocellular carcinomas. An exon 3 mutation was also observed in 15% (2/13) of hepatocellular carcinomas with patchy glutamine synthetase staining. The results show a modest correlation between diffuse glutamine synthetase immunostaining and exon 3 β-catenin mutations in hepatocellular adenoma and hepatocellular carcinoma with discrepancy rates exceeding 50% in both hepatocellular adenoma and hepatocellular carcinoma. The interpretation of β-catenin activation based on glutamine synthetase staining should be done with caution, and the undetermined significance of various glutamine synthetase patterns should be highlighted in pathology reports.
Introduction
β-catenin activation in hepatocellular tumors is generally due to missense mutation or small in-frame insertion or deletions in exon 3 of the CTNNB1 gene, which results in cytoplasmic and nuclear accumulation of β-catenin. Nuclear β-catenin accumulation leads to transcriptional activation of several downstream target genes, one of which is GLUL, which codes for glutamine synthetase (1, 2). In normal liver, glutamine synthetase expression is confined to hepatocytes surrounding terminal hepatic venules (3), whereas the majority of neoplastic hepatocytes express glutamine synthetase in β-catenin activated tumors (4). Nuclear β-catenin staining is often focal in hepatocellular adenoma and is an unreliable marker in detecting β-catenin activation, whereas diffuse glutamine synthetase staining has greater sensitivity and better correlation with β-catenin activation (4).
The 2010 World Health Organization classification subdivides hepatocellular adenomas into four categories: β-catenin activated, hepatocyte nuclear factor 1α inactivated, inflammatory, and unclassified (4–6). The β-catenin activated hepatocellular adenoma subset poses a major diagnostic challenge as these tumors often show overlapping features with well-differentiated hepatocellular carcinoma and are frequently associated with hepatocellular carcinoma at the time of initial diagnosis or on follow-up (4, 7, 8). In initial studies, the β-catenin activated subtype accounted for 10–15% of total hepatocellular adenomas (4, 6, 7).
Aberrant activation of the Wnt signaling pathway also occurs in hepatocellular carcinoma with a frequency varying from 8–42% (9–17). Hepatocellular carcinoma with β-catenin mutations are typically more well-differentiated, occur in non-cirrhotic liver, infrequently have TP53 mutations, have a lower level of chromosomal instability and a relatively better prognosis (15, 18, 19). A higher association of β-catenin mutations with hepatocellular carcinoma arising in hepatitis has also been reported (14, 20).
It has been proposed that hepatocellular neoplasms with diffuse glutamine synthetase expression are β-catenin activated, irrespective of whether nuclear β-catenin staining is present (4), and this has become standard practice in the diagnosis of hepatocellular tumors. However, the proposed strong correlation between diffuse glutamine synthetase staining and β-catenin activation is based on a limited number of studies (4, 21, 22), likely represents an oversimplification of a more complex relationship. Further, the immunohistochemical patterns of glutamine synthetase expression show wide variability leading to interobserver variability, even among experienced pathologists (23). Standardized criteria for interpretation of these diverse patterns and the relationship of each pattern with β-catenin mutations have not been established. The current study examines the correlation between different staining patterns of glutamine synthetase and β-catenin mutations in hepatocellular adenoma and hepatocellular carcinoma.
Materials and Methods
Study cases
The institutional review board of the University of California, San Francisco, approved this study. Study cases were selected from the pathology files of the University of California, San Francisco and from the Southern California Permanente Medical Group. The majority of the hepatocellular adenoma and atypical hepatocellular neoplasm cases included in this study had previously been classified by immunohistochemistry according to the 2010 World Health Organization guidelines (24). Case selection was aimed at including cases with a range of glutamine synthetase staining patterns. Hematoxylin and eosin stained sections and reticulin stains were reviewed in all cases, which were assigned to the categories enumerated below:
Typical hepatocellular adenoma (n=15; 12 resections, 3 biopsies): Tumors that occurred in women 15–50 years of age and showed typical histologic features of hepatocellular adenoma with no morphologic atypia.
Atypical hepatocellular neoplasm (n=5; 4 resections, 1 biopsy): Tumors that occurred in men (any age), women >50 years and/or showed focal atypical morphological features (<5% of the tumor) such as small cell change, pseudoacinar architecture and thick cell plates that were insufficient for definite diagnosis of hepatocellular carcinoma (25).
Hepatocellular carcinoma (n=60; 59 resections, 1 biopsy): Tumors with obvious cytoarchitectural features of hepatocellular carcinoma such as thick cell plates, small cell change and reticulin loss in most areas of the tumor.
Hepatocellular carcinomas were graded as well, moderate and poorly differentiated tumors according to the 2010 World Health Organization classification (26), and clinical data on background liver disease and degree of fibrosis was obtained for each case.
Immunohistochemistry
Using formalin-fixed paraffin-embedded tissue, immunohistochemistry was performed for β-catenin and glutamine synthetase on all hepatocellular carcinomas and hepatocellular lesions. Serum amyloid A and liver fatty acid binding protein had previously been performed on all hepatocellular adenomas and atypical hepatocellular neoplasms as previously described (24). The sections obtained on Superfrost Plus slides were heated at 65–70°C for 60 minutes, deparaffinized and hydrated to distilled water. Slides were washed in running distilled water, and then incubated with blocking solution (3% hydrogen peroxide, 0.05% Tween-20, 0.1% sodium azide in calcium-magnesium-free phosphate-buffered saline) for 10 minutes. Following two washes in deionized water and washing with 0.2% Tween-20 in phosphate-buffered saline, antigen retrieval was performed by incubating slides with the ENVISION+ system (DAKO) for 15 minutes (β-catenin, glutamine synthetase), 30 minutes (serum amyloid A) or by using Bond Epitope Retrieval (Leica) for 20 minutes (liver fatty acid binding protein, C-reactive protein), followed by incubation with the primary antibodies for 30 minutes at room temperature. The slides were washed again with 0.2% Tween-20 in phosphate-buffered saline, incubated for 10 minutes at room temperature in DAB+ liquid (DAKO), and washed in water. Slides were counterstained with hematoxylin for 10 seconds, washed in tap water, incubated in phosphate-buffered saline for 30 seconds, washed in tap water, dehydrated in graded ethanols, cleared in xylene, and cover-slipped.
Interpretation of immunohistochemistry
In all tumors, the glutamine synthetase staining pattern was classified into one of the following three patterns: (a) diffuse homogeneous: moderate to strong cytoplasmic staining in more than 90% of lesional cells, without a map-like pattern, (b) diffuse heterogeneous: moderate to strong staining in 50–90% of lesional cells, without a map-like pattern, and (c) patchy: moderate to strong staining in <50% of lesional cells (often in a perivascular distribution), or weak staining irrespective of extent, and all other staining patterns (including negative cases). Hepatocellular adenomas with diffuse homogeneous or heterogeneous glutamine synthetase staining were considered β-catenin activated, irrespective of mutation status or nuclear β-catenin immunohistochemical staining as per 2010 World Health Organization recommendations and studies by Bioulac-Sage et al (4, 6). Based on immunohistochemistry that has been performed as part of the diagnostic work-up, hepatocellular adenomas with loss of liver fatty acid binding protein were considered as hepatocyte nuclear factor 1α inactivated, while adenomas were classified as inflammatory when serum amyloid A and/or C-reactive protein was positive – even in the absence of typical inflammatory adenoma morphologic features (i.e., inflammation and sinusoidal dilatation). Nuclear β-catenin immunohistochemical staining was interpreted as positive irrespective of number of positive cells. Membranous and/or cytoplasmic staining was not considered positive.
DNA extraction, polymerase chain reaction and sequencing analysis
Sanger sequencing analysis of exon 3 of the CTNNB1 gene was performed in all cases. Formalin-fixed paraffin-embedded tumor tissue was scraped from unstained sections and deparaffinized in xylene. Then genomic DNA was purified using the QIAamp DNA FFPE Tissue Kit (Cat. No. 56404), according to the manufacturer’s instructions (Qiagen, Valencia, CA, USA).
Exon 3 of CTNNB1 was amplified using the following primers: forward 5’-TCCAATCTACTAATGCTAATACTGTTTCGTA-3’; reverse 5’-CATTCTGACTTTCAGTAAGGCAATG-3’ (27). Approximately 50 ng of genomic DNA from tumor samples was denatured at 95°C for 5 min and then cycled 40× at 95°C for 30 seconds, 62°C for 45 seconds, 72°C for 45 seconds, with a final extension at 72°C for 20 min. Sanger sequencing of polymerase chain reaction products was performed using BigDye v3.1 (Applied Biosystems, Foster City, CA, USA) and the same primers used for amplification. Sequencing reactions were run on an ABI Prism 310 Genetic Analyzer (Applied Biosystems). Traces were analyzed using Mutation Surveyor (Softgenetics, State College, PA, USA) with NM_001098209 as a reference.
In select cases where a small insertion/deletion was suspected based on this analysis, polymerase chain reaction products were purified and cloned into a pDrive cloning vector (Qiagen polymerase chain reaction Cloning Kit, Cat. No. 23112) according to the manufacturer’s protocol. An individual clone was isolated, and the insert was subsequently sequenced using an M13F primer.
Results
Clinical features
The clinical features of hepatocellular adenoma and atypical hepatocellular neoplasm are summarized in Table 1. A history of oral contraceptive use was noted in 2 hepatocellular adenoma cases. Each hepatocellular adenoma and atypical hepatocellular neoplasm was further subclassified based on the 2010 World Health Organization classification (6), as previously described (Table 1) (24). Of the 60 hepatocellular carcinomas, there were 16 well-differentiated, 32 moderately differentiated, and 12 poorly differentiated cases. A background of cirrhosis was present in 31 (52%) hepatocellular carcinoma cases. The underlying etiology for cirrhosis was hepatitis C (n=19), hepatitis B (n=5) and non-alcoholic steatohepatitis (n=2); the etiology was not known in 5 cases. Of the hepatocellular carcinomas in noncirrhotic liver, 4 patients had hepatitis C and 3 patients had hepatitis B.
Table 1.
Clinicopathologic characteristics of typical hepatocellular adenomas and atypical hepatocellular neoplasms.
| Diagnosis | Female gender (%) |
Mean Age (range) |
Subtype |
|---|---|---|---|
| Typical hepatocellular adenoma (n=15) |
15 (100) | 38 (23–46) | Inflammatory (n=2) Inflammatory with β-catenin activation (n=3) Hepatocyte nuclear factor1α inactivated (n=4) Hepatocyte nuclear factor1αinactivated with β-catenin activation (n=1) β-catenin activated (n=2) Unclassified (n=3) |
| Atypical hepatocellular neoplasm(n=5) |
2 (40) | 33 (13–53) | Inflammatory with β-catenin activation(n=3) Hepatocyte nuclear factor1α inactivated with β-catenin activation (n=1) Unclassified (n=1) |
Glutamine synthetase and β-catenin staining
Diffuse glutamine synthetase staining (homogeneous or heterogeneous) was seen in 6/15 (40%) of typical hepatocellular adenoma cases and therefore all of these cases were considered β-catenin activated. Four hepatocellular adenomas had diffuse heterogeneous glutamine synthetase staining: inflammatory hepatocellular adenoma with β-catenin activation (n=1), hepatocyte nuclear factor 1α inactivated with β-catenin activation (n=1), β-catenin activated hepatocellular adenoma without expression or loss of other adenoma markers (n=2). The two hepatocellular adenomas with diffuse homogeneous staining also expressed inflammatory markers. Nuclear β-catenin staining was present in 2/15 (13%) typical hepatocellular adenoma cases, both of which showed diffuse heterogeneous glutamine synthetase staining.
Among atypical hepatocellular neoplasms, diffuse glutamine synthetase staining was seen in 4/5 (80%) cases (1 homogeneous, 3 heterogeneous). None of the atypical hepatocellular neoplasm cases showed nuclear β-catenin staining.
Among hepatocellular carcinomas, diffuse glutamine synthetase staining was seen in 47/60 (78%) cases (19 diffuse homogeneous, 28 diffuse heterogeneous). Nuclear β-catenin staining was identified in 18 (38%) hepatocellular carcinomas, all of which showed diffuse glutamine synthetase staining (14 diffuse homogeneous, 4 diffuse heterogeneous).
β-catenin exon 3 mutation
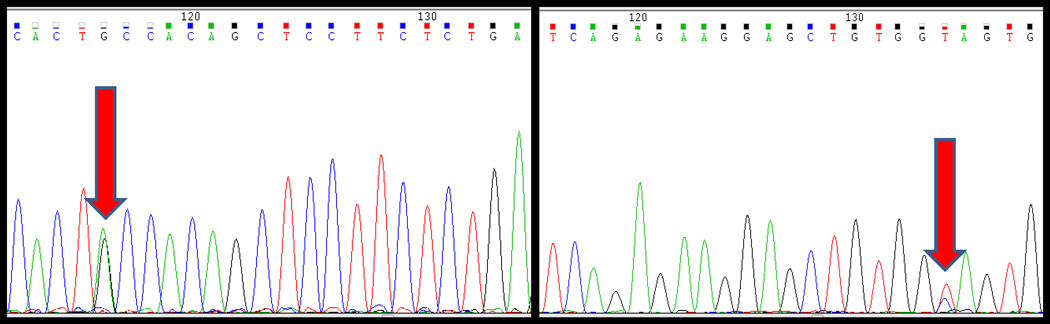
Exon 3 β-catenin mutations (Table 2) were detected in 2/15 (13%) hepatocellular adenomas; both mutations were T41A missense mutations. Of the 5 atypical hepatocellular neoplasms, 3 (60%) showed β-catenin mutations: two with T41A missense mutation (Figure 1), and one with a heterozygous in-frame insertion of AAT at codon 35.
Table 2.
Clinical and immunohistochemical characteristics of cases with a β-catenin mutation.
| Diagnosis | Age/gender | Procedure | β-catenin mutation |
GS staining pattern | β-catenin staining pattern |
|---|---|---|---|---|---|
| Hepatocellular adenoma, inflammatory with β-catenin activation |
33/F | Resection | T41A | Diffuse homogeneous | Nuclear |
| Hepatocellular adenoma, inflammatory with β-catenin activation |
32/F | Resection | T41A | Diffuse heterogeneous | Membranous |
| Atypical hepatocellular neoplasm | 26/M | Biopsy | T41A | Diffuse homogeneous | Membranous |
| Atypical hepatocellular neoplasm | 37/M | Resection | S35insN | Diffuse heterogeneous | Membranous |
| Atypical hepatocellular neoplasm | 36/M | Resection | T41A | Diffuse heterogeneous | Membranous |
| Hepatocellular carcinoma, non-cirrhotic |
71/M | Resection | S45P | Diffuse homogeneous | Nuclear |
| Hepatocellular carcinoma, cirrhotic | 65/M | Resection | S33C | Diffuse homogeneous | Nuclear |
| Hepatocellular carcinoma, cirrhotic | 59/M | Resection | T41A | Diffuse homogeneous | Nuclear |
| Hepatocellular carcinoma, cirrhotic | 63/M | Resection | T41A | Diffuse homogeneous | Nuclear |
| Hepatocellular carcinoma, non-cirrhotic |
36/M | Resection | T41A | Patchy | Membranous |
| Hepatocellular carcinoma, non-cirrhotic |
50/M | Biopsy | T41A | Diffuse heterogeneous | Membranous |
| Hepatocellular carcinoma, non-cirrhotic |
78/M | Resection | T41A | Diffuse heterogeneous | Nuclear |
| Hepatocellular carcinoma, cirrhotic | 66/M | Resection | S35insI | Diffuse homogeneous | Nuclear |
| Hepatocellular carcinoma, non-cirrhotic |
45/F | Resection | T41A | Diffuse heterogeneous | Nuclear |
| Hepatocellular carcinoma, cirrhotic | 59/F | Resection | S35_T41del | Patchy | Membranous |
Figure 1.
Bidirectional sequence traces over exon 3 of the CTNNB1 gene in inflammatory atypical hepatocellular neoplasm, demonstrating an A>G missense mutation causing a threonine to alanine substitution at amino acid 41.
In hepatocellular carcinomas, β-catenin mutations were seen in 10 (17%) cases (Table 2): T41A (n=6), S45P (n=1), and S33C (n=1), heterozygyous in-frame insertion of ATT at codon 35 (n=1), and heterozygous in-frame deletion of codons 35–41 (n=1). This last in-frame deletion was confirmed by cloning the polymerase chain reaction product and sequencing 10 individual subclones, of which 5 demonstrated this in-frame deletion seen on initial Sanger sequencing. There was no correlation between β-catenin mutation status and histologic grade, presence of cirrhosis, or etiology of cirrhosis (data not shown).
Correlation between β-catenin mutation and glutamine synthetase staining
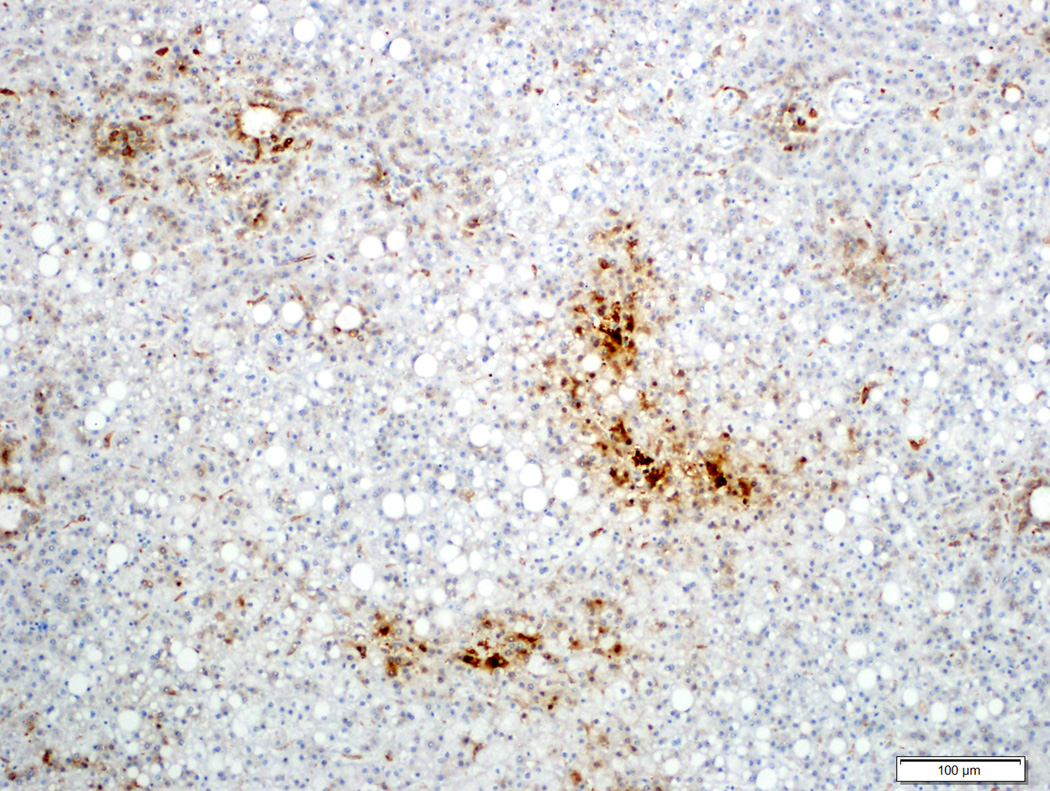
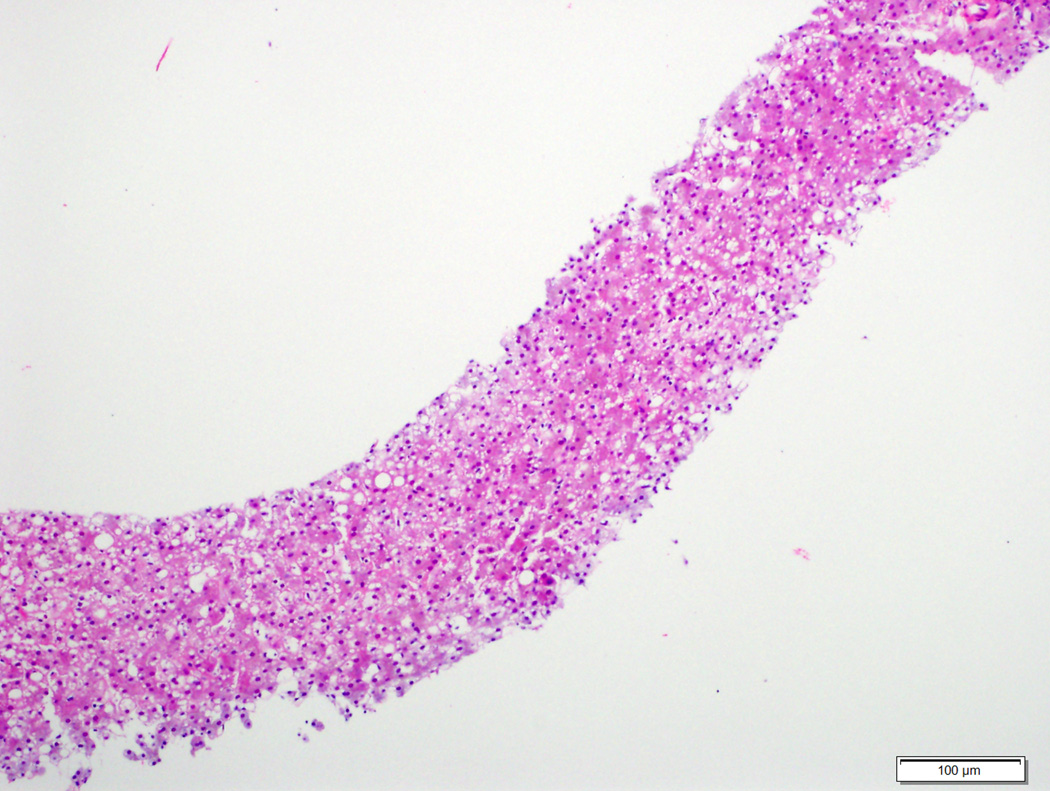
Of the 6 typical hepatocellular adenomas with diffuse staining (2 diffuse homogeneous, 4 diffuse heterogeneous), a β-catenin mutation was detected in 2 (33%) cases (both T41A), one each with diffuse homogeneous and heterogeneous glutamine synthetase staining. Nuclear β-catenin staining was seen in one of the mutated cases. Both of these cases also expressed inflammatory markers. No mutations were detected in the other typical hepatocellular adenoma with diffuse homogeneous glutamine synthetase staining. Figures 2–4 demonstrate examples of glutamine synthetase staining patterns in hepatocellular adenoma and atypical hepatocellular neoplasms, and the correlation with β-catenin mutations. Of the 4 atypical hepatocellular neoplasms with diffuse glutamine synthetase staining, β-catenin mutation was identified in 3 (75%) cases (1 diffuse homogeneous, 2 diffuse heterogeneous). β-catenin point mutations or insertions/deletions were not identified in any hepatocellular adenoma or atypical hepatocellular neoplasm with patchy glutamine synthetase staining (Table 3).
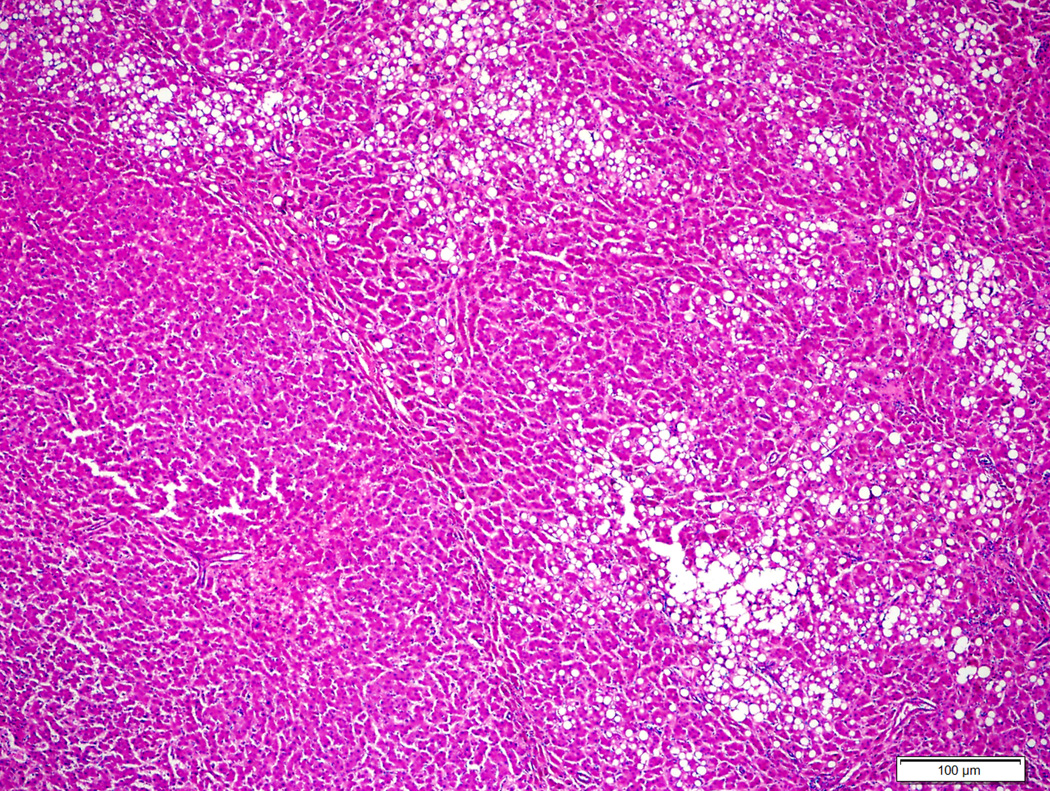
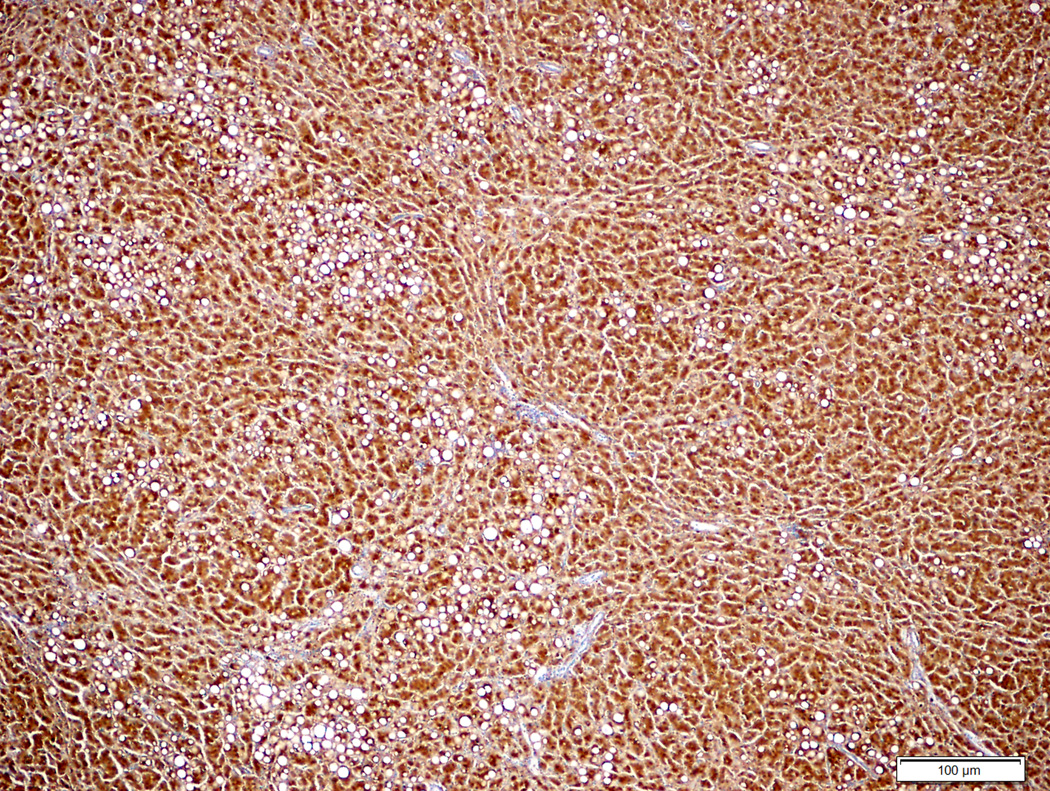
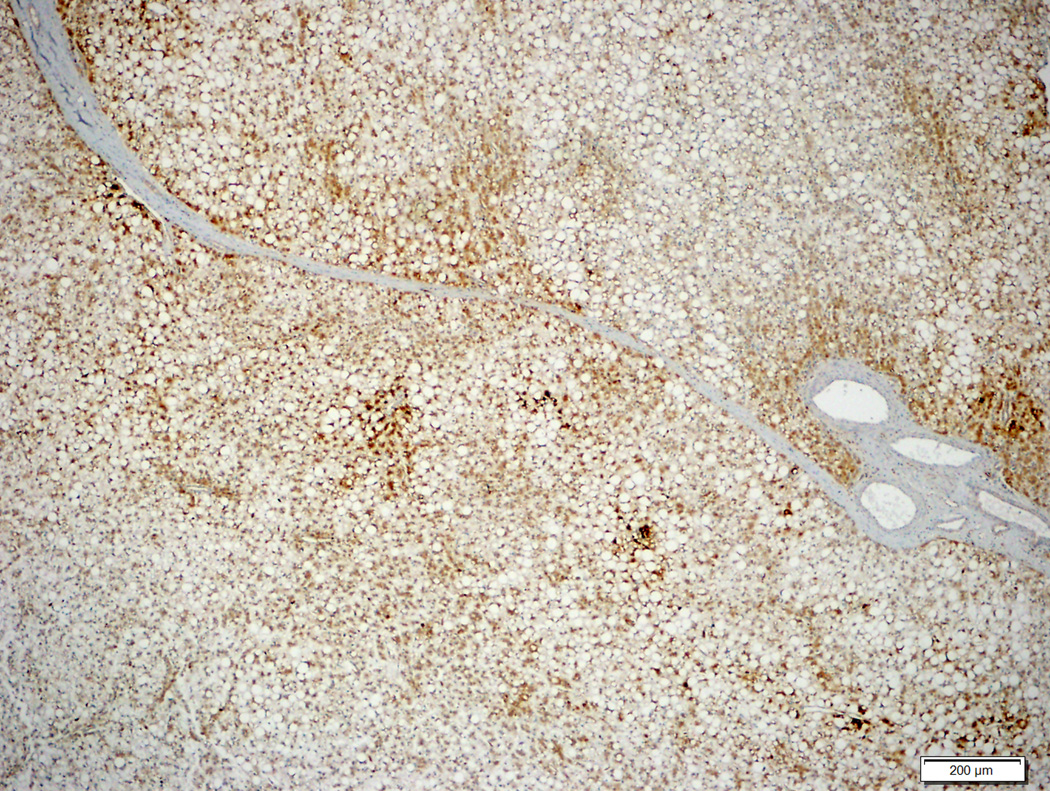
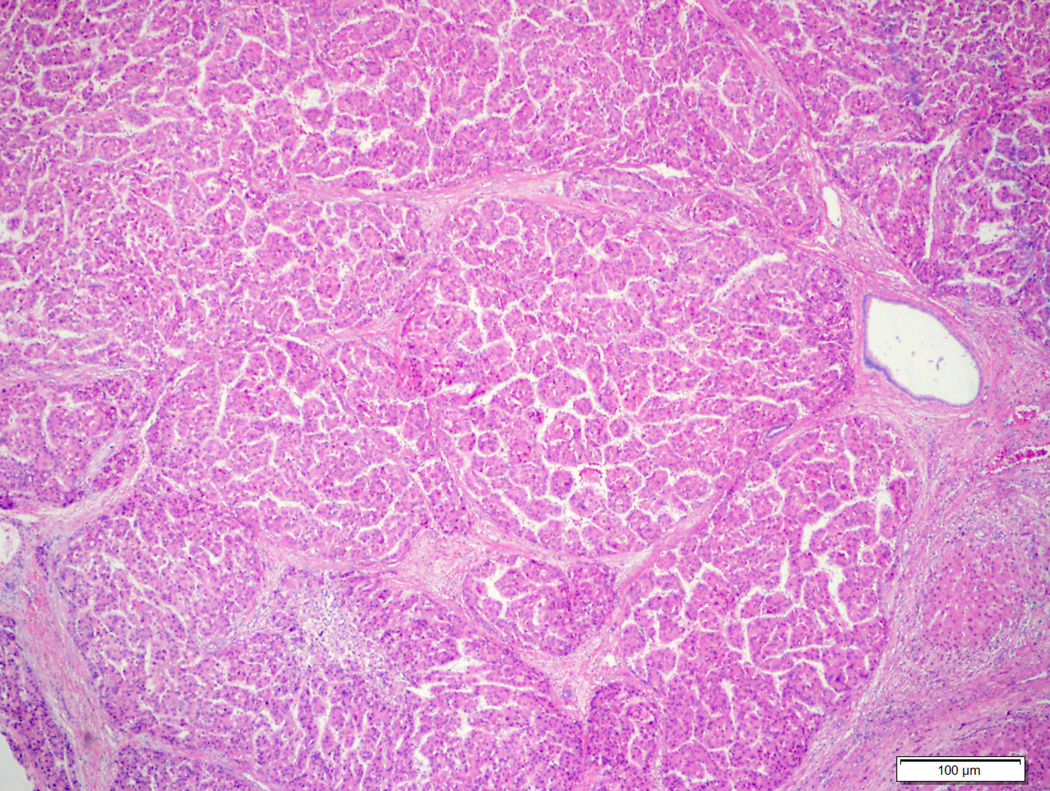
Figure 2.
Hepatocellular adenoma with no specific histologic characteristics and no β-catenin mutation (2A, H&E stain, 4X) showing diffuse homogeneous glutamine synthetase staining (2B, 4X).
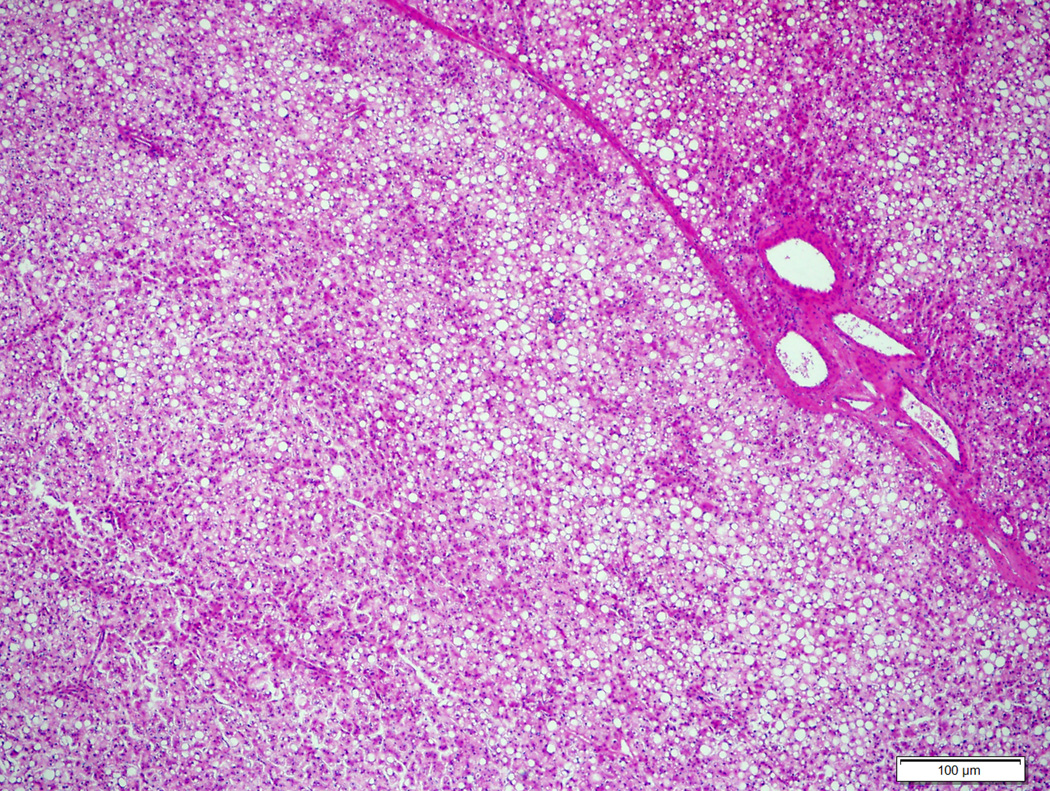
Figure 4.
Atypical hepatocellular neoplasm with diffuse steatosis, liver fatty acid binding protein loss and heterozygous missense mutation at codon 41 (T41A) (4A, H&E stain, 4X) with diffuse heterogeneous glutamine synthetase staining (4B, 4X).
Table 3.
Correlation between β-catenin mutation and β-catenin nuclear staining with the glutamine synthetase staining pattern in hepatocellular tumors.
| Glutamine synthetase staining |
Typical hepatocellular adenoma (n=15) |
Atypical hepatocellular neoplasm (n=5) |
Hepatocellular carcinoma (n=60) |
|||
|---|---|---|---|---|---|---|
| β-catenin mutated (%) |
β-catenin nuclear (%) |
β-catenin mutated (%) |
β-catenin nuclear (%) |
β-catenin mutated (%) |
β-catenin nuclear (%) |
|
|
Patchy glutamine synthetase |
0/9 | 0/9 | 0/1 | 0/1 | 2/13 (15) | 0/13 |
|
Diffuse glutamine synthetase-all |
2/6 (33) | 2/6 (33) | 3/4 (75) | 0/4 | 8/47 (17) | 18/47 (38) |
|
Diffuse glutamine synthetase - homogeneous pattern only |
0/2 | 0/2 | 1/1 (100) | 0/1 | 5/19 (26) | 14/19 (74) |
|
Diffuse glutamine synthetase - heterogeneous pattern only |
2/4 (50) | 2/4 (50) | 2/3 (67) | 0/3 | 3/28 (11) | 4/28 (14) |
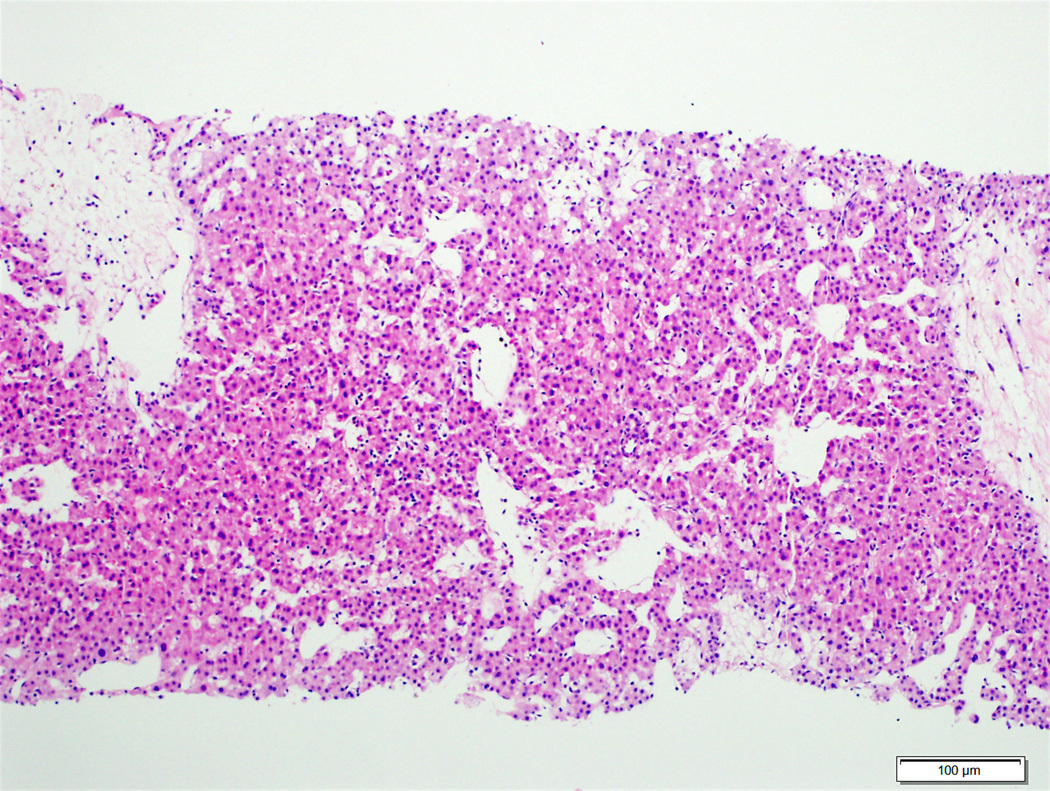
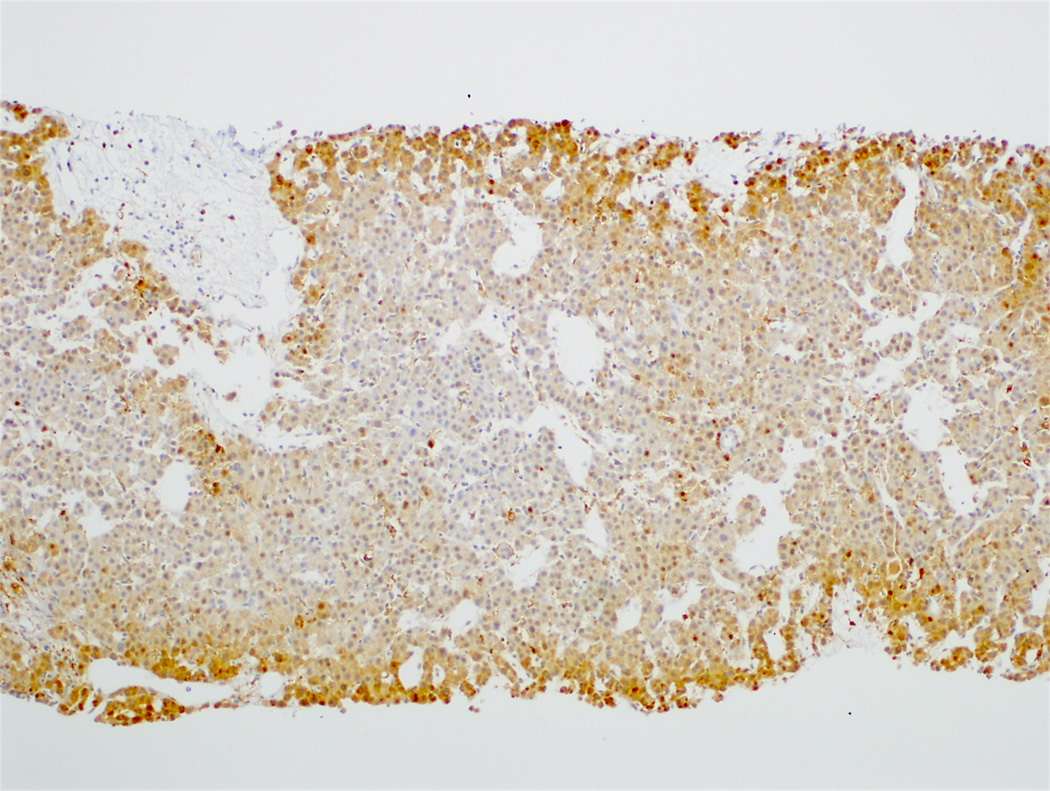
Of the 47 hepatocellular carcinoma cases with diffuse glutamine synthetase staining, β-catenin mutations were identified in 8 (17%) cases (5 diffuse homogeneous, 3 diffuse heterogeneous) (Table 3). Examples of glutamine synthetase staining patterns in hepatocellular carcinoma and their correlation with β-catenin mutations are demonstrated in Figures 5–7. A β-catenin mutation was not detected in 14 of the 19 hepatocellular carcinoma cases with diffuse homogeneous staining (Figure 5). All 5 hepatocellular carcinomas with diffuse homogeneous glutamine synthetase staining and β-catenin mutations also had nuclear β-catenin staining, while 2 of the 3 cases with diffuse heterogeneous glutamine synthetase staining and β-catenin mutations had nuclear β-catenin expression. Of the 13 hepatocellular carcinomas with patchy glutamine synthetase staining, 2 (15%) had a β-catenin mutation. None of the patchy glutamine synthetase staining hepatocellular carcinomas had nuclear β-catenin staining (Table 3). Of the 18 hepatocellular carcinoma cases that showed nuclear β-catenin staining, 7 (39%) showed β-catenin mutations (Table 3).
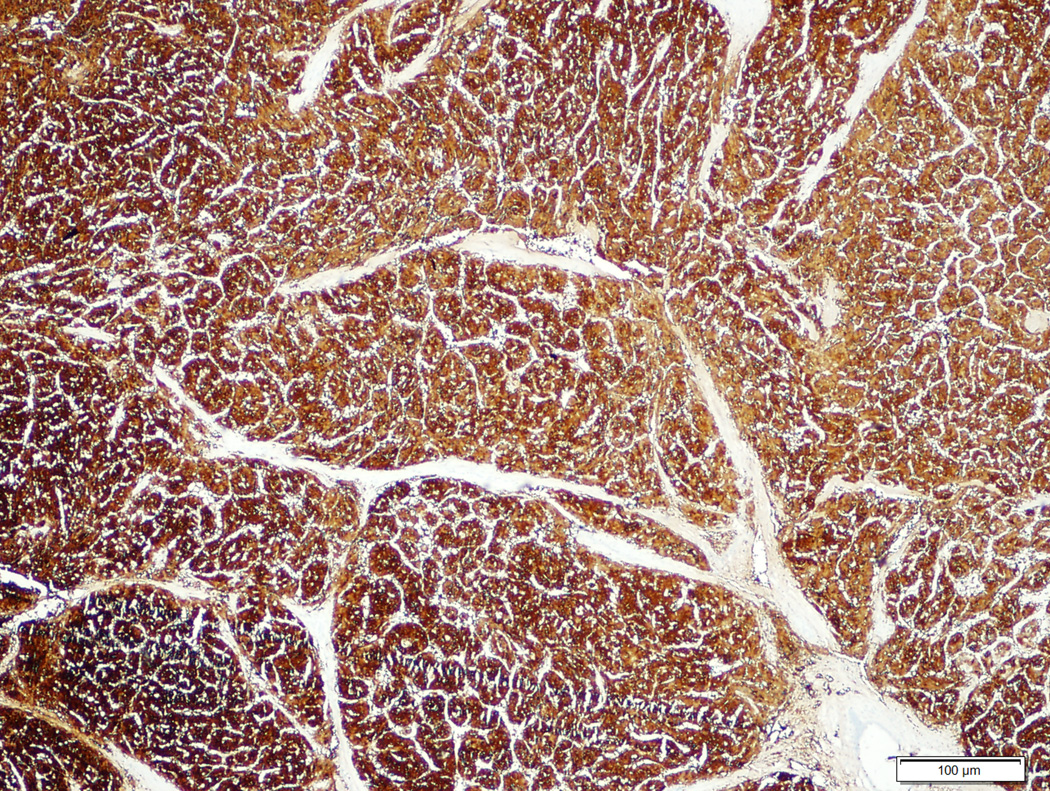
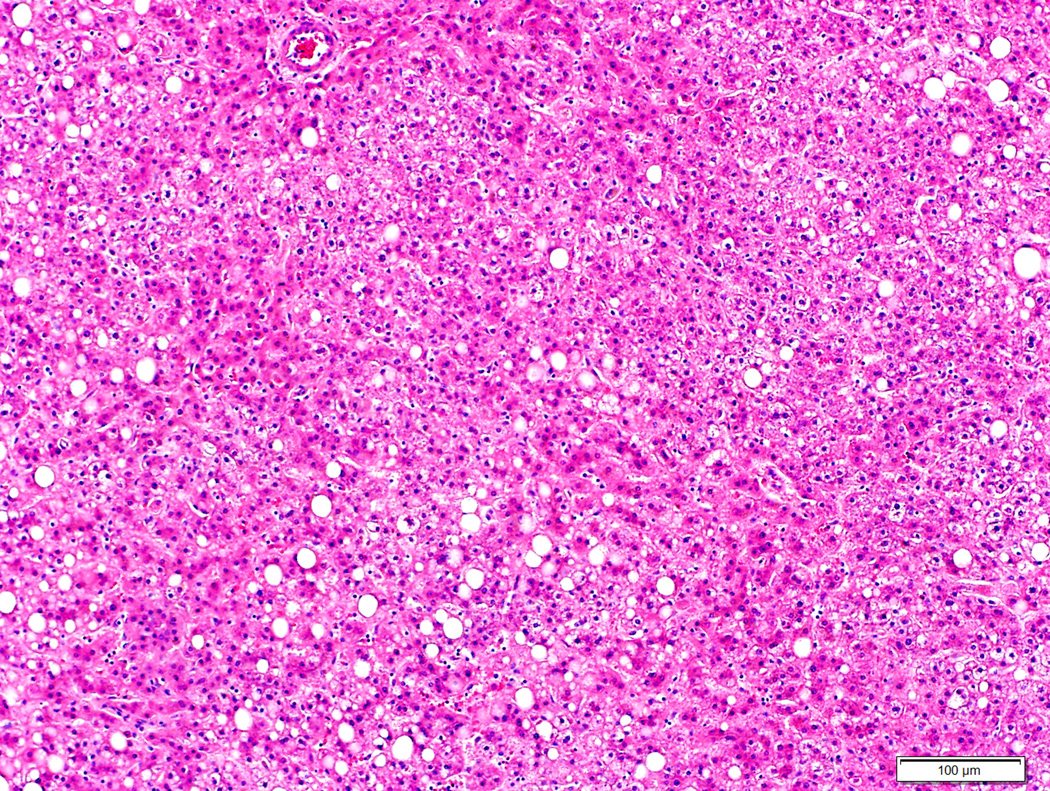
Figure 5.
Moderately differentiated hepatocellular carcinoma with no β-catenin mutation (5A, H&E stain, 4X) and diffuse homogeneous glutamine synthetase staining (5B, 4X).
Figure 7.
Well-differentiated hepatocellular carcinoma with heterozygous missense mutation at codon 41 (T41A) (7A, H&E stain, 4X) and patchy glutamine synthetase staining (7B, 4X).
There was no correlation between the type of mutation and a specific glutamine synthetase staining pattern. All three glutamine synthetase staining patterns were observed in the 6 hepatocellular carcinoma cases with T41A mutation: 2 diffuse homogeneous, 3 diffuse heterogeneous (Figure 6), 1 patchy (Figure 7). One case each of S45P and S33C mutation had diffuse homogeneous glutamine synthetase staining. An insertion was identified in one hepatocellular carcinoma case with diffuse homogeneous staining. The hepatocellular carcinoma case with a deletion had patchy glutamine synthetase staining. A similar lack of correlation was also observed in hepatocellular adenoma and atypical hepatocellular neoplasm.
Figure 6.
Well-differentiated hepatocellular carcinoma with heterozygous missense mutation at codon 41 (T41A), (6A, H&E stain, 10X) and diffuse heterogeneous glutamine synthetase staining (6B, 10X).
There was no significant difference between resection specimens and biopsies with respect to the mutation status in hepatocellular adenoma, atypical hepatocellular neoplasm and hepatocellular carcinoma cases.
Discussion
Activation of β-catenin is a high-risk feature in hepatocellular adenomas, and these tumors are frequently associated with hepatocellular carcinoma (4, 28). Our previous studies have shown that careful morphologic review of these tumors can enable the diagnosis of hepatocellular carcinoma in most cases (24, 25, 28). It has been recommended that tumors with β-catenin activation should be classified as atypical hepatocellular neoplasms or hepatocellular neoplasms with uncertain malignant potential (29, 30). Due to the low sensitivity of β-catenin immunohistochemistry, diffuse glutamine synthetase staining has been recommended as a surrogate marker for β-catenin activation (4), and this has become a standard practice in diagnostic pathology (6). However, the staining patterns observed with glutamine synthetase immunohistochemistry are diverse, and the correlation between the various patterns and β-catenin mutations is not well established.
In this study, mutations in exon 3 of the CTNNB1 gene were detected in 33% of typical hepatocellular adenomas and 75% of atypical hepatocellular neoplasms with diffuse glutamine synthetase staining. This is in contrast to the earlier reported 100% correlation between diffuse glutamine synthetase staining and β-catenin mutation in hepatocellular adenomas (4). In our series, exon 3 CTNNB1 gene mutations were detected in 21% of hepatocellular carcinomas with diffuse glutamine synthetase staining. The reported correlation between diffuse glutamine synthetase staining and β-catenin mutations varies widely in hepatocellular carcinoma among reported series, ranging from 30–100% (4, 9, 21, 22, 31) (Table 4). The discrepancy in glutamine synthetase staining and β-catenin mutation in liver neoplasms, especially hepatocellular carcinoma is not surprising. Diffuse glutamine synthetase staining has been described in 60–70% of hepatocellular carcinoma, while β-catenin mutation occurs in only 15–35% of hepatocellular carcinoma (16, 32, 33). This indicates that mechanisms other than β-catenin mutation can lead to overexpression of glutamine synthetase in hepatocellular carcinoma, which may be related to alterations in blood flow, oxygenation, nutritional status, cholestasis and fibrosis/cirrhosis (34–36).
Table 4.
Correlation between β-catenin mutation and diffuse glutamine synthetase staining in different studies.
| Total number of cases |
Correlation in hepatocellular carcinoma cases |
Correlation in hepatocellular adenoma cases |
|
|---|---|---|---|
| Bioulac-Sage, 2007 | 93 | not done | 100% |
| Zucman-Rossi, 2007 | 45 | 100% | not done |
| Audard, 20071 | 190 | 90% | not done |
| Austinat, 2008 | 40 | 25% | not done |
| Cieply, 2009 | 32 | 28% | not done |
| Hale, 2015 | 80 | 17% | 50%2 |
This study included cases with homogeneous glutamine synthetase staining only.
Includes typical adenomas and atypical neoplasms.
Other reasons may also contribute to discordance between diffuse glutamine synthetase staining and β-catenin mutations in hepatocellular adenoma and hepatocellular carcinoma. Mutation analysis of exon 3 of CTNNB1 in our study was performed using paraffin-embedded tissue, which may have missed large deletions in exon 3. These have been reported in 12% of hepatocellular adenomas (7). β-catenin deletions tend to be associated with diffuse homogeneous glutamine synthetase staining (22). Hence, some of our cases with diffuse homogeneous glutamine synthetase staining and no evidence of β-catenin mutation may have deletions that were not detected due to use of paraffin-embedded tissue. However, this would likely account for only a small number of discrepant cases in our series. Mutations in other genes involved in the Wnt signaling pathway can also lead to β-catenin activation and diffuse glutamine synthetase staining in the absence of β-catenin mutation. Inactivation of the AXIN1 gene has been reported in 5–15% of hepatocellular carcinoma (19, 37). Diffuse glutamine synthetase staining has been observed in AXIN1-mutated hepatocellular carcinoma cases (22), and can account for a small number of discrepant cases. Mutations in other exons of the CTNNB1 gene such as exons 7 and 8 have recently been reported in approximately 10% of hepatocellular adenomas (38); however, these cases typically show weak glutamine synthetase expression and are unlikely to explain the discrepancy between CTNNB1 exon 3 mutations and diffuse glutamine synthetase staining.
Just as the correlation between diffuse glutamine synthetase staining and β-catenin mutation is not perfect, patchy glutamine synthetase staining does not necessarily signify absence of β-catenin mutation. We observed an exon 3 CTNNB1 gene mutation in 15% of hepatocellular carcinomas with patchy glutamine synthetase staining. Exon 3 mutations with patchy or weak glutamine synthetase staining have been observed in a small number of cases in other studies (21, 31). There were no cases of hepatocellular adenoma or atypical hepatocellular neoplasm in our series in which patchy glutamine synthetase staining was associated with β-catenin mutation. Mutation in exon 45 of CTNNB1 gene has been reported in hepatocellular adenoma, which was not associated with diffuse glutamine synthetase staining in many areas of the tumor (39).
The mutations detected in our study (T41A, S33C, S45P) are phosphorylation sites for glycogen synthase kinase 3β. T41A was the most common mutation in this series, accounting for two-thirds of the cases. In the study by Zucman-Rossi (22), T41A mutations were seen in only 11% of cases, while mutations at serine position 45 (33%) and serine at position 33 (22%) were the most common. The reason for this difference is not clear. It may be surmised that the different glutamine synthetase staining patterns may result from specific mutations in exon 3 of the CTNNB1 gene. However, this correlation was not observed in our study. A similar lack of correlation of diffuse homogeneous or heterogeneous glutamine synthetase staining and specific β-catenin mutations has been reported in another study (22), although the deletions tended to be associated with diffuse homogeneous glutamine synthetase staining.
Similar to other studies (21, 40) we found a low correlation between β-catenin mutations and nuclear expression using immunohistochemistry. In our study, 70% (7/10) of hepatocellular carcinomas and 20% (1/5) of hepatocellular adenomas with β-catenin mutation showed nuclear β-catenin expression by immunohistochemistry. Higher correlations approximating 100% have been reported in earlier studies, which is likely related to cytoplasmic staining being considered positive in the absence of nuclear staining (31). Due to the possibility of nonspecific background cytoplasmic staining, it is recommended that β-catenin staining should be considered positive only in the presence of nuclear staining (4).
Another interesting finding is the detection of a T41A mutation in an atypical hepatocellular neoplasm with loss of liver fatty acid binding protein staining. Although the French studies have generally classified hepatocyte nuclear factor 1α inactivated hepatocellular adenomas as benign neoplasms with very low or negligible risk of transformation to hepatocellular carcinoma, a recent series demonstrates the loss of liver fatty acid binding protein in approximately 30% (6/20) hepatocellular carcinomas (41); the series included 2 cases of hepatocellular carcinoma that arose in an hepatocyte nuclear factor 1α inactivated hepatocellular adenoma. These findings and our study suggest that these adenomas may not be considered entirely benign neoplasms.
Our results challenge the near perfect correlation that has been reported between β-catenin mutation status and diffuse glutamine synthetase staining (4, 42), and suggest that the use of glutamine synthetase staining pattern for determining the histologic subtype and management of hepatocellular adenoma needs to be revisited. It is likely that diffuse homogeneous glutamine synthetase staining has a high correlation with β-catenin mutation (including deletions). This pattern is relatively easy to recognize and hepatocellular adenoma with this pattern of staining can be regarded as high-risk tumors. However, the significance of diffuse heterogeneous or patchy glutamine synthetase staining is less certain. We recommend that these patterns of glutamine synthetase staining should be considered indeterminate for β-catenin activation, especially in needle biopsies. Our results show that analysis of paraffin embedded tissue can demonstrate missense mutations and small deletions, and hence provide definite evidence of β-catenin activation in a subset of cases with borderline glutamine synthetase staining results. A negative result obtained by mutation testing in paraffin-embedded tissue is not informative, as it does not exclude β-catenin activation due to large deletions, mutations in exons other than exon 3, and mutations in other genes of the Wnt signaling pathway. Other modalities such as TERT promoter mutations and cytogenetic abnormalities may help to stratify risk in cases with borderline glutamine synthetase staining patterns. TERT promoter mutations are associated with β-catenin mutations (43) and have been reported in 17% of atypical hepatocellular neoplasms and 56% of hepatocellular adenoma with malignant progression (38). Similarly, chromosomal abnormalities such as gains of chromosomes 1q, 7q and 8q, have been shown to occur in a subset of atypical hepatocellular neoplasm and most of hepatocellular carcinoma cases, but not in typical hepatocellular adenoma, suggesting that these changes would suggest high-risk behavior and possible malignant potential in tumors with borderline features (38, 44). Additional work-up with mutation analysis, TERT promoter mutation and cytogenetics may be helpful to determine the risk status in indeterminate cases.
In summary, this study reveals that there is a modest correlation between diffuse glutamine synthetase immunostaining and mutations in exon 3 of the CTNNB1 gene in hepatocellular adenoma and hepatocellular carcinoma using paraffin-embedded tissue. The interpretation of β-catenin activation based on glutamine synthetase staining should be performed with caution in clinical practice, and the undetermined significance of various staining patterns should be emphasized in the pathology report. Mutation testing for exon 3 of β-catenin can be accomplished from paraffin-embedded tissue and can provide definite evidence of β-catenin activation in a subset of cases.
Supplementary Material
Figure 3.
Inflammatory atypical hepatocellular neoplasm with heterozygous missense mutation at codon 41 (T41A) (3A, H&E stain, 10X) and diffuse homogeneous glutamine synthetase staining (3B, 10X). The inflammatory designation was based on strong serum amyloid A staining.
Acknowledgments
This work is in part supported by NIH grant (R01CA136606) to X.C.
Footnotes
Disclosure/conflict of Interest
The authors declare no conflict of interest.
References
- 1.Chen YW, Jeng YM, Yeh SH, Chen PJ. P53 gene and Wnt signaling in benign neoplasms: beta-catenin mutations in hepatic adenoma but not in focal nodular hyperplasia. Hepatology. 2002;36:927–935. doi: 10.1053/jhep.2002.36126. [DOI] [PubMed] [Google Scholar]
- 2.Cadoret A, Ovejero C, Terris B, et al. New targets of beta-catenin signaling in the liver are involved in the glutamine metabolism. Oncogene. 2002;21:8293–8301. doi: 10.1038/sj.onc.1206118. [DOI] [PubMed] [Google Scholar]
- 3.Gebhardt R, Baldysiak-Figiel A, Krugel V, et al. Hepatocellular expression of glutamine synthetase: an indicator of morphogen actions as master regulators of zonation in adult liver. Prog Histochem Cyto. 2007;41:201–266. doi: 10.1016/j.proghi.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Bioulac-Sage P, Rebouissou S, Thomas C, et al. Hepatocellular adenoma subtype classification using molecular markers and immunohistochemistry. Hepatology. 2007;46:740–748. doi: 10.1002/hep.21743. [DOI] [PubMed] [Google Scholar]
- 5.Bioulac-Sage P, Blanc JF, Rebouissou S, et al. Genotype phenotype classification of hepatocellular adenoma. World J Gastroenterol. 2007;13:2649–2654. doi: 10.3748/wjg.v13.i19.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bioulac-Sage P, Balabaud C, Wanless I. Focal nodular hyperplasia and hepatocellular adenoma. In: Bosman F, Carneiro F, Hruban R, Theise ND, editors. WHO Classification of Tumours of the Digestive System. 4th. Lyon, France: IARC Press; 2010. pp. 198–204. [Google Scholar]
- 7.Zucman-Rossi J, Jeannot E, Nhieu JT, et al. Genotype-phenotype correlation in hepatocellular adenoma: new classification and relationship with HCC. Hepatology. 2006;43:515–524. doi: 10.1002/hep.21068. [DOI] [PubMed] [Google Scholar]
- 8.Micchelli ST, Vivekanandan P, Boitnott JK, et al. Malignant transformation of hepatic adenomas. Mod Pathol. 2008;21:491–497. doi: 10.1038/modpathol.2008.8. [DOI] [PubMed] [Google Scholar]
- 9.Cieply B, Zeng G, Proverbs-Singh T, et al. Unique phenotype of hepatocellular cancers with exon-3 mutations in beta-catenin gene. Hepatology. 2009;49:821–831. doi: 10.1002/hep.22695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui J, Zhou X, Liu Y, et al. Wnt signaling in hepatocellular carcinoma: analysis of mutation and expression of beta-catenin, T-cell factor-4 and glycogen synthase kinase 3-beta genes. J Gastroenterol Hepatol. 2003;18:280–287. doi: 10.1046/j.1440-1746.2003.02973.x. [DOI] [PubMed] [Google Scholar]
- 11.Yamamoto Y, Sakamoto M, Fujii G, et al. Overexpression of orphan G-protein-coupled receptor, Gpr49, in human hepatocellular carcinomas with beta-catenin mutations. Hepatology. 2003;37:528–533. doi: 10.1053/jhep.2003.50029. [DOI] [PubMed] [Google Scholar]
- 12.Taniguchi K, Roberts LR, Aderca IN, et al. Mutational spectrum of beta-catenin, AXIN1, and AXIN2 in hepatocellular carcinomas and hepatoblastomas. Oncogene. 2002;21:4863–4871. doi: 10.1038/sj.onc.1205591. [DOI] [PubMed] [Google Scholar]
- 13.Devereux TR, Stern MC, Flake GP, et al. CTNNB1 mutations and beta-catenin protein accumulation in human hepatocellular carcinomas associated with high exposure to aflatoxin B1. Mol Carcinog. 2001;31:68–73. doi: 10.1002/mc.1041. [DOI] [PubMed] [Google Scholar]
- 14.Huang H, Fujii H, Sankila A, et al. Beta-catenin mutations are frequent in human hepatocellular carcinomas associated with hepatitis C virus infection. Am J Pathol. 1999;155:1795–1801. doi: 10.1016/s0002-9440(10)65496-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Legoix P, Bluteau O, Bayer J, et al. Beta-catenin mutations in hepatocellular carcinoma correlate with a low rate of loss of heterozygosity. Oncogene. 1999;18:4044–4046. doi: 10.1038/sj.onc.1202800. [DOI] [PubMed] [Google Scholar]
- 16.de La Coste A, Romagnolo B, Billuart P, et al. Somatic mutations of the beta-catenin gene are frequent in mouse and human hepatocellular carcinomas. Proc Natl Acad Sci USA. 1998;95:8847–8851. doi: 10.1073/pnas.95.15.8847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Terris B, Pineau P, Bregeaud L, et al. Close correlation between beta-catenin gene alterations and nuclear accumulation of the protein in human hepatocellular carcinomas. Oncogene. 1999;18:6583–6588. doi: 10.1038/sj.onc.1203051. [DOI] [PubMed] [Google Scholar]
- 18.Laurent-Puig P, Zucman-Rossi J. Genetics of hepatocellular tumors. Oncogene. 2006;25:3778–3786. doi: 10.1038/sj.onc.1209547. [DOI] [PubMed] [Google Scholar]
- 19.Laurent-Puig P, Legoix P, Bluteau O, et al. Genetic alterations associated with hepatocellular carcinomas define distinct pathways of hepatocarcinogenesis. Gastroenterology. 2001;120:1763–1773. doi: 10.1053/gast.2001.24798. [DOI] [PubMed] [Google Scholar]
- 20.Hsu HC, Jeng YM, Mao TL, et al. Beta-catenin mutations are associated with a subset of low-stage hepatocellular carcinoma negative for hepatitis B virus and with favorable prognosis. Am J Pathol. 2000;157:763–770. doi: 10.1016/s0002-9440(10)64590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Audard V, Grimber G, Elie C, et al. Cholestasis is a marker for hepatocellular carcinomas displaying beta-catenin mutations. J Pathol. 2007;212:345–352. doi: 10.1002/path.2169. [DOI] [PubMed] [Google Scholar]
- 22.Zucman-Rossi J, Benhamouche S, Godard C, et al. Differential effects of inactivated Axin1 and activated beta-catenin mutations in human hepatocellular carcinomas. Oncogene. 2007;26:774–780. doi: 10.1038/sj.onc.1209824. [DOI] [PubMed] [Google Scholar]
- 23.Hale G, Kim G, Ferrell L, Kakar S. Correlation of B-catenin exon 3 mutations with glutamine synthetase staining patterns in hepatocellular carcinoma [Abstract] Mod Pathol. 2015;28:419A. doi: 10.1038/modpathol.2016.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shafizadeh N, Genrich G, Ferrell L, Kakar S. Hepatocellular adenomas in a large community population, 2000 to 2010: reclassification per current World Health Organization classification and results of long-term follow-up. Hum Pathol. 2014;45:976–983. doi: 10.1016/j.humpath.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 25.Kakar S, Chen X, Ho C, et al. Chromosomal abnormalities determined by comparative genomic hybridization are helpful in the diagnosis of atypical hepatocellular neoplasms. Histopathology. 2009;55:197–205. doi: 10.1111/j.1365-2559.2009.03343.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirohashi S, Ishak KG, Kojiro M, et al. Hepatocellular carcinoma. In: Bosman FT, Carneiro F, Hruban RH, Theise ND, editors. WHO classification of tumours of the digestive system. 4th. Lyon, France: IARC Press; 2010. pp. 159–172. [Google Scholar]
- 27.Le Guellec S, Soubeyran I, Rochaix P, et al. CTNNB1 mutation analysis is a useful tool for the diagnosis of desmoid tumors: a study of 260 desmoid tumors and 191 potential morphologic mimics. Mod Pathol. 2012;25:1551–1558. doi: 10.1038/modpathol.2012.115. [DOI] [PubMed] [Google Scholar]
- 28.Kakar S, Grenert JP, Paradis V, et al. Hepatocellular carcinoma arising in adenoma: similar immunohistochemical and cytogenetic features in adenoma and hepatocellular carcinoma portions of the tumor. Mod Pathol. 2014;27:1499–1509. doi: 10.1038/modpathol.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bedossa P, Burt AD, Brunt EM, et al. Well-differentiated hepatocellular neoplasm of uncertain malignant potential: proposal for a new diagnostic category. Hum Pathol. 2014;45:658–660. doi: 10.1016/j.humpath.2013.09.020. [DOI] [PubMed] [Google Scholar]
- 30.Kakar S, Evason KJ, Ferrell LD. Well-differentiated hepatocellular neoplasm of uncertain malignant potential: proposal for a new diagnostic category--reply. Hum Pathol. 2014;45:660–661. doi: 10.1016/j.humpath.2013.09.019. [DOI] [PubMed] [Google Scholar]
- 31.Austinat M, Dunsch R, Wittekind C, et al. Correlation between beta-catenin mutations and expression of Wnt-signaling target genes in hepatocellular carcinoma. Mol Cancer. 2008;7:21. doi: 10.1186/1476-4598-7-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guichard C, Amaddeo G, Imbeaud S, et al. Integrated analysis of somatic mutations and focal copy-number changes identifies key genes and pathways in hepatocellular carcinoma. Nat Genet. 2012;44:694–698. doi: 10.1038/ng.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prange W, Breuhahn K, Fischer F, et al. Beta-catenin accumulation in the progression of human hepatocarcinogenesis correlates with loss of E-cadherin and accumulation of p53, but not with expression of conventional WNT-1 target genes. J Pathol. 2003;201:250–259. doi: 10.1002/path.1448. [DOI] [PubMed] [Google Scholar]
- 34.Arnason T, Fleming KE, Wanless IR. Peritumoral hyperplasia of the liver: a response to portal vein invasion by hypervascular neoplasms. Histopathology. 2013;62:458–464. doi: 10.1111/his.12032. [DOI] [PubMed] [Google Scholar]
- 35.Ueberham E, Arendt E, Starke M, et al. Reduction and expansion of the glutamine synthetase expressing zone in livers from tetracycline controlled TGF-beta1 transgenic mice and multiple starved mice. J Hepatol. 2004;41:75–81. doi: 10.1016/j.jhep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- 36.Paxian M, Rensing H, Geckeis K, et al. Perflubron emulsion in prolonged hemorrhagic shock: influence on hepatocellular energy metabolism and oxygen-dependent gene expression. Anesthesiology. 2003;98:1391–1399. doi: 10.1097/00000542-200306000-00014. [DOI] [PubMed] [Google Scholar]
- 37.Satoh S, Daigo Y, Furukawa Y, et al. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet. 2000;24:245–250. doi: 10.1038/73448. [DOI] [PubMed] [Google Scholar]
- 38.Pilati C, Letouze E, Nault JC, et al. Genomic profiling of hepatocellular adenomas reveals recurrent FRK-activating mutations and the mechanisms of malignant transformation. Cancer Cell. 2014;25:428–441. doi: 10.1016/j.ccr.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 39.Berry RS, Gullapalli RR, Wu J, et al. Diffuse glutamine synthetase overexpression restricted to areas of peliosis in a beta-catenin-activated hepatocellular adenoma: a potential pitfall in glutamine synthetase interpretation. Virchows Archiv. 2014;465:241–245. doi: 10.1007/s00428-014-1620-8. [DOI] [PubMed] [Google Scholar]
- 40.Torbenson M, Lee JH, Choti M, et al. Hepatic adenomas: analysis of sex steroid receptor status and the Wnt signaling pathway. Mod Pathol. 2002;15:189–196. doi: 10.1038/modpathol.3880514. [DOI] [PubMed] [Google Scholar]
- 41.Cho SJ, Ferrell LD, Gill RM. Expression of liver fatty acid binding protein in hepatocellular carcinoma. Hum Pathol. 2016;50:135–139. doi: 10.1016/j.humpath.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bioulac-Sage P, Laumonier H, Couchy G, et al. Hepatocellular adenoma management and phenotypic classification: the Bordeaux experience. Hepatology. 2009;50:481–489. doi: 10.1002/hep.22995. [DOI] [PubMed] [Google Scholar]
- 43.Nault JC, Mallet M, Pilati C, et al. High frequency of telomerase reverse-transcriptase promoter somatic mutations in hepatocellular carcinoma and preneoplastic lesions. Nat Commun. 2013;4:2218. doi: 10.1038/ncomms3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evason KJ, Grenert JP, Ferrell LD, Kakar S. Atypical hepatocellular adenoma-like neoplasms with beta-catenin activation show cytogenetic alterations similar to well-differentiated hepatocellular carcinomas. Hum Pathol. 2013;44:750–758. doi: 10.1016/j.humpath.2012.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.