Abstract
Signal recognition particle (SRP) plays a central role in the delivery of classical secretory and membrane proteins to the endoplasmic reticulum (ER). All nascent chains studied to date dissociate from SRP once released from the ribosome, thereby supporting a strictly cotranslational mode of action for eukaryotic SRP. We now report a novel post-translational function for SRP in the targeting of tail-anchored (TA) proteins to the ER. TA proteins possess a hydrophobic membrane insertion sequence at their C-terminus such that it can only emerge from the ribosome after translation is terminated. We show that SRP can associate post-translationally with this type of ER-targeting signal, and deliver newly synthesised TA proteins to the ER membrane by a pathway dependent upon GTP and the SRP receptor. We find that dependency upon this SRP-dependent route is precursor specific, and propose a unifying model to describe the biogenesis of TA proteins in vivo.
Keywords: ER targeting, membrane protein biogenesis, SRP, tail-anchored protein
Introduction
Tail-anchored (TA) proteins display their hydrophilic N-termini to the cytosol and are widely distributed throughout the membranes of eukaryotic organelles (Wattenberg and Lithgow, 2001; Borgese et al, 2003). Their principal sites of membrane integration are the endoplasmic reticulum (ER) and mitochondrial outer membrane (Kutay et al, 1995; Borgese et al, 2001), and they ultimately function in many key cellular processes including vesicle trafficking (SNAREs), protein translocation (Sec61β and Sec61γ) and apoptosis (Bcl-2 family).
The C-terminal location of the sole ER-targeting signal present in TA proteins poses a unique challenge to the cellular targeting and membrane integration machineries, since it only emerges from the ribosome and becomes available to cytosolic targeting factors post-translationally (Kutay et al, 1995). At the ER membrane, authentic integration of TA proteins is strictly dependent upon proteinaceous factors (Kutay et al, 1995), although their precise identity remains controversial and incompletely defined (Kutay et al, 1995; Steel et al, 2002; Abell et al, 2003; Yabal et al, 2003). The post-translational integration of TA proteins in vitro is ATP dependent, consistent with a role of cytosolic chaperones in maintaining the precursor in an insertion-competent state (Kutay et al, 1995; Kim et al, 1999; Steel et al, 2002). However, no cytosolic components responsible for the organelle-specific targeting of TA proteins have been identified to date (Kutay et al, 1995; Kim et al, 1999; Borgese et al, 2001).
The in vitro integration of an archetypal TA protein, cytochrome b5 (Cytb5), can occur independently of the signal recognition particle (SRP), consistent with a post-translational membrane insertion pathway (Anderson et al, 1983). In contrast, the insertion of most membrane proteins at the ER depends upon SRP binding to their loosely conserved N-terminal signal sequences and internal signal anchors. A key feature of these signals is a contiguous stretch of 7–15 hydrophobic residues that interacts with the 54 kDa subunit of SRP (SRP54) (Keenan et al, 2001). Furthermore, SRP specifically associates with the ribosome at the exit tunnel, thereby allowing it to bind signal sequences immediately upon their emergence (Pool et al, 2002; Halic et al, 2004). Once SRP has bound a signal sequence, it promotes cotranslational membrane insertion by attenuating translation until the ribosome-nascent chain complex (RNC) has been received by the SRP receptor (Egea et al, 2004; Focia et al, 2004) at the ER membrane and transferred to the Sec61 protein translocation channel (Song et al, 2000). The strictly cotranslational nature of SRP action is supported by the observation that SRP dissociates completely from nascent secretory proteins immediately upon their release from the ribosome (Wiedmann et al, 1994; Plath and Rapoport, 2000).
In order to identify cellular factors responsible for the ER targeting of TA proteins, we pursued a crosslinking approach using polypeptides that had been synthesised in reticulocyte lysate. Given previous studies (Anderson et al, 1983; Kutay et al, 1995), we were initially surprised to discover that TA proteins form adducts with SRP54. However, using the well-characterised ER-targeted TA proteins synaptobrevin 2 (Syb2) and Sec61β, we show that SRP can specifically associate with TA proteins in a strictly post-translational manner, and via an interaction that is mediated by the proteins' hydrophobic insertion sequences. At a functional level, we find that TA proteins can be targeted to ER membranes via a previously undefined post-translational pathway that is dependent upon SRP, the SRP receptor and GTP. The efficient operation of this pathway requires the presence of SRP during nascent chain release from the ribosome, but ribosomes play no role in the targeting of the polypeptide to the ER membrane. We find that the capacity to utilise this SRP-dependent route is precursor specific, and, while this pathway accounts for a significant proportion of Syb2 and Sec61β integration, Cytb5 insertion is entirely independent of this route. On the basis of these findings, we propose a unifying model to describe the pathways responsible for TA protein biosynthesis in vivo.
Results
TA proteins were synthesised in rabbit reticulocyte lysate using mRNAs lacking stop codons, thereby enabling the release of the polypeptides from the ribosome to be synchronised by puromycin treatment (Gilmore et al, 1991). Associations between the newly released TA proteins and cytosolic factors present in the complete cell-free system were then assessed by crosslinking. As a benchmark for SRP binding, we used the N-terminal 86 residues of the secretory protein preprolactin (PPL86) in its cycloheximide-stabilised, ribosome-bound, form. Consistent with previous studies (Wiedmann et al, 1994), SRP54 was one of several cytosolic components crosslinked to PPL86 (Figure 1A, lanes 1–3). Surprisingly, Syb2 chains that had been released from the ribosome by puromycin treatment also crosslinked to SRP54 (Figure 1A, lanes 5–7), as did puromycin-released Sec61β chains (Figure 1A, lanes 10–12). Hence, TA proteins can associate with the SRP54 subunit in a post-translational manner. In contrast, we found no evidence of crosslinking between these TA proteins and members of the Hsc70 chaperone family (data not shown), although these components are estimated to be present at a 500-fold molar excess over SRP (Siegel and Walter, 1988; Nollen et al, 2000). Several of the other adducts observed during this analysis were found to reflect in vitro ubiquitination of the precursor proteins (Figure 1A, lanes 4, 8 and 13), as previously seen during studies of preproα-factor (Plath and Rapoport, 2000).
Figure 1.
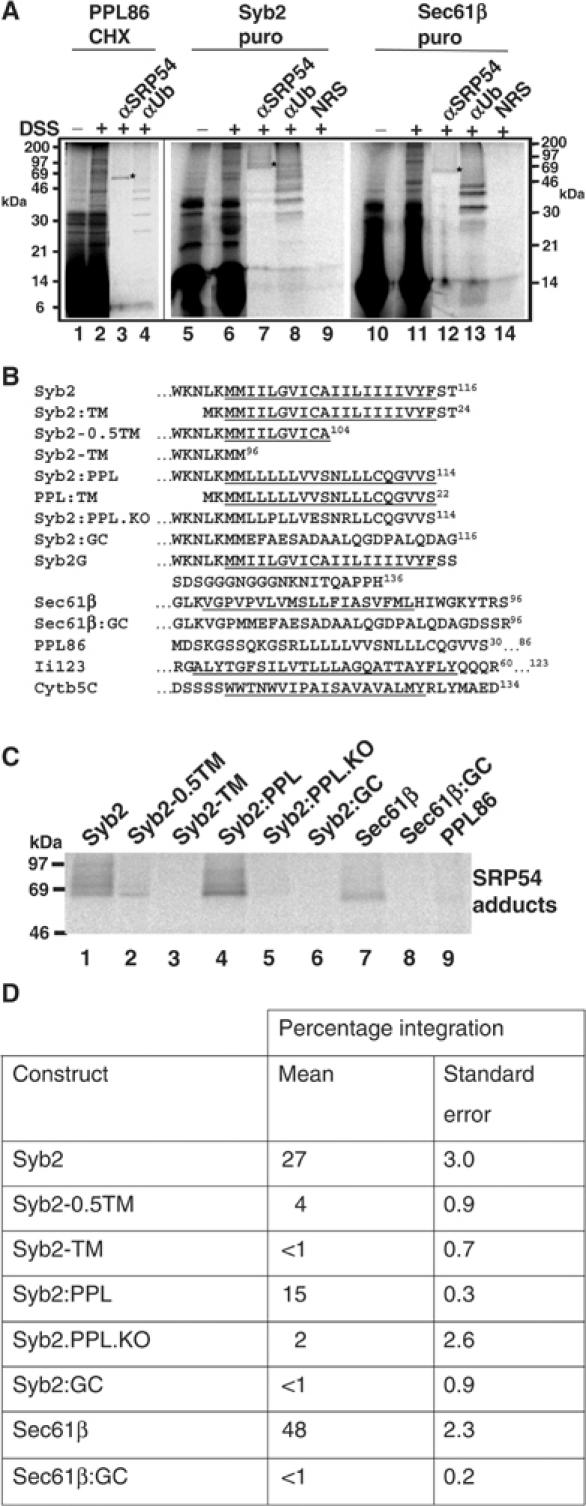
TA proteins associate with SRP54 post-translationally. (A) Stop codon minus RNA encoding full-length Syb2 (lanes 5–9), full-length Sec61β (lanes 10–14) or the first 86 residues of preprolactin (lanes 1–4) was translated for 20 min, and synthesis then terminated by the addition of cycloheximide (CHX) or puromycin (puro) as shown. Crosslinking was induced with DSS as indicated. Total products were analysed directly (lanes 1, 2, 5, 6, 10 and 11) or following immunoprecipitation with antisera for SRP54 (αSRP54), ubiquitin (αUb) or a nonrelated serum (NRS). (B) Amino-acid sequences of the hydrophobic ER-targeting signals from the polypeptides used in this study. Potential TM domains are underlined, while dots indicate regions where hydrophilic domains of the polypeptide extend beyond the sequence presented. Numbers in superscript show the total length of the various polypeptides used. Where two numbers are given, the first is the limit of the sequence presented and the second the length of the polypeptide studied. (C) Cell-free translations of the various precursors were terminated with puromycin and the samples treated with DSS. In order to avoid any variability resulting from differences in translation efficiency, a fraction of each sample was first analysed by SDS–PAGE and the relative amount of each nascent chain determined by quantitative phosphorimaging (data not shown). On the basis of this analysis, equivalent amounts of each DSS-treated radiolabelled precursor were then used for the immunoprecipitation of adducts with SRP54. (D) Stop codon minus RNA was translated for 20 min in the presence or absence of canine pancreatic microsomes, and then incubated with puromycin for a further 20 min to enable membrane integration. The membrane fraction was isolated by centrifugation through a high-salt sucrose cushion. The resulting pellet was then resuspended in alkaline sodium carbonate solution and the membrane pellet re-isolated by centrifugation. For each precursor, the translation reaction lacking any exogenously added membranes was processed in parallel to provide an estimate of the amount of each protein that was recovered in the final pellet fraction independent of membrane integration. The amount of precursor present in each of the fractions was determined by SDS–PAGE and quantitative phosphorimaging, and was expressed as a percentage of the total protein synthesised in that reaction. The final value for membrane integration is the percentage of the total protein synthesised, which was specifically recovered with the isolated microsomal membranes (see Supplementary data). These values are the means of two or more independent experiments, and standard errors are indicated.
SRP54 normally binds to the hydrophobic core of ER signal sequences (Keenan et al, 2001), and we therefore investigated the validity of the interaction between SRP54 and the TA proteins by manipulating their hydrophobic tail anchors (see Figure 1B). When the Syb2 tail anchor is truncated from 20 to 10 residues (Syb2-0.5TM), its association with SRP54 is substantially reduced, while upon its complete removal (Syb2-TM) no association with SRP54 is detected (Figure 1C, cf. lanes 1–3). The hydrophobic core from the PPL signal sequence could replace the authentic Syb2 tail anchor (Syb2:PPL) and facilitate a strong interaction with SRP54 (Figure 1C, lane 4). However, a nonfunctional version of the PPL signal sequence (Syb2:PPL.KO; see Luirink et al, 1992) gave no adducts with SRP54 (Figure 1C, lane 5). When the tail-anchor regions of Syb2 and Sec61β were replaced with hydrophilic sequences (Syb2:GC and Sec61β:GC, respectively), no crosslinking to SRP54 was observed (Figure 1C, lanes 6 and 8). Thus, the binding of cytosolic SRP to TA proteins requires an intact, hydrophobic, ER-targeting signal. The efficacy of the crosslinking to SRP54 observed in this assay was further investigated by analysing the PPL86 polypeptide after puromycin treatment to release it from the ribosome. In agreement with previous studies (Wiedmann et al, 1987), no adducts of PPL86 with SRP54 were observed (Figure 1C, lane 9; cf. Figure 1A, lane 3), confirming the efficiency of the puromycin release and the specificity of adduct formation between SRP54 and the TA proteins.
If the binding of SRP to the C-terminal hydrophobic targeting sequences of TA proteins was functionally significant, a defect in SRP binding should result in a defect in membrane integration, and we therefore investigated this possibility. Ribosome-bound versions of the polypeptides were generated in the presence of canine pancreatic microsomes, and the precursors then released from the ribosome by puromycin treatment. Parallel control reactions were carried out in the absence of any membranes, and integration was quantitatively assessed by determining the resistance of the membrane-associated precursor proteins to extraction with alkaline sodium carbonate solution (see Kutay et al, 1995; Whitley et al, 1996; cf. Figure 4A). As previously observed (cf. Kutay et al, 1995; Whitley et al, 1996), when no membranes were present in the incubations, only background levels of the TA precursors were recovered in the final pellet (see Supplementary data). Likewise, precursors with disrupted or deleted TA sequences showed very low levels of membrane integration (Figure 1D). In contrast, authentic TA proteins that could recruit SRP (Figure 1C) were efficiently membrane integrated (e.g. 27% of Syb2 and 48% of Sec61β; see Figure 1D). Given the role of the TA sequence in mediating stable membrane insertion, the reductions in integration observed upon perturbation of the hydrophobic TA sequences are likely to reflect defects in both targeting and membrane insertion. Nevertheless, taken together, these data are consistent with the hypothesis that the binding of SRP to TA proteins might play an important role in their delivery to the ER membrane.
Figure 4.
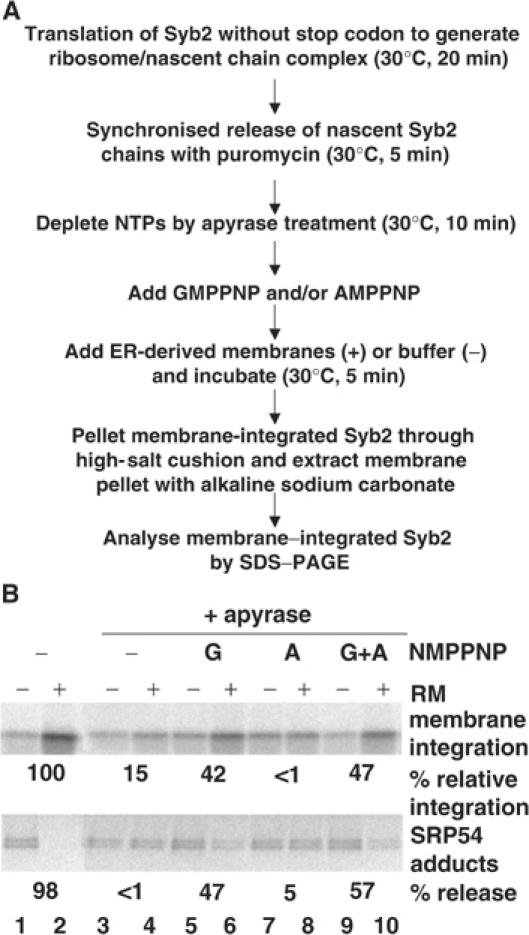
Membrane insertion of Syb2 is GTP dependent. (A) Flow diagram of the assay used to investigate the nucleotide dependence of Syb2 integration following apyrase treatment. (B) Syb2 RNA was translated for 20 min and treated with puromycin. Samples were depleted of nucleotide di- and triphosphates by treatment with apyrase, or mock treated. Aliquots were supplemented with combinations of AMPPNP, GMPPNP and RMs. After a 5 min incubation, the extent of membrane integration was assessed using sodium carbonate extraction, and SRP release assessed by DSS crosslinking and immunoprecipitation with αSRP54. Relative membrane integration (see the upper panel) was calculated by quantifying the increase in sodium carbonate-resistant material upon the addition of RMs (+), relative to a control sample with no added RMs (−), cf. Supplementary data. Mock-treated membranes were set at 100% relative membrane integration (lane 2). SRP release (lower panel) indicates the reduction in the amount of SRP54 adduct caused by the addition of RMs.
We examined the precise relationship between SRP binding and the state of the TA polypeptide, focusing on the question of whether the interaction was strictly post-translational. To this end, we analysed the association of SRP with Syb2 when it was present as either an artificially generated ribosome/nascent chain complex (RNC) or a fully released polypeptide. The ribosome attachment of the nascent chain was specifically monitored by analysing the crosslinking of Syb2 to NACα, a component that is known to interact with nascent chains in a sequence-independent, but strictly ribosome-dependent, manner (Wiedmann et al, 1994). In the case of PPL86, the bulk of the nascent chains and all their adducts with both SRP54 and NACα co-isolated with the ribosomal pellet after stabilisation with cycloheximide (Figure 2, lane 2). After puromycin treatment, the PPL86 nascent chains were recovered primarily in the supernatant, and adducts with both SRP54 and NACα were absent (Figure 2, lane 3). Hence, PPL86 will only associate with SRP54 and NACα as a nascent, ribosome-attached, polypeptide chain (Wiedmann et al, 1994; Plath and Rapoport, 2000). Syb2 RNCs responded to both cycloheximide and puromycin treatment in the same fashion as PPL86 RNCs (Figure 2, Syb2 nascent chain, lanes 1–4), and the Syb2-NACα adducts were similarly restricted to the ribosome-attached form of the polypeptides (Figure 2, lane 2). In contrast, the Syb2 adduct with SRP54 was seen exclusively in the ribosome-free supernatant and primarily after puromycin release (Figure 2, lane 3). A small fraction of Syb2 chains were lost from the ribosome even with cycloheximide stabilisation (Figure 2, Syb 2 nascent chain, cf. lanes 1 and 2), resulting in the appearance of some SRP54 adducts under these conditions (Syb2, αSRP54, lane 1). We conclude that SRP binds to TA proteins in a strictly post-translational manner.
Figure 2.
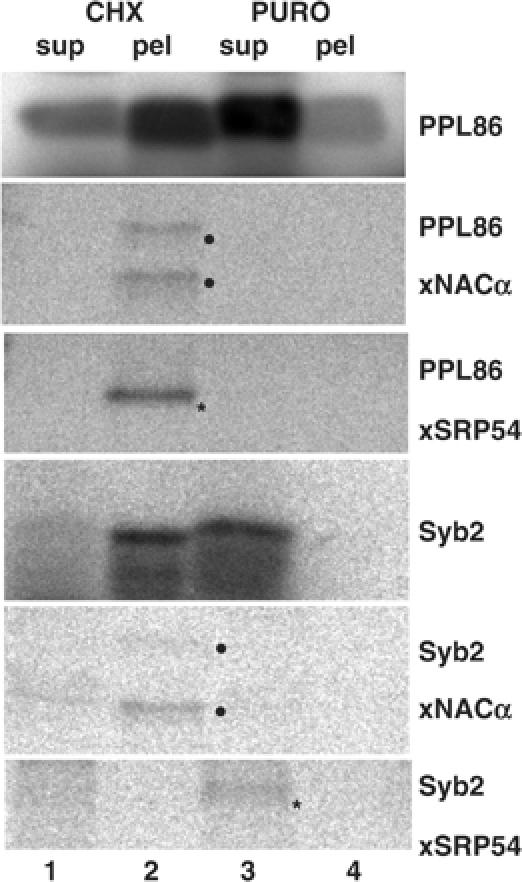
TA protein association with SRP54 is strictly post-translational. Syb2 or PPL86 RNA was translated for 5 min, treated with cycloheximide or puromycin, and crosslinked using DSS. RNCs were pelleted through a high-salt cushion (pel), leaving free polypeptides in the supernatant (sup). A fraction of each sample was analysed prior to crosslinking (PPL85 and Syb2), while the remainder was used for the immunoprecipitation of adducts with NACα and SRP54 as indicated.
To determine the region of the TA protein that binds to SRP54, we performed a site-specific crosslinking analysis using four precursor proteins that each contains a single, naturally occurring, cysteine residue within its hydrophobic tail-anchor sequence. When crosslinking was mediated by a short (6 Å), bifunctional, cysteine reactive reagent (o-PDM), distinct adducts with SRP54 were detected using both the full-length proteins (Figure 3, lanes 1 and 3, o-PDM), or their hydrophobic tail-anchor regions alone (Figure 3, lanes 5 and 7, o-PDM). As an additional control, point mutants of each of these four polypeptides, where the single cysteine was altered to a leucine residue, were analysed in parallel. In this case, no significant o-PDM-dependent crosslinking to SRP54 was detected (Figure 3, lanes 2, 4, 6 and 8), confirming the authenticity of the adducts seen with the cysteine-containing polypeptides. In some cases, trace levels of o-PDM-dependent adducts were obtained with derivatives lacking any cysteine residues (Figure 3, lanes 4 and 8). This probably reflects the low-efficiency modification of amino groups by o-PDM. When the same series of polypeptides was analysed using a bifunctional crosslinking reagent specific for the several amino groups present in each polypeptide, adducts with SRP54 were observed with all of the precursors (Figure 3, lanes 1–8, DSS). Taken together, the ability of SRP54 to be crosslinked to a single cysteine residue located near the centre of their ER-targeting signals strongly suggests that SRP associates directly with the hydrophobic regions of TA proteins.
Figure 3.
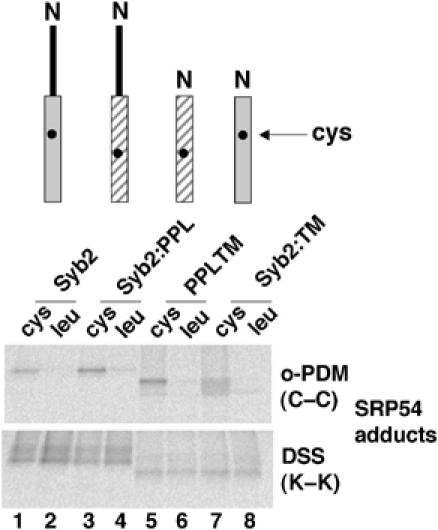
SRP associates with the hydrophobic domain of TA proteins. Stop codon minus RNAs encoding the indicated polypeptides with either a single cysteine probe (upper panel) or a leucine at the equivalent location (lower panel) was translated for 15 min. The resulting RNCs were then isolated through a low-salt cushion to separate the precursors from DTT in the translation reaction and facilitate crosslinking with the bifunctional maleimide o-PDM. Nascent chains were released from ribosomes with puromycin, treated with DSS or o-PDM, and adducts with SRP54 recovered by immunoprecipitation. The relative efficiency with which each precursor was synthesised was established by quantitative phophorimaging prior to immunoprecipitation, and equivalent amounts of radiolabelled polypeptides were used for subsequent immunoisolation as for Figure 1C.
We next investigated the biological significance of the specific interaction between SRP and TA proteins that we had defined. A hallmark of the SRP-dependent targeting pathway is its dependency upon GTP (Keenan et al, 2001). This GTP requirement can be substituted with nonhydrolysable analogues such as GMPPNP in the context of typical in vitro assays, where a single round of SRP-dependent targeting is normally sufficient to promote significant membrane integration (Connolly et al, 1991; High et al, 1991). Under these conditions, the lack of SRP recycling from the ER membrane that results from the absence of GTP hydrolysis has little effect (Connolly et al, 1991). As a read-out for protein targeting, we determined the efficiency of Syb2 membrane integration (cf. Figure 1D), in this case following various pretreatments of the cell-free translation reaction (outlined in Figure 4A). In the control reaction, we found that ∼7% of total Syb2 was specifically integrated into ER membranes during a short, 5 min, post-treatment incubation (Figure 4B, cf. lanes 1 and 2, upper panel). This level of efficiency is on a par with previous studies where quantification has been carried out (Whitley et al, 1996; Kim et al, 1999; Lan et al, 2000), and reflects the relatively short incubation period used for this insertion assay (cf. Figure 1D, where a 20 min incubation was used).
As previously observed in other studies, the substantial depletion of NTPs from the cell-free system prior to membrane addition largely abolished the post-translational integration of Syb2 into ER-derived membranes (Figure 4B, cf. lanes 2 and 4, upper panel; see Kutay et al, 1995). However, we made the striking observation that the nonhydrolysable GTP analogue GMPPNP could restore Syb2 membrane integration to 42% of the original level (Figure 4B, cf. lanes 5 and 6, upper panel). In contrast, the addition of the ATP analogue AMPPNP caused no increase in membrane integration (Figure 4B, lanes 7 and 8, upper panel), confirming that the stimulation was specific for GTP analogues. Likewise, a combination of GMPPNP and AMPPNP gave the same stimulation as GMPPNP alone (Figure 4B, cf. lanes 5 and 6 with 9 and 10, upper panel). If the addition of GMPPNP were enabling a biologically productive role for SRP in promoting the authentic membrane integration of Syb2, one would expect a reciprocal release of SRP from the TA proteins upon the addition of ER membranes bearing the SRP receptor. Consistent with this proposal, we find that the addition of GMPPNP and membranes results in a significant reduction of Syb2-SRP54 adduct formation to a level that mirrors the stimulation of membrane insertion observed (Figure 4B, lanes 3–6, 9 and 10, lower panel). As with integration, the addition of AMPPNP in combination with microsomal membranes causes no significant reduction in SRP54 adduct formation. We conclude that Syb2 can become membrane integrated via a GTP-dependent pathway that correlates with the release of SRP from the polypeptide.
To further investigate the role of SRP in the targeting of TA proteins, we manipulated the SRP receptor (SR), which is sensitive to low concentrations of trypsin that specifically remove the SRα subunit but leave the SRβ subunit largely intact (Andrews et al, 1989; Miller et al, 1995). In this case, we found that ∼8% of the total Syb2 present in the incubation was specifically membrane integrated in the control reaction (Figure 5A, cf. lanes 1 and 2). If the ER-derived membranes were first treated with trypsin, we found that their capacity for integrating Syb2 was reduced to 23% of the original level, consistent with a role for specific ER proteins in mediating targeting/integration (Figure 5A, lane 4; cf. Kutay et al, 1995). However, when these trypsinised membranes were specifically reconstituted with recombinant SR (Fulga et al, 2001), a substantial recovery of Syb2 membrane integration to 65% of the original level was obtained (Figure 5A, lane 5). In contrast, the addition of recombinant SR in the absence of trypsin treatment had only a marginal effect on Syb2 integration (Figure 5A, lanes 2 and 3). To confirm the specificity of this reconstitution assay, we determined the efficiency of targeting of the α subunit of SR (SRα). SRα is a peripheral ER membrane protein that binds directly to the integral membrane β subunit via a mechanism that is independent of SRP (Andrews et al, 1989). In contrast to the effect upon Syb2 integration, SRα was relatively unaffected by trypsin treatment and SR reconstitution, with ∼7% of the total input being specifically recovered with the membrane fraction under all conditions (Figure 5B, cf. lanes 2–5).
Figure 5.
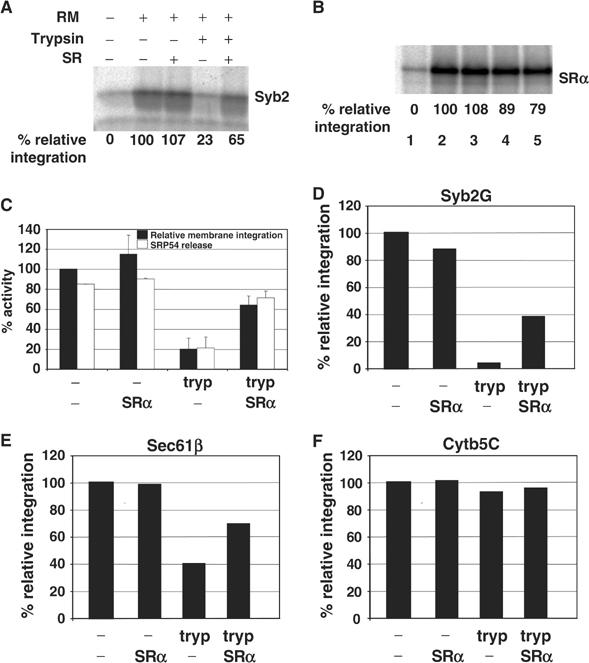
Membrane insertion of Syb2 is dependent on the SRP receptor. (A) Syb2 RNA lacking a stop codon was translated for 20 min and then treated with puromycin. RMs were treated with trypsin or mock treated as indicated, re-isolated by centrifugation, and added to the translation mix with or without soluble SR. After a 5 min incubation, membrane integration was assessed by resistance to sodium carbonate extraction. Samples were separated by SDS–PAGE. Quantification of membrane integration was performed as described for Figure 4B, with mock-treated RM set to 100%. Lane 1 shows the level of background signal obtained in the absence of any added membranes. This contribution was subtracted from the signals obtained in the presence of membranes during quantification, and is therefore labelled as 0% relative integration. (B) SRα RNA was translated in vitro and the membrane association of the resulting protein analysed as described in (A). (C) The membrane integration of Syb2 into various RM preparations was performed as described in (A), except that the RMs were reconstituted with in vitro-synthesised SRα in place of the recombinant protein used in panels A and B. SRP54 release indicates the percentage reduction in SRP54 adduct formation caused by the addition of the various RM preparations. In this case, the mean and standard error of two independent experiments are shown. (D) The membrane integration of Syb2G into various RM preparations was established as described in (C), except that the level of N-glycosylation was assayed in place of the amount of material resistant to sodium carbonate extraction. (E) The membrane integration of Sec61β was determined as described in (C). (F) The membrane integration of Cytb5C was determined as described in (C).
In vitro-synthesised SRα is also known to rescue the targeting defect resulting from the trypsin treatment of ER membranes (Andrews et al, 1989). As with the recombinant protein, we found that the addition of in vitro-synthesised SRα resulted in a significant recovery of specific membrane integration after trypsinisation (20–64%; Figure 5C). This increase in membrane integration was accompanied by a comparable increase in the release of SRP from the Syb2 polypeptides, as judged by the accompanying reduction in its crosslinking to the SRP54 subunit (21–71%; Figure 5C).
To further verify that we were observing an SR-dependent stimulation in authentic membrane insertion, we exploited a derivative of Syb2 with a site for N-linked glycosylation at its C-terminus (Syb2G; see Figure 1B). The efficiency of Syb2G N-glycosylation (∼5% of total input) was comparable to that seen in previous studies (Kutay et al, 1995), and we used the quantity of glycosylated product as a specific measure of complete membrane insertion. As with Syb2, we found that, when trypsinised pancreatic microsomes were repopulated with in vitro synthesised SRα, an ∼40% recovery in the levels of N-glycosylated material was obtained (Figure 5D, cf. tryp and tryp plus SRα samples). Hence, Syb2G responds to the re-addition of SRα in the same manner as native Syb2, and we conclude that SR stimulates the topologically correct insertion of Syb2 at the ER membrane.
In order to establish whether the SR-dependent integration of TA proteins was a general effect, we investigated two other precursors, Sec61β, which we had found to crosslink SRP54 (Figure 1A), and a point mutant of cytochrome b5 (Cytb5C), which can integrate into the ER independently of SRP (Anderson et al, 1983). In the case of Sec61β, while the residual integration observed after trypsin treatment was higher than that for Syb2 or Syb2G (cf. Figure 5C–E), an ∼30% stimulation in membrane integration was observed upon repopulation of trypsinised membranes with in vitro synthesised SRα (Figure 5E). In contrast, the integration of Cytb5C was essentially unaffected by either trypsinisation or subsequent re-addition of SRα (Figure 5F). We therefore conclude that some TA proteins can be membrane integrated in an SR-dependent manner, while others, such as cytochrome b5, are efficiently integrated independently of this pathway (cf. Anderson et al, 1983).
In order to investigate directly the role of SRP in TA protein targeting, we studied the effect of purified SRP on the integration of newly synthesised Syb2. To separate Syb2 polypeptides from SRP and other cellular components present in the reticulocyte lysate (cf. Figure 1A), we isolated Syb2 RNCs through a high-salt sucrose cushion. This process specifically removes SRP that is bound only to ribosomes, but does not deplete the more tightly associated SRP bound to RNCs bearing an exposed signal sequence (Hauser et al, 1995). Following their isolation, the Syb2 nascent chains were released from their ribosomes by puromycin treatment in the presence or absence of SRP and GTP. The membrane insertion of Syb2 polypeptides prepared under these conditions was tested using high-salt-washed microsomes, which are free of endogenous SRP (Walter and Blobel, 1983a). The presence of both SRP and GTP during the release of Syb2 from RNCs stimulated its subsequent membrane insertion by more than three-fold, such that ∼9% of the total precursor was specifically membrane integrated (Figure 6A, upper panel, cf. lanes 2 and 5). The specificity of this effect was demonstrated by the failure of either SRP or GTP to provide any significant stimulation alone (Figure 6A, upper panel, lanes 2–4). The inability of GTP alone to stimulate membrane insertion confirms that the Syb2 RNCs are unable to recruit SRP in a high-salt-resistant manner. This is entirely consistent with buried tail-anchor sequences being unavailable for SRP binding when present in ribosome-bound chains (cf. Figure 7).
Figure 6.
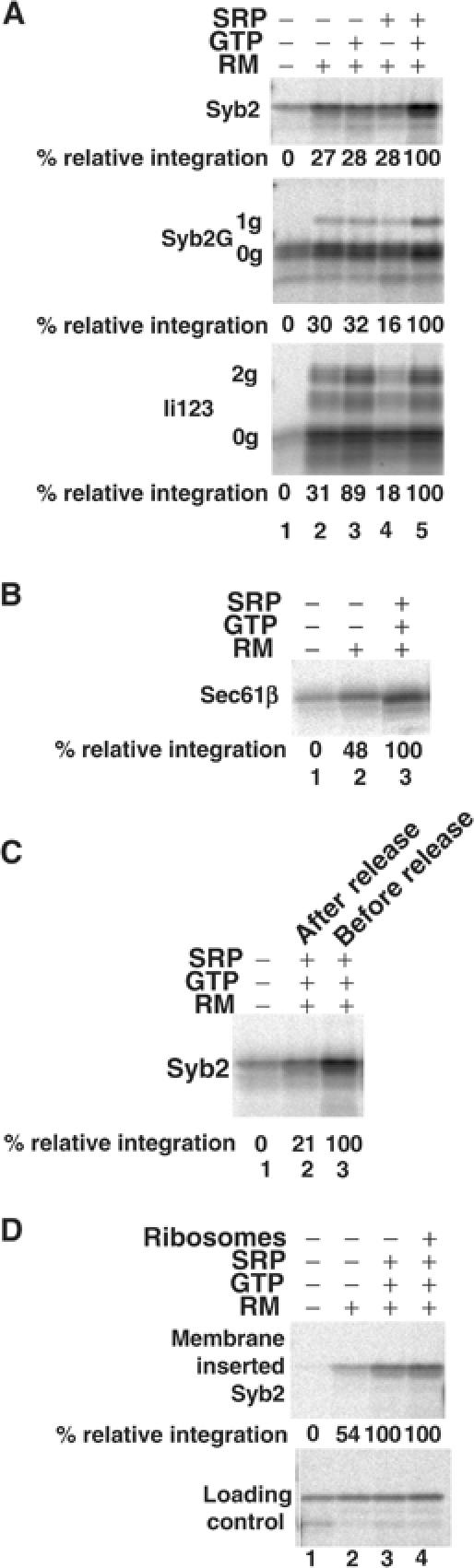
Membrane insertion of Syb2 is SRP dependent. (A) Syb2 or Syb2G was released from isolated RNCs by puromycin treatment in the presence or absence of GTP and SRP, and then incubated with SRP-depleted membranes for 10 min. With Ii123, the isolated RNCs were incubated with SRP-depleted membranes for 10 min before the puromycin treatment, and the incubation continued for a further 10 min after puromycin treatment in order to mimic cotranslational targeting. Membrane integration was assessed by extraction with sodium carbonate solution and quantification of Syb2 (upper panel), Syb2-1g product (middle panel) or Ii123-2g product (lower panel). Lane 1 shows the level of background signal obtained in the absence of any added membranes, and is set at 0% relative integration, as indicated in the legend to Figure 5A. (B) Sec61β was released from isolated RNCs by puromycin treatment in the presence or absence of GTP and SRP, and then incubated with SRP-depleted membranes for 10 min. Membrane integration was determined as described for Syb2 in (A). (C) Syb2 integration was assayed as for (A), except that the ‘after-release' sample was treated with puromycin before the addition of SRP and GTP. (D) Syb2 integration (upper panel) was assayed as for (A), except that ribosomes were removed by centrifugation prior to incubation with membranes. In one case (lane 4), an equivalent amount of purified, nonprogrammed, ribosomes was added back. To allow for potential losses of material during centrifugation, a fraction of the total products wase analysed and used to normalise the quantification of inserted Syb2 (loading control).
Figure 7.
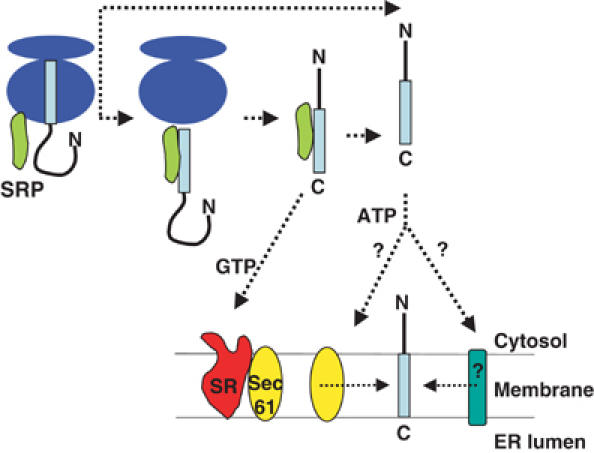
Unifying model for TA protein targeting to the ER membrane. Upon termination of translation, the insertion sequence of the TA protein emerges from the ribosomal exit tunnel, where an interaction with SRP is favoured by its proximity. If SRP remains bound, the TA protein can be targeted to SR at the ER membrane, allowing the possibility of its coordinated transfer to the Sec61 translocon for membrane insertion. Alternatively, where SRP fails to bind the TA protein, or binds and then becomes dissociated, a complementary ATP-dependent pathway can deliver the precursor to the ER membrane. The ATP pathway may deliver the TA protein to the Sec61 complex and/or novel component(s) for membrane insertion.
As with the analysis of SR function, we again exploited the N-glycosylated Syb2G derivative. When tested in the same assay, the membrane insertion of Syb2G, as assessed by the level of its N-glycosylation, was also stimulated over three-fold by the addition of SRP plus GTP, but not by either SRP or GTP alone (Figure 6A, middle panel, cf. lanes 2–5, see 1g product). Hence, Syb2G responds to SRP and GTP in the same manner as native Syb2, confirming authentic membrane insertion.
We compared the insertion characteristics of Syb2 with those of the 123-residue N-terminal fragment of invariant chain (Ii123), a polypeptide known to insert at the ER in a cotranslational and SRP-dependent manner (High et al, 1991). Ii123 possesses a single-membrane span, displays its N-terminus to the cytosol, and its membrane insertion can be monitored by the glycosylation of two asparagines in its C-terminus. Since Ii123 is only capable of membrane integration when present as a ribosome-bound precursor, the purified RNCs were added to ER microsomes before puromycin treatment in order to mimic a cotranslational targeting assay (cf. High et al, 1991). In this case, we found that the membrane insertion of Ii123 was stimulated three-fold by the addition of GTP, in either the presence or absence of additional SRP, to result in a total of ∼23% of the input being fully N-glycosylated (Figure 6A, bottom panel, cf. lanes 2, 3 and 5, see 2g product). Significantly, the addition of SRP alone failed to have any effect upon Ii123 membrane integration (Figure 6A, bottom panel, cf. lanes 2 and 4, 2g product). This characteristic confirms previous studies showing that SRP can bind to the exposed signal-anchor region of Ii to yield a high-salt-stable RNC–SRP complex (High et al, 1991). This contrasts with the behaviour of Syb2, which we find is unable to recruit SRP in a high-salt-resistant form when present as an RNC, with SRP binding only taking place once the insertion sequence is released from the ribosomal exit tunnel by the termination of translation. Our analysis of SR dependency had indicated that Sec61β could also utilise the SRP-dependent targeting route (Figure 5E), and we therefore analysed its membrane integration using this SRP-depleted system. In this case, we found a two-fold stimulation of Sec61β integration upon the addition of both SRP and GTP (Figure 6B, cf. lanes 2 and 3), confirming that Sec61β can also exploit the SRP-dependent targeting (cf. Figure 5E).
We investigated the coordination of the SRP interaction with Syb2 by adding SRP either during or after the puromycin-mediated release of Syb2 chains from the ribosome. The maximal stimulatory effect of SRP addition (∼10% of total input being membrane integrated; Figure 6C, lane 3) was only observed if it was present during the release of Syb2 (Figure 6C, lanes 2 and 3), suggesting that SRP recruitment occurs shortly after the release of the polypeptide from the ribosome. Finally, we determined whether there was any potential role for ribosomes in the subsequent targeting of the Syb2–SRP complex to the ER membrane. However, we found that when ribosomes were removed from the reaction mixture after the release of the nascent chains from their RNCs, SRP was still able to stimulate membrane insertion (Figure 6D, lanes 1–3). Furthermore, the re-addition of purified ribosomes had no effect on insertion efficiency, which in this case remained at a level of ∼19% of the input (Figure 6D, lane 4). Hence, any potential role of the ribosome is restricted to promoting the initial interaction between Syb2 and SRP. Taken together, these data suggest that SRP scans the Syb2 polypeptide as it emerges from the ribosome, and can bind its tail anchor during the release of the nascent chain, thereby forming a complex that is competent for ER targeting.
Discussion
We have shown that SRP can associate post-translationally with the hydrophobic membrane insertion sequences of some TA proteins, and deliver these polypeptides to the ER membrane by a pathway requiring the SRP receptor and GTP. We find that SRP must be present during the release of the TA protein from the ribosome, consistent with the ability of SRP to bind at the ribosomal exit tunnel and scan the emerging nascent chain for appropriate signal sequences (Pool et al, 2002; Halic et al, 2004). However, the action of SRP in the targeting of TA proteins differs fundamentally from its strictly cotranslational function in the targeting of classical membrane proteins. While SRP forms a targeting complex with the ribosome-bound nascent chains of cotranslational precursors, the SRP–TA protein complex targets to membranes independently of ribosomes. This novel post-translational function for SRP appears to be unique to TA proteins, and is facilitated by the ability of SRP to remain bound to newly synthesised TA proteins after their release from the ribosome. This capacity for post-translational association is clearly not a specific feature of tail-anchor sequences per se, since the well-characterised ‘cotranslational' PPL signal sequence can functionally replace the native Syb2 tail anchor. A plausible explanation for this behaviour is that the ability of SRP to bind polypeptides after release from the ribosome is dependent upon the folding of regions flanking the ER-targeting signal.
Based on the levels at which we can reconstitute the SRP-dependent membrane integration of Syb2 (see Figures 4, 5A, C and D) and Sec61β (Figure 5E), our data indicate that this pathway can account for between one-third and one-half of their membrane insertion observed in vitro. Interestingly, with Cytb5, we find no requirement for an intact SR, suggesting that this TA protein is integrated exclusively by an alternative route. This is entirely consistent with a previous study of Cytb5 biosynthesis (Anderson et al, 1983), and indicates that this precursor uses only a poorly characterised ATP-dependent pathway for membrane integration (cf. Yabal et al, 2003). In the case of Syb2 and Sec61β, we propose that these precursors can utilise both the SRP-dependent route characterised in this study and the alternative ATP-dependent pathway (Kutay et al, 1995; Steel et al, 2002; Yabal et al, 2003).
On the basis of the work presented here and in previous studies, we propose a unifying model to describe the molecular basis for the biogenesis of TA proteins. Hence, the ATP-dependent targeting route would act as the primary pathway for the targeting of precursors such as Cytb5, but as a complementary pathway for proteins such as Syb2 and Sec61β that can exploit the SRP-dependent route (Figure 7). A precedent for the use of multiple targeting pathways by individual proteins has been established in Saccharomyces cerevisiae, where different precursors exhibit varying preferences for either a cotranslational, SRP-dependent pathway or a post-translational, SRP-independent pathway (Ng et al, 1996). We observe a similar phenomenon with the post-translational integration of TA proteins, and find that different precursors display distinct capacities to use the SRP-dependent targeting route (cf. Figure 7).
In the case of a TA protein, the cotranslational recognition of the ER-targeting signal by SRP is prevented by the signal's inaccessibility within the ribosome. Once the nascent chain is released, the signal sequence may only be transiently available for SRP binding due to sequestration by rapid folding or by the binding of other cellular factors. Consistent with this suggestion, we observe a limited window of opportunity for SRP to bind productively to TA proteins after their release from the ribosome. Alternatively, the released TA proteins might aggregate in the absence of SRP, which may play some role in maintaining their solubility (cf. Sanz and Meyer, 1988). In any case, the ability of SRP to bind at the exit site on the ribosome is likely to be central to its ability to exploit the limited opportunity for binding TA proteins. There is a substantial increase in the affinity of SRP for ribosomes that are engaged in translation, suggesting that the majority of SRP will be associated with ribosomes synthesising polypeptides (Flanagan et al, 2003). While this provides a potential opportunity for SRP to scan emerging TA proteins, the limited concentration of SRP relative to that of ribosomes (Flanagan et al, 2003) implies that there is a significant probability of a newly synthesised TA protein failing to bind to SRP. This scenario would obviously favour the presence of an alternative ER-targeting route for TA proteins (cf. Figure 7).
For cotranslational membrane integration, the interaction between SRP and its receptor is the first step in a highly coordinated mechanism that leads to the transfer of the RNC to the Sec61 translocon (Song et al, 2000; Fulga et al, 2001; Helmers et al, 2003). Hence, the ability of TA proteins such as Syb2 to be targeted to the ER via an SRP-dependent pathway is entirely consistent with such precursors being integrated via the classical ER translocon (Figure 7, cf. Abell et al, 2003). The ER components responsible for the integration of precursors such as Cytb5 that are delivered via the ATP-dependent pathway remain uncertain (cf. Yabal et al, 2003). While a post-translational role for SRP in the mammalian cytosol is unprecedented, a paradigm is provided by the stromal SRP of chloroplasts. In this case, several precursors are inserted into the internal thylakoid membrane in a strictly post-translational process that is dependent upon stromal SRP (reviewed by Robinson et al, 2001). Our data show that the post-translational action of SRP is more widespread and generic than previously believed.
Materials and methods
Antibodies
Rabbit polyclonal antibodies recognising SRP54 and NACα were gifts from B Dobberstein, ZMBH, Heidelberg, Germany, and M Wiedmann, Sloan-Kettering Cancer Center, New York, USA, respectively. The anti-ubiquitin antibody was from Stressgen.
Transcription
Rat synaptobrevin 2 was in pBluescript, and human Sec61β and human invariant chain in pSPUTK (Abell et al, 2003). Preprolactin and canine SRα were in pGEM4. The coding region of human cytochrome b5 (IMAGE ID: 4701789) was subcloned into pSPUTK. Cytb5C is a point mutant with the wild-type leucine at residue 81 replaced by a cysteine. Transcription templates for TA proteins were prepared by PCR and all lacked a stop codon (see Figure 1B for relevant sequences). The DNA templates for PPL86 and SRα were generated by digestion with PvuII and EcoRI, respectively. Transcripts were synthesised using T7 RNA polymerase for PPL86 and SRα, T3 RNA polymerase for Syb2-derived constructs and SP6 RNA polymerase for all other templates, according to the manufacturer's instructions (New England Biolabs).
Translation and membrane insertion
Proteins were synthesised using rabbit reticulocyte lysate (Promega) prespun at 200 000 g for 10 min to remove any contaminating membranes. Incubations were at 30°C in the presence of [35S]-methionine, according to the manufacturer's instructions. Cycloheximide was used at 2.5 mM and puromycin at 1 mM, with subsequent incubation at 30°C for 5 min. Microsomes were prepared from canine pancreas (Walter and Blobel, 1983a) and added to a final concentration of 1.5–2.0 OD280/ml. Microsomes were analysed for TA protein insertion by centrifugation through 100 μl HSC (500 mM sucrose, 500 mM KOAc, 5 mM Mg(OAc)2, 50 mM Hepes–KOH pH 7.9) at 132 000 g for 5 min. For sodium carbonate extraction, the resulting pellet was resuspended in 100 μl of cold 0.1 M Na2CO3, incubated on ice for 10 min, and repelleted.
RNC isolation
RNCs were recovered by layering 30–50 μl translations over 150 μl LSC (250 mM sucrose, 100 mM KOAc, 5 mM (MgOAc)2, 50 mM Hepes–KOH, pH 7.9) or HSC, as appropriate, followed by centrifugation at 213 000 g for 20 min.
Crosslinking and immunoprecipitation
Samples in lysate or LSC were incubated at 30°C for 10 min with either 1 mM disuccinimidyl suberate (DSS) (Pierce) or 1 mM ortho-phenylenedimaleimide (o-PDM) (Sigma), diluted from a 20 mM stock in DMSO. Crosslinking was stopped with 50 mM glycine (DSS) or 10 mM 2-mercaptoethanol (o-PDM). Denaturing immunoprecipitation was performed as described previously (Abell et al, 2003).
Apyrase treatment
In all, 1 U of type III apyrase (Sigma) was added per 30 μl translation and incubated at 30°C for 10 min. The reaction was supplemented with microsomes or microsome buffer (50 mM Hepes, pH 7.8, 250 mM sucrose, 1 mM dithiothreitol (DTT)), 0.5 mM Na3GMPPNP (Sigma) or 1.5 mM NaOAc and 0.5 mM Li4AMPPNP (Roche) or H2O, and incubated at 30°C for a further 10 min.
SR knockout and reconstitution
Microsomes were incubated on ice with 5 μg/ml trypsin for 60 min, phenylmethylsulphonyl fluoride was added to 2 mM, and the microsomes were then layered onto HSC buffer and recovered by centrifugation at 132 000 g for 5 min. The resulting membrane pellet was resuspended in microsome buffer (Walter and Blobel, 1983a). Mock-treated microsomes were prepared in parallel, omitting the trypsin treatment. Soluble SR is comprised of full-length SRα and an N-terminally deleted SRβ lacking its transmembrane domain. It was prepared as previously described (Fulga et al, 2001) and added to incubations at a final concentration of 75 nM (Pool et al, 2002). Alternatively, SRα was reconstituted onto trypsinised (or mock-treated) microsomes by pretranslation with the SRα transcript, where [35S]-methionine was substituted with 20 μM nonlabelled methionine (Andrews et al, 1989). The resulting membranes were reisolated by pelleting through HSC and resuspended in microsome buffer.
Reconstitution of SRP-dependent targeting
RNCs were pelleted twice through HSC as described above with 2.5 mM cycloheximide and 1 mM DTT in all buffers. The final RNC pellet was resuspended in LSC and 1 mM DTT. Ribosomes were prepared from reticulocyte lysate using the same method and added to reactions at a level corresponding to that in translations. GTP was added at 0.5 mM and SRP was added at ∼12.5 nM. Treatment with 1 mM puromycin for 5 min at 30°C was performed either before or after the addition of SRP and GTP, and then followed by a 10 min incubation at 30°C with high-salt-washed, SRP-depleted RMs (final concentration 1.5–2.0 OD280/ml). In cotranslational assays for Ii123, the puromycin treatment was performed after the incubation with membranes, and was extended to 10 min. Ribosome depletion was performed by centrifugation of samples at 213 000 g for 20 min. SRP and K-RMs were prepared as previously described (Walter and Blobel, 1983a, 1983b).
Gel electrophoresis
Samples were heated to 70°C for 10 min in SDS/PAGE sample buffer and the solubilised material was resolved on 16% polyacrylamide Tris–glycine gels under denaturing conditions. Gels were fixed, dried and then exposed to phosphorimage plates, which were read using a Fuji BAS-1800 phosphorimager. Radiolabelled products separated by SDS–PAGE were quantified using Aida software.
Supplementary Material
Supplementary data
Acknowledgments
This work was supported by funding from the Biotechnology and Biological Sciences Research Council (BBSRC), by the award of a BBSRC Professorial Fellowship to SH, and by grants from the European Union and the Deutsche Forschungs-gemeinschaft. We thank Bernhard Dobberstein for his support, Martin Wiedmann for the NACα antisera and our colleagues for their invaluable comments during the preparation of the manuscript.
References
- Abell BM, Jung M, Oliver JD, Knight BC, Tyedmers J, Zimmermann R, High S (2003) Tail-anchored and signal-anchored proteins utilize overlapping pathways during membrane insertion. J Biol Chem 278: 5669–5678 [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Mostov KE, Blobel G (1983) Mechanisms of integration of de novo-synthesized polypeptides into membranes: signal-recognition particle is required for integration into microsomal membranes of calcium ATPase and of lens MP26 but not of cytochrome b5. Proc Natl Acad Sci USA 80: 7249–7253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews DW, Lauffer L, Walter P, Lingappa VR (1989) Evidence for a two-step mechanism involved in assembly of functional signal recognition particle receptor. J Cell Biol 108: 797–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, Colombo S, Pedrazzini E (2003) The tale of tail-anchored proteins: coming from the cytosol and looking for a membrane. J Cell Biol 161: 1013–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese N, Gazzoni I, Barberi M, Colombo S, Pedrazzini E (2001) Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways. Mol Biol Cell 12: 2482–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T, Rapiejko PJ, Gilmore R (1991) Requirement of GTP hydrolysis for dissociation of the signal recognition particle from its receptor. Science 252: 1171–1173 [DOI] [PubMed] [Google Scholar]
- Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM (2004) Substrate twinning activates the signal recognition particle and its receptor. Nature 427: 215–221 [DOI] [PubMed] [Google Scholar]
- Flanagan JJ, Chen JC, Miao Y, Shao Y, Lin J, Bock PE, Johnson AE (2003) Signal recognition particle binds to ribosome-bound signal sequences with fluorescence-detected subnanomolar affinity that does not diminish as the nascent chain lengthens. J Biol Chem 278: 18628–18637 [DOI] [PubMed] [Google Scholar]
- Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM (2004) Heterodimeric GTPase core of the SRP targeting complex. Science 303: 373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulga TA, Sinning I, Dobberstein B, Pool MR (2001) SRbeta coordinates signal sequence release from SRP with ribosome binding to the translocon. EMBO J 20: 2338–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore R, Collins P, Johnson J, Kellaris K, Rapiejko P (1991) Transcription of full-length and truncated mRNA transcripts to study protein translocation across the endoplasmic reticulum. In Methods in Cellular Biology, Tartakoff AM (ed), Vol. 34, pp 223–239. New York: Academic Press [DOI] [PubMed] [Google Scholar]
- Halic M, Becker T, Pool MR, Spahn CM, Grassucci RA, Frank J, Beckmann R (2004) Structure of the signal recognition particle interacting with the elongation-arrested ribosome. Nature 427: 808–814 [DOI] [PubMed] [Google Scholar]
- Hauser S, Bacher G, Dobberstein B, Lutcke H (1995) A complex of the signal sequence binding protein and the SRP RNA promotes translocation of nascent proteins. EMBO J 14: 5485–5493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmers J, Schmidt D, Glavy JS, Blobel G, Schwartz T (2003) The beta-subunit of the protein-conducting channel of the endoplasmic reticulum functions as the guanine nucleotide exchange factor for the beta-subunit of the signal recognition particle receptor. J Biol Chem 278: 23686–23690 [DOI] [PubMed] [Google Scholar]
- High S, Flint N, Dobberstein B (1991) Requirements for the membrane insertion of signal-anchor type proteins. J Cell Biol 113: 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keenan RJ, Freymann DM, Stroud RM, Walter P (2001) The signal recognition particle. Annu Rev Biochem 70: 755–775 [DOI] [PubMed] [Google Scholar]
- Kim PK, Hollerbach C, Trimble WS, Leber B, Andrews DW (1999) Identification of the endoplasmic reticulum targeting signal in vesicle-associated membrane proteins. J Biol Chem 274: 36876–36882 [DOI] [PubMed] [Google Scholar]
- Kutay U, Ahnert-Hilger G, Hartmann E, Wiedenmann B, Rapoport TA (1995) Transport route for synaptobrevin via a novel pathway of insertion into the endoplasmic reticulum membrane. EMBO J 14: 217–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L, Isenmann S, Wattenberg BW (2000) Targeting and insertion of C-terminally anchored proteins to the mitochondrial outer membrane is specific and saturable but does not strictly require ATP or molecular chaperones. Biochem J 349: 611–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luirink J, High S, Wood H, Giner A, Tollervey D, Dobberstein B (1992) Signal sequence recognition by an E. coli ribonucleoprotein particle. Nature 359: 741–743 [DOI] [PubMed] [Google Scholar]
- Miller JD, Tajima S, Lauffer L, Walter P (1995) The beta subunit of the signal recognition particle receptor is a transmembrane GTPase that anchors the alpha subunit, a peripheral membrane GTPase, to the endoplasmic reticulum membrane. J Cell Biol 128: 273–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng DT, Brown JD, Walter P (1996) Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J Cell Biol 134: 269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen EA, Brunsting JF, Song J, Kampinga HH, Morimoto RI (2000) Bag1 functions in vivo as a negative regulator of Hsp70 chaperone activity. Mol Cell Biol 20: 1083–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Rapoport TA (2000) Spontaneous release of cytosolic proteins from posttranslational substrates before their transport into the endoplasmic reticulum. J Cell Biol 151: 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool MR, Stumm J, Fulga TA, Sinning I, Dobberstein B (2002) Distinct modes of signal recognition particle interaction with the ribosome. Science 297: 1345–1348 [DOI] [PubMed] [Google Scholar]
- Robinson C, Thompson SJ, Woolhead C (2001) Multiple pathways used for the targeting of thylakoid proteins in chloroplasts. Traffic 2: 245–251 [DOI] [PubMed] [Google Scholar]
- Sanz P, Meyer DI (1988) Signal recognition particle (SRP) stabilizes the translocation-competent conformation of presecretory proteins. EMBO J 7: 3553–3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel V, Walter P (1988) The affinity of signal recognition particle for presecretory proteins is dependent on nascent chain length. EMBO J 7: 1769–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Raden D, Mandon EC, Gilmore R (2000) Role of Sec61alpha in the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell 100: 333–343 [DOI] [PubMed] [Google Scholar]
- Steel GJ, Brownsword J, Stirling CJ (2002) Tail-anchored protein insertion into yeast ER requires a novel posttranslational mechanism which is independent of the SEC machinery. Biochemistry 41: 11914–11920 [DOI] [PubMed] [Google Scholar]
- Walter P, Blobel G (1983a) Preparation of microsomal membranes for cotranslational protein translocation. Meth Enzymol 96: 84–93 [DOI] [PubMed] [Google Scholar]
- Walter P, Blobel G (1983b) Signal recognition particle: a ribonucleoprotein required for cotranslational translocation of proteins, isolation and properties. Methods Enzymol 96: 682–691 [DOI] [PubMed] [Google Scholar]
- Wattenberg B, Lithgow T (2001) Targeting of C-terminal (tail)-anchored proteins: understanding how cytoplasmic activities are anchored to intracellular membranes. Traffic 2: 66–71 [DOI] [PubMed] [Google Scholar]
- Whitley P, Grahn E, Kutay U, Rapoport TA, von Heijne G (1996) A 12-residue-long polyleucine tail is sufficient to anchor synaptobrevin to the endoplasmic reticulum membrane. J Biol Chem 271: 7583–7586 [DOI] [PubMed] [Google Scholar]
- Wiedmann B, Sakai H, Davis TA, Wiedmann M (1994) A protein complex required for signal-sequence specific sorting and translocation. Nature 370: 434–440 [DOI] [PubMed] [Google Scholar]
- Wiedmann M, Kurzchalia TV, Bielka H, Rapoport TA (1987) Direct probing of the interaction between the signal sequence of nascent preprolactin and the signal recognition particle by specific cross-linking. J Cell Biol 104: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabal M, Brambillasca S, Soffientini P, Pedrazzini E, Borgese N, Makarow M (2003) Translocation of the C terminus of a tail-anchored protein across the endoplasmic reticulum membrane in yeast mutants defective in signal peptide-driven translocation. J Biol Chem 278: 3489–3496 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data


