Abstract
The frequency of polymyxin-resistant pathogenic Gram-negative bacteria appearing in the clinic is increasing, and the consequences are largely mediated by modification of lipopolysaccharide (LPS) in the outer membrane. As polymyxins exert their antibacterial effect by binding to LPS, understanding their mode of binding will prove highly valuable for new antibiotic discovery. In this study, we assess the potential of MIPS-9451, a fluorescent polymyxin analogue designed for imaging studies, as a fluorescent reporter molecule, titrating it against 17 different Gram-negative species and/or strains of LPS. MIPS-9451 bound to the various species and/or strains of LPS with a dissociation constant ranging between 0.14 ± 0.01 µM (Escherichia coli) and 0.90 ± 0.42 µM (Porphyromonas gingivalis) (mean ± SE). Furthermore, we assessed the applicability of MIPS-9451 to assess affinities of polymyxin B to different LPS species in a displacement assay which yielded inhibition constants of 6.2 µM ± 33%, 7.2 µM ± 30% and 0.95 µM ± 13% for Klebsiella pneumoniae, Pseudomonas aeruginosa and Salmonella enterica, respectively (mean ± CV). The results from this study are concordant with those observed with similarly structured polymyxin probes, confirming the potential for MIPS-9451 for quantitation of polymyxin-LPS affinities in discovery programs of novel polymyxin antibiotics.
Keywords: Fluorescence spectroscopy, In vitro models, Structure-activity relationship, Antiinfectives, Peptides, Drug resistance
1 INTRODUCTION
The manifestation of multidrug-resistant (MDR) Gram-negative bacteria has been widely acknowledged as a major health concern.1–3 The rapidity with which the resistance to antibiotics is forming is alarming, with clinicians reporting that depending on the pathogen of interest, currently available antibiotics may only provide a defensive barrier for a timespan measuring decades, or potentially only years.4 The clinically available polymyxins (polymyxin B and colistin, the latter also known as polymyxin E) are one of the last remaining families of antibiotics still providing activity against problematic Gram-negative pathogens.5,6 Whilst the mechanism of antibacterial action of polymyxins is yet to be fully elucidated, it is known that a key initial interaction involves binding of the polymyxins to lipopolysaccharide (LPS),7 a large and complex molecule which makes up approximately 90% of the Gram-negative outer membrane.8 It is generally considered that upon binding to LPS, the polymyxins cause disruption and hyper-permeability of the bacterial outer membrane.9 This hyper-permeability is considered to result in an osmotic imbalance between the microbial interior and its external environment, leading eventually to bacterial cell death.10
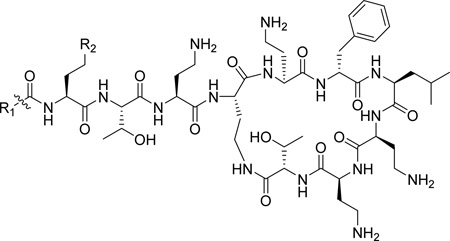
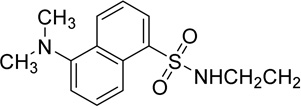
Given that the binding of polymyxins to LPS represents a key driving component of antibiotic activity, it follows that polymyxin analogues which possess enhanced affinity for LPS may potentially provide more efficacy against increasingly problematic Gram-negative pathogens. However, the non-protein nature and inherent heterogeneity render LPS11 systems less amenable to commonly used ligand/target affinity methods used in drug discovery programs. Thus the past few decades have seen increasing efforts to expand the available toolbox for assessing polymyxin/LPS structure-activity relationships (SAR), beginning with a novel displacement assay using a polydansylated polymyxin probe system introduced by Moore et al in 1986.12 The dansyl moiety (shown in Figure 1) exhibits a low level of fluorescence when occupying polar environments, and fluorescence is significantly enhanced upon interacting with hydrophobic structures, such as that provided by LPS.13 Whilst Moore’s novel assay represented an important step forward, the extent and positioning of dansylation upon the polymyxin scaffold was largely uncontrolled, limiting precision and confidence in the data produced therefrom. To address this, Soon et al developed the first fully synthetic mono-dansylated polymyxin probe, [dansyl-Lys]1polymyxin B3 (abbreviated to DPmB3 and shown in Table 1), which was utilised successfully to determine inhibition constants for the binding of colistin and polymyxin B with several species of LPS.13 The DPmB3 probe proved beneficial for the subsequent exploration of SARs within the polymyxin scaffold. The position of the dansyl group within the DPmB3 was chosen for the facilitation of synthesis, with 1 of the 5 positively charged dab residues normally found within a polymyxin structure (see position R2 on DPmB3 in Table 1) being replaced with the dansyl moiety.
Figure 1.
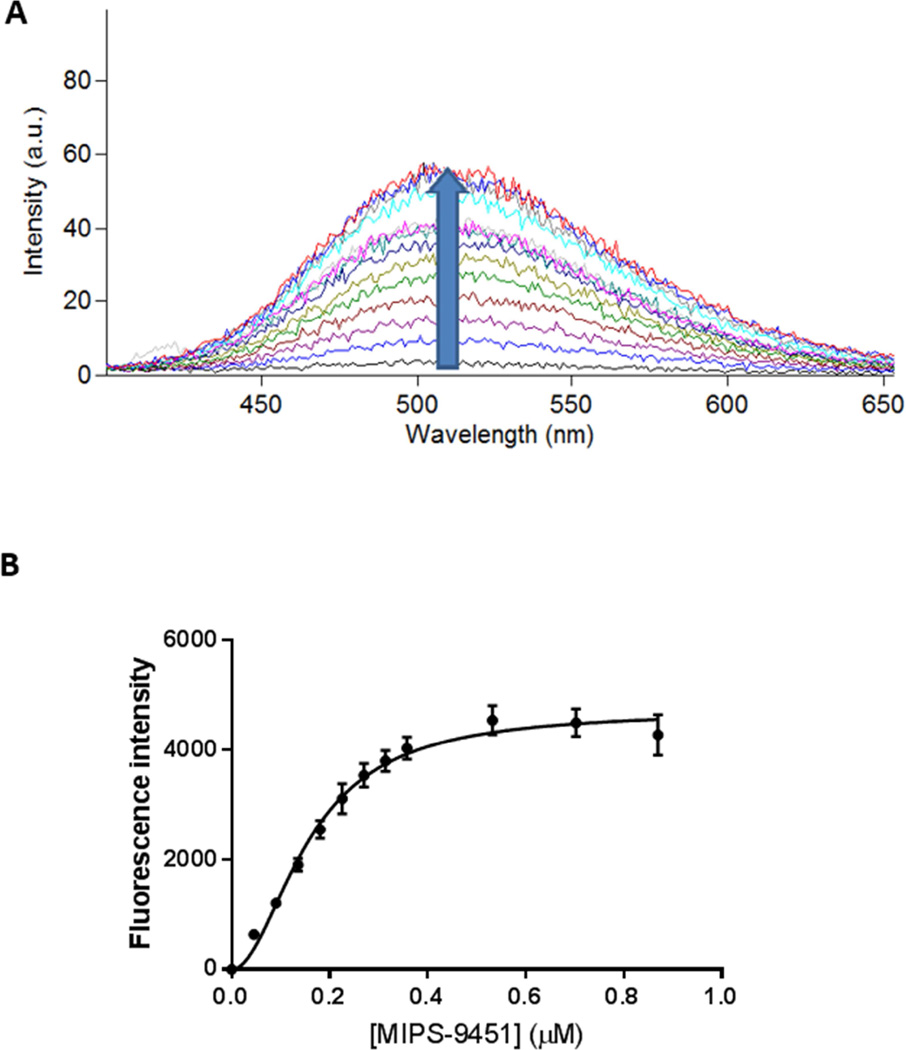
A) Representative emission spectra illustrating fluorescence enhancement upon the titration of increasing MIPS-9451 concentration into S. enterica (typhimurium) LPS, where the upward arrow indicates a rise in the curve upon successive titrations and B) area under curve of each titration plotted as a function of MIPS-9451 concentration. Data are presented as mean ± SD (n = 3) and are fitted to a Hill-slope augmented one-site specific binding model.
Table 1.
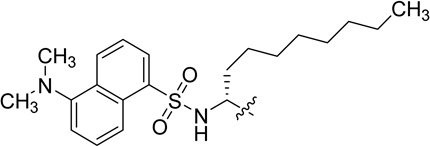
The chemical structures of polymyxin B1, polymyxin B2, DPmB3 and MIPS-9541.
Deris et al recently designed a new fully synthetic polymyxin probe, MIPS-9451, which was successfully utilized to image in real time, the physiological mechanism of polymyxins when interacting with the Gram-negative pathogen, Klebsiella pneumoniae.14 In MIPS-9451, the dansyl group is incorporated into the N-terminus of the polymyxin scaffold through attachment to an octylglycine residue, which serves to mimic the N-terminal fatty acyl groups found in polymyxin B peptides, thereby retaining all of the fundamental features of the polymyxin scaffold (see position R1 on MIPS-9451 in Table 1). This can be considered an iterative improvement in the physiological relevance of the molecule, when compared to previous versions of probes used in polymyxin fluorescence displacement assays.12,13 However, whether this modified dansylated probe exhibits similar or higher affinity for LPS, can discriminate between different LPS species and can be employed to assess displacement of polymyxins from LPS is yet to be determined. Therefore, the aims of this study were to determine the dissociation constants of MIPS-9451 binding to LPS from a number of species and/or strains of Gram-negative bacteria; and to investigate the suitability of MIPS-9451 as a probe in a fluorescent displacement assay to yield quantitative data (inhibition constants) on polymyxin-LPS interactions using polymyxin B (a mixture of polymyxin B1 and B2) as a model polymyxin antibiotic.
2 MATERIALS AND METHODS
2.1 Materials
LPS from the bacterial species, Escherichia coli serotype: 0111:B4, E. coli serotype: 026:B6, Klebsiella pneumoniae, Pseudomonas aeruginosa, Salmonella enterica serotype: abortus equi, S. enterica serotype: enteritidis, S. enterica serotype: Minnesota R595 (rough strain), S. enterica, serotype: Minnesota (smooth strain), S. enterica, serotype: typhimurium, S. typhosa and S. marcescens, all purified by phenol extraction, and 4-(2-hydroxyethyl)-1-piperazineethanesulphonic acid (HEPES buffer) were purchased from Sigma Aldrich (St. Louis, MO). LPS from the bacterial species Campylobacter jejuni, Proteus mirabilis, Proteus vulgaris, all purified by phenol extraction, and Helicobacter pylori and Porphyromonas gingivalis, purified by ultracentrifugation, were purchased from Wako chemicals, Osaka, Japan. Polymyxin B was purchased from BetaPharma (Shanghai, China). The synthesis of MIPS-9451 has been previously described elsewhere.14
2.2 Fluorometric assay: Binding of MIPS-9451 to various strains and/or species of LPS
The binding of MIPS-9451 to the various strains and/or species of LPS tested was executed using previously described assay methods.13 Briefly, 1 mL of a solution containing 5 mM HEPES buffer (pH 7.10) and 3 mg/L of LPS was dispensed into a 1.4 mL quartz cuvette with a path length of 10 mm (Starna, Baulkham Hills, New South Wales, Australia). After mixing the cuvette was then placed into a Carey Eclipse fluorescence spectrophotometer (Varian, Mulgrave, Victoria, Australia) and allowed to thermally equilibrate for 10 min within the internal cell block, which was set to 37°C. The excitation wavelength was set to 340 nm, with a slit width of 5 nm, and the emission spectrum was collected from 400 – 650 nm, with a slit width of 10 nm. Aliquots of the fluorescent probe, MIPS-9451, were titrated into the cuvette at 2 min intervals, until the fluorescence intensity reached an observable plateau. Each experiment was repeated 3 times, and the emission maximum was found to vary between different types of LPS, but always ranged between 490 – 520 nm. The fluorescence intensity was determined by integration of the area under the entire 400 – 650 nm spectrum. Titrations as described above were also conducted 3 times in the absence of LPS (control), to detect any non-specific fluorescence, and the average of these values was calculated and subtracted from the experimental data points. The data were corrected for dilution before being fit to a 1-site binding model (Equation 1), using GraphPad Prism 6 software (La Jolla, CA):
| (Equation 1) |
where ΔF is the specific fluorescence enhancement upon addition of MIPS-9451, ΔFmax is the maximum specific fluorescence enhancement at saturation of the LPS/MIPS-9451 complex, KD is the dissociation constant for the interaction, [L] is the total MIPS-9451 concentration in the mixture and h is the hill-slope coefficient.
2.3 Displacement of MIPS-9451 from LPS by polymyxin B
The displacement of MIPS-9451 from 3 different species of LPS by polymyxin B was also executed using methods described previously.13 Briefly, a 1 mL solution containing 5 mM HEPES buffer (pH 7.10), 3 mg/L of each LPS species, and MIPS-9451 at the concentration corresponding to 95% occupancy of the plateau identified above (0.9, 0.55 and 1.5 µM for K. pneumoniae, S. enterica and P. aeruginosa, respectively), was added to a quartz cuvette within the temperature controlled fluorescent spectrophotometer for 10 min to facilitate thermal equilibration. Into this solution, increasing concentrations of polymyxin B were titrated at 2 min intervals, and the resulting decrease in fluorescence was measured at each concentration, using the same instrumentation and experimental parameters detailed above. An additional control was included in this study, whereby the experiment was conducted 3 times in the absence of MIPS-9451. The averages for each of these data points were calculated, and these values were subtracted from experimental data points, to eliminate the effects of polymyxin light interference caused by micellular Rayleigh scattering. Next the fluorescence was corrected for dilution, and plotted as a function of the polymyxin B concentration in the mixture. The data were fit to a sigmoidal dose-response curve (Equation 2), using the same software as described above and the IC50 (the concentration of polymyxin B required to reduce fluorescence emission by 50%) was calculated from Equation 2:
| (Equation 2) |
where Fmin is the fluorescent signal of MIPS-9451 when unbound to LPS, Fmax is the fluorescent signal of MIPS-9451 when binding to LPS was saturated and [L] is the concentration of polymyxin B. The IC50 was then applied to the Cheng Prusoff equation,15 from which an inhibition constant, or Ki value, was derived (Equation 3):
| (Equation 3) |
where [MIPS-9451] is the concentration of probe in the cuvette solution (0.75 µM) and KD (MIPS-9451) is the KD of the probe.
3 RESULTS
3.1 Binding of MIPS-9451 to LPS from multiple strains and/or species
Figure 1A shows a representative titration of MIPS-9451 into an S. enterica (serotype typhimurium) LPS solution. As with all species and/or strains of LPS assessed, a progressive increase in fluorescence emission occurred with increasing concentration of MIPS-9451, before eventually reaching a plateau. When titrated against S. enterica the probe had an emission wavelength maximum of 513 nm. However with other species and/or strains of LPS the maximum emission wavelengths ranged between 490 and 520 nm. The plotting of fluorescence enhancement versus probe concentration when MIPS-9451 was titrated against S. enterica is shown in Figure 1B. Each of the binding profiles of the 17 species and/or strains of LPS tested, conformed well to a Hill slope 1-site binding model. Summarised in Table 2 are the parameters obtained for the LPS from all strains/species examined in this study, including the dissociation constant (KD), magnitude of signal return (Bmax), and Hill-slope coefficient (h). MIPS-9451 bound to all of the LPS target molecules with similar affinity, ranging between 0.14 ± 0.01 µM for E. coli; serotype: 026:B6 to 0.90 ± 0.42 µM for P. gingivalis.
Table 2.
The dissociation constant (KD) of MIPS-9451 against a 3 mg/L solution of LPS from various strains and/or species of LPS, and the magnitude of signal return (Bmax) and Hill-slope coefficient obtained following titration of MIPS-9451 under the same conditions. Data are presented as mean ± SEM (n = 3).
| LPS species and strain | KD (µM) | Bmax (arbitrary units) |
Hill-slope coefficient |
|---|---|---|---|
| Bordetella pertussis | 0.32 ± 0.01 | 19140 | 2.69 ± 0.22 |
| Campylobacter jejuni | 0.41 ± 0.01 | 19012 | 2.32 ± 0.13 |
|
Escherichia coli, serotype: 0111:B4 |
0.29 ± 0.02 | 11397 | 2.54 ± 0.37 |
|
Escherichia coli, serotype: 026:B6 |
0.14 ± 0.01 | 6677 | 2.18 ± 0.21 |
| Helicobacter pylori | 0.18 ± 0.01 | 5765 | 1.82 ± 0.18 |
| Klebsiella pneumoniae | 0.29 ± 0.01 | 13594 | 2.66 ± 0.26 |
|
Porphyromonas gingivalis |
0.90 ± 0.42 | 3841 | 1.03 ± 0.14 |
| Proteus mirabilis | 0.38 ± 0.01 | 11750 | 2.04 ± 0.13 |
| Proteus vulgaris | 0.40 ± 0.02 | 16172 | 2.27 ± 0.22 |
|
Pseudomonas aeruginosa |
0.54 ± 0.03 | 18446 | 2.92 ± 0.49 |
|
Salmonella enterica, Serotype: abortus equi |
0.25 ± 0.01 | 9639 | 2.65 ± 0.21 |
|
Salmonella enterica, Serotype: enteritidis |
0.28 ± 0.01 | 12903 | 2.42 ± 0.29 |
|
Salmonella enterica, Serotype: Minnesota R595 (rough strain) |
0.52 ± 0.04 | 14427 | 2.00 ± 0.26 |
|
Salmonella enterica, Serotype: Minnesota (smooth strain) |
0.23 ± 0.01 | 6974 | 2.55 ± 0.29 |
|
Salmonella enterica, Serotype: typhimurium |
0.16 ± 0.01 | 4718 | 1.96 ± 0.16 |
| Salmonella typhosa | 0.30 ± 0.01 | 12592 | 1.83 ± 0.10 |
| Serratia marcescens | 0.27 ± 0.01 | 11504 | 2.14 ± 0.11 |
3.2 Displacement of MIPS-9451 from LPS by polymyxin B
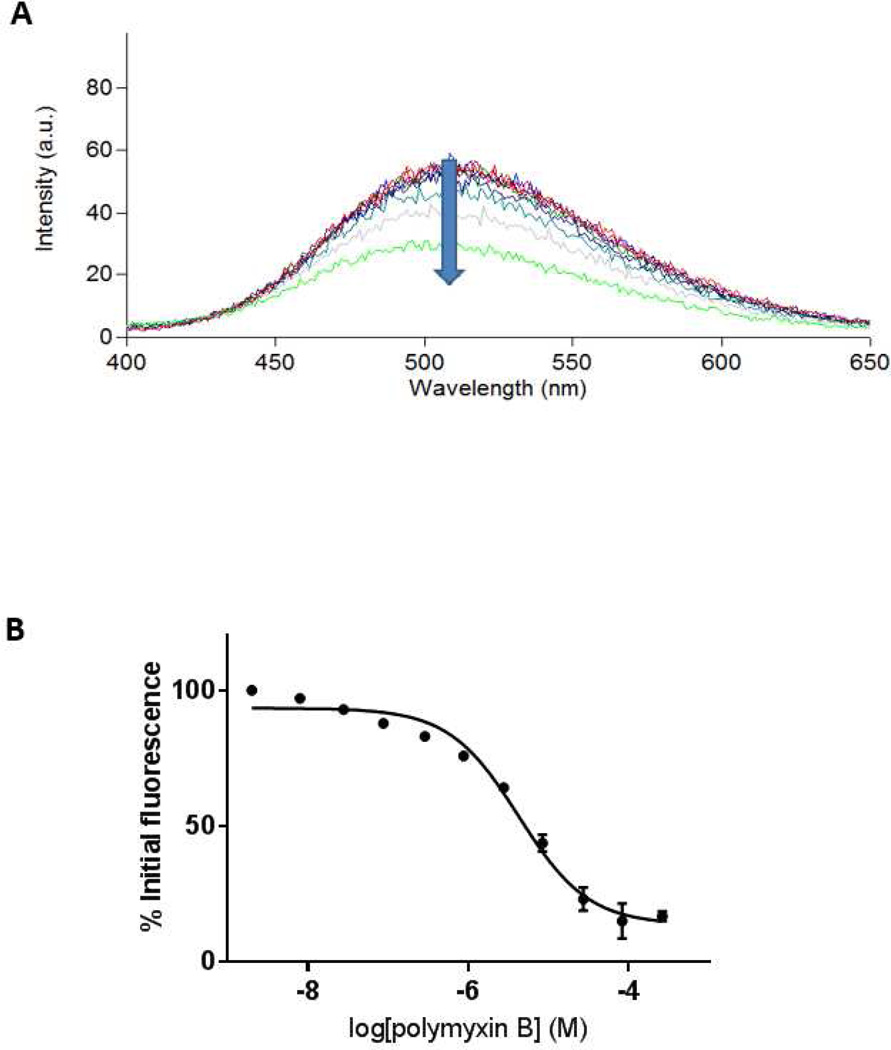
As shown in Figure 2A, titration of increasing concentrations of polymyxin B into a solution containing LPS and MIPS-9451 (the latter corresponding to 95% probe occupancy of S. enterica LPS binding sites) resulted in a progressive decrease in fluorescence emission. This displacement (Figure 2B) also approached a plateau, corresponding to near 0% probe occupancy of the LPS binding sites. The plotting of fluorescence reduction as a function of polymyxin B concentration is shown in Figure 2B, where the y-axis is expressed as a percentage of initial fluorescence. The titration of polymyxin B against LPS from three Gram-negative species (K. pneumoniae, P. aeruginosa and S. enterica) in the presence of MIPS-9451, and subsequent calculation of inhibition constants via the Cheng/Prusoff equation, yielded the values of 6.2, 7.2 and 0.95 µM, respectively (Table 3).
Figure 2.
A) Representative emission spectra illustrating fluorescence attenuation upon titration of increasing polymyxin B concentration against S. enterica LPS with the direction of the arrow indicating increasing polymyxin B concentration; and B) area under the curve of each titration plotted as a function of polymyxin B concentration. Data are presented as mean ± SD (n = 3) and fit to a competitive one-site binding model. Note: some data points were associated with very small error, thus the error bars are not visible.
Table 3.
Experimentally determined inhibition constants (Ki) of polymyxin B when titrated against 3 different species of LPS. Data are presented as mean ± coefficient of variation (CV), (n = 3).
| LPS species | Ki (µM) ± CV |
|---|---|
| Klebsiella pneumoniae | 6.2 ± 33% |
| Pseudomonas aeruginosa | 7.2 ± 30% |
|
Salmonella enterica (typhimurium) |
0.95 ± 13% |
4 DISCUSSION
Given that MIPS-9451 was recently utilized as an imaging agent, and that the structure of the molecule represents an incremental improvement in the physiological relevance over previous polymyxin probe molecules,12–14,16 the aim of this study was to assess the potential of MIPS-9451 as a fluorescent reporter molecule for LPS binding studies. The availability of a robust and sensitive displacement assay using MIPS-9451 could then be applied to polymyxin discovery settings.
MIPS-9451 appears to possess the properties required for functioning as a reporter molecule in a fluorescence displacement assay targeting LPS, namely; intrinsically low fluorescence in solution; enhanced and saturable fluorescence upon binding to its LPS target; and a suitable level of affinity such that displacement is possible by polymyxins in ranges below their critical micelle concentration. The KD of MIPS-9451 for all of the LPS strains tested fell within the range of 0.14 – 0.90 µM. Soon et al assessed the affinity of DPmB3 for 4 separate species of LPS, and the values derived ranged between 0.5 – 1.2 µM.13 Each of these 4 species was also tested in this study, and MIPS-9451 produced a KD which was approximately two to three-fold lower than that of DPmB3 in each case. This modest but consistent improvement in affinity is likely due to the reinstatement of the Dab residue in the R1 position (see Table 1), which facilitates the ionic interactions with the negatively charged phosphate groups of the lipid A component of LPS, thus mildly enhancing the affinity of MIPS-9451 for LPS molecules.
Since MIPS-9451 did exhibit similar affinity to each of LPS species tested, this suggests that this probe may be relevant as a screening tool for assessing binding of polymyxins intended to target a number of Gram-negative species and/or strains. Notably, for each species and/or strain of LPS tested, the emission wavelength maximum remained constant throughout the experiment, but tended to experience a slight red-shift, or broadening of the right-hand tail of the curve upon binding saturation. This effect could potentially be an artefact of changes in the local electronic environments which immediately surround the probe, where upon binding saturation of LPS aggregates by MIPS-9451, water molecules may be displaced by the steric considerations of the much larger probe molecule.
Having demonstrated that MIPS-9451 possessed suitable binding properties whilst interacting with LPS, we then tested its ability to perform in a proof-of-concept displacement type assay in three different LPS species with polymyxin B as a model polymyxin. When the inhibition constants for polymyxin B produced by the new probe assay are compared with values previously reported in the literature, a high degree of similarity is observed. Soon et al, using as DPmB3 the displacing reporter molecule, reported the affinity of polymyxin B for K. pneumoniae, P. aeruginosa and S. enterica to be 0.5, 3.5 and 0.4 µM respectively.13 As shown in Table 3, the values obtained from titrating polymyxin B against LPS from the same 3 species of bacteria but using MIPS-9451 as the displacing reporter molecule were 6.2, 7.2 and 0.95 µM, respectively. Whilst direct comparison of these inhibition constants reveals some minor variations, for the most part they are consistent (within the same order of magnitude) with those inhibition constants generated by Soon et al. These studies demonstrate that MIPS-9451 performs appropriately as a fluorescent probe in a displacement assay in addition to being a suitable imaging agent,14 being a probe molecule that can assist in elucidation of both mechanism of polymyxin activity and affinities of polymyxin-LPS interactions, and therefore useful in antimicrobial drug discovery programs.
5 CONCLUSION
This study demonstrates that MIPS-9451, originally developed as an analogue of polymyxin for imaging interactions between Gram-negative bacteria and polymyxins, can also function as a suitable probe in a fluorometric displacement assay to quantify polymyxin-LPS affinities. MIPS-9451 may therefore provide useful insight in future polymyxin drug discovery programs by identifying novel compounds with high affinity to LPS, the antimicrobial target of polymyxins.
Acknowledgments
This study is supported by a research grant to J.L and T.V. from the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (R01 AI111965). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health. J. L. is an Australian National Health and Medical Research Council (NHMRC) Senior Research Fellow, and T. V. is an Australian NHMRC Industry Career Development Level 2 Research Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Choudhury R, Panda S, Singh DV. Emergence and dissemination of antibiotic resistance: a global problem. Indian J Med Microbiol. 2012;30:384–390. doi: 10.4103/0255-0857.103756. [DOI] [PubMed] [Google Scholar]
- 2.Rossolini GM, Arena F, Pecile P, Pollini S. Update on the antibiotic resistance crisis. Curr Opin Pharmacol. 2014;18C:56–60. doi: 10.1016/j.coph.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 3.Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, Scheld WM, Bartlett JG, Edwards J, Jr the Infectious Diseases Society of America. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the infectious diseases society of America. Clin Infect Dis. 2008;46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- 4.Wellington EMH. The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect Dis. 2013;13:155–165. doi: 10.1016/S1473-3099(12)70317-1. [DOI] [PubMed] [Google Scholar]
- 5.Biswas S, Brunel JM, Dubus JC, Reynaud-Gaubert M, Rolain JM. Colistin: an update on the antibiotic of the 21st century. Expert Rev Anti Infect Ther. 2012;10:917–934. doi: 10.1586/eri.12.78. [DOI] [PubMed] [Google Scholar]
- 6.Falagas ME, Grammatikos AP, Michalopoulos A. Potential of old-generation antibiotics to address current need for new antibiotics. Expert Rev Anti Infect Ther. 2008;6:593–600. doi: 10.1586/14787210.6.5.593. [DOI] [PubMed] [Google Scholar]
- 7.Pristovsek P, Kidric J. Solution structure of polymyxins B and E and effect of binding to lipopolysaccharide: an NMR and molecular modeling study. J Med Chem. 1999;42:4604–4613. doi: 10.1021/jm991031b. [DOI] [PubMed] [Google Scholar]
- 8.Rosenfeld Y, Shai Y. Lipopolysaccharide (Endotoxin)-host defense antibacterial peptides interactions: role in bacterial resistance and prevention of sepsis. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2006;1758:1513–1522. doi: 10.1016/j.bbamem.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 9.Velkov T, Thompson PE, Nation RL, Li J. Structure-activity relationships of polymyxin antibiotics. J Med Chem. 2010;53:1898–1916. doi: 10.1021/jm900999h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock RE, Chapple DS. Peptide antibiotics. Antimicrob Agents Chemother. 1999;43:1317–1323. doi: 10.1128/aac.43.6.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jann B, Reske K, Jann K. Heterogeneity of lipopolysaccharides. Analysis of polysaccharide chain lengths by sodium dodecylsulfate-polyacrylamide gel electrophoresis. Eur J Biochem. 1975;60:239–246. doi: 10.1111/j.1432-1033.1975.tb20996.x. [DOI] [PubMed] [Google Scholar]
- 12.Moore RA, Bates NC, Hancock RE. Interaction of polycationic antibiotics with Pseudomonas aeruginosa lipopolysaccharide and lipid A studied by using dansyl-polymyxin. Antimicrob Agents Chemother. 1986;29:496–500. doi: 10.1128/aac.29.3.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soon RL, Velkov T, Chiu F, Thompson PE, Kancharla R, Robers K, Larson I, Nation RL, Li J. Design, synthesis, and evaluation of a new fluorescent probe for measuring polymyxin-lipopolysaccharide binding interactions. Anal Biochem. 2011;409:273–283. doi: 10.1016/j.ab.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deris ZZ, Swarbrick JD, Roberts KD, Azad MA, Akter J, Horne AS, Nation RL, Rogers KL, Thompson PE, Velkov T, Li J. Probing the penetration of antimicrobial polymyxin lipopeptides into gram-negative bacteria. Bioconjug Chem. 2014;25:750–760. doi: 10.1021/bc500094d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng YC, Prusoff WH. Relationship between the inhibition constant (Ki) and the concentration of inhibitor which causes 50 percent inhibition of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 16.Azad MA, Yun B, Roberts KD, Nation RL, Thompson PE, Velkov T, Li J. Measuring polymyxin uptake by renal tubular cells: is BODIPY-polymyxin B an appropriate probe? Antimicrob Agents Chemother. 2014;58:6337–6338. doi: 10.1128/AAC.03733-14. [DOI] [PMC free article] [PubMed] [Google Scholar]