Abstract
Pseudomonas syringae relies on type III secretion system to deliver effector proteins into the host cell for parasitism. Type III genes are induced in planta, but host factors affecting the induction are poorly understood. Here we report on the identification of an Arabidopsis mutant, att1 (for aberrant induction of type three genes), that greatly enhances the expression of bacterial type III genes avrPto and hrpL. att1 plants display enhanced disease severity to a virulent strain of P. syringae, suggesting a role of ATT1 in disease resistance. ATT1 encodes CYP86A2, a cytochrome P450 monooxygenase catalyzing fatty acid oxidation. The cutin content is reduced to 30% in att1, indicating that CYP86A2 plays a major role in the biosynthesis of extracellular lipids. att1 has a loose cuticle membrane ultrastructure and shows increased permeability to water vapor, demonstrating the importance of the cuticle membrane in controlling water loss. The enhanced avrPto-luc expression is specific to att1, but not another cuticle mutant, wax2. The results suggest that certain cutin-related fatty acids synthesized by CYP86A2 may repress bacterial type III gene expression in the intercellular spaces.
Keywords: Arabidopsis, cutin, disease resistance, Pseudomonas, type III secretion system
Introduction
The ability of plant-associated microbes to sense the host environment is crucial for their successful establishment in plants. Plant-sensing triggers virulence/pathogenicity gene expression, enabling the microbes to suppress, tolerate, or evade host defenses and exploit nutrients for their multiplication (Lee and Camilli, 2000). Conversely, plant extracellular factors affecting bacterial gene expression, either positively or negatively, may play important roles in disease susceptibility or resistance.
Many Gram-negative plant bacterial pathogens, including Pseudomonas syringae, Xanthomonas, Erwinia, and Ralstonia solanacearum, use the type III secretion system (TTSS) to deliver a repertoire of effector proteins into their host cells for parasitism (Galan and Collmer, 1999; He and Jin, 2003). These bacteria sense host-derived signals to activate coordinately genes encoding TTSS and effectors. However, host signals and bacterial mechanisms that regulate these genes are poorly understood.
Certain carbon sources and amino acids are known to affect type III gene expression in culture media (Huynh et al, 1989; Arlat et al, 1992; Rahme et al, 1992; Wei et al, 1992). However, it is unclear if these truly mimic the apoplast environment of plants, and little is known about biochemical and physical features of the apoplast involved in type III gene regulation. It is therefore imperative to use living plants for host signal studies.
Animal bacterial type III genes are activated upon contact with host cells (Francis et al, 2002), first described in Yersinia pseudotuberculosis (Pettersson et al, 1996) and later in Pseudomonas aeruginosa (Vallis et al, 1999), Yersinia enterocolitica (Stainier et al, 1997), and Shigella flexneri (Demers et al, 1998). In Y. pseudotuberculosis, the contact with HeLa cells is believed to trigger the release of LcrQ, a TTSS secreted protein that negatively regulates other type III gene expression (Pettersson et al, 1996). Plant bacterial type III genes are also induced by the host. However, the molecular basis of the contact-dependent gene induction remains largely unknown.
A series of recent studies elegantly demonstrated that R. solanacearum type III genes are induced when the bacterium is in direct contact with the plant cell (Aldon et al, 2000). The induction is mediated by PrhA, a membrane protein with significant similarities with siderophore receptors (Marenda et al, 1998). The prhA mutation does not affect the induction by an inducing medium, suggesting distinct signaling mechanisms in the perception of signals derived from host cell and culture medium. Additional components acting downstream of PrhA are PrhR, PrhI, PrhJ, HrpG, and HrpB, which function in a sequential order (Brito et al, 1999, 2002). The plant signal perceived by PrhA is likely a nondiffusible wall component that is resistant to protease and heat (Aldon et al, 2000), but the chemical nature of the signal remains unknown.
The cuticle envelops the aerial parts of the plant. It serves as a protective barrier that prevents water and solute losses (Jenks, 2002) and pathogen penetration (Jenks et al, 1994). The cuticle is composed of two basic layers, an outermost epicuticular wax layer and an underlying cuticle membrane layer (Jeffree, 1996). In addition to the external epidermal surface, the cuticle membrane extends into the substomatal chamber (Esau, 1977). This notion is potentially interesting, because the substomatal chamber is where bacteria reside immediately after entering the stomatal opening. The cuticle membrane is primarily an insoluble polyester of hydroxyl and epoxy hydroxyl fatty acids (Kolattukudy, 1981), whereas the epicuticular waxes and intracuticular waxes (embedded in the cutin matrix) are primarily soluble long-chain aliphatic molecules. Cutin content has been analyzed in a number of plant species (Kolattukudy, 2001), but not Arabidopsis. Typical cutin monomers reported in the literature are 10,16-dihydroxy hexadecanoic acid, 18-hydroxy-9,10 epoxy octadecanoic acid, and 9,10,18-trihydroxy octadecanoic acid. Diacids, monohydroxy diacids, and unsaturated acids have also been reported (Holloway, 1982). While biochemical studies have established the cutin biosynthetic pathway, genes encoding the key enzymes have not been unequivocally identified (Kolattukudy, 2001; Nawrath, 2002).
Here we describe the use of a genetic approach to investigate host signals involved in the regulation of Pseudomonas type III gene expression. We developed a highly sensitive reporter assay for P. syringae type III gene expression in plants. As a first step to understand host factors affecting bacterial type III gene expression, we identified a genetic locus in Arabidopsis, named ATT1 (for Aberrant induction of Type Three genes), that negatively regulates the expression of type III effector gene avrPto and regulatory gene hrpL. The latter encodes an ECF family alternate σ factor that recognizes the ‘hrp box' motif conserved in the promoters of type III genes including avrPto (Salmeron and Staskawicz, 1993; Xiao et al, 1994). ATT1 encodes CYP86A2, a cytochrome P450 monooxygenase catalyzing fatty acid oxidation. att1 contains only 30% of the cutin in the wild-type plants and has a loosely structured cuticle membrane. The results led us to suggest that CYP86A2 is involved in the biosynthesis of hydroxylated fatty acids that function in both cuticle development and repression of bacterial type III gene expression.
Results
Isolation of att1 mutant by using avrPto-luc reporter gene
The avrPto gene of P. syringae pv. tomato contains a typical hrp box in the promoter and is induced strongly in planta (Salmeron and Staskawicz, 1993). P. syringae pv. phaseolicola and P. syringae pv. tomato DC3000 strains carrying avrPto-luc were constructed to monitor nonintrusively the activation of a type III gene promoter in planta. The reporter strains displayed strong luciferase activity when infiltrated into Arabidopsis plants, which can be detected as luminescence by a cooled charge-coupled device (CCD). P. syringae pv. phaseolicola is a nonhost strain, whereas DC3000 is a virulent strain on Arabidopsis. Both strains displayed similar induction of avrPto-luc, consistent with the finding that the host signal(s) for type III genes are not determinants of host specificity (Rahme et al, 1992).
In an attempt to identify host factors involved in type III gene induction, we screened for Arabidopsis mutants displaying altered induction of luc. The use of P. syringae pv. phaseolicola (avrPto-luc), which is unable to multiply in Arabidopsis, avoids potential confusion between bacterial growth with avrPto-luc transcription. From approximately 4000 ethyl methanesulfonate (EMS)-mutagenized Arabidopsis Col-gl M2 plants, a single mutant that hyperinduced the avrPto-luc reporter gene was isolated (Figure 1A). This mutant is named att1-1 (for aberrant induction of a type three gene). att1-1 plants are morphologically identical to wild-type plants when grown in the growth chamber. At 12 h after infiltration, avrPto-luc was 10 times more active in att1-1 than in wild-type plants. In contrast, the constitutive reporter gene trp-luc was not affected in this mutant (Figure 1B), suggesting that the effect of att1-1 was specific to the type III gene.
Figure 1.
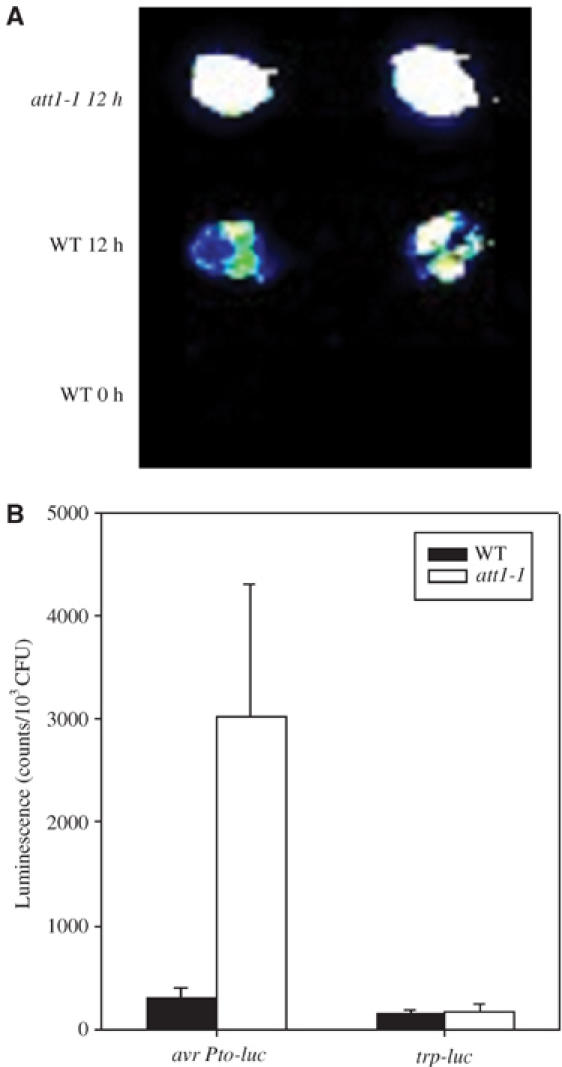
att1-1 specifically enhances type III gene expression. (A) CCD image of avrPto-luc expression in wild-type Col-gl (WT) and att1-1 leaves. (B) Activity of avrPto-luc and trp-luc in wild-type Col-gl (WT) and att1-1 leaves. Plants were inoculated with P. syringae pv. phaseolicola carrying avrPto-luc or trp-luc, and leaves were detached at 0 or 12 h for luciferase assay. Error bars indicate standard errors. The experiments were repeated three times with similar results.
We next examined the kinetics of avrPto-luc induction in wild-type and att1-1 plants. Figure 2A shows that while the wild-type plants transiently activated avrPto-luc in the bacterium, the att1-1 mutant enabled a much greater and prolonged induction of avrPto-luc. The observed values (normalized to leaf bacterial numbers) reflect the reporter gene expression, because bacteria did not grow in either wild-type or att1-1 plants (Supplementary Figure S1; data not shown). We also determined if att1-1 affected the expression of hrpL-luc in the plant. Similar to avrPto-luc, hrpL-luc was strongly induced in wild-type plants, and the induction was dramatically enhanced and long lasting in att1-1 (Figure 2B). The differential induction in att1-1 and wild-type plants was also observed in DC3000 (data not shown).
Figure 2.
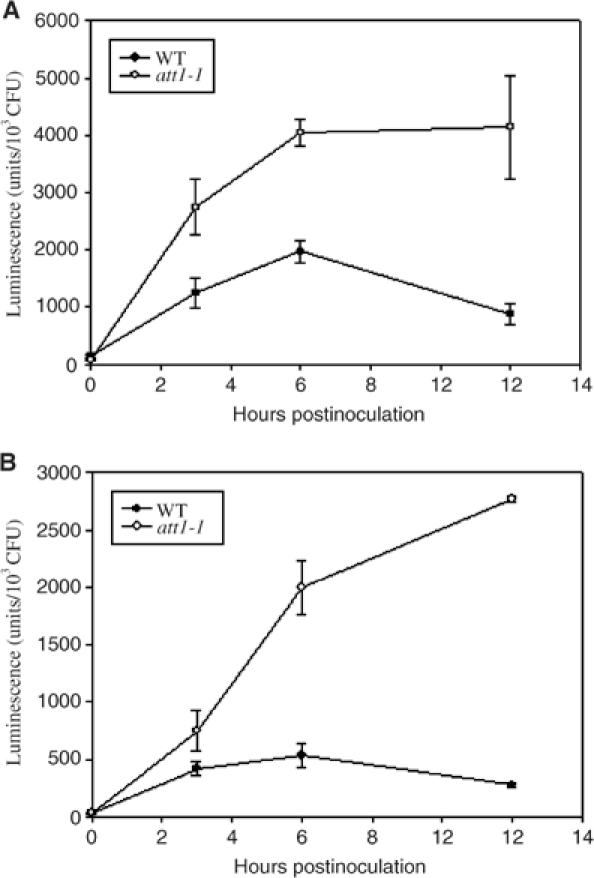
Kinetics of type III gene expression in wild-type Col-gl (WT) and att1-1 leaves. Plants were inoculated with P. syringae pv. phaseolicola carrying avrPto-luc (A) or hrpL-luc (B), and leaves were detached at the indicated times for luciferase assay. Error bars indicate standard errors. The experiments were repeated three times with similar results.
att1 enhances disease severity in plants
To determine if the increased type III gene induction altered disease susceptibility, we dip-inoculated plants with DC3000 (Figure 3A). Typical disease symptoms were observed in wild-type plants, but they were much more severe in att1-1 plants. A small but reproducible increase of bacterial growth (∼5-fold) was observed in att1-1 plants 2–4 days after dipping inoculation (Figure 3B). However, the increased bacterial growth was not observed when plants were inoculated by syringe infiltration (Supplementary Figure S1). The cause of different bacterial growth results between dipping and infiltration inoculations is not understood, but similar results have also been reported for strains of DC3000 lacking the phytotoxin coronatine (Mittal and Davis, 1995).
Figure 3.
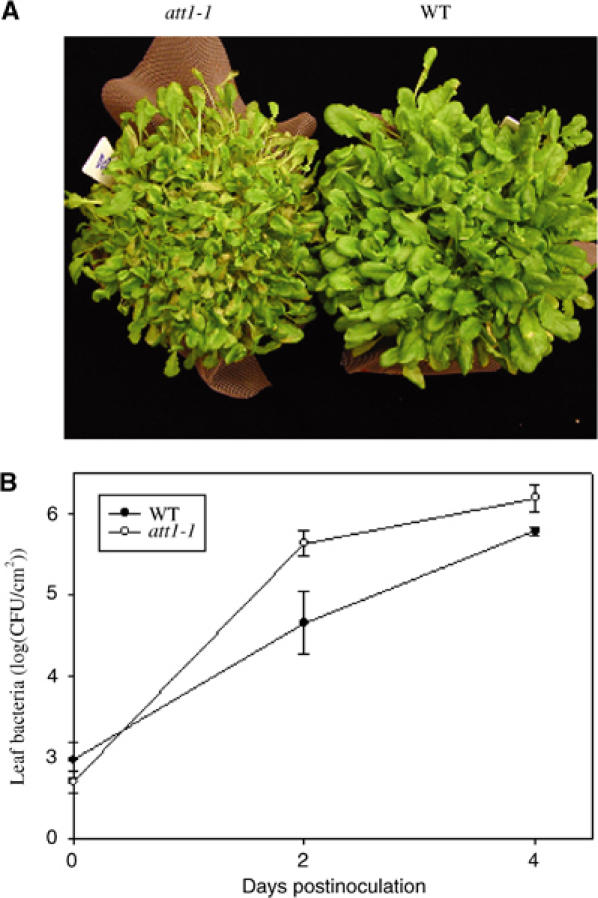
att1 displays enhanced disease severity. (A) Disease symptoms. (B) Bacterial growth assay. Wild-type Col-gl (WT) and att1-1 plants were dip-inoculated with DC3000 at 1 × 108 CFU/ml. Disease symptoms were photographed 7 days after inoculation. Leaf bacterial number was determined at 0 and 4 days after inoculation. The experiment was repeated twice with similar results.
We also inoculated att1-1 plants with the nonhost strain P. syringae pv. phaseolicola and the incompatible strain DC3000 (avrB) by infiltration. No difference in bacterial growth (Supplementary Figure S1) or hypersensitive response was observed between att1-1 and wild-type plants, indicating that the enhanced type III gene induction by itself does not convert the plant from resistant to susceptible. This is consistent with a previous report that the expression level of type III gene does not determine host range (Rahme et al, 1992).
Isolation of ATT1
To determine the nature of mutation, the att1-1 mutant was back-crossed with wild-type Col-0 plants. All five F1 plants tested supported normal avrPto-luc induction. F2 plants segregated in a 1:3 ratio (17 mutant to 55 wild-type, χ2=0.07), indicative of a single recessive mutation. The att1-1 mutant was crossed with ecotype Nossen to generate an F2 mapping population. Simple sequence-length polymorphism (SSLP) markers located ATT1 to the top of chromosome IV. Further mapping using approximately 1000 F2 plants placed ATT1 on the BAC clone F5I10 in a 50 kb interval containing six genes (Figure 4A). Sequence analysis of the six genes in the att1-1 mutant revealed a single nucleotide substitution (C to T at nucleotide 157 143 of chromosome IV) in the At4g00360 gene, causing a single amino-acid substitution (R309C). No mutation was found in the remaining five genes. At4g00360 contains a single intron and encodes a protein of 553 amino acids. To further confirm if this mutation is responsible for the att1-1 phenotype, we identified a T-DNA insertion line, Salk_005826, from the Salk T-DNA database. This line contained a T-DNA insertion disrupting the second exon of At4g00360 and supported identical avrPto-luc induction compared to att1-1 (Figure 4B), providing further evidence that At4g00360 was the ATT1 gene. The allele in the T-DNA line is thus named att1-2. To assign unequivocally At4g00360 as ATT1, we introduced an 8 kb PvuII fragment containing At4g00360 into att1-1 by Agrobacterium-mediated transformation. Four transgenic plants were tested for the avrPto-luc induction in planta, and all displayed normal expression of avrPto-luc, demonstrating that At4g00360 is ATT1 (Figure 4C).
Figure 4.
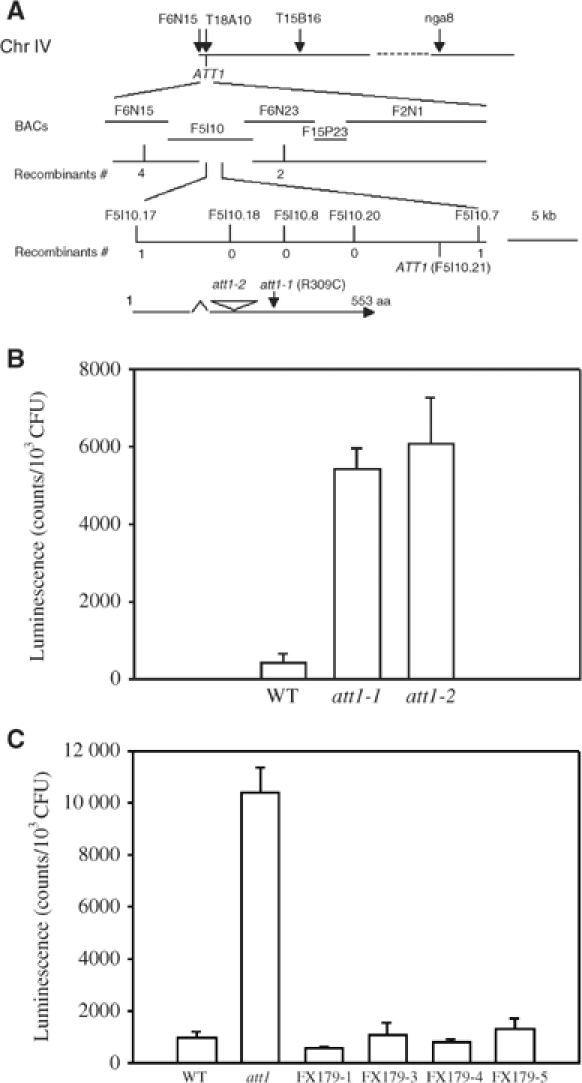
Map-based cloning of ATT1. (A) Physical mapping of ATT1. ATT1 was mapped to the upper arm of chromosome IV and positioned on the BAC clone F5I10. (B) T-DNA insertion in At4g00360 results in att1 phenotype. Col-gl (WT), att1-1, and the T-DNA line (Salk-005826; att1-2) were infiltrated with P. syringae pv. phaseolicola carrying the avrPto-luc reporter. Relative LUC activity was measured 12 h after inoculation. Error bars represent standard error. (C) At4g00360 complements the att1-1 mutation. An 8 kb PvuII fragment containing At4g00360 was mobilized from the BAC clone F5I10 to pBI121 (by replacing the 35S-GUS fragment) and transformed into att1-1 mutant plants. Four of the five putative primary transgenic plants tested contained the transgene. These plants (FX179-1, -3, -4, -5) were tested for avrPto-luc activation and compared with wild-type and untransformed att1-1 plants. Error bars indicate standard error.
ATT1 mRNA is repressed by bacterial infection
We next determined if the ATT1 gene expression is affected by bacterial infection. Northern analyses indicated that the ATT1 transcripts were nonspecifically induced at 3 h by buffer infiltration (Figure 5). However, inoculation with P. syringae, regardless of whether compatible, incompatible, or nonhost strains were used, repressed the gene expression at later time points.
Figure 5.
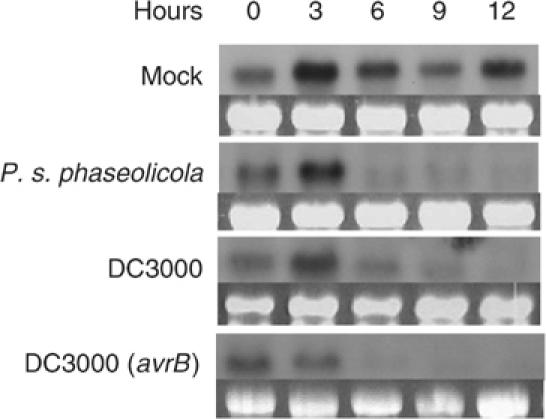
Bacteria repress ATT1 expression. Wild-type plants were infiltrated with water (mock) or the indicated bacterial strains at a concentration of 2 × 106 CFU/ml, and tissues were harvested at the indicated times for RNA isolation. RNA blots were hybridized with a radiolabeled ATT1 probe. The rRNA gel pictures indicate equal loading of RNA in lanes. The Northern analysis was repeated three times with similar results.
ATT1 encodes a putative fatty acid hydroxylase
At4g00360 encodes the cytochrome P450 monooxygenase CYP86A2. For consistency, the ATT1 gene is referred to as CYP86A2 in the rest of the paper. Arabidopsis contains 246 cytochrome P450 genes, which are divided into different families based on sequence and function (Werck-Reichhart et al, 2002). The Arabidopsis CYP86 family is highly homologous to the animal and fungal P450 families CYP52 and CYP4, which catalyze the metabolism of fatty acids and alkanes through ω-hydroxylation (Werck-Reichhart et al, 2002; Figure 6). CYP86A2 shows 87% amino-acid sequence identity with CYP86A8 and 73% with CYP86A1. Both CYP86A1 and CYP86A8 catalyze the ω-hydroxylation of C12 to C18 saturated and unsaturated fatty acids in vitro (Beneviste et al, 1998; Wellesen et al, 2001). Thus these results suggest that CYP86A2 is a fatty acid ω-hydroxylase.
Figure 6.

Sequence alignment of ATT1 with other CYP86A subfamily members. Sequences underlined with a solid line, a dashed line, and a double line indicate oxygen binding and activation motif, ERR triad, and heme binding motif, respectively.
The cyp86a2-1 (R309C) is phenotypically indistinguishable from cyp86a2-2, a likely null mutant because of an early truncation caused by the T-DNA insertion. R309 is a variant residue in the oxygen binding/activation motif of P450 proteins (AGRDTS; Werck-Reichhart et al, 2002), but is conserved among members of the CYP86A subfamily. This suggests that R309 plays a critical role specific to the CYP86A subfamily.
CYP86A2 is required for the biosynthesis of cutin and cuticle development
ω-Hydroxy fatty acids are exported to outside of plant cells through an unknown mechanism. The most abundant form of plant extracellular lipids in leaves is cutin, which is formed upon interesterification of ω-hydroxy fatty acids (Kolattukudy, 2001). We therefore tested if CYP86A2 is involved in the synthesis of hydroxy fatty acids in vivo by examining leaf cuticle membrane structure of cyp86a2-1 using transmission electron microscopy. Figure 7A and B shows that the cyp86a2-1 epidermis had a cuticle membrane that was less osmiophilic, as indicated by the reduced electron density, but more than twice as thick compared with that of wild type. Osmium tetroxide stains lipid components. The lower electron density in cyp86a2-1 suggests a reduction of fatty acids in the cuticle membrane. Interestingly, we found that the Arabidopsis cuticle membrane covered cells surrounding the entire substomatal chamber including mesophyll cells, although it was much thinner than that on the epidermal surface. This mesophyll cuticle membrane of cyp86a2 was more than twice as thick as that in the wild-type plants (Figure 7C and D), indicating that the defects are not limited to epidermal cells. Similar observations were made in cyp86a2-2 (data not shown).
Figure 7.
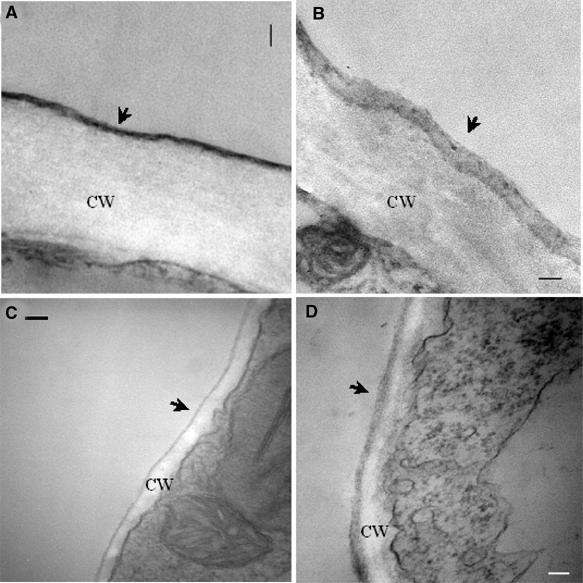
cyp86a2-1 displays altered cuticle membrane ultrastructure. Ultrastructure of the wild-type Col-gl (A, C) and the cyp86a2-1 mutant (B, D) cuticle membrane from an epidermal cell (A, B) and substomatal chamber mesophyll cell (C, D). The cuticle (arrowhead) and cell wall (CW) are shown. Note the lower electron density of the cyp86a2-1 cuticle membrane, which was found in all samples examined, compared with the dense, compact cuticle in the wild-type plant. Bar equals 200 nm.
We next determined cutin composition in the wild-type and cyp86a2-1 plants. The cuticle membrane consists of both hydrolyzable cutin polymer and nonhydrolyzable fraction, named cutan. Our studies revealed that leaf cutin was altered in the cyp86a2-1 plants. However, a protocol for isolation of Arabidopsis leaf cutin in amounts needed for complete cutin monomer analysis has yet to be developed. Therefore, the more abundant cutin of Arabidopsis inflorescence stem was examined. Table I shows that the most abundant cutin monomer in wild-type Arabidopsis stem was hexadecane-1,16-dioic acid (49.3%) followed by 10,16-dihydroxy hexadecanoic acid (16.4%), octadecane-1,18-dioic acid (11.8%), 7-hydroxy hexadecane-1,16-dioic acid (11.3%), 10(9)-hydroxy heptadecanoic acid (3.9%), 16-hydoxy hexadecanoic acid (3.2%), and 9-hydroxy pentadecanoic acid (2.6%). In cyp86a2-1, the total cutin monomers per surface area were reduced by ∼70% compared with that in the wild-type plants. Except for octadecadien-1,18-dioic acid, all monomers were significantly reduced in cyp86a2 (31–78% less than the wild type). The results indicated a broad effect of cyp86a2-1 on cutin biosynthesis. The ratio of cutin/cutan largely remained unchanged, suggesting that both cutin and cutan were proportionally reduced in cyp86a2-1 (Table II).
Table 1.
Stem cutin monomer amount (μg/dm2±s.d.) and percent cutin monomers (%±s.d.)
| Columbia |
att1-1 |
|||
|---|---|---|---|---|
| Cutin monomer | Amount | Percent | Amount | Percent |
| 9-Hydroxy pentadecanoic acid | 1.3±0.4 | 2.6±0.3 | 0.7±0.1 | 4.3±1.0 |
| 10(9)-Hydroxy heptadecanoic acid | 1.9±1.1 | 3.9±2.3 | 0.7±0.3 | 4.3±1.9 |
| 16-Hydroxy hexadecanoic acid | 1.6±0.3 | 3.2±0.2 | 1.1±0.3 | 6.4±1.6 |
| 10,16(9,16)-Dihydroxy hexadecanoic acid | 8.1±2.6 | 16.4±3.5 | 3.5±1.1 | 21.3±5.3 |
| Hexadecane-1,16-dioic acid | 24.1±3.5 | 49.3±1.2 | 5.4±0.1 | 33.6±2.6 |
| 7(8)-Hydroxy hexadecane-1,16-dioic | 5.5±0.7 | 11.3±0.6 | 2.2±0.3 | 13.7±2.7 |
| Octadecane-1,18-dioic acid | 5.8±1.1 | 11.8±0.3 | 1.8±0.1 | 11.1±1.5 |
| Octadecadien-1,18-dioic acid | 0.7±0.1 | 1.4±0.1 | 0.8±0.1 | 5.3±0.7 |
| Total |
49.1±8.2 |
|
16.1±1.1 |
|
| Cuticle was enzymatically isolated from the first three internodes of the main stem of 35-day-old plants (three replicates). Each replicate includes 12–14 individual plants pooled together. Isomers are listed in parentheses. Position of the two double bonds in octadecadien-1,18-dioic acid was not determined. | ||||
Table 2.
Cutin and cutan as a percent (%) of total weight of dried, dewaxed cuticle membrane
| Col-0 | cyp86a2-1 | |
|---|---|---|
| % cutin | 35.8±2.5 | 41.8±5.0 |
| % cutan | 64.2±2.5 | 58.2±5.0 |
cyp86a2 mutants showed greater sensitivity to dehydration. Detached leaves from the cyp86a2-1 mutant wilted much faster than did wild-type leaves, indicating a higher epidermal permeability of cyp86a2-1 to water vapor than that of wild-type (data not shown). Whole-plant in-pot transpiration analysis showed that cyp86a2-1 plants had higher transpiration rates than did wild-type plants in both light and dark environments (Figure 8). The cyp86a2-1 mutant had normal stomatal opening and closing responses to photoperiod. However, cyp86a2-1 had ∼2-fold higher water loss rate relative to wild-type both in the light and dark, when stomata were closed, indicating that a higher permeability of cuticle rather than stomatal pore was the primary cause for the increase in the transpiration rate.
Figure 8.
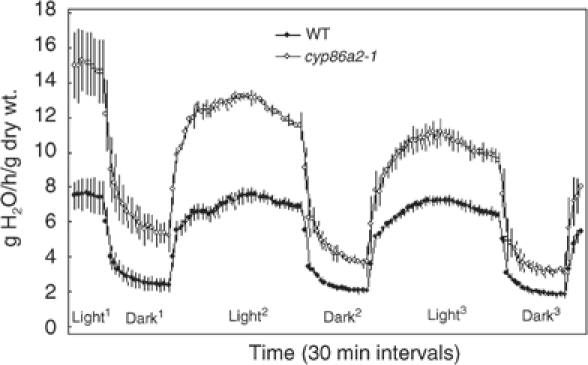
cyp86a2-1 has a higher transpiration rate. Whole flowering plant in-pot transpiration rates were measured for consecutive 16 h light and 8 h dark photoperiods. Error bars indicate standard deviation.
The wax2 mutant does not show enhanced avrPto-luc expression
The defects of cyp86a2 cuticle membrane prompted us to test the potential role of cuticle structure in bacterial type III gene expression. Bacterial avrPto-luc expression was examined in another Arabidopsis mutant, wax2. Like att1, wax2 also shows a thick but translucent cuticle membrane (Chen et al, 2003). However, wax2 only supported normal avrPto-luc expression similar to the wild-type plants (Figure 9), indicating that the enhanced type III gene induction was specific to cyp86a2 mutants.
Figure 9.
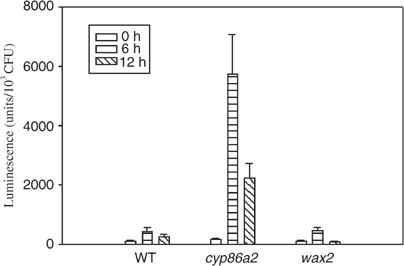
wax2 mutant shows normal induction of avrPto-luc. Col-0 (WT), cyp86a2-2, and wax2 plants were infiltrated with P. syringae pv. phaseolicola (avrPto-luc), and the luciferase activity was measured at 0, 6, and 12 h after inoculation. Bars indicate standard errors.
To further determine the role of cuticle in avrPto induction, we constructed avrPto-gfp and examined its expression from bacteria in the intercellular spaces and at the leaf surface. P. syringae pv. phaseolicola bacteria expressed avrPto-gfp only in the intercellular spaces, primarily in the substomatal chamber, whereas no expression was detected at the leaf surface of either wild-type or cyp86a2 plants (Figure 10). The avrPto-gfp expression in the intercellular spaces was much stronger in cyp86a2-1 than in wild-type plants.
Figure 10.
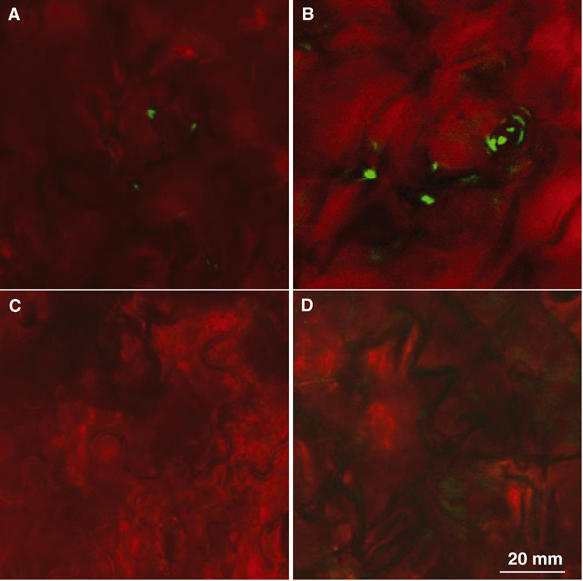
Induction of avrPto-gfp in the intercellular spaces. avrPto-gfp expression in the intercellular spaces (A, B) or at the leaf surface (C, D) of the wild-type (A, C) and cyp86a2-2 (B, D) plants. Leaves were inoculated with P. syringae pv. phaseolicola (avrPto-gfp) for 6 h before being examined under a confocal microscope. The images are representative of multiple plants in two experiments.
Discussion
The work described here assigns two important biological functions to CYP86A2: normal development of cuticle and repressing the in planta expression of bacterial type III genes avrPto and hrpL. The latter is a master regulatory gene for P. syringae genes containing the hrp box. At least when dip-inoculated, the cyp86a2-1 mutant displays enhanced disease severity to the virulent bacteria P. syringae pv. tomato, indicating a positive role of CYP86A2 in disease resistance. CYP86A2 belongs to a family of fatty acid hydroxylases and is involved in the biosynthesis of extracellular lipids, as indicated by the severely reduced cutin content in att1 plants.
Cutin monomers are synthesized intracellularly through multiple hydroxylation and epoxidation reactions catalyzed by cytochrome P450-dependent enzymes (Kolattukudy, 2001). These hydroxyl fatty acids are then exported and polymerized extracellularly. Several genes encoding cytochrome P450 monooxygenases that catalyze internal and ω-hydroxylation of fatty acids have been isolated, and their in vitro enzymatic activities demonstrated (Pinot et al, 1992, 1998; Beneviste et al, 1998; Cabello-Hurtado et al, 1998; Tijet et al, 1998). However, no cytochrome P450 monooxygenase genes have been demonstrated to function in cutin biosynthesis in vivo.
To date, genetic studies have suggested three proteins involved in the biosynthesis of cutin. These include CYP86A8 (Wellesen et al, 2001), WAX2, a lipid desaturase-like metabolic enzyme (Chen et al, 2003), and LACS2 (long-chain acyl-CoA synthetase; Schnurr et al, 2004). Mutations in CYP86A8 and WAX2 lead to postgenital organ fusion, a phenotype known for plants expressing a fungal cutinase (Sieber et al, 2000). However, cutin compositions in these mutants remain unknown. Consequently, the role of these proteins in cutin biosynthesis remains to be established. Our results demonstrate that CYP86A2 is required for the accumulation of the majority of cutin monomers and normal cuticle membrane development in vivo, suggesting that CYP86A2 is a major cutin biosynthetic enzyme. Unlike other mutants, cyp86a2 mutants are morphologically normal and do not show postgenital fusions. The cuticle membrane of cyp86a2 mutants has a ‘puffy' appearance, which may reflect reduced polyester bonding between cutin monomers, a consequence of reduced fatty acid hydroxylation.
The finding that cyp86a2 mutants have much greater water permeability is highly significant. One of the major functions of the cuticle is to prevent water loss. However, the relative contribution of the wax layer and the cuticle membrane has been unclear. A causal relationship between wax load and dehydration tolerance has not been convincingly established (Jenks, 2002). Our results show that the cuticle membrane plays a large role in preventing water loss by reducing the permeability to water vapor.
CYP86A2 may have multiple functions, a condition that might explain the reduction of all cutin monomers of the cyp86a2-1 mutant. A comparable P450, CYP94A5, was shown in Nicotiana tabacum to both hydroxylate the terminal methyl group of 9,10-expoxy octadecanoic acid and then oxidize the hydroxy end group to a carboxyl in vitro (Le Bouquin et al, 2001). The Arabidopsis CYP86A family contains five members, CYP86A1, CYP86A2, CYP86A4, CYP86A7, and CYP86A8 (LCR). The cyp86a2 mutants are not completely abolished in cuticle development, suggesting that other family members may account for the synthesis of the remaining cutin.
While the biochemical function of CYP86A2 readily explains the defects in cutin composition and cuticle ultrastructure in the cyp86a2 mutants, it remains to be elucidated how the mutants enable a stronger type III gene induction in the bacterium. The enhanced avrPto-luc gene induction occurs in cyp86a2 but not the wax2 mutants. Furthermore, the avrPto-luc expression does not occur epiphytically in either wild-type or cyp86a2 plants. These results indicate that the alteration of cuticle membrane structure, by itself, does stimulate type III gene activation. CYP86A2 must repress bacterial type III gene expression from the intercellular spaces. This appeared puzzling at first, because cuticle is known as a surface structure. However, our findings that cells in the substomatal chambers are covered with a continuous cuticle membrane and that this cuticle membrane is also altered in cyp86a2 plants argue for a functional relevance of cutin in type III gene expression. The substomatal chamber cuticle membrane may repress bacterial type III gene expression, either by blocking the access of a positive host factor or active repression of bacterial gene expression. The less compact structure of the cyp86a2 substomatal chamber cuticle membrane may lead to the leakage of this host factor or enable better attachment of the bacterial cell to the host cell in the intercellular spaces, which in turn stimulates type III gene expression.
It is also possible that certain lipids may repress type III gene expression from the intercellular spaces, thus serving to discourage bacterial infection. These lipids could be either cutin monomers or other fatty acids related to cutin monomers that are synthesized by CYP86A2. Consistent with a role of extracellular lipids in plant–pathogen interaction, lipid transfer protein genes are often induced during defense responses. A recent report showed that the apoplast lipid transfer protein DIR1 is required for systemic acquired resistance (Maldonado et al, 2002), although the interacting lipids have yet to be identified. Interestingly, both DIR1 and ATT1 transcripts are downregulated following P. syringae inoculation, suggesting a functional link between the two genes. While a role of fatty acids in bacterial type III gene regulation has not been reported, free fatty acids derived from cutin are known to function as a signal for plant fungal pathogens (Woloshuk and Kolattukudy, 1986; Gilbert et al, 1996; Dickman et al, 2003). In support of this hypothesis, we have tested a variety of commercial fatty acids and found that certain cutin-related fatty acid species are capable of repressing avrPto-luc but not trp-luc expression (Y Xiao and JM Zhou, unpublished results). A sensitive assay for leaf cutin and other extracellular lipids remains to be developed to determine if these fatty acid species are responsible for type III gene repression in planta. Regardless of the nature of the host factor involved, the predicted enzymatic function of CYP86A2 and the reduction of cutin in cyp86a2 mutants strongly suggest that lipids are host factors modulating bacterial gene expression in the intercellular spaces.
In summary, our results demonstrate that CYP86A2 plays a major role in cutin biosynthesis and is required for normal development of cuticle membrane. CYP86A2 negatively modulates the expression of P. syringae type III genes. The predicted biochemical function of CYP86A2 raises the possibility that plant lipids may act as host factors modulating bacterial virulence gene expression.
Materials and methods
Bacterial strains and plants
The bacterial strains used in this study were P. syringae pv. phaseolicola NPS3121 (Lindgren et al, 1986) and P. syringae pv. tomato DC3000 (Davis et al, 1991). Reporter gene constructs were made as follows: for avrPto-luc, the luc gene was PCR-amplified from pSP-luc (Promega, Madison, WI) with a forward primer (5′-AGAATTCGGATCCGAAGACGCCAAAA ACATAAAG-3′) and the T7 primer. The PCR product was cloned into pGEM7Z (Promega, Madison, WI) between EcoRI and XbaI for sequencing confirmation. The resulting plasmid was digested with XbaI and BamHI, and the luc fragment was inserted into the corresponding sites of pPTE6 (Ronald et al, 1992) to replace the avrPto open reading frame (ORF). This gave rise to avrPto-luc. For the hrpL-luc construct, the hrpL promoter was PCR-amplified from DC3000 genomic DNA with a forward primer (5′-AGGAATTCTGTGCAGATCAGCATTGG GAAG-3′) and a reverse primer (5′-TTGGATCCCATGGGCTTACCCTGATT TAGTG-3′) and inserted into the BamHI and EcoRI sites of avrPto-luc to replace the avrPto promoter. To construct the trp-luc construct, luc was first excised from pSP-luc with XbaI and BglII and inserted between the XbaI and BamHI sites of pBluscript SK (−) (Stratagene, La Jolla, CA) to create pBS-luc. A HindIII fragment containing the Salmonella typhimurium trp promoter (Gage et al, 1996) was inserted into the HindIII site of pBS-luc to create pBS-trp-luc. The trp-luc cassette was then excised with XbaI and KpnI and inserted between the XbaI and KpnI sites of pPTE6 (Ronald et al, 1992). For avrPto-gfp construct, the ORF of gfp was amplified from pCKGFP S65C containing gfp (Reichel et al, 1996) with the following primers: 5′-TTGGATCCGGTAAAGGAGAAGAACTT TTC-3′, and 5′-TTTCTAGATTATTTGTATAGTTCATC CATG-3′. The PCR product was digested with BamHI and XbaI and replaced the luc gene in avrPto-luc.
All Arabidopsis thaliana ecotypes (Col-0, Col-gl, and No-0) and EMS-mutagenized M2 seeds (Col-gl background) were obtained from Lehle Seeds (Tucson, AZ). Plants were grown in growth chambers or greenhouse at 20°C at night and 22°C during the day with a 10 h/day photoperiod.
Luminescence and fluorescence imaging assay
For luminescence imaging, plants were inoculated with P. syringae pv. phaseolicola carrying a luc construct at 108 CFU/ml, and leaves were detached at the indicated times, sprayed with a solution containing 1 mM luciferin and 0.01% Triton X-100, and kept in the dark for 5 min before imaging by using a cooled CCD (Roper Scientific, Trenton, NJ). The relative LUC activity was normalized to leaf bacterial numbers. Each data point represents an average of 3–4 replicates.
For assaying the bacterial expression of avrPto-gfp in the intercellular spaces, plants were infiltrated with 5 × 108 CFU/ml P. syringae pv. phaseolicola containing avrPto-gfp. For assaying bacterial expression of avrPto-gfp at the leaf surface, leaves were coated with the same bacteria suspension in the presence of 0.02% silwet L-77. Leaves were then examined for green fluorescence under a confocal microscope 6 h after inoculation.
Mutant screening and cloning of ATT1
Col-gl M2 plants were grown in the greenhouse for 5 weeks. Fully expanded leaves were hand-inoculated with P. s. pv. phaseolicola (avrPto-luc) at 4 × 108 CFU/ml. The luciferase activity was examined 8 h after injection by CCD imaging. Plants showing high luminescence were verified in the M3 generation.
For genetic mapping, the att1-1 mutant was crossed with wild-type No-0 plants. The resulting F1 plants were self-pollinated to produce F2 populations. F2 plants were screened for individuals with the att1 phenotype. Genomic DNA from these plants was extracted and analyzed with SSLP markers spanning the five chromosomes. For fine mapping, a single nucleotide polymorphism (SNP) was developed for every 5 kb of the chromosome by comparing a 1 kb PCR-amplified genomic DNA fragment from No-0 with that of Col-0 in the database. DNA fragments spanning the SNPs were then PCR-amplified from putative recombinant plants and sequenced to determine the heterozygosity of the marker.
Disease and bacterial growth assay
Bacterial culture was grown in King's B medium (King et al, 1954) with appropriate antibiotics, and the inoculum was prepared in sterile water. Plants (5–6 weeks old) were dipped in a suspension containing 108 CFU/ml bacteria and 0.04% silwet L-77 (OSI, Danbury, CT), and leaf bacterial number was determined by plating bacteria on King's B agar plates containing appropriate antibiotics.
Transpiration assay
For whole-plant transpiration measurements, 3-inch pots containing one 35-day-old plant were enclosed in polyethylene terephthalate film and bound under the rosette to prevent water loss except through the exposed plant surfaces. Allowing 8 h for plant transpiration rates to stabilize, transpiration rates of three wild-type and three cyp86a2-1 replicates were monitored instantaneously using six electronic balances and WinWedge data acquisition software (TalTech, Philadelphia, PA). To increase water loss amounts to improve measurement accuracy, two plants placed on one balance constituted one replicate. Measurements were performed in complete darkness (when stomata were assumed to be generally closed) and in the light (when stomata were assumed to be generally open). A 400 W metal halide lamp provided illumination (120 mol m2 s). Temperature during the study ranged from 22 to 24°C, and RH was between 25 and 35%.
Agrobacterium-mediated transformation
An 8 kb PvuII genomic DNA fragment containing At4g00360 was subcloned from the BAC clone F5I10, blunt-ended by Klenow fill-in, and inserted between the HindIII and EcoRI sites (also blunt-ended by Klenow fill-in) of the binary vector pBI121 (Clontech, Palo Alto, CA). att1-1 inflorescences were dipped with Agrobacterium tumafaciens strain LBA4404 carrying the transgene construct as described (http://plantpath.wisc.edu/%7Eafb/protocol.html).
Northern blot analysis
Plants were vacuum-infiltrated with bacteria, and leaves were sampled at indicated times for RNA isolation. Total RNA (10 μg/lane) was fractionated with formaldehyde gel, transferred to a nitrocellulose membrane, and hybridized to 32P-radiolabeled At4g00360 genomic DNA fragment.
Transmission electron microscopy
Mid-blade sections of fully expanded leaves were dissected into 1 × 1 mm squares and immediately placed in 100 mM sodium cacodylate buffer (pH 7.0) containing 2.5% (w/v) glutaraldehyde and 4% (w/v) paraformaldehyde. Vacuum was applied until samples were fully submerged. Tissues were then fixed for 3 h at room temperature, washed three times with 100 mM sodium cacodylate buffer (pH 7.0), and postfixed in 100 mM sodium cacodylate buffer (pH 7.0) containing 1% (w/v) osmium tetroxide overnight. Samples were subsequently washed with 100 mM sodium cacodylate buffer (pH 7.0), dehydrated in an acetone series (30–100% by 10% steps, 10 min each step), and infiltrated with Spurr's (Spurr, 1969) epoxy resin (1:2, 1:1, 2:1 resin:acetone, and pure resin for 4 h, overnight, 3 h, and 5 h, respectively). Infiltrated tissues were placed in molds, and the resin was cured at 60°C for 2 days.
Embedded materials were thin-sectioned using an ultramicrotome (Leica Inc., Deerfield, IL). Sections were collected onto 200-mesh copper grids, stained with 2% (w/v) uranyl acetate for 15 min, and rinsed for 30 min, before being viewed and photographed with a transmission electron microscope (FEI, Hillsboro, OR).
Cutin monomer analysis
The cuticle was enzymatically isolated as per Schönherr and Riederer (1986) from first to third internodes of main stems of 35-day-old plants grown under greenhouse conditions as stated previously. Stem areas were measured, and then stem segments from 12–14 individual plants were pooled per sample. The cuticle was refluxed in chloroform:methanol (1:1) for 24 h and then depolymerized with 14% BF3 in anhydrous methanol (Sigma) to produce methyl esters, which were then extracted with diethyl ether as per Riederer and Schönherr (1986). Extracts were derivatized with N,O-bis(trimethylsily)trifluoroacetamide and incubated for 30 min at 70°C. After derivatization, samples were analyzed with a Hewlett-Packard 5890 series II gas chromatograph (GC) equipped with a flame ionization detector. The GC was equipped with a 12 m, 0.2 mm HP-1 capillary column with helium as the carrier gas. The GC was programmed with an initial temperature of 80°C, and then increased at 15°C/min to 200°C and at 2°C/min to 280°C. Quantification was based on flame ionization detector peak areas and the internal standard eicosane added prior to depolymerization. Specific correction factors were developed from multilevel calibration curves (as for wax analysis; Chen et al, 2003) developed from external standards hexadecanoic acid (Aldrich), hexadecane-1,16-dioic acid (Aldrich), and 16-hydroxy hexadecanoic acid (Sigma). All values shown in tables represent the average of three replicate samples. Selected subsamples were analyzed in a GC–mass spectrometer (FinniganMAT/Thermospray Corporation, San Jose, CA). Percentage of cutin and cutan (nonsaponable fraction) was determined by weighing cuticle before and after depolymerization (Table II).
Cutin monomer composition was also analyzed using whole-organ samples. Results of three replicates were statistically equivalent (P<0.05) to the results obtained from enzymatically isolated cuticles, as determined by the Student's t-test. Palmitate and stearate had amounts 4- to 8-fold greater in whole than the enzymatically isolated samples, and were likely derived in large part from internal lipid pools. For this reason, we did not include these acid compounds in the cutin monomer profiles presented.
All cutin monomers were identified from electron impact mass spectra of the methyl ester TMS derivatives on the basis of the published spectra (Eglinton and Hunneman, 1968; Holloway, 1982), retention indexes (Holloway, 1984), and retention times of the external standards.
Methyl ester monomers identified in this study are listed with retention times, diagnostic fragment ions, and relative abundances. Minor isomers are in parentheses.
Methyl hexadecanoate, retention time (RT) 8.305 min, molecular mass (MM) 270; m/z 270, 5; m/z 241, 2; m/z 239, 3; m/z 227, 7; m/z 87, 60; m/z 74, 100.
Methyl 9-TMS pentadecanoate, RT 9.658 min, MM 344; m/z 329, 4; m/z 313, 5; m/z 297, 3; m/z 259, 4; m/z 230, 5; m/z 187, 90; m/z 75, 32; m/z 73, 100.
Methyl octadecanoate, RT 10.144 min, MM 298; m/z 298, 32; m/z 269, 11; m/z 267, 10; m/z 255, 18; m/z 87, 68; m/z 74, 100.
Dimethyl hexadecane-1,16-dioate, RT 11.597 min, MM 314; m/z 283, 32; m/z 250, 10; m/z 241, 21; m/z 222, 3; m/z 209,18; m/z 191, 14; m/z 112, 32; m/z 98, 100; m/z 84, 45; m/z 74, 56.
Methyl 16-TMS hexadecanoate, RT 12.298 min, MM 358; m/z 343, 32; m/z 327, 5; m/z 311, 100; m/z 159, 9; m/z 146, 10; m/z 75, 45.
Methyl 10(9)-TMS heptadecanoic acid, RT 13.225 min, MS 372; m/z 357, 4; m/z 325, 4; m/z 273, 68; m/z 259, 9; m/z 244, 10; m/z 215, 5; m/z 201, 29; m/z 81, 37; m/z 75, 50; m/z 73, 100; m/z 55, 60.
Dimethyl octadecadiene-1,18-dioate, RT 13.996 min, MM 338; m/z 306, 45; m/z 274, 34; m/z 264, 5; m/z 246, 8; m/z 95, 40; m/z 94, 50; m/z 93, 44; m/z 81, 62; m/z 67, 100; m/z 55, 96.
Dimethyl octadecane-1,18-dioate, RT 14.996 min, MM 342; m/z 311, 22; m/z 269, 18; m/z 250, 8; m/z 237, 11; m/z 219, 10; m/z 112, 42; m/z 98, 90; m/z 74, 54; m/z 55, 100.
Dimethyl 7(8)-TMS hexadecane-1,16-dioate, RT 15.099 min, MM 402; m/z 387, 6; m/z 371, 2; m/z 355, 3; m/z 273, 82; m/z 259, 36; m/z 245, 35; m/z 244, 12; m/z 231, 98; m/z 230, 5; m/z 216, 5; m/z 202, 12; m/z 73, 100.
Methyl 10,16(9,16)-bisTMS hexadecanoate, RT 16.024 min, MM 446; m/z 431, 5; m/z 415, 3; m/z 399, 3; m/z 309, 9; m/z 289, 2; m/z 275, 44; m/z 273, 100; m/z 259, 3; m/z 244, 16; m/z 230, 1; m/z 73, 63.
Two non-cutin phenolic acids esterified to carbohydrates (Holloway, 1982) were identified.
Methyl m-coumarate, RT 7.239 min, MM 250; m/z 250, 100; m/z 235, 64; m/z 219, 34; m/z 203, 40; m/z 75, 75; m/z 73, 39.
Methyl ferulate, RT 7.409 min, MM 280; m/z 280, 74; m/z 265, 45; m/z 250, 100; m/z 219, 35; m/z 191, 8; m/z 73, 20.
Supplementary Material
Supplementary Figure
Acknowledgments
We thank Dr Marty Dickman for providing cutin monomers, Drs Frank White and Scot Hulbert for critical reading of the manuscript, Dr Daniel Boyle for assistance in electron and confocal microscopy, and Dr Karl Wood for help in identifying cutin monomer spectra. The work was supported by Kansas Agriculture Experimental Station (contribution #04-225-J) and an NIH grant (P20 RR16443-01) to XT.
References
- Aldon D, Brito B, Boucher C, Genin S (2000) A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes. EMBO J 19: 2304–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlat M, Gough CL, Zischek C, Barberis PA, Trigalet A, Boucher CA (1992) Transcriptional organization and expression of the large hrp gene cluster of Pseudomonas solanacearum. Mol Plant Microbe Interact 5: 187–193 [DOI] [PubMed] [Google Scholar]
- Beneviste I, Tijet N, Adas F, Philipps G, Salaun JP, Durst F (1998) CYP86A1 from Arabidopsis thaliana encodes a cytochrome P450-dependent fatty acid omega-hydroxylase. Biochem Biophys Res Commmun 243: 688–693 [DOI] [PubMed] [Google Scholar]
- Brito B, Aldon D, Barberis P, Boucher C, Genin S (2002) A signal transfer system through three compartments transduces the plant cell contact-dependent signal controlling Ralstonia solanacearum hrp genes. Mol Plant Microbe Interact 15: 109–119 [DOI] [PubMed] [Google Scholar]
- Brito B, Marenda M, Barberis P, Boucher C, Genin S (1999) prhJ and hrpG, two new components of the plant signal-dependent regulatory cascade controlled by PrhA in Ralstonia solanacearum. Mol Microbiol 31: 237–251 [DOI] [PubMed] [Google Scholar]
- Cabello-Hurtado F, Batard Y, Salaun J-P, Durst F, Pinot F, Werck-Reichhart D (1998) Cloning, expression in yeast, and functional characterization of CYP81B1, a plant cytochrome P450 that catalyzes in-chain hydroxylation of fatty acids. J Biol Chem 273: 7260–7267 [DOI] [PubMed] [Google Scholar]
- Chen X, Goodwin M, Boroff VL, Liu X, Jenks MA (2003) Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 15: 1170–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis KR, Schott E, Ausubel FM (1991) Virulence of selected phytopathogenic pseudomonads in Arabidopsis thaliana. Mol Plant Microbe Interact 4: 477–488 [Google Scholar]
- Demers B, Sansonetti PJ, Parsot C (1998) Induction of type III secretion in Shigella flexneri is associated with differential control of transcription of genes encoding secreted proteins. EMBO J 17: 2894–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickman MB, Ha Y-S, Yang Z, Adams B, Huang C (2003) A protein kinase from Colletotrichum trifolii is induced by plant cutin and is required for appressorium formation. Mol Plant Microbe Interact 16: 411–421 [DOI] [PubMed] [Google Scholar]
- Eglinton G, Hunneman DH (1968) Gas chromatographic-mass spectrometric studies of long chain hydroxy acids—I. Phytochemistry 7: 313–322 [Google Scholar]
- Esau K (1977) Anatomy of Seed Plants, 2nd edn. New York: Wiley [Google Scholar]
- Francis MS, Wolf-Watz H, Forsberg A (2002) Regulation of type III secretion systems. Curr Opin Microbiol 5: 166–172 [DOI] [PubMed] [Google Scholar]
- Gage DJ, Bobo T, Long SR (1996) Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa). J Bacteriol 178: 7159–7166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, Collmer A (1999) Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284: 1322–1328 [DOI] [PubMed] [Google Scholar]
- Gilbert RD, Johnson AM, Dean RA (1996) Chemical signals responsible for appressorium formation in the rice blast fungus Magnaporthe grisea. Physiol Mol Plant Pathol 48: 335–346 [Google Scholar]
- He SY, Jin Q (2003) The Hrp pilus: learning from flagella. Curr Opin Microbiol 6: 15–19 [DOI] [PubMed] [Google Scholar]
- Holloway PJ (1982) The chemical constitution of plant cutins. In The Plant Cuticle. Linnaean Society Symposium Series, Cutler D, Alvin K, Price CE (eds) pp 45–85. London: Academic Press [Google Scholar]
- Holloway PJ (1984) Cutins and suberins, the polymeric plant lipids. In CRC Handbook of Chromatography: Lipids, Mangold HK, Zweig G, Sherma J (eds) Vol I, pp 321–345. Boca Raton, FL: CRC Press [Google Scholar]
- Huynh TV, Dahlbeck D, Staskawicz BJ (1989) Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science 245: 1374–1377 [DOI] [PubMed] [Google Scholar]
- Jeffree CE (1996) Structure and ontogeny of plant cuticles. In Plant Cuticles, Kerstiens G (ed) pp 33–82. Oxford: BIOS Scientific Publishers Ltd [Google Scholar]
- Jenks MA (2002) Critical issues with the plant cuticle's function in drought tolerance. In Biochemical & Molecular Responses of Plants to the Environment, Wood AJ (ed) pp 97–127. Kerala, India: Research Signpost Press ISBN: 81-7736-167-8 [Google Scholar]
- Jenks MA, Joly RA, Rich PJ, Peters PJ, Axtell JD, Ashworth EN (1994) Chemically-induced cuticle mutation affecting epidermal conductance to water vapor and disease susceptibility in Sorghum bicolor (L.) Moench. Plant Physiol 105: 1239–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluoresin. J Lab Clin Med 44: 301–307 [PubMed] [Google Scholar]
- Kolattukudy PE (1981) Structure, biosynthesis, and biodegradation of cutin and suberin. Annu Rev Plant Physiol 32: 539–567 [Google Scholar]
- Kolattukudy PE (2001) Polyesters in higher plants. In Advances in Biochemical Engineering/Biotechnology, Scheper Th (ed) Vol 71. Berlin, Heidelberg: Springer-Verlag [DOI] [PubMed] [Google Scholar]
- Le Bouquin R, Skrabs M, Kahn R, Benveniste I, Salaün J, Schreiber L, Durst F, Pinot F (2001) CYP94A5, a new cytochrome P450 from Nicotiana tabacum is able to catalyze the oxidation of fatty acids to the ω-alcohol and to the corresponding diacid. Eur J Biochem 268: 3083–3090 [DOI] [PubMed] [Google Scholar]
- Lee SH, Camilli A (2000) Novel approaches to monitor bacterial gene expression in infected tissue and host. Curr Opin Microbiol 3: 97–101 [DOI] [PubMed] [Google Scholar]
- Lindgren PB, Peet RC, Panopoulos NJ (1986) Gene cluster of Pseudomonas syringae pv. ‘phaseolicola' controls pathogenicity of bean plants and hypersensitivity on nonhost plants. J Bacteriol 168: 512–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado AM, Doerner P, Dixon RA, Lamb CJ, Cameron RK (2002) A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419: 399–403 [DOI] [PubMed] [Google Scholar]
- Marenda M, Brito B, Callard D, Genin S, Barberis P, Boucher C, Arlat M (1998) PrhA controls a novel regulatory pathway required for the specific induction of Ralstonia solanacearum hrp genes in the presence of plant cells. Mol Microbiol 27: 437–453 [DOI] [PubMed] [Google Scholar]
- Mittal S, Davis KR (1995) Role of the phytotoxin coronatine in the infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Mol Plant Microbe Interact 8: 165–171 [DOI] [PubMed] [Google Scholar]
- Nawrath C (2002) The biopolymers cutin and suberin. In The Arabidopsis Book, Somerville C, Meyerowitz E (eds) American Society of Plant Biologists (http://www.aspb.org/publications/arabidopsis) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson J, Nordfelth R, Dubinina E, Bergman T, Gustafsson M, Magnusson KE, Wolf-Watz H (1996) Modulation of virulence factor expression by pathogen target cell contact. Science 273: 1231–1233 [DOI] [PubMed] [Google Scholar]
- Pinot F, Benveniste I, Salaun JP, Durst F (1998) Methyl jasmonate induces lauric acid omega-hydroxylase activity and accumulation of CYP94A1 transcripts but does not affect epoxide hydrolase activities in Vicia sativa. Plant Physiol Biochem 35: 103–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinot F, Salaun J-P, Bosch H, Lesot A, Minoskowski C, Durst F (1992) Omega-hydroxylation of Z9-octadecenoic, Z9,10-epoxystearic and 9,10-dihydroxystearic acids by microsomal cytochrome P450 systems from Vicia sativa. Biochem Biophys Res Commun 184: 183–193 [DOI] [PubMed] [Google Scholar]
- Rahme LG, Mindrinos MN, Panopoulos NJ (1992) Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syrinagae pv. phaseolicola. J Bacteriol 174: 3499–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichel C, Mathur J, Eckes P, Langenkemper K, Koncz C, Schell J, Reiss B, Maas C (1996) Enhanced green fluorescence by the expression of an Aequorea victoria green fluorescent protein mutant in mono- and dicotyledonous plant cells. Proc Natl Acad Sci USA 93: 5888–5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riederer M, Schönherr J (1986) Quantitative gas chromatographic analysis of methyl esters of hydroxy fatty acids derived from plant cutin. J Chromatogr 360: 151–161 [Google Scholar]
- Ronald PC, Salmeron JM, Carland FM, Staskawicz BJ (1992) The cloned avirulence gene avrPto induces disease resistance in tomato cultivars containing the Pto resistance gene. J Bacteriol 174: 1604–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmeron JM, Staskawicz BJ (1993) Molecular characterization and hrp dependence of the avirulence gene avrPto from Pseudomonas syringae pv. tomato. Mol Gen Genet 239: 6–16 [DOI] [PubMed] [Google Scholar]
- Schnurr J, Shockey J, Browse J (2004) The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 16: 629–642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönherr J, Riederer M (1986) Plant cuticles sorb lipophilic compounds during enzymatic isolation. Plant Cell Environ 9: 459–466 [Google Scholar]
- Sieber P, Schorderet M, Ryser U, Buchala A, Kolattukudy P, Metraux J-P, Nawrath C (2000) Transgenic Arabidopsis plants expressing a fungal cutinase show alterations in the structure and properties of the cuticle and postgenital organ fusion. Plant Cell 12: 721–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurr AR (1969) A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res 261: 31–43 [DOI] [PubMed] [Google Scholar]
- Stainier I, Iriarte M, Cornelis GR (1997) YscM1 and YscM2, two Yersinia enterocolitica proteins causing downregulation of yop transcription. Mol Microbiol 26: 833–843 [DOI] [PubMed] [Google Scholar]
- Tijet N, Helvig C, Pinot F, Le Bouquin R, Lesot A, Durst F, Salaun JP, Beneviste I (1998) Functional expression in yeast and characterization of a clofibrate-inducible plant cytochrome P450 (CYP94A1) involved in cutin monomer synthesis. Biochem J 332: 583–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallis AJ, Yahr TL, Barbieri JT, Frank DW (1999) Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect Immun 67: 914–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei ZM, Sneath BJ, Beer SV (1992) Expression of Erwinia amylovora hrp genes in response to environmental stimuli. J Bacteriol 174: 1875–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellesen K, Durst D, Pinot F, Benveniste I, Nettesheim K, Wisman E, Steiner-Lange S, Saedler H, Yephremov A (2001) Functional analysis of the LACERATA gene of Arabidopsis provides evidence for different roles of fatty acid ω-hydroxylation in development. Proc Natl Acad Sci USA 98: 9694–9699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werck-Reichhart D, Bak S, Paquette S (2002) Cytochrome P450. In The Arabidopsis Book, Somerville C, Meyerowitz (eds) American Society of Plant Biologists (http://www.aspb.org/publications/arabidopsis) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woloshuk CP, Kolattukudy P (1986) Mechanism by which contact with plant cuticle triggers cutinase gene expression in the spores of Fusarium solani f. sp. Pisi. Proc Natl Acad Sci USA 83: 1704–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Heu S, Yi J, Lu Y, Hutcheson SW (1994) Identification of a putative alternate sigma factor and characterization of a multicomponent regulatory cascade controlling the expression of Pseudomonas syringae pv. syringae Pss61 hrp and hrmA genes. J Bacteriol 176: 1025–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure


