Abstract
The major histocompatibility complex (MHC) class I chain-related A (MICA) is the most polymorphic non-classical MHC class I gene in humans. It encodes a ligand for NKG2D (NK group 2, member D), an activating natural killer (NK) receptor that is expressed mainly on NK cells and CD8+ T cells. The single-nucleotide polymorphism (SNP) rs1051792 causing a valine (Val) to methionine (Met) exchange at position 129 of the MICA protein is of specific interest. It separates MICA into isoforms that bind NKG2D with high (Met) and low affinities (Val). Therefore, this SNP has been investigated for associations with infections, autoimmune diseases, and cancer. Here, we systematically review these studies and analyze them in view of new data on the functional consequences of this polymorphism. It has been shown recently that the MICA-129Met variant elicits a stronger NKG2D signaling, resulting in more degranulation and IFN-γ production in NK cells and in a faster costimulation of CD8+ T cells than the MICA-129Val variant. However, the MICA-129Met isoform also downregulates NKG2D more efficiently than the MICA-129Val isoform. This downregulation impairs NKG2D-mediated functions at high expression intensities of the MICA-Met variant. These features of the MICA-129Met/Val dimorphism need to be considered when interpreting disease association studies. Particularly, in the field of hematopoietic stem cell transplantation, they help to explain the associations of the SNP with outcome including graft-versus-host disease and relapse of malignancy. Implications for future disease association studies of the MICA-129Met/Val dimorphism are discussed.
Keywords: NK cells, T cells, activating NK receptor, costimulation, single-nucleotide polymorphism, autoimmune diseases, cancer, hematopoietic stem cell transplantation
Introduction
The major histocompatibility complex (MHC) class I chain-related A (MICA) is the most polymorphic non-classical MHC class I gene in humans, and 105 alleles are known encoding for 82 protein variants (http://www.ebi.ac.uk/imgt/hla/, release 3.25.0). MICA is encoded within the human leukocyte antigen (HLA) complex close to HLA-B (1, 2). The protein structure is similar to classical class I molecules, but MICA is not associated with β2-microglobulin and does not present peptides. MICA is constitutively expressed only on a few cell types, including gastrointestinal epithelium, but is induced due to cellular and genotoxic stress (3, 4), malignant transformation, or virus infection (5, 6). MICA is a ligand for NKG2D (NK group 2, member D), an activating natural killer (NK) receptor encoded by the KLRK1 gene (7). NKG2D is expressed on most human NK cells, CD8+ αβ T cells, γδ T cells, iNKT cells, and subsets of effector or memory CD4+ T cells (8, 9). On NK cells, NKG2D signaling elicits killing of target cells (10) and secretion of IFN-γ (11). On CD8+ αβ T cells, NKG2D provides a costimulatory signal to activate naïve cytotoxic T lymphocytes (12). NKG2D contributes to the elimination of tumor cells (13) and plays a role in the defense against pathogens (14, 15). In addition to MICA, MICB and the UL16-binding proteins (ULBP) encoded by the retinoic acid early transcript 1 (RAET1) family function as ligands for NKG2D. MICB is also very polymorphic with 42 alleles encoding 28 protein variants (http://www.ebi.ac.uk/imgt/hla/, release 3.25.0). The RAET1 gene family is localized on chromosome 6 outside the HLA complex and six loci encode functional proteins (16). RAET1 genes are less polymorphic than MICA and MICB.
Polymorphisms of MICA have been investigated for their role in infections, autoimmune diseases, and cancer (17–21). The single-nucleotide polymorphism (SNP) rs1051792 (G/A) causing a valine (Val) to methionine (Met) exchange at position 129 in the α2 domain of the MICA protein has gained specific interest. It separates MICA alleles into two groups (22). MICA isoforms containing a methionine at position 129 bind NKG2D with high affinity, whereas those with a valine bind NKG2D with low affinity. High-affinity alleles include MICA*001, *002, *007, and *017; among the low-affinity alleles are MICA*004, *006, *008, *009, and *010 (23). Due to its functional consequences, the MICA-129Met/Val dimorphism has been investigated in several disease association studies. Here, we review these studies in view of new data on the functional consequences of this amino acid variation elicited after binding to NKG2D.
MICA-129Met/Val Disease Association Studies
In September 2016, we searched Pubmed for MICA-129Met/Val disease association studies using the key words rs1051792, MICA-129, MICA AND polymorphism AND Met, and MICA AND polymorphism AND Val. Moreover, we exchanged polymorphism by SNP, Met by methionine, and Val by valine. We identified 17 publications, in which an association of the MICA-129Met/Val dimorphism with a disease or disease complication has been investigated. One study in Chinese language (24) appeared to be not independent of a larger study published in English (25). Thus, we analyzed 16 independent studies published between 2005 and 2015 (Table S1 in Supplementary Material). Three studies are small with less than 100 cases. All others are of a medium size with more than 100 but less than 1,000 patients included, and most studies used a case–control design.
Eight studies investigated associations with autoimmune diseases, i.e., ankylosing spondylitis (AS) (26), rheumatoid arthritis (RA) (27–29), inflammatory bowel disease (IBD) (25, 30) [including ulcerative colitis (UC) and Crohn’s disease], systemic lupus erythematosus (SLE) (28), type I diabetes (31), latent autoimmune diabetes in adults (LADA) (31), and psoriasis (32). In one study, the MICA-129 SNP has not been determined directly. Instead, the SNP rs1051794 was typed and reported to be in complete linkage disequilibrium with the rs1051792 (27). Five studies reported on malignancies, i.e., nasopharyngeal cancer (33), hepatitis B virus (HBV)-induced hepatocellular carcinoma (HCC) (34), cutaneous malignant melanoma (35), and relapse of malignancy after hematopoietic stem cell transplantation (HSCT) (36, 37). Three studies investigated infections or their complications, i.e., HBV infection and HBV-induced HCC (34), left ventricular systolic dysfunction (LVSD) in chronic Chargas heart disease (38), and ocular toxoplasmosis (39). One study investigated an association of the MICA-129Met/Val dimorphism with recurrent miscarriage (40). The two studies on HSCT (36, 37) investigated besides relapse also other outcomes including graft-versus-host disease (GVHD).
Three studies, on recurrent miscarriage (40), ocular toxoplasmosis (39), and malignant melanoma (35), failed to demonstrate an association with the SNP. Thus, 81% of the studies showed an association at least for a subgroup, e.g., juvenile AS, whereas in all patients with AS, the association was dependent on HLA-B27 (26), or a sub-phenotype, e.g., severe LVSD (38). However, we must assume that other negative association studies have not been published. In seven studies, a MICA-129 allele and the corresponding homozygous genotype were both associated with a disease risk (25, 28, 29, 31, 32, 34, 38). The odds ratio (OR) was then always higher for the genotype than the allele. In six studies, the Met allele and/or the Met/Met genotype were found to be associated with a risk, including autoimmune diseases [juvenile AS (26), UC (30), SLE (28), and psoriasis (32)], a malignancy (HBV-induced HCC) (34), and a complication of an infection (severe LVSD in chronic Chargas disease) (38). In three studies, the Val allele and/or the Val/Val genotype has been identified to confer a risk for autoimmune diseases [including RA (27), UC (25), and diabetes (31)] and for nasopharyngeal carcinoma (NPC) (33). Moreover, rheumatoid factor (RF) positivity in RA patients has been associated with the Val allele and the Val/Val genotype (29). In the studies on HSCT, different outcomes showed different associations. In one study (36), the Met/Met genotype was associated with an increased risk of relapse and the Val/Val genotype with an increased risk of chronic GVHD. In our recent study (37), the Met/Met genotype conferred a risk of acute GVHD, whereas having Met alleles reduced the risk to die from acute GVHD. Overall, the Val allele was associated with a higher mortality after HSCT (37).
The results of these disease association studies do not allow for a simple unifying interpretation, such as the high-affinity MICA-129Met variant being associated with an activation of the immune system resulting in a lower risk of infections and cancer but higher risk of autoimmunity (Figures 1A,B). Autoimmune diseases are associated with both variants even within the same disease entity. UC, e.g., has been associated with the Met/Met genotype in a small study from Spain (30) but with the Val allele and Val/Val genotype in a larger study from China (25). RA has been associated with the Val allele in a study from France and Germany (27), but no association was found in cohorts from Japan (28) and Tunisia (29). Notably, a role of the NKG2D pathway has been reported for the pathogenesis of RA (41) and SLE (42), although this has not been linked to polymorphisms. Juvenile AS has been associated in a small study with the Met/Met genotype (26), and a larger sequencing study identified the MICA*007:01 allele that encodes a methionine at position 129 as a risk allele for AS in both Caucasian and Han Chinese populations (43). However, MICA*019, encoding a valine-129, has been identified as the major risk allele in Han Chinese (43). Malignancies were found to be associated with Val/Val genotype in the case of NPC (33) but with the Met/Met genotype in the case of relapse after HSCT (36). These different associations could suggest that the observed associations are random or dependent on the population studied. However, since the MICA-129Met/Val dimorphism is functional, it could also indicate that we need to better understand this function to predict its consequences in the pathophysiology of different diseases in various populations, which might be exposed to different interfering environmental factors. This assumption is supported by genome-wide association studies (GWAS), which have assigned disease risks for NPC (44), HCC (45, 46), cervical cancer (47, 48), and asthma (49) or advantages, such as HIV long-term non-progression (50) to the MICA gene region in an unbiased manner.
Figure 1.
Reported associations of the homozygous MICA-129 genotypes (A) and the MICA-129 alleles (B) with health risks [odds ratio (OR) > 1] or advantages (OR < 1). Shown are ORs with 95% confidence intervals (CI) or hazard ratios reported by Boukouaci et al. (36) and Isernhagen et al. (37) (overall survival) in event-time data. The number of patients analyzed in the studies is indicated at the y-axis. Studies reporting on autoimmune diseases are shown by open and closed blue symbols and malignancies by red symbols; studies reporting complications of infections (LVSD, Chargas disease; HCC, hepatitis B virus infection) are shown by green frames, and others are displayed by black symbols. The investigated diseases or complications and the references for the studies are indicated. (A) MICA-129Met/Met genotype effects are directly displayed. For studies that reported MICA-129Val/Val genotype effects [chronic GVHD (36), NPC (33), UC (25), RA (27), T1D, and LADA (31), indicated by brown font], the graph displays the corresponding effect of the pooled MICA-129 Met/Met and MICA-129Met/Val genotypes to allow for a direct comparison. (B) MICA-129Met allele effects are directly displayed. For studies that reported MICA-129Val allele effects [UC (25), T1D, and LADA (31), indicated by brown font], the graph displays the corresponding effect of the MICA-129Met allele; ORMet = 1/ORVal and 95%-CIMet = (1/CIVal, upper, 1/CIVal, lower). Abbreviations: CPs, cutaneous psoriasis; GVHD, graft-versus-host disease; HCC, hepatocellular carcinoma; JAS, juvenile ankylosing spondylitis; LADA, latent autoimmune diabetes in adults; LVSD, left ventricular systolic dysfunction; NPC, nasopharyngeal carcinoma; PsA, psoriatic arthritis; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; T1D, type 1 diabetes; UC, ulcerative colitis.
Functional Consequences of the MICA-129Met/Val Dimorphism
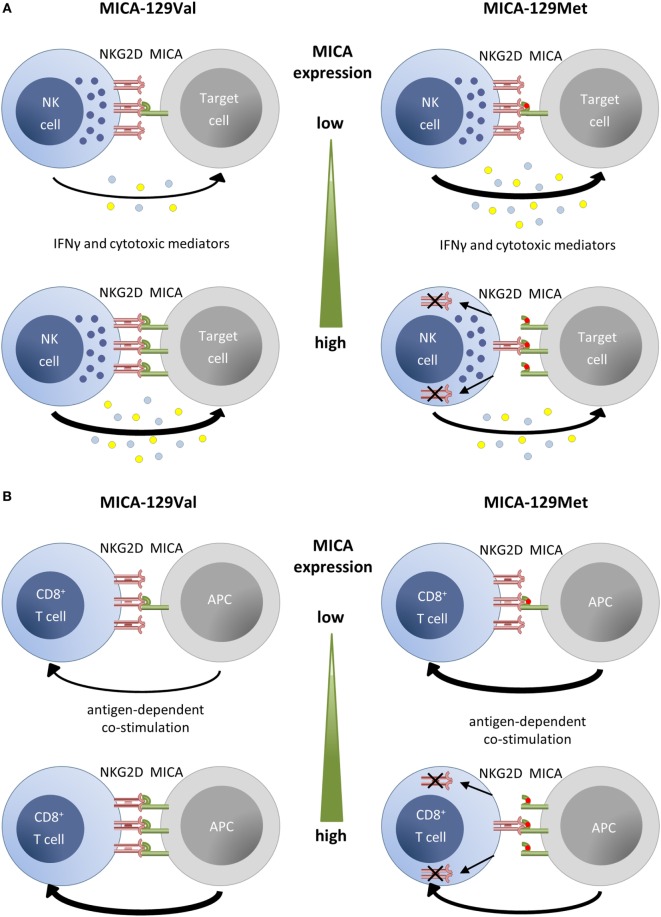
It has been shown by Steinle and colleagues that MICA-129Met isoforms bind NKG2D with high affinity in contrast to MICA-129Val isoforms that bind with low affinity (22). Yoshida and colleagues combined the MICA-129Met variant with the A9 variant of a microsatellite polymorphism in the transmembrane (TM) region and the MICA-129Val variant with the A5-TM variant in GST-fusion proteins (28). NK92MI cells showed a reduced NKG2D expression and killed K562 cells less efficiently when exposed to the MICA-129Met-A9-TM variant, but IFN-γ production was increased (28). We recently studied the consequences of binding of the two MICA-129 variants to NKG2D on primary NK cells and CD8+ T cells using cell lines transfected with expression constructs and recombinant Fc-fusion proteins differing only in amino acid 129 (37, 51). The recombinant MICA-129Met variant stimulated a stronger phosphorylation of SRC family kinases in NK cells than the MICA-129Val variant. Subsequently, the MICA-129Met ligand triggered more degranulation and IFN-γ production than the MICA-129Val ligand (Figure 2A). We then exposed NK cells to target cells expressing different amounts of the MICA-129 variants. The extent of degranulation and IFN-γ secretion correlated positively with the MICA expression intensity on the target cells but only for the MICA-129Val isoform. The expression intensity of the MICA-129Met isoform, in contrast, had either none or even a negative effect on the extent of degranulation, target cell killing, and IFN-γ release (37). On CD8+ T cells, the MICA-129Met isoform induced an earlier costimulatory activation than the MICA-129Val isoform (Figure 2B). Importantly, the MICA-129Met ligand induced also a stronger downregulation of NKG2D on both NK and CD8+ T cells than the MICA-129Val ligand. This downregulation of NKG2D impaired the capability of NK and CD8+ T cells to receive signals via NKG2D (37). Thus, MICA-129Met ligands, which elicit strong NKG2D responses, stimulate in parallel a robust negative feedback signal by downregulation of NKG2D that limits the initially stronger effects of MICA-129Met ligands. These data show that the biological effect of the MICA-129Met/Val dimorphism changes with the MICA expression intensity. Variant MICA-129Met triggers more NKG2D signals at low expression intensities, whereas variant MICA-129Val elicits more NKG2D effects at high expression, at which the MICA-129Met variant already downregulates NKG2D leading to impaired function. Thus, the biological effect of the SNP can hardly be predicted without information on the expression intensity of MICA.
Figure 2.
Summary of functional effects of MICA-129 variants depending on expression intensity. (A) For target cells expressing the MICA-129Val variant, the degree of natural killer (NK) cell cytotoxicity and IFN-γ production increases steadily with the MICA expression intensity. Augmented expression of the high-affinity MICA-129Met isoform, in contrast, has none or even a negative effect on these NK cell functions due to a rapid downregulation of NKG2D on NK cells. (B) Antigen-dependent costimulation of CD8+ T cells with the MICA-129Met variant allows for an earlier antigen-dependent activation than costimulation with the MICA-129Val variant. However, the downregulation of NKG2D in response to MICA-129Met ligands impairs any subsequent NKG2D-dependent costimulation and T cell activation. The downregulation of NKG2D on CD8+ T cells is augmented with MICA-129Met expression intensity. The figure is reproduced from Isernhagen et al. (37).
It is known that expression intensities vary for certain MICA alleles (52, 53). The G allele of the SNP at -1878 (rs2596542) in the promoter region of the MICA gene region, e.g., was found to have a higher transcriptional activity (54). Biological effects of the MICA-129Met/Val dimorphism can be expected to be modified by polymorphisms affecting MICA gene expression. We have investigated whether the Met/Val dimorphism itself affects MICA expression. In transfected cells, more of the MICA-129Met variant was retained in intracellular compartments (51). A similar alteration of the intracellular transport has been described for MICA-A5.1 variants (55). Thus, the combination of polymorphisms affecting transcription and intracellular transport of MICA could modify the effect of the Met/Val dimorphism.
Another important aspect of MICA is the generation of soluble MICA (sMICA) by proteolytic shedding. sMICA can induce NKG2D downregulation (56, 57) resulting in tumor immune escape (58). Some MICA polymorphisms have been reported to affect the amounts of sMICA in sera of patients including the SNP at -1878 (rs2596542) in the promoter region (34, 45, 59) that affects transcription (54), a microsatellite in exon 5 encoding the TM region (60, 61), and the MICA-129Met/Val dimorphism. In patients with UC, the MICA-129Val/Val genotype was associated with higher sMICA serum levels (25), and the MICA-129Val allele was also associated with higher sMICA serum levels in HBV patients and controls (34). In transfected cells, we found that the MICA-129Met isoform was more susceptible to shedding than the MICA-129Val isoform (51). However, due to the intracellular retention of the MICA-129Met variant (51), less sMICA might appear in sera (25, 34). Notably, intracellular retention and preferred shedding both appear to limit the expression of the high-affinity MICA-129Met isoform at the plasma membrane.
MICA-129Met/Val Disease Associations in View of Biological Functions
Recent data on the MICA-129Met/Val variation demonstrate the complexity of the functional consequences of this exchange of a single amino acid (37, 51). There are several layers of this complexity, which are as follows: (1) the function of the variant is not constant but dynamic (37); it depends on the MICA expression intensity, and the direction of the biological effect can invert for the MICA-129Met variant at higher expression. (2) Epistatic effects must be expected for this SNP as polymorphisms affecting the expression of MICA will modify the functional effects of the MICA-129Met/Val isoforms. Moreover, the expression intensity of NKG2D can be modified by SNPs in the KLRK1 gene (62) and those might interact with the MICA-129 variants. Other genes within the NKG2D pathway including other ligands might also show epistatic effects (63). (3) MICA can target NKG2D on several cell types, and biological effects on different cell types might be synergistic or antagonistic. An activation of NK cells and a costimulation of CD8+ T cells both can promote antitumor immunity. By contrast, a strong activation of NK cells might polarize an immune response to a Th1 reaction and reduce the risk to develop a Th2-mediated autoimmune disease. (4) Additional factors, such as sMICA or anti-MICA antibodies (36) that might neutralize sMICA, have been shown to be functionally important and have been determined in some of the disease association studies (25, 34, 36).
Currently, we mostly have not sufficient clinical and biological information to interpret the MICA-129Met/Val disease association studies in view of the complex function of this polymorphism. However, the two HSCT studies do provide more information and illustrate the clinical effects of the MICA-129Met/Val dimorphism as explained previously in detail (37). In our study (37), the homozygous carriers of Met alleles had an increased risk to experience acute GVHD, possibly due to immediate strong effects of MICA-129Met variants on NKG2D signaling. Having at least one Met allele reduced the risk to die from acute GVHD likely due to a rapid downregulation of NKG2D on alloreactive CD8+ T cells mediated by engagement of a high-affinity MICA-129Met variant. Carrying a MICA-129Met allele increased in consequence the chance of survival in all patients and in patients receiving a MICA-129-matched graft (37). Boukouaci and colleagues reported an increased risk of chronic GVHD for recipients with the Val/Val genotype, whereas the Met/Met genotype was associated with the risk of relapse (36). Sustained NKG2D-mediated activation of alloreactive CD8+ T cells would be expected if only MICA-129Val variants are present that fail to efficiently downregulate NKG2D, and this could increase the risk of chronic GVHD but reduce the risk of relapse. Thus, the different risk associations reported in the two studies are not arguing against the relevance of the MICA-129 dimorphism for the outcome of HSCT. The principal relevance of the NKG2D pathway for HSCT is further emphasized by studies showing an effect of the genotype of the NKG2D ligand RAET1L (64) and NKG2D itself (65) on the survival of patients. Moreover, matching for MICA alleles (66–69) and specifically for the MICA-129 polymorphism (70) is beneficial in HSCT. The huge effect of MICA-129 matching appears hardly explainable solely by the avoidance of a potential minor histocompatibility antigen. A “tuning” of the threshold of NKG2D signaling toward the affinity of NKG2D ligands present in an individual (52) and disturbance of this balance by mismatching could be considered as an alternative explanation.
Despite the functional relevance of the MICA-129 SNP, it cannot be excluded that some of the associations reported are random or caused by linkage disequilibrium with classical HLA genes. The association of MICA-129 with psoriasis (32) has been disproven in large GWAS cohorts (71). However, associations with NPC (33) and HCC (34) are supported by GWAS data pointing to the MICA gene region (44–46).
Conclusion
Information on functional consequences of a polymorphism is indispensable for understanding disease associations. The variation in the disease associated allele or genotype of MICA-129 in the published studies must not indicate random associations. For MICA-129, the biological function can change with expression intensity, epistatic interactions can be expected, the effect on different lymphocytes can vary, and modifying factors, such as sMICA, have to be considered. Notably, as expected for a functional SNP with a minor allele (MICA-129Met) frequency ranging from 48% in Africans to 30% in Asians (72), and being even the major allele reported in one of the analyzed studies (26), both alleles appear to confer advantages and disadvantages in specific situations suggesting balancing evolution of the MICA alleles. Since the MICA-129 dimorphism is considered as decisive for distinguishing low- and high-affinity variants (22), the frequency of alleles encoding high-affinity MICA variants is expected to match the frequency of the MICA-129Met variant. However, other MICA polymorphisms and their interaction need to be studied further (73).
In future studies, the MICA-129Met/Val dimorphism should be analyzed in larger cohorts. Detailed clinical information would help to understand why associations might differ in cohorts. Additional biological information should be obtained in parallel to genetic data. Most important would be data on MICA expression intensities in relevant tissues at relevant time points. Due to the complexity of MICA-129Met/Val effects, this polymorphism is unlikely to become a simple genetic biomarker for prediction of disease risks. However, it still may provide highly important information. We found that Val/Val genotype carriers undergoing HSCT specifically profited from a treatment with antithymocyte globulin to deplete T cells (37). This might be explained by a lack of a high-affinity MICA variant that efficiently downregulates NKG2D on alloreactive donor CD8+ T cells. Moreover, the MICA-129 dimorphism might be relevant when considering therapies aiming at upregulation of MICA on tumor cells to sensitize them for NK cells (74, 75). Increasing the expression of MICA-129Met variants could result in opposite effects than intended.
Author Contributions
RD searched the literature; RD and AI interpreted the functional data; RD and DM interpreted the genetic association data; RD drafted the manuscript; AI, DM, and HB commented the draft; and all the authors approved the final version.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank all the colleagues who contributed to their studies on the MICA-129Met/Val dimorphism.
Funding
The authors’ work was supported by the Deutsche Forschungsgemeinschaft (GRK 1034 and SFB 1002, TP C05) and the European Union Grant FP7-PEOPLE-2012-ITN-315963 (CELLEUROPE). They acknowledge support by the Open Access Publication Funds of the Göttingen University.
Supplementary Material
The Supplementary Material for this article can be found online at http://journal.frontiersin.org/article/10.3389/fimmu.2016.00588/full#supplementary-material.
References
- 1.Bahram S, Bresnahan M, Geraghty DE, Spies T. A second lineage of mammalian major histocompatibility complex class I genes. Proc Natl Acad Sci U S A (1994) 91:6259–63. 10.1073/pnas.91.14.6259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leelayuwat C, Townend DC, Degli-Esposti MA, Abraham LJ, Dawkins RL. A new polymorphic and multicopy MHC gene family related to nonmammalian class I. Immunogenetics (1994) 40:339–51. 10.1007/BF01246675 [DOI] [PubMed] [Google Scholar]
- 3.Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Cell stress-regulated human major histocompatibility complex class I gene expressed in gastrointestinal epithelium. Proc Natl Acad Sci U S A (1996) 93:12445–50. 10.1073/pnas.93.22.12445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature (2005) 436:1186–90. 10.1038/nature03884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Champsaur M, Lanier LL. Effect of NKG2D ligand expression on host immune responses. Immunol Rev (2010) 235:267–85. 10.1111/j.0105-2896.2010.00893.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Raulet DH, Gasser S, Gowen BG, Deng W, Jung H. Regulation of ligands for the NKG2D activating receptor. Annu Rev Immunol (2013) 31:413–41. 10.1146/annurev-immunol-032712-095951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glienke J, Sobanov Y, Brostjan C, Steffens C, Nguyen C, Lehrach H, et al. The genomic organization of NKG2C, E, F, and D receptor genes in the human natural killer gene complex. Immunogenetics (1998) 48:163–73. 10.1007/s002510050420 [DOI] [PubMed] [Google Scholar]
- 8.Zafirova B, Wensveen FM, Gulin M, Polic B. Regulation of immune cell function and differentiation by the NKG2D receptor. Cell Mol Life Sci (2011) 68:3519–29. 10.1007/s00018-011-0797-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanier LL. NKG2D receptor and its ligands in host defense. Cancer Immunol Res (2015) 3:575–82. 10.1158/2326-6066.CIR-15-0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Billadeau DD, Upshaw JL, Schoon RA, Dick CJ, Leibson PJ. NKG2D-DAP10 triggers human NK cell-mediated killing via a Syk-independent regulatory pathway. Nat Immunol (2003) 4:557–64. 10.1038/ni929 [DOI] [PubMed] [Google Scholar]
- 11.Andre P, Castriconi R, Espeli M, Anfossi N, Juarez T, Hue S, et al. Comparative analysis of human NK cell activation induced by NKG2D and natural cytotoxicity receptors. Eur J Immunol (2004) 34:961–71. 10.1002/eji.200324705 [DOI] [PubMed] [Google Scholar]
- 12.Groh V, Rhinehart R, Randolph-Habecker J, Topp MS, Riddell SR, Spies T. Costimulation of CD8alphabeta T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat Immunol (2001) 2:255–60. 10.1038/85321 [DOI] [PubMed] [Google Scholar]
- 13.Guerra N, Tan YX, Joncker NT, Choy A, Gallardo F, Xiong N, et al. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity (2008) 28:571–80. 10.1016/j.immuni.2008.02.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang M, Lanier LL, Sigal LJ. A role for NKG2D in NK cell-mediated resistance to poxvirus disease. PLoS Pathog (2008) 4:e30. 10.1371/journal.ppat.0040030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wesselkamper SC, Eppert BL, Motz GT, Lau GW, Hassett DJ, Borchers MT. NKG2D is critical for NK cell activation in host defense against Pseudomonas aeruginosa respiratory infection. J Immunol (2008) 181:5481–9. 10.4049/jimmunol.181.8.5481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Radosavljevic M, Cuillerier B, Wilson MJ, Clement O, Wicker S, Gilfillan S, et al. A cluster of ten novel MHC class I related genes on human chromosome 6q24.2-q25.3. Genomics (2002) 79:114–23. 10.1006/geno.2001.6673 [DOI] [PubMed] [Google Scholar]
- 17.Choy MK, Phipps ME. MICA polymorphism: biology and importance in immunity and disease. Trends Mol Med (2010) 16:97–106. 10.1016/j.molmed.2010.01.002 [DOI] [PubMed] [Google Scholar]
- 18.Chen D, Gyllensten U. MICA polymorphism: biology and importance in cancer. Carcinogenesis (2014) 35:2633–42. 10.1093/carcin/bgu215 [DOI] [PubMed] [Google Scholar]
- 19.Goto K, Kato N. MICA SNPs and the NKG2D system in virus-induced HCC. J Gastroenterol (2015) 50:261–72. 10.1007/s00535-014-1000-9 [DOI] [PubMed] [Google Scholar]
- 20.Ji M, Wang J, Yuan L, Zhang Y, Zhang J, Dong W, et al. MICA polymorphisms and cancer risk: a meta-analysis. Int J Clin Exp Med (2015) 8:818–26. [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Zhou X. Associations of MICA polymorphisms with inflammatory rheumatic diseases. Open Rheumatol J (2015) 9:94–100. 10.2174/1874312901409010094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steinle A, Li P, Morris DL, Groh V, Lanier LL, Strong RK, et al. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics (2001) 53:279–87. 10.1007/s002510100325 [DOI] [PubMed] [Google Scholar]
- 23.Stastny P. Introduction: MICA/MICB in innate immunity, adaptive immunity, autoimmunity, cancer, and in the immune response to transplants. Hum Immunol (2006) 67:141–4. 10.1016/j.humimm.2006.02.019 [DOI] [PubMed] [Google Scholar]
- 24.Zhao J, Jiang Y, Lei Y, Chen LP, Yi FM, Wang CG, et al. [The relationship between major histocompatibility complex class I chain-related antigens A (MICA)-129 gene polymorphism, soluble MICA level and ulcerative colitis]. Zhonghua Nei Ke Za Zhi (2011) 50:311–5. [PubMed] [Google Scholar]
- 25.Zhao J, Jiang Y, Lei Y, Zou K, Wang C, Huang S, et al. Functional MICA-129 polymorphism and serum levels of soluble MICA are correlated with ulcerative colitis in Chinese patients. J Gastroenterol Hepatol (2011) 26:593–8. 10.1111/j.1440-1746.2010.06524.x [DOI] [PubMed] [Google Scholar]
- 26.Amroun H, Djoudi H, Busson M, Allat R, El Sherbini SM, Sloma I, et al. Early-onset ankylosing spondylitis is associated with a functional MICA polymorphism. Hum Immunol (2005) 66:1057–61. 10.1016/j.humimm.2005.09.004 [DOI] [PubMed] [Google Scholar]
- 27.Kirsten H, Petit-Teixeira E, Scholz M, Hasenclever D, Hantmann H, Heider D, et al. Association of MICA with rheumatoid arthritis independent of known HLA-DRB1 risk alleles in a family-based and a case control study. Arthritis Res Ther (2009) 11:R60. 10.1186/ar2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoshida K, Komai K, Shiozawa K, Mashida A, Horiuchi T, Tanaka Y, et al. Role of the MICA polymorphism in systemic lupus erythematosus. Arthritis Rheum (2011) 63:3058–66. 10.1002/art.30501 [DOI] [PubMed] [Google Scholar]
- 29.Achour Y, Kammoun A, Ben Hamad M, Mahfoudh N, Chaabane S, Marzouk S, et al. Association study of MICA gene polymorphisms with rheumatoid arthritis susceptibility in south Tunisian population. Int J Immunogenet (2014) 41:486–92. 10.1111/iji.12146 [DOI] [PubMed] [Google Scholar]
- 30.Lopez-Hernandez R, Valdes M, Lucas D, Campillo JA, Martinez-Garcia P, Salama H, et al. Association analysis of MICA gene polymorphism and MICA-129 dimorphism with inflammatory bowel disease susceptibility in a Spanish population. Hum Immunol (2010) 71:512–4. 10.1016/j.humimm.2010.02.003 [DOI] [PubMed] [Google Scholar]
- 31.Raache R, Belanteur K, Amroun H, Benyahia A, Heniche A, Azzouz M, et al. Association of major histocompatibility complex class 1 chain-related gene a dimorphism with type 1 diabetes and latent autoimmune diabetes in adults in the Algerian population. Clin Vaccine Immunol (2012) 19:557–61. 10.1128/CVI.05473-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pollock RA, Chandran V, Pellett FJ, Thavaneswaran A, Eder L, Barrett J, et al. The functional MICA-129 polymorphism is associated with skin but not joint manifestations of psoriatic disease independently of HLA-B and HLA-C. Tissue Antigens (2013) 82:43–7. 10.1111/tan.12126 [DOI] [PubMed] [Google Scholar]
- 33.Douik H, Ben Chaaben A, Attia Romdhane N, Romdhane HB, Mamoghli T, Fortier C, et al. Association of MICA-129 polymorphism with nasopharyngeal cancer risk in a Tunisian population. Hum Immunol (2009) 70:45–8. 10.1016/j.humimm.2008.10.008 [DOI] [PubMed] [Google Scholar]
- 34.Tong HV, Toan NL, Song LH, Bock CT, Kremsner PG, Velavan TP. Hepatitis B virus-induced hepatocellular carcinoma: functional roles of MICA variants. J Viral Hepat (2013) 20:687–98. 10.1111/jvh.12089 [DOI] [PubMed] [Google Scholar]
- 35.Campillo JA, Lopez-Hernandez R, Martinez-Banaclocha H, Bolarin JM, Gimeno L, Mrowiec A, et al. MHC class I chain-related gene a diversity in patients with cutaneous malignant melanoma from southeastern Spain. Dis Markers (2015) 2015:831864. 10.1155/2015/831864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boukouaci W, Busson M, Peffault De Latour R, Rocha V, Suberbielle C, Bengoufa D, et al. MICA-129 genotype, soluble MICA, and anti-MICA antibodies as biomarkers of chronic graft-versus-host disease. Blood (2009) 114:5216–24. 10.1182/blood-2009-04-217430 [DOI] [PubMed] [Google Scholar]
- 37.Isernhagen A, Malzahn D, Viktorova E, Elsner L, Monecke S, von Bonin F, et al. The MICA-129 dimorphism affects NKG2D signaling and outcome of hematopoietic stem cell transplantation. EMBO Mol Med (2015) 7:1480–502. 10.15252/emmm.201505246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ayo CM, Oliveira AP, Camargo AV, Mattos CC, Bestetti RB, Mattos LC. Association of the functional MICA-129 polymorphism with the severity of chronic Chagas heart disease. Clin Infect Dis (2015) 61:1310–3. 10.1093/cid/civ540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ayo CM, Camargo AV, Frederico FB, Siqueira RC, Previato M, Murata FH, et al. MHC class I chain-related gene a polymorphisms and linkage disequilibrium with HLA-B and HLA-C alleles in ocular toxoplasmosis. PLoS One (2015) 10:e0144534. 10.1371/journal.pone.0144534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hizem S, Mtiraoui N, Massaoudi S, Fortier C, Boukouaci W, Kahina A, et al. Polymorphisms in genes coding for the NK-cell receptor NKG2D and its ligand MICA in recurrent miscarriage. Am J Reprod Immunol (2014) 72:577–85. 10.1111/aji.12314 [DOI] [PubMed] [Google Scholar]
- 41.Groh V, Bruhl A, El-Gabalawy H, Nelson JL, Spies T. Stimulation of T cell autoreactivity by anomalous expression of NKG2D and its MIC ligands in rheumatoid arthritis. Proc Natl Acad Sci U S A (2003) 100:9452–7. 10.1073/pnas.1632807100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dai Z, Turtle CJ, Booth GC, Riddell SR, Gooley TA, Stevens AM, et al. Normally occurring NKG2D+CD4+ T cells are immunosuppressive and inversely correlated with disease activity in juvenile-onset lupus. J Exp Med (2009) 206:793–805. 10.1084/jem.20081648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou X, Wang J, Zou H, Ward MM, Weisman MH, Espitia MG, et al. MICA, a gene contributing strong susceptibility to ankylosing spondylitis. Ann Rheum Dis (2014) 73:1552–7. 10.1136/annrheumdis-2013-203352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Low JS, Chin YM, Mushiroda T, Kubo M, Govindasamy GK, Pua KC, et al. A genome wide study of copy number variation associated with nasopharyngeal carcinoma in Malaysian Chinese identifies CNVs at 11q14.3 and 6p21.3 as candidate loci. PLoS One (2016) 11:e0145774. 10.1371/journal.pone.0145774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar V, Kato N, Urabe Y, Takahashi A, Muroyama R, Hosono N, et al. Genome-wide association study identifies a susceptibility locus for HCV-induced hepatocellular carcinoma. Nat Genet (2011) 43:455–8. 10.1038/ng.809 [DOI] [PubMed] [Google Scholar]
- 46.Chen K, Shi W, Xin Z, Wang H, Zhu X, Wu X, et al. Replication of genome wide association studies on hepatocellular carcinoma susceptibility loci in a Chinese population. PLoS One (2013) 8:e77315. 10.1371/journal.pone.0077315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen D, Juko-Pecirep I, Hammer J, Ivansson E, Enroth S, Gustavsson I, et al. Genome-wide association study of susceptibility loci for cervical cancer. J Natl Cancer Inst (2013) 105:624–33. 10.1093/jnci/djt051 [DOI] [PubMed] [Google Scholar]
- 48.Chen D, Cui T, Ek WE, Liu H, Wang H, Gyllensten U. Analysis of the genetic architecture of susceptibility to cervical cancer indicates that common SNPs explain a large proportion of the heritability. Carcinogenesis (2015) 36:992–8. 10.1093/carcin/bgv083 [DOI] [PubMed] [Google Scholar]
- 49.Nieuwenhuis MA, Siedlinski M, Van Den Berge M, Granell R, Li X, Niens M, et al. Combining genomewide association study and lung eQTL analysis provides evidence for novel genes associated with asthma. Allergy (2016) 71(12):1712–20. 10.1111/all.12990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Clerc S, Delaneau O, Coulonges C, Spadoni JL, Labib T, Laville V, et al. Evidence after imputation for a role of MICA variants in nonprogression and elite control of HIV type 1 infection. J Infect Dis (2014) 210:1946–50. 10.1093/infdis/jiu342 [DOI] [PubMed] [Google Scholar]
- 51.Isernhagen A, Schilling D, Monecke S, Shah P, Elsner L, Walter L, et al. The MICA-129Met/Val dimorphism affects plasma membrane expression and shedding of the NKG2D ligand MICA. Immunogenetics (2016) 68:109–23. 10.1007/s00251-015-0884-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shafi S, Vantourout P, Wallace G, Antoun A, Vaughan R, Stanford M, et al. An NKG2D-mediated human lymphoid stress surveillance response with high interindividual variation. Sci Transl Med (2011) 3:113ra124. 10.1126/scitranslmed.3002922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi C, Li H, Couturier JP, Yang K, Guo X, He D, et al. Allele specific expression of MICA variants in human fibroblasts suggests a pathogenic mechanism. Open Rheumatol J (2015) 9:60–4. 10.2174/1874312901409010060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lo PH, Urabe Y, Kumar V, Tanikawa C, Koike K, Kato N, et al. Identification of a functional variant in the MICA promoter which regulates MICA expression and increases HCV-related hepatocellular carcinoma risk. PLoS One (2013) 8:e61279. 10.1371/journal.pone.0061279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashiru O, Lopez-Cobo S, Fernandez-Messina L, Pontes-Quero S, Pandolfi R, Reyburn HT, et al. A GPI anchor explains the unique biological features of the common NKG2D-ligand allele MICA*008. Biochem J (2013) 454:295–302. 10.1042/BJ20130194 [DOI] [PubMed] [Google Scholar]
- 56.Groh V, Wu J, Yee C, Spies T. Tumour-derived soluble MIC ligands impair expression of NKG2D and T-cell activation. Nature (2002) 419:734–8. 10.1038/nature01112 [DOI] [PubMed] [Google Scholar]
- 57.Salih HR, Rammensee HG, Steinle A. Cutting edge: down-regulation of MICA on human tumors by proteolytic shedding. J Immunol (2002) 169:4098–102. 10.4049/jimmunol.169.8.4098 [DOI] [PubMed] [Google Scholar]
- 58.El-Gazzar A, Groh V, Spies T. Immunobiology and conflicting roles of the human NKG2D lymphocyte receptor and its ligands in cancer. J Immunol (2013) 191:1509–15. 10.4049/jimmunol.1301071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kumar V, Yi Lo PH, Sawai H, Kato N, Takahashi A, Deng Z, et al. Soluble MICA and a MICA variation as possible prognostic biomarkers for HBV-induced hepatocellular carcinoma. PLoS One (2012) 7:e44743. 10.1371/journal.pone.0044743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tamaki S, Kawakami M, Yamanaka Y, Shimomura H, Imai Y, Ishida J, et al. Relationship between soluble MICA and the MICA A5.1 homozygous genotype in patients with oral squamous cell carcinoma. Clin Immunol (2009) 130:331–7. 10.1016/j.clim.2008.09.004 [DOI] [PubMed] [Google Scholar]
- 61.Jiang X, Zou Y, Huo Z, Yu P. Association of major histocompatibility complex class I chain-related gene A microsatellite polymorphism and hepatocellular carcinoma in South China Han population. Tissue Antigens (2011) 78:143–7. 10.1111/j.1399-0039.2011.01693.x [DOI] [PubMed] [Google Scholar]
- 62.Hayashi T, Imai K, Morishita Y, Hayashi I, Kusunoki Y, Nakachi K. Identification of the NKG2D haplotypes associated with natural cytotoxic activity of peripheral blood lymphocytes and cancer immunosurveillance. Cancer Res (2006) 66:563–70. 10.1158/0008-5472.CAN-05-2776 [DOI] [PubMed] [Google Scholar]
- 63.Isernhagen A, Malzahn D, Monecke S, Schilling D, Shah P, Multhoff G, et al. Functional consequences of genetic polymorphisms in the NKG2D receptor signaling pathway and putative gene interactions. Receptors Clin Investig (2016) 3:e1269. 10.14800/rci.1269 [DOI] [Google Scholar]
- 64.Antoun A, Vekaria D, Salama RA, Pratt G, Jobson S, Cook M, et al. The genotype of RAET1L (ULBP6), a ligand for human NKG2D (KLRK1), markedly influences the clinical outcome of allogeneic stem cell transplantation. Br J Haematol (2012) 159:589–98. 10.1111/bjh.12072 [DOI] [PubMed] [Google Scholar]
- 65.Espinoza JL, Takami A, Onizuka M, Sao H, Akiyama H, Miyamura K, et al. NKG2D gene polymorphism has a significant impact on transplant outcomes after HLA-fully-matched unrelated bone marrow transplantation for standard risk hematologic malignancies. Haematologica (2009) 94:1427–34. 10.3324/haematol.2009.008318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kitcharoen K, Witt CS, Romphruk AV, Christiansen FT, Leelayuwat C. MICA, MICB, and MHC beta block matching in bone marrow transplantation: relevance to transplantation outcome. Hum Immunol (2006) 67:238–46. 10.1016/j.humimm.2006.02.012 [DOI] [PubMed] [Google Scholar]
- 67.Parmar S, Del Lima M, Zou Y, Patah PA, Liu P, Cano P, et al. Donor-recipient mismatches in MHC class I chain-related gene A in unrelated donor transplantation lead to increased incidence of acute graft-versus-host disease. Blood (2009) 114:2884–7. 10.1182/blood-2009-05-223172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Askar M, Sun Y, Rybicki L, Zhang A, Thomas D, Kalaycio M, et al. Synergistic effect of major histocompatibility complex class I-related chain a and human leukocyte antigen-DPB1 mismatches in association with acute graft-versus-host disease after unrelated donor hematopoietic stem cell transplantation. Biol Blood Marrow Transplant (2014) 20:1835–40. 10.1016/j.bbmt.2014.07.019 [DOI] [PubMed] [Google Scholar]
- 69.Carapito R, Jung N, Kwemou M, Untrau M, Michel S, Pichot A, et al. Matching for the nonconventional MHC-I MICA gene significantly reduces the incidence of acute and chronic GVHD. Blood (2016) 128:1979–86. 10.1182/blood-2016-05-719070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fuerst D, Neuchel C, Niederwieser D, Bunjes D, Gramatzki M, Wagner E, et al. Matching for the MICA-129 polymorphism is beneficial in unrelated hematopoietic stem cell transplantation. Blood (2016). 10.1182/blood-2016-05-716357 [DOI] [PubMed] [Google Scholar]
- 71.Okada Y, Han B, Tsoi LC, Stuart PE, Ellinghaus E, Tejasvi T, et al. Fine mapping major histocompatibility complex associations in psoriasis and its clinical subtypes. Am J Hum Genet (2014) 95:162–72. 10.1016/j.ajhg.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cunningham F, Amode MR, Barrell D, Beal K, Billis K, Brent S, et al. Ensembl 2015. Nucleic Acids Res (2015) 43:D662–9. 10.1093/nar/gku1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carapito R, Bahram S. Genetics, genomics, and evolutionary biology of NKG2D ligands. Immunol Rev (2015) 267:88–116. 10.1111/imr.12328 [DOI] [PubMed] [Google Scholar]
- 74.Kato N, Tanaka J, Sugita J, Toubai T, Miura Y, Ibata M, et al. Regulation of the expression of MHC class I-related chain A, B (MICA, MICB) via chromatin remodeling and its impact on the susceptibility of leukemic cells to the cytotoxicity of NKG2D-expressing cells. Leukemia (2007) 21:2103–8. 10.1038/sj.leu.2404862 [DOI] [PubMed] [Google Scholar]
- 75.Diermayr S, Himmelreich H, Durovic B, Mathys-Schneeberger A, Siegler U, Langenkamp U, et al. NKG2D ligand expression in AML increases in response to HDAC inhibitor valproic acid and contributes to allorecognition by NK-cell lines with single KIR-HLA class I specificities. Blood (2008) 111:1428–36. 10.1182/blood-2007-07-101311 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.