Summary
Actin plays a critical role during the early stages of pathogenic microbe internalization by immune cells. In this study, we identified a key mechanism of actin filament tethering and stabilization to the surface of phagosomes in human dendritic cells. We found that the actin-binding protein SWAP70 is specifically recruited to nascent phagosomes by binding to the lipid phosphatidylinositol (3,4)-bisphosphate. Multi-color super-resolution stimulated emission depletion (STED) microscopy revealed that the actin cage surrounding early phagosomes is formed by multiple concentric rings containing SWAP70. SWAP70 colocalized with and stimulated activation of RAC1, a known activator of actin polymerization, on phagosomes. Genetic ablation of SWAP70 impaired actin polymerization around phagosomes and resulted in a phagocytic defect. These data show a key role for SWAP70 as a scaffold for tethering the peripheral actin cage to phagosomes.
Keywords: phagocytosis; dendritic cell; actin; cytoskeleton; phosphoinositides; phosphatidylionositol 3,4-bisphosphate; SWAP70; rho-GTPases; Rac1; STED microscopy
Graphical Abstract
Highlights
-
•
SWAP70 is transiently recruited to the nascent cup of emerging phagosomes
-
•
Phagosomal recruitment of SWAP70 is driven by RAC1 and phosphoinositides
-
•
SWAP70 tethers concentric rings of F-actin on the phagosomal membrane
-
•
Absence of SWAP70 results in phagocytic defects
Baranov et al. find a key role for SWAP70 in tethering concentric rings of F-actin to emerging phagosomes. Specific phagosomal recruitment of SWAP70 is achieved by interactions with the GTPase RAC1 and phosphatidylinositol (3,4)-bisphosphate. Absence of SWAP70 leads to disruption of phagosomal actin and impaired phagocytosis.
Introduction
Phagocytosis is an evolutionarily conserved mechanism by which immune cells take up foreign particles for degradation, such as microbial pathogens and tumor cells. Phagocytosis, thereby, plays an essential role in the defense against disease. Actin executes a pivotal function in phagocytosis with its precise role depending on the stimuli and receptors involved (Freeman and Grinstein, 2014, May et al., 2000, Rohatgi et al., 2001, Swanson, 2008). F-actin-rich protrusions facilitate the initial capture of phagocytic targets. Actin is polymerized around the nascent phagosome forming a cage that helps wrapping of the membrane for particle engulfment and membrane fission. The actin cytoskeleton also exerts physical forces for pulling the pathogen inside the cell from the outside environment (May et al., 2000). Although the precise course of actin cytoskeleton assembly varies for different receptors (Allen and Aderem, 1996, Caron and Hall, 1998, Kuiper et al., 2008), actin polymerization is mediated by small GTPases of the RHO family: CDC42, RHOA, and RAC1 (Caron and Hall, 1998, May et al., 2000, Swanson, 2008). Within minutes after internalization, actin is depolymerized, a step that is required for the unhindered fusion of the maturing phagosome with endomembranes (Ferrari et al., 1999, Liebl and Griffiths, 2009). The maturation process that succeeds phagosome formation can be accompanied by successive waves of actin polymerization and depolymerization, which can facilitate or impede additional fusion events with endolysosomal compartments (Anes et al., 2003, Greenberg et al., 1991, Kjeken et al., 2004, Yam and Theriot, 2004). Despite phagocytosis being a well-studied phenomenon, the molecular mechanisms for F-actin cage tethering to the phagocytic surface are not clear. The structure of the phagosomal actin cage has also not been resolved, as F-actin is densely packed around the phagosome, and there is a lack of high resolution data.
In this study, we report that the actin cage in dendritic cells is connected to phagosomes by the 69 kDa scaffolding protein SWAP70. SWAP70 is expressed in many cell types, including mast cells (Gross et al., 2002), fibroblasts (Shinohara et al., 2002), dendritic cells (Oberbanscheidt et al., 2007), monocytes, and macrophages (Hilpelä et al., 2003). In these cells, the absence of SWAP70 leads to defects in actin polymerization and impairs cell migration, adhesion, polarization, and morphology (Bahaie et al., 2012, Chopin et al., 2010, Garbe et al., 2012, Murugan et al., 2008, Ocaña-Morgner et al., 2011, Pearce et al., 2006, Ripich and Jessberger, 2011, Shinohara et al., 2002, Sivalenka and Jessberger, 2004). SWAP70 can directly bind, bundle, and stabilize actin filaments by means of its C-terminal actin binding domain (Chacón-Martínez et al., 2013, Gomez-Cambronero, 2012, Hilpelä et al., 2003, Ihara et al., 2006, Murugan et al., 2008, Pearce et al., 2011, Shinohara et al., 2002). SWAP70 regulates cellular actin dynamics and organization via activation of RHOA and RAC1 (Dwyer et al., 2015, Oberbanscheidt et al., 2007, Ocana-Morgner et al., 2009, Ocaña-Morgner et al., 2011, Shinohara et al., 2002, Sivalenka and Jessberger, 2004, Sivalenka et al., 2008). Binding of SWAP70 to phosphoinositides is required for this activation of RHO-GTPases (Murugan et al., 2008, Shinohara et al., 2002). We found that SWAP70 was transiently recruited to nascent phagosomes in human monocyte-derived dendritic cells. SWAP70 remained associated to these phagosomes shortly after phagocytic cup closure by specific binding of its Pleckstrin homology (PH-) domain to phosphatidylinositol (3,4)-bisphosphate (PI(3,4)P2) and to a lesser extent to phosphatidylinositol (3,4,5)-trisphosphate (PI(3,4,5)P3). Knockdown of SWAP70 by small interfering RNA (siRNA) significantly decreased RAC1 activation, actin polymerization, and phagocytic capability. Multi-color super-resolution STED microscopy showed that SWAP70 overlapped with RAC1 and aligned with parallel F-actin filaments and concentric rings surrounding the phagosome. These F-actin structures were not observable upon siRNA knockdown of SWAP70. Our findings show that SWAP70 is a scaffolding protein that promotes particle internalization by enabling the formation of the phagosomal actin cage.
Results
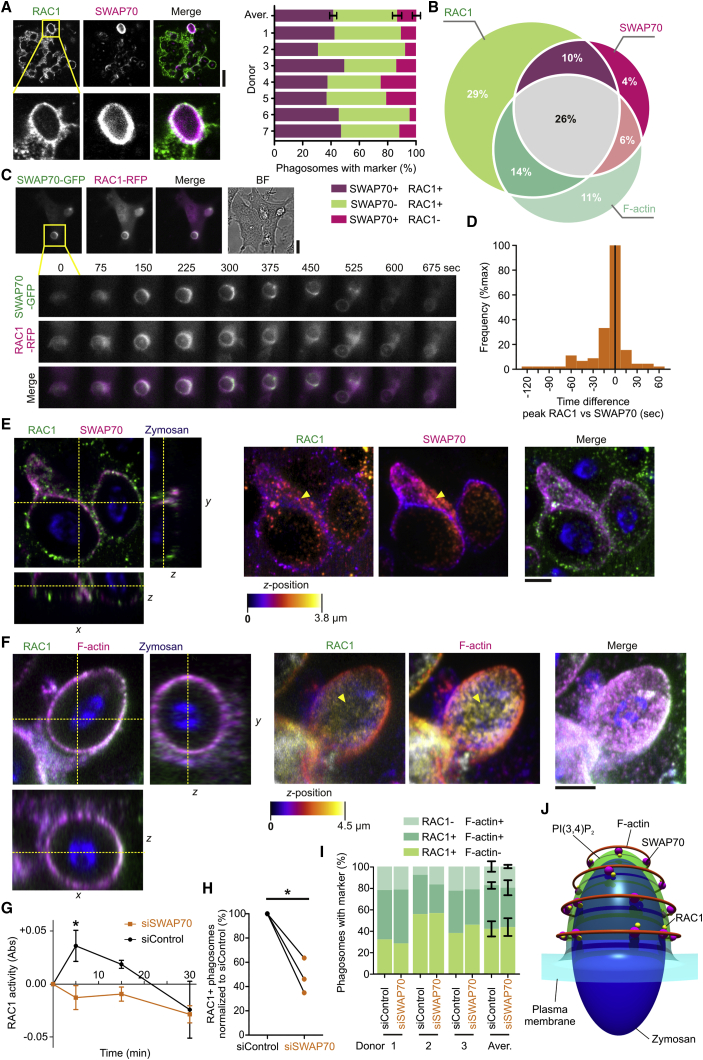
SWAP70 Aligns with F-Actin Filaments on the Surface of Early Phagosomes
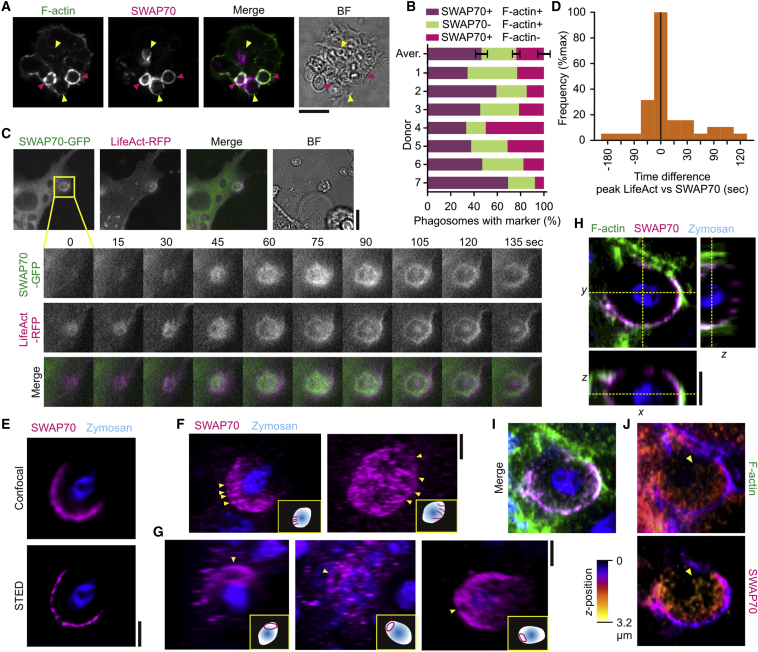
Because SWAP70 can bind directly to F-actin via its C-terminal region (Chacón-Martínez et al., 2013, Hilpelä et al., 2003, Ihara et al., 2006) and can regulate cellular actin dynamics and organization via activation of RHOA and RAC1 (Dwyer et al., 2015, Oberbanscheidt et al., 2007, Ocana-Morgner et al., 2009, Ocaña-Morgner et al., 2011, Shinohara et al., 2002, Sivalenka and Jessberger, 2004, Sivalenka et al., 2008), we hypothesized that SWAP70 would have an important role in the formation of the phagocytic actin cage. To prove this hypothesis, we first investigated whether SWAP70 is recruited to phagosomes. Dendritic cells derived from monocytes isolated from blood of healthy volunteers were pulsed with zymosan particles, a cell wall preparation of the yeast Saccharomyces cerevisiae. Zymosan signals through a number of pattern recognition receptors, including Toll-like receptor 2 and the C-type lectin CLEC7A (Dectin-1). Zymosan is taken up by CLEC7A, which is the main phagocytic receptor for live fungi (Goodridge et al., 2009). Cells were immunolabeled for SWAP70 and F-actin was stained with fluorescently labeled phalloidin. A substantial portion of SWAP70 localized to F-actin-rich phagosomes that often (35%–70%, depending on the donor) were positive for SWAP70 (Figures 1A and 1B). Live cell experiments with dendritic cells overexpressing SWAP70 fused to GFP revealed transient recruitment of SWAP70 to phagosomes simultaneously with the F-actin binding probe LifeAct-RFP (Riedl et al., 2008) (Figures 1C and 1D; Movie S1). We performed three-dimensional super-resolution stimulated emission depletion (STED) microscopy to visualize the ultrastructure of SWAP70 on the surface of the phagosomes. The cores of the zymosan particles were visualized by their auto-fluorescence in the GFP channel. Filamentous structures of SWAP70 were distinguishable by STED but not by conventional confocal microscopy (Figure 1E). Interestingly, we frequently observed that SWAP70 was arranged in parallel arched structures that aligned with the membrane of the phagosome (Figure 1F; Movies S2, S3, and S4). We also often observed concentric ring-like structures of SWAP70 (Figure 1G; Movies S5 and S6). This observation raised the possibility that SWAP70 would align with the F-actin filaments previously described to form around the phagocytic cup following uptake of non-opsonized zymosan (Goodridge et al., 2012, Huang et al., 2014, Liebl and Griffiths, 2009). We performed multi-color STED microscopy to address this possibility and observed perfect alignment of the F-actin filaments with SWAP70 (Figures 1H–1J; Movie S7). These results suggest a role for SWAP70 in the organization and/or formation of peripheral actin filaments on the phagosome. We next investigated when and under what conditions SWAP70 was recruited to phagosomes.
Figure 1.
SWAP70 Aligns with Actin Filaments on the Phagosomal Surface
(A) Confocal micrograph of zymosan pulsed dendritic cells immunostained for SWAP70 (magenta in merge) and F-actin (green). Pink arrowheads, double-positive phagosomes; yellow arrowheads, phagosomes only positive for SWAP70; BF, bright field.
(B) Quantification of (A). Aver., average ± SEM (∼100 cells/donor).
(C) Live cell imaging of zymosan-pulsed dendritic cells expressing SWAP70-GFP (green in merge) and LifeAct-RFP (magenta). The inset shows a time series during zymosan uptake. See also Movie S1.
(D) Quantification from (C). The histogram shows the time difference of peak recruitment of SWAP70-GFP and LifeAct-RFP based on fluorescence intensities (40 phagosomes). Negative values indicate that LifeAct was recruited prior to SWAP70 and positive values later than SWAP70.
(E) Confocal (top) and super-resolution 3D-STED (bottom) microscopy of dendritic cells pulsed with zymosan (blue) and immunostained for SWAP70 (magenta).
(F and G) 3D-reconstructions of the STED from (E). Often, parallel arches (F) (yellow arrowheads and depicted in the insets) and rings (G) of SWAP70 were visible. This is more clear in the 3D-rotations (Movies S2, S3, S4, S5, and S6).
(H–J) Same as (F) and (G) but now with co-staining for F-actin (green). Shown are cross-section and orthogonal sections (H; indicated by dashed yellow lines), the maximum intensity surface projection (I), and maximum intensity height maps (J). Note the overlap of SWAP70 and F-actin on the surface of the phagosome (yellow arrowhead). Scale bars, 10 μm (A and C); 2 μm (E–H).
See also Movie S7.
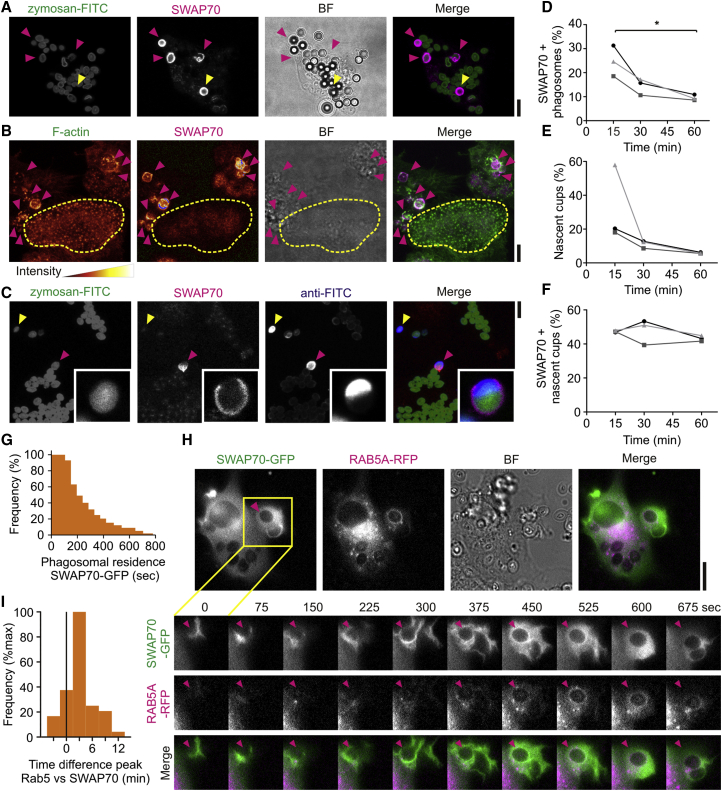
The recruitment of SWAP70 to phagosomes was not dependent on the type of phagocytic receptor, as it was broadly observed for different phagocytic cargoes, such as IgG-opsonized and naked latex beads (taken up by integrin αMβ2) (Freeman and Grinstein, 2014) (Figures 2A and S1A). Remarkably, the recruitment of SWAP70 to phagosomes was much stronger than to other actin-rich structures, such as podosomes (>5-fold based on fluorescence intensity; Figure 2B). SWAP70 has been shown to locate to podosomes in mouse dendritic cells, but its role there is not clear, as it is not required for podosome formation or turnover (Götz and Jessberger, 2013). Although SWAP70 has been detected in nuclear fractions of B cells (Borggrefe et al., 1999) and mouse dendritic cells (Ocaña-Morgner et al., 2013), we did not detect endogenous or overexpressed SWAP70 in the nucleus of human monocyte-derived dendritic cells. In order to determine when SWAP70 was recruited to phagosomes, dendritic cells were pulsed with fluorescein isothiocyanate (FITC)-labeled zymosan followed by immunolabeling with an antibody directed against FITC (i.e., without permeabilization). Zymosan particles that were completely taken up by the dendritic cells are inaccessible to antibody, allowing for selective labeling of free zymosan and nascent phagocytic cups. SWAP70 was recruited to sealed phagosomes (i.e., no anti-FITC labeling) and to nascent cups (Figure 2C; Movie S8). SWAP70 was only transiently recruited to phagosomes, as the total fraction of all SWAP70-positive phagosomes readily decreased in time (Figure 2D). As expected, the fraction of nascent cups compared to total phagosomes also readily decreased in time as more and more zymosan particles were fully internalized (Figure 2E). Approximately 50% of all nascent cups were positive for SWAP70 at all time points tested (Figure 2F). SWAP70 resided on phagosomes for ∼1–3 min after zymosan uptake based on the time-lapse imaging of dendritic cells expressing SWAP70-GFP, although we sometimes observed considerable longer residence times (up to 10–15 min; Figure 2G). We performed co-immunofluorescence labeling experiments to determine whether SWAP70 was still present on early phagosomes defined by the presence of the early endosomal protein EEA1 (Figures S1B and S1C). We could not find a single phagosome positive for both EEA1 and SWAP70, indicating that SWAP70 dissociated from the phagosomes prior to EEA1 recruitment. We also observed complete exclusion of SWAP70 with the late endosomal/lysosomal protein LAMP1 (Figures S1D and S1E). In agreement with these findings, live cell imaging revealed that SWAP70-GFP was recruited ∼3 min prior to mRFP-tagged RAB5A (Vonderheit and Helenius, 2005), an early endosomal small GTPase that binds EEA1 (Figures 2H and 2I; Movie S9). These results mirror the transient recruitment of SWAP70 to macropinosomes in mouse dendritic cells prior to RAB5A (Oberbanscheidt et al., 2007). Thus, in most cases, SWAP70 is already recruited to the nascent cup during phagosome formation and disappears within minutes after cup closure prior to the recruitment of EEA1.
Figure 2.
SWAP70 Is Recruited to Nascent Phagosomes
(A) Human dendritic cells were pulsed with a combination of FITC-labeled zymosan (zymosan-FITC; green in merge) and latex beads followed by immunostaining for SWAP70 (magenta). SWAP70 was recruited to both zymosan (pink arrowheads) and latex beads (yellow arrowhead). BF, bright field. See also Figure S1A.
(B) Maximum intensity z projection of zymosan-pulsed dendritic cells immunostained for SWAP70 (magenta) and F-actin (green). F-actin and SWAP70 are shown in the fire look-up-table to emphasize the low recruitment of SWAP70 to podosomes (podosome region encircled in yellow) compared to phagosomes (pink arrowheads).
(C) Identification of nascent cups by labeling of zymosan-FITC (green) with an antibody (blue; anti-FITC) in absence of permeabilization. Only free zymosan particles (yellow arrowhead) and nascent cups (pink) were accessible to anti-FITC labeling. Inset: magnification of a nascent cup. See also Movie S8.
(D–F) Phagosomes in dendritic cells from three different donors over time of uptake (∼70 cells/donor/time point) were analyzed for the fraction of SWAP70-positive phagosomes (D), the fraction of nascent cups to total phagosomes (E), and the fraction of SWAP70-positive nascent cups to all SWAP70-positive phagosomes (F).
(G) Residence time of SWAP70-GFP on phagosomes quantified based on live cell imaging (150 phagosomes).
(H) Live cell imaging of dendritic cells expressing SWAP70-GFP (green in merge) and RAB5A-RFP (magenta). The inset shows a time series during zymosan uptake (arrowhead). See also Figures S1B–S1E and Movie S9.
(I) Quantification from (H). The histogram shows the time difference of peak recruitment of SWAP70-GFP and RAB5A-RFP based on fluorescence intensities (50 phagosomes). Negative values indicate that RAB5A was recruited prior to SWAP70 and positive values later than SWAP70. Scale bars, 10 μm.
Phagosomal Recruitment of SWAP70 Is Promoted by PI(3,4)P2
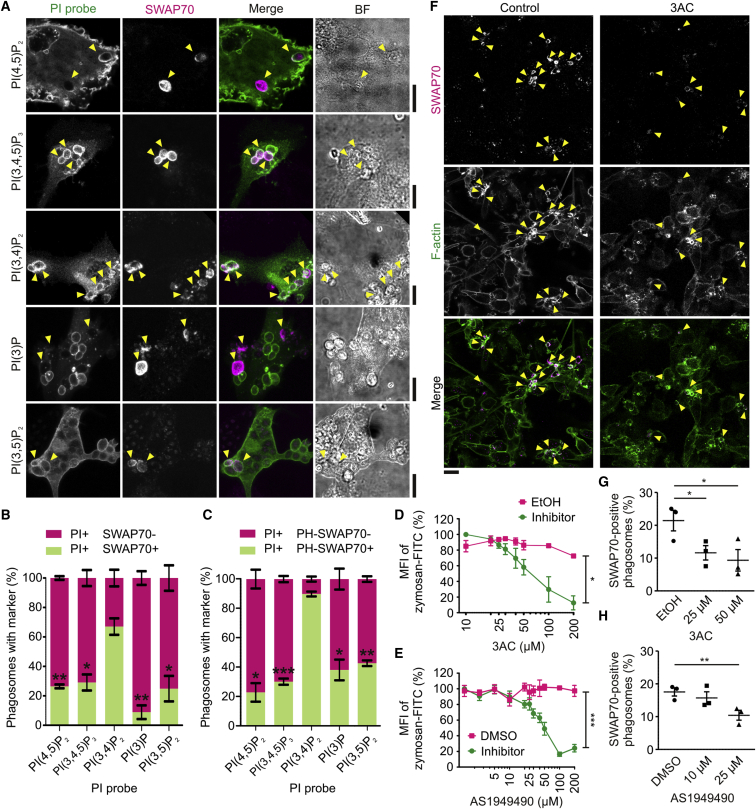
The phosphoinositide composition of phagosomes changes progressively during phagosome formation and maturation. Phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2), which is the predominant phosphoinositide at the plasma membrane (van den Bogaart and ter Beest, 2014), converts into PI(3,4,5)P3 by the action of PI3-kinases (Greenberg and Grinstein, 2002, Gu et al., 2003, Hoppe and Swanson, 2004, Terebiznik et al., 2002). PI(3,4,5)P3 is then dephosphorylated by SHIP1 (INPP5D) and SHIP2 (INPPL1), yielding PI(3,4)P2 (Brooks et al., 2010, Drobek et al., 2015, Fuhler et al., 2012), which in turn is converted into PI(3)P by PI 4-phosphatases. Both SHIP1 and 2 are expressed by human monocyte-derived dendritic cells (Figure S2A). As EEA1 is known to be recruited to early phagosomes by binding to phosphatidylinositol 3-phosphate (PI(3)P) (Levin et al., 2015, Simonsen et al., 1998), we speculated that SWAP70 is recruited to the phagosome through its known binding partners PI(3,4)P2 and PI(3,4,5)P3 (Hilpelä et al., 2003, Shinohara et al., 2002). To assess this, we overexpressed in dendritic cells GFP-tagged probes with specificity for several phosphoinositide species: the PLCδ1 (PLCD1) PH-domain specific for PI(4,5)P2 (Lemmon et al., 1997, Stauffer et al., 1998), the PH-domain of AKT specific for PI(3,4,5)P3 and less for PI(3,4)P2 (Klippel et al., 1997, Rosen et al., 2012), the PH-domain of TAPP2 (PLEKHA2) specific for PI(3,4)P2 (Marshall et al., 2002), the PX-domain of p40phox (NCF4) specific for PI(3)P (Kanai et al., 2001), and the N-terminal sequence of MCOLN1 specific for PI(3,5)P2 (Li et al., 2013). We then tested the co-localization of endogenous SWAP70 (i.e., by immunostaining) with these phosphoinositide-probes (Figures 3A and 3B). Of all probes, co-localization of SWAP70 on phagosomes was strongest with the probe sensing PI(3,4)P2 (∼70% of all PI(3,4)P2 positive phagosomes). Live cell imaging of dendritic cells co-expressing the GFP-tagged probes together with SWAP70 fused to mCherry showed simultaneous recruitment of SWAP70 and the probe for PI(3,4)P2 to phagosomes (Figures S2B and S2C; Movie S10). The probes for PI(4,5)P2 and PI(3,4,5)P3 were mostly present on phagosomes before SWAP70-mCherry was recruited and the probe for PI(3)P was recruited later than SWAP70-mCherry (Figures S2D–S2I; Movies S11, S12, and S13). Experiments with co-expression of the mCherry-tagged PH-domain of SWAP70 (amino acid residues 210–306) with our GFP-tagged phosphoinositide probes demonstrated that PI(3,4)P2 binding was mediated by the PH-domain of SWAP70 (Figures 3C, S3A, and S3B). Phagosome binding was not observed for the PH-domain carrying mutations R223E and R224E (Figure S3C), which is unable to bind to phosphoinositides in vitro (Hilpelä et al., 2003).
Figure 3.
Phagosomal SWAP70 Is Mainly Recruited by PI(3,4)P2
(A) Confocal images of zymosan-pulsed dendritic cells expressing GFP-tagged phosphoinositide-probes (PI probe; green in merge) and immunolabeled for SWAP70 (magenta). The following PI probes were used: the PH-domain of PLCδ1 for PI(4,5)P2, the PH-domain of AKT for PI(3,4,5)P3, the PH-domain of TAPP2 for PI(3,4)P2, the PX-domain of NCF4 for PI(3)P, and the N-terminal sequence of MCOLN1 for PI(3,5)P2. Yellow arrowheads, phagosomes positive for SWAP70; BF, bright field. See also Figure S2 and Movies S10, S11, S12, and S13.
(B) The percentages of phagosomes positive for the phosphoinositide biomarkers and endogenous SWAP70 (mean ± SEM for three donors; >60 phagosomes/condition).
(C) Same as (B) but now with the mCherry-tagged PH-domain of SWAP70 (PH-SWAP70; mean ± SEM for three donors). See also Figures S3A–S3C.
(D) Phagocytosis capacity with the SHIP1 inhibitor 3AC by flow cytometry. MFI, mean fluorescence intensity of the zymosan-FITC signal (mean ± SEM for three donors). EtOH, ethanol solvent control. See also Figure S3D.
(E) Same as (D) but now for the SHIP2 inhibitor AS1949490. DMSO, solvent control. See also Figure S3E.
(F) Confocal micrographs of dendritic cells pulsed with zymosan in the presence or absence of 25 μM 3AC. Yellow arrowheads, phagosomes positive for SWAP70.
(G and H) Quantification of SWAP70-positive phagosomes after 30 min internalization for the 3AC (G) and AS1949490 (H) concentrations indicated (mean ± SEM for three donors; >80 phagosomes/donor/condition). Scale bars, 10 μm (A), 20 μm (F).
See also Figures S3F–S3K and S4.
To further validate the role of PI(3,4)P2 in SWAP70 recruitment, we cultured dendritic cells in the presence of the specific SHIP1 inhibitor 3-a-aminocholestane (3AC) or the specific SHIP2 inhibitor AS1949490 (Brooks et al., 2010). We selected concentrations of 25 and 50 μM for 3AC and 10 and 25 μM for AS1949490; just below and above the concentrations where cellular viability and phagocytosis capacity were affected (Figures 3D, 3E, S3D, and S3E). These concentrations of 3AC and AS1949490 resulted in a lower recruitment of the PI(3,4)P2 probe to phagosomes compared to the solvent controls, indicating a reduced presence of PI(3,4)P2 at the phagosomes (Figures S3F–S3H). We also observed ∼50% reduction of phagosomes containing SWAP70 with 3AC or AS1949490 (Figures 3F–3H), supporting a role for PI(3,4)P2 in phagosomal recruitment of SWAP70. This reduced recruitment of SWAP70 was not due to a general trafficking defect, because the presence of gp91phox (CYBB, an integral membrane component of the NADPH oxidase NOX2) at the phagosome was not affected by 3AC nor AS1949490 (Figures S3I–S3K).
PI(3,4)P2 is not the only factor recruiting SWAP70 to the phagosome, as the PH-domain alone showed considerably higher cytoplasmic background compared to endogenous SWAP70 (Figure S3a) and to overexpressed full-length SWAP70 fused to GFP (Figure S4). This is also indicated by the finding that a GFP-tagged full-length SWAP70 mutant unable to bind to phosphoinositides (R223E and R224E (Hilpelä et al., 2003)) was still recruited to phagosomes, albeit less efficiently than wild-type SWAP70 (Figure S4). We tested GFP-tagged N- and C-terminal truncation mutants of SWAP70 to test which domains contribute to SWAP70 binding to phagosomes. Truncation mutants of SWAP70 lacking its N-terminal fragment containing the EF-hand motif or its C-terminal fragment containing the putative Dbl-homology domain (DH) (Shinohara et al., 2002) and actin-binding domain were recruited to phagosomes less efficiently compared to full-length SWAP70 (Figure S4), indicating that not only the PH-domain but also the N- and C-terminal regions contribute to phagosomal recruitment of SWAP70. These results compare well with the reported domain requirement for binding of SWAP70 to macropinosomes (Oberbanscheidt et al., 2007), except that the PH-domain alone is recruited to phagosomes (Figure S3a, S4) but not to macropinosomes (Oberbanscheidt et al., 2007). Possibly, phagosomes have prolonged presence and/or higher levels of PI(3,4)P2 than macropinosomes. F-actin is not a major contributor for phagosomal SWAP70 recruitment, as mutants lacking part of or the entire C-terminal actin-binding domain (Ihara et al., 2006) were recruited to phagosomes with similar efficiency as full-length SWAP70 (Figure S4).
SWAP70 Controls Phagocytosis via F-Actin Polymerization
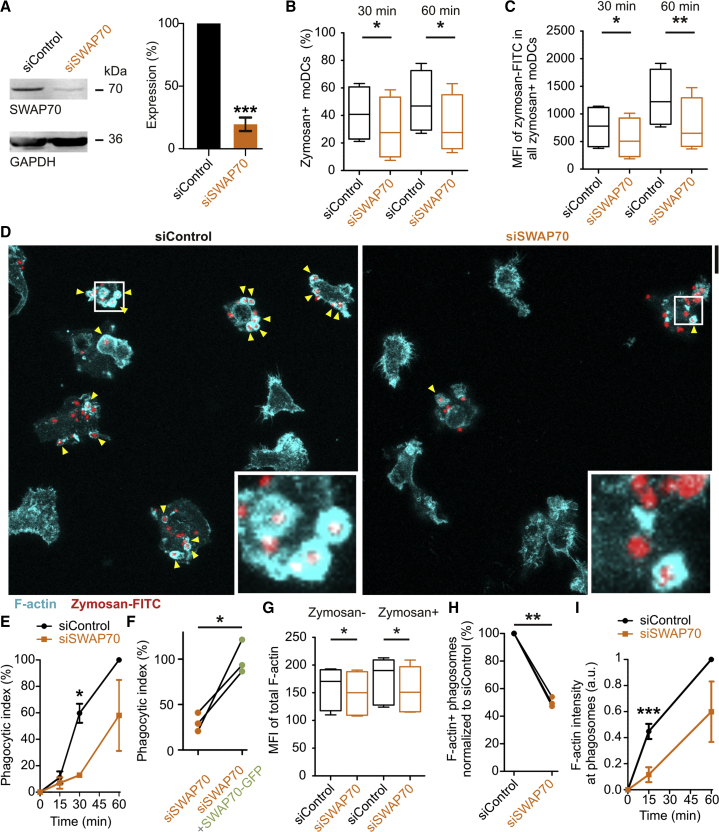
We then addressed the functional role of SWAP70 in phagocytosis. We first evaluated how siRNA knockdown of SWAP70 (∼80% knockdown efficiency; Figure 4A) affected uptake of FITC-labeled zymosan by flow cytometry (Figure S5A). For all donors tested, SWAP70 knockdown reduced the phagocytosis capacity of dendritic cells. The total fraction of cells that ingested zymosan was significantly reduced upon SWAP70 knockdown (Figures 4B and S5B). In this fraction of cells that took up zymosan, the total uptake was also significantly reduced (Figures 4C, S5C, and S5D). Lower zymosan uptake upon SWAP70 knockdown was also apparent from confocal imaging (Figures 4D and 4E). Zymosan uptake could be rescued by combining SWAP70 knockdown with overexpression of SWAP70-GFP with altered codon usage to protect it from siRNA targeting (Figures 4F, S5E, and S5F). These results contrast a study where TAT-mediated transduction of human granulocytes with a dominant-negative form of SWAP70 did not (or only somewhat) affect phagocytosis of gonococci via CEACAM3 (Schmitter et al., 2007). We also tested the effect of SWAP70 knockdown on pinocytosis, because SWAP70 is recruited to pinosomes (Oberbanscheidt et al., 2007) and S1P-induced endocytosis is reduced in dendritic cells from SWAP70−/− mice (Ocaña-Morgner et al., 2011). Similar to our findings with zymosan particles, we observed a significant reduction (∼25%) in uptake of BSA labeled with Alexa Fluor 488 (Figure S5G).
Figure 4.
SWAP70 Regulates Phagocytosis and Phagosomal F-Actin
(A) Knockdown of dendritic cells with control (black, siControl) or SWAP70 siRNA (orange, siSWAP70) quantified by western blot (mean ± SEM). GAPDH, loading control.
(B) Dendritic cells with siSWAP70 were pulsed with FITC-labeled zymosan. Zymosan-positive cells were quantified by FACS after 30 and 60 min for at least four donors. See also Figures S5A and S5B.
(C) The total zymosan-FITC signal for the zymosan-positive cells from (B). MFI, mean fluorescence intensity. See also Figures S5C and S5D.
(D) Confocal micrographs of FITC-labeled zymosan (red) pulsed dendritic cells with siControl (left) or siSWAP70 (right). F-actin was stained with phalloidin (cyan). Yellow arrowheads, F-actin positive phagosomes. Scale bar, 20 μm.
(E) Quantification of phagocytosis from (D) (mean ± SEM; >40 cells/donor/condition).
(F) Rescue experiment of phagocytosis for dendritic cells with siSWAP70 and SWAP70-GFP (individual donors shown). See also Figures S5E and S5F.
(G) The MFI of phalloidin labeling in the zymosan-positive and -negative populations of cells from (B).
(H) Percentage of phalloidin-positive phagosomes from (D).
(I) Quantification of the phalloidin signals of F-actin-positive phagosomes from (D).
See also Figures S5G, S5H, and S6.
In our flow cytometry experiments, we stained F-actin with fluorescently labeled phalloidin. This allowed quantification of the amount of F-actin per cell. Knockdown of SWAP70 led to a small (∼10%–20%) but significant reduction of F-actin (Figures 4G and S5H). This result is in line with the reduced F-actin levels in macrophages from SWAP70−/− mice (Chacón-Martínez et al., 2013). Confocal imaging showed that the percentage of F-actin-positive phagosomes was reduced by ∼50% upon SWAP70 knockdown and the phalloidin intensity at these F-actin-positive phagosomes was also reduced by ∼50% (Figures 4H and 4I). We no longer observed the parallel arches or rings of F-actin on the surface of the phagosomes by STED microscopy upon SWAP70 knockdown, whereas these structures were readily observable in the non-targeting siRNA control (Figures S6A and S6B). Phagosomes of SWAP70 knockdown dendritic cells still contained PI(4,5)P2, PI(3,4,5)P3, and PI(3,4)P2 based on experiments with the GFP-tagged phosphoinositide probes (Figure S6C). Thus, knockdown of SWAP70 leads to a phagocytotic defect and reduced actin filament formation around phagosomes. Our data suggest that SWAP70 acts as a scaffolding protein, coordinating the tethering and stabilization of actin filaments on the phagosome surface. We then investigated the role of RAC1 in this process.
SWAP70 Promotes Phagosomal RAC1 Activity
SWAP70 can bind to RHO-GTPases, including the activated form of RAC1 (Ihara et al., 2006, Oberbanscheidt et al., 2007, Ocana-Morgner et al., 2009, Sivalenka and Jessberger, 2004). RAC1 activity is promoted by its binding to SWAP70 (Dwyer et al., 2015, Murugan et al., 2008, Shinohara et al., 2002, Sivalenka et al., 2008). RAC1 stimulates actin polymerization on phagosomes by Arp2/3 through activation of WAVE (Swanson, 2008). In its inactive state, RAC1 is associated to RHO-GDI that keeps the protein in the cytosol. Dissociation of RHO-GDI from RAC1 results in membrane association of RAC1 via insertion of its geranyl-geranyl moiety (Bustelo et al., 2007, van de Donk et al., 2005). RAC1 is transiently recruited to phagosomes and disappears within minutes after phagocytic cup closure (Hoppe and Swanson, 2004, Oberbanscheidt et al., 2007). By confocal microscopy, we estimated that the majority (60%–90%, depending on the donor) of SWAP70-positive zymosan phagosomes contained RAC1 and ∼70% of these also contained F-actin (Figures 5A and 5B). RAC1 not only localized to phagosomes but also to the plasma membrane, as reported previously for lymphocytes (Cernuda-Morollón et al., 2010). In live cell imaging experiments, overexpressed RAC1 fused to RFP (Hoppe and Swanson, 2004) also localized to phagosomes simultaneously with SWAP70-GFP (Figures 5C and 5D; Movie S14). By multi-color three-dimensional STED microscopy, we observed strong overlap between RAC1 and SWAP70 on the surface of phagosomes (Figure 5E; Movie S15). As expected, based on the high overlap between SWAP70 and F-actin, RAC1 also overlapped with F-actin on the surface of phagosomes (Figure 5F; Movie S16).
Figure 5.
SWAP70 Promotes Phagosomal RAC1 Activity
(A) Confocal micrographs and quantification of RAC1 (green in merge) and SWAP70 (magenta) positive phagosomes for seven donors (∼100 cells/donor). Aver., average ± SEM.
(B) Venn diagram showing phagosomal distribution of RAC1, F-actin, and SWAP70.
(C) Live cell imaging of dendritic cells expressing SWAP70-GFP (green in merge) and RAC1-RFP (magenta). The inset shows a time series during zymosan uptake. BF, bright field. See also Movie S14.
(D) Quantification from (C). The histogram shows the time difference of peak recruitment of SWAP70-GFP and RAC1-RFP based on fluorescence intensities (88 phagosomes). Negative values indicate that RAC1 was recruited prior to SWAP70 and positive values later than SWAP70.
(E and F) Multicolor 3D-STED microscopy of zymosan-pulsed dendritic cells immunostained for RAC1 (green) and SWAP70 (magenta; E) or F-actin (F). Left: cross-section and orthogonal sections (indicated by dashed yellow lines). Middle: maximum intensity height maps. Right: maximum intensity surface projection. Yellow arrowheads, overlap of RAC1 with SWAP70 (E) or F-actin (F) on the surface of the phagosomes. See also Movies S15 and S16.
(G) RAC1 activation upon control (siControl) and SWAP70 siRNA (siSWAP70) after zymosan addition by G-LISA assay (Abs, absorbance units at 490 nm; mean ± SEM). The background (t = 0) is from cells without zymosan.
(H) RAC1-positive phagosomes upon siControl and siSWAP70 counted by microscopy (>20 cells/donor/condition analyzed; individual donors shown).
(I) Phagosomes positive for F-actin and/or RAC1 with siControl or siSWAP70.
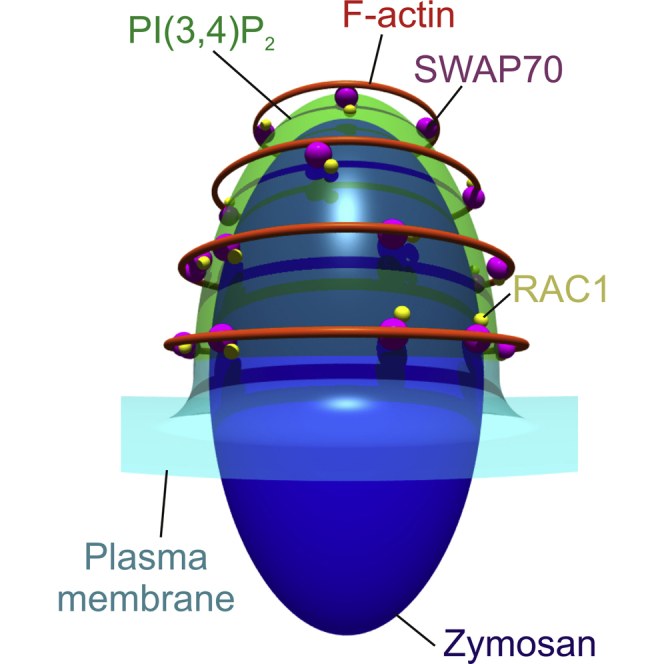
(J) Model of F-actin cage formation by SWAP70. SWAP70 is recruited to phagosomes by coincidence detection. Scale bars: 10 μm (A and C), 2 μm (E and F).
We then decided to test how knockdown of SWAP70 would affect RAC1 activation by measuring activated (GTP-bound) RAC1 by a colorimetric assay. Although such global methods are not optimal to detect localized changes, SWAP70 knockdown reduced the zymosan-induced activation of RAC1 almost completely (Figure 5G). The recruitment of total RAC1 to phagosomes was also reduced upon SWAP70 knockdown by ∼60% (Figure 5H). We showed that phagosomal F-actin was also reduced upon SWAP70 knockdown (Figures 4H and 4I). The residual phagosomes that contained RAC1 in the SWAP70 knockdown samples had similar levels of F-actin as the RAC1-positive phagosomes in the control samples (Figure 5I). This indicates that SWAP70 affects both phagosomal RAC1 and F-actin equally. The residual RAC1 and F-actin recruitment could be due to functional redundancy (i.e., another scaffolding protein) and/or to residual SWAP70. Our results are reminiscent of the reduced levels of activated RAC1 in Mast cells isolated from SWAP70−/− mice (Sivalenka and Jessberger, 2004) and of the reduced RAC1 recruitment to actin-rich areas in dendritic cells from SWAP70−/− mice (Ocaña-Morgner et al., 2011). Thus, SWAP70 promotes the presence of F-actin and RAC1 on phagosomes.
Discussion
SWAP70 is a multitasking protein, harboring multiple domains that can bind to DNA, phosphoinositides, RHO-GTPases, and F-actin. SWAP70 is implicated in many diseases, including cancer (Chiyomaru et al., 2011, Fukui et al., 2007, Murugan et al., 2008, Shu et al., 2013), various autoimmune diseases (Bahaie et al., 2012, Biswas et al., 2012, Erdağ et al., 2012, Türkoğlu et al., 2014, Vural et al., 2009), and HIV (Kimbara et al., 2006). In this study, we show that SWAP70 is recruited to the surface of the nascent phagocytic cup in dendritic cells. SWAP70 remains associated with phagosomes for ∼1–3 min but up to 15 min after cup closure, reminiscent of the transient localization of SWAP70 to early macropinosomes in mouse dendritic cells and NIH/3T3 cells (Oberbanscheidt et al., 2007). Knockdown of SWAP70 reduces phagosomal F-actin and RAC1 and impairs phagocytosis. SWAP70 localization to phagosomes was much stronger than to other F-actin-rich structures such as podosomes (Götz and Jessberger, 2013). Our results indicate that SWAP70 is recruited to phagosomes via association of its PH-domain to PI(3,4)P2. This confirms previous phosphoinositide dot-blot results indicating preferential binding of SWAP70 to PI(3,4)P2 regulates the actin cytoskeleton for cell protrusion (Hilpelä et al., 2003). SWAP70 was also identified as a PI(3,4)P2 binding protein in a quantitative phosphoinositide-binding proteomics study in HeLa cells (Jungmichel et al., 2014). Finally, binding of SWAP70 to PI(3,4)P2 was observed in competitive pull-down experiments, although in this case SWAP70 bound stronger to PI(3,4,5)P3 (Shinohara et al., 2002). Thus, it seems reasonable to conclude that the PH-domain of SWAP70 preferentially binds to PI(3,4)P2, although it may also bind to PI(3,4,5)P3.
Phosphoinositides recruit factors promoting the polymerization and tethering of actin fibers to phagosomes (Levin et al., 2015). Especially, PI(4,5)P2 is well-understood in these processes via the ERM proteins, CDC42 and WASP (Bretscher et al., 2002, Hoppe and Swanson, 2004, Rohatgi et al., 2001, Yonemura et al., 2002). PI(3,4)P2 has not yet been described to play a role in phagosomal actin and was long believed to be merely a transition product in the conversion from PI(3,4,5)P3 to PI(3)P (Li and Marshall, 2015). In fact, the action of SHIP1 was even reported to negatively affect both CR3 and Fcγ receptor-mediated phagocytosis, as it decreased levels of PI(3,4,5)P3 on the nascent phagosome (Horan et al., 2007). However, it is now increasingly clear that PI(3,4)P2 is a signaling phosphoinositide itself with unique roles in phagocytosis and pinocytosis (Li and Marshall, 2015, Maekawa et al., 2014, Welliver and Swanson, 2012). Several proteins specifically bind to PI(3,4)P2, such as TAPP1 (PLEKHA1) or TAPP2 (PLEKHA2), which are both involved in actin remodeling via their interaction with syntrophins (Hogan et al., 2004). PI(3)P is the main phosphoinositide of early endosomes/phagosomes, and the RAB5A effector protein EEA1 is recruited to PI(3)P by binding of its FYVE-domain (Levin et al., 2015, Simonsen et al., 1998). The finding that SWAP70 does not bind to PI(3)P is in accordance with our observations that SWAP70 is recruited to phagosomes prior to EEA1 and RAB5A and with the reported finding that SWAP70 recruitment to macropinosomes occurs prior to RAB5A (Oberbanscheidt et al., 2007).
Our super-resolution STED-microscopy showed that SWAP70 aligned with F-actin and RAC1 on the surface of phagosomes, forming parallel arches and rings. Upon knockdown of SWAP70, these structures were no longer observable. The binding of the C-terminal domain of SWAP70 to non-muscle F-actin is well-established (Gomez-Cambronero, 2012, Hilpelä et al., 2003, Ihara et al., 2006, Murugan et al., 2008, Shinohara et al., 2002). SWAP70 can oligomerize and thereby bundle actin filaments in both a parallel and anti-parallel fashion and this delays dilution-induced F-actin depolymerization (Chacón-Martínez et al., 2013). This bundling of actin by SWAP70 likely explains the filamentous-like organization of SWAP70 on the phagosome surface revealed by STED. We also observed overlap between SWAP70 and RAC1 and impairment of RAC1 activation upon SWAP70 knockdown. Although SWAP70 was initially shown to be a guanine nucleotide exchange factor (GEF) for RAC1 (Shinohara et al., 2002), this is no longer believed (Oberbanscheidt et al., 2007) because mammalian SWAP70 (and also plant SWAP70) (Yamaguchi et al., 2012) binds to activated RHOA and RAC1, but not to GDP-bound or nucleotide-free RAC (Ihara et al., 2006, Murugan et al., 2008, Oberbanscheidt et al., 2007, Ocana-Morgner et al., 2009). Likely, RAC1 is activated by GEFs such as DOCK180 (DOCK1) and VAV (Rossman et al., 2005, Swanson, 2008) and stabilized in its active form by SWAP70. Our findings are in accordance with the reduced activation of RAC1 and decreased recruitment to actin-rich cellular regions in SWAP70−/− mice (Shinohara et al., 2002, Sivalenka et al., 2008, Ocaña-Morgner et al., 2011). Moreover, the aberrant activation of RAC seen in Kaposi’s sarcoma requires SWAP70 (Dwyer et al., 2015). Our data and these studies all support a role for SWAP70 in regulation of RHO-GTPases and hence we conclude that SWAP70 does not only bind and stabilize the phagocytic actin cage, but also promotes their formation via binding-activated RAC1 (Figure 5J).
What is the sequence of events of SWAP70, RAC1, and F-actin recruitment to the phagocytic cup? Our data show that phosphoinositide binding by the PH-domain of SWAP70 clearly contributes to specific phagosomal recruitment. This result corroborates previous findings that a SWAP70 mutant impaired in PI(3,4,5)P3 binding failed to locate to F-actin rich plasma membrane ruffles (Fukui et al., 2007, Shinohara et al., 2002, Wakamatsu et al., 2006). The binding of SWAP70 to RAC1 likely also contributes to phagosomal recruitment, as our data show that phagosomal recruitment is supported by both the N-terminal fragment (containing the EF-hand motif) and the putative Dbl-homology domain (DH) (Shinohara et al., 2002) of SWAP70, but not by its C-terminal actin-binding region, similar to previous findings with macropinosomes (Oberbanscheidt et al., 2007). In accordance with this notion, RAC1 is recruited to the extending pseudopods of the nascent cup of phagosomes (Hoppe and Swanson, 2004), and SWAP70 is recruited after RAC1 to macropinosomes (Oberbanscheidt et al., 2007). However, we did not observe such a clear time-dependency for phagosomes in dendritic cells and instead saw simultaneous recruitment of SWAP70 and RAC1 and reduced RAC1 recruitment upon SWAP70 knockdown. Because SWAP70 binds to activated RAC1 (Ihara et al., 2006, Murugan et al., 2008, Ocana-Morgner et al., 2009), it may well be that SWAP70 and RAC1 are recruited to nascent phagosomes by a positive feedback loop, where binding of SWAP70 to PI(3,4)P2 results in recruitment of activated RAC1 and F-actin that in turn recruits more SWAP70 (Figure 5J). PI(3,4)P2 may also directly recruit RAC1 itself and promote RAC-GEF activation, as is well-established for PI(3,4,5)P2 (Fleming et al., 2000, Hill and Welch, 2006, Missy et al., 1998). The notion that SWAP70 is recruited to phagosomes by multiple binding partners is supported by the finding that the N-terminal region, the PH-domain, and the putative Dbl-homology domain (DH) all contribute to binding of SWAP70 to phagosomes. Such a synergistic engagement of multiple binding partners is called coincidence detection (Carlton and Cullen, 2005) and is a well-understood mechanism to achieve highly restricted localizations as the one we report here for SWAP70 at phagosomes.
Our data show that SWAP70 plays a role in the assembly of the F-actin cage on phagosomes that persists well after cup-closure and depends on PI(3,4)P2. This is not only a novel role for PI(3,4)P2 in the organization of actin on phagosomes, but it also refutes the long-standing believe that F-actin on early phagosomes is primarily depending on PI(4,5)P2 and PI(3,4,5)P3, but not on other 3-phosphoinositides (Bretscher et al., 2002, Hoppe and Swanson, 2004, Rohatgi et al., 2001, Yonemura et al., 2002). Likely, the formation of an F-actin cage by SWAP70 promotes the remodeling of the membrane and provides a mechanic pulling force for ingestion of the phagocytic load (May et al., 2000). F-actin surrounding newly internalized phagosomes is known to rapidly depolymerize upon the conversion of PI(4,5)P2 to PI(3,4,5)P3, which leaves the phagosome membrane available to fuse with endosomes and lysosomes (May et al., 2000, Schlam et al., 2015). Retaining the F-actin cage on the phagocytic surface by SWAP70 likely delays this phagosomal maturation, which could well explain the maturation defects and reduced T cell activation observed in SWAP70-deficient dendritic cells (Ocana-Morgner et al., 2009, Ocaña-Morgner et al., 2013). Thereby, SWAP70 could potentially lead to preservation or rerouting of the phagocytic cargo for specific processing of antigen for naive T cell activation.
Experimental Procedures
Antibodies and Reagents
The following primary antibodies were used: rabbit polyclonal anti-SWAP70 (Novus Biologicals; cat. no NBP1-82979) at 1:200 dilution (v/v) for immunofluorescence (IF) and 1:500 for western blot; mouse IgG1 anti-EEA1 (BD Biosciences; cat. no. 610457) at 1:100; mouse IgG1 anti-LAMP1 (Biolegend; 328601) at 1:200; mouse monoclonal anti-RAC1 (Cell Biolabs; 240106) at 1:100; mouse IgG1 anti-FITC Alexa Fluor 647 (Jackson ImmunoResearch; 200-602-037) at 1:200; rabbit polyclonal anti-actin (Sigma; A5060) at 1:100; and mouse IgG1 anti-CYBB (MBL; D162-3) at 1:200. The following secondary antibodies were used: goat-anti-mouse Alexa Fluor 488 (Life Technologies; A11029); goat-anti-rabbit Alexa Fluor 568 or 647 (Life Technologies; A11036 and A21245); goat-anti-mouse STAR635 (Abberior; 2-0002-002-0); sheep-anti-mouse KK114 (Abberior Red); and goat-anti-rabbit Alexa Fluor 594 (Abcam; ab150084). For immunoblotting goat-anti-rabbit IRDye 800CW (Li-Cor; 926-32211) and goat-anti-mouse IRDye 680CW (Li-Cor; 926-32220) were used. Alexa Fluor 633-labeled phalloidin was from ThermoFisher (A22284) and used at 1:200 (v/v). mCLING labeled with ATTO647N was from Synaptic Systems (710006AT1) and used at 1:200 (v/v). The RAC1 activation G-LISA kit was from Cytoskeleton (BK128-S).
Cells and Transfection
Buffy coats from healthy donors (consent obtained and according to national ethics guidelines and approved by the institutional review board [Radboud UMC]) were used to isolate peripheral monocytes as described (de Vries et al., 2002). Constructs for full-length human SWAP70, the PH-domain (residues 210–306), the PH-domain and full-length SWAP70 with R223E and R224E mutations, and C- and N-terminal truncations (residues 205–585, 1–313, 1–525, and 1–572) were generated as codon-optimized synthetic genes (Genscript) in the EcoRI/BamHI sites of pEGFP-C1 and pmCherry-C1 (Clontech). RAB5A-mRFP was a gift from Ari Helenius (Addgene plasmid #14437) (Vonderheit and Helenius, 2005). YFP-RAC1 was a gift from Joel Swanson (Addgene plasmid #11391) (Hoppe and Swanson, 2004) and was re-cloned into mCherry-C1. LifeAct-RFP was a gift from Michael Sixt (Max Planck Institute of Biochemistry, Martinsried). GFP-C1-PLCD1-PH was a gift from Tobias Meyer (Addgene plasmid #21179) (Stauffer et al., 1998). The GFP-tagged PH-domain of AKT is described (de Keijzer et al., 2011). NCF4-PX-EGFP was a gift from Michael Yaffe (Addgene plasmid #19010) (Kanai et al., 2001). The GFP-tagged residues 179–311 of TAPP2 are described (de Keijzer et al., 2011, Haugh et al., 2000). Mouse MCOLN1 residues 1–68 were generated as a synthetic gene in the XhoI/BamHI sites of pEGFP-C1. Transfection was performed as described (Baranov et al., 2014, Dingjan et al., 2016) and zymosan uptake and imaging experiments were performed 8–12 hr post-transfection.
Microscopy
Samples were imaged with a Leica SP8 confocal microscope fitted with a 63× 1.2 NA water immersion objective. For live cell microscopy, cells were imaged 3–5 hr post-transfection with IgG-opsonized zymosan at 1:10 cell-to-particle ratio. Imaging was performed in Hank’s balanced salt solution (HBSS) (ThermoFisher, 14025050) supplemented with 10 mM HEPES at pH 7.4. Samples were imaged with a Leica DMI6000 epi-fluorescence microscope fitted with a 63× 1.4 NA oil immersion objective, a metal halide EL6000 lamp for excitation, a DFC365FX CCD camera, and GFP and DsRed filter sets (all from Leica). Focus was kept stable with the adaptive focus control from Leica.
The STED microscope was from Abberior Instruments. Emission of two fluorescent markers (excitation [Ex.]: 560/640 nm, emission [Em.]: 580–630 nm/640–720 nm) was depleted with a single STED-laser operating at 775 nm with 800 picosecond (ps) long pulses at 40 MHz. The STED-laser point-spread-function was shaped by a spatial light modulator. STED-laser power in the back-focal-plane was 180 mW, with 65% of that power used for the 2D-donut and 35% for the 3D-donut. Zymosan particles were detected by excitation at 485 nm and collecting autofluorescence at 500–550 nm. Excitation power of the three excitation lasers 485 nm/560 nm/640 nm were 4 μW/10 μW/8 μW in the back-focal-plane of an 100× 1.4 NA oil immersion objective (Olympus).
Fluorescence-Activated Cell Sorting
For fluorescence-activated cell sorting (FACS), cells were incubated with FITC-labeled zymosan at 1:10 cell-to-zymosan ratio or 100 μg mL−1 BSA-AF488 for 5 min to 2 hr. Incubation was 1 hr for the uptake experiments with 3AC and AS1949490. Cells were fixed in 4% PFA (w/v) and F-actin was labeled with Alexa Fluor 633 phalloidin in S-PBS (see the Supplemental Information). Data were acquired on a FACS-Calibur cytometer (BD Biosciences).
Statistical Analysis
All data were analyzed using Student’s two-tailed paired t tests. A p value < 0.05 was regarded as statistically significant (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
Author Contributions
M.V.B., M.t.B., and G.v.d.B. designed the experiments and wrote the paper. A.H. and R.M. performed STED experiments. N.H.R., I.D., and M.t.B helped with experiments and cloning. M.V.B. performed all other experiments. All authors contributed to writing the manuscript.
Acknowledgments
We thank Rolf Jessberger for SWAP70 constructs, Tobias Meyer for PH-PLCD1, Michael Sixt for LifeAct-RFP, Joel Swanson for YFP-RAC1, Ari Helenius for RAB5A-mRFP, and Michael Yaffe for NCF4-PX constructs. This work was supported by a Starting Grant from the European Research Council (ERC) under the European Union’s Seventh Framework Programme (Grant Agreement 336479). G.v.d.B. is funded by a Hypatia fellowship from the Radboud University Medical Center, a Career Development Award from the Human Frontier Science Program, the NWO Gravitation Programme 2013 (ICI-024.002.009), and a Vidi grant from the Netherlands Organization for Scientific Research (NWO-ALW VIDI 864.14.001). N.H.R. is funded by an EMBO Long-Term Fellowship (ALTF 232-2016) and a Veni grant from NWO-ALW (016.Veni.171.097).
Published: November 1, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, six figures, and sixteen movies and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2016.10.021.
Supplemental Information
Time-lapse epi-fluorescence imaging of dendritic cells transiently expressing SWAP70-GFP (green) and LifeAct-RFP (magenta) and pulsed with IgG-opsonized zymosan. Images were acquired every 20 sec. BF: bright field. Scale bar, 5 μm.
The animated reconstruction from Figure 1F shows concentric F-actin rings on the phagosomal surface. Dendritic cells were pulsed with zymosan (blue; autofluorescent core) and immunostained for SWAP70 (magenta). Total membranes were labeled with the fixable endocytic marker mCLING (green; Revelo et al., 2014). The bright mCLING-labeled structures surrounding the phagosome are endo/lysosomal compartments. The movies show 3D-rotations of single SWAP70-positive phagosomes. Note the alignment of SWAP70 in concentric fibers (movies 2–4) and rings (movies 5–6) on the surface of the phagosomes. The ventral membrane is positioned on the surface of the microscope cover glass; the dorsal membrane is facing away from it. Scale bars, approximately 2 μm.
The animated reconstruction from Figure 1F shows concentric F-actin rings on the pagosomal surface. Dendritic cells were pulsed with zymosan (blue; autofluorescent core) and immunostained for SWAP70 (magenta). Total membranes were labeled with the fixable endocytic marker mCLING (green; Revelo et al., 2014). The bright mCLING-labeled structures surrounding the phagosome are endo/lysosomal compartments. The movies show 3D-rotations of single SWAP70-positive phagosomes. Note the alignment of SWAP70 in concentric fibers (movies 2–4) and rings (movies 5–6) on the surface of the phagosomes. The ventral membrane is positioned on the surface of the microscope cover glass; the dorsal membrane is facing away from it. Scale bars, approximately 2 μm.
The animated reconstruction from Figure 1F shows concentric F-actin rings on the phagosomal surface. Dendritic cells were pulsed with zymosan (blue; autofluorescent core) and immunostained for SWAP70 (magenta). Total membranes were labeled with the fixable endocytic marker mCLING (green; Revelo et al., 2014). The bright mCLING-labeled structures surrounding the phagosome are endo/lysosomal compartments. The movies show 3D-rotations of single SWAP70-positive phagosomes. Note the alignment of SWAP70 in concentric fibers (movies 2–4) and rings (movies 5–6) on the surface of the phagosomes. The ventral membrane is positioned on the surface of the microscope cover glass; the dorsal membrane is facing away from it. Scale bars, approximately 2 μm.
The animated reconstruction from Figure 1F shows concentric F-actin rings on the phagosomal surface. Dendritic cells were pulsed with zymosan (blue; autofluorescent core) and immunostained for SWAP70 (magenta). Total membranes were labeled with the fixable endocytic marker mCLING (green; Revelo et al., 2014). The bright mCLING-labeled structures surrounding the phagosome are endo/lysosomal compartments. The movies show 3D-rotations of single SWAP70-positive phagosomes. Note the alignment of SWAP70 in concentric fibers (movies 2–4) and rings (movies 5–6) on the surface of the phagosomes. The ventral membrane is positioned on the surface of the microscope cover glass; the dorsal membrane is facing away from it. Scale bars, approximately 2 μm.
The animated reconstruction from Figure 1F shows concentric F-actin rings on the phagosomal surface. Dendritic cells were pulsed with zymosan (blue; autofluorescent core) and immunostained for SWAP70 (magenta). Total membranes were labeled with the fixable endocytic marker mCLING (green; Revelo et al., 2014). The bright mCLING-labeled structures surrounding the phagosome are endo/lysosomal compartments. The movies show 3D-rotations of single SWAP70-positive phagosomes. Note the alignment of SWAP70 in concentric fibers (movies 2–4) and rings (movies 5–6) on the surface of the phagosomes. The ventral membrane is positioned on the surface of the microscope cover glass; the dorsal membrane is facing away from it. Scale bars, approximately 2 μm.
Dendritic cells were pulsed with zymosan (blue; autofluorescent core) and immunostained for SWAP70 (magenta) a1nd actin (green). The movie shows a 3D-rotation of a single SWAP70-positive phagosome. Note the alignment of SWAP70 and F-actin on the surface of the phagosomes. Yellow lines indicate the orientation of F-actin and SWAP70-positive filamentous structures. Scale bar, approximately 2 μm.
3D-confocal stack of a nascent phagosome. FITC-labeled zymosan (green) was phagocytosed by dendritic cells. Uninternalized zymosan and nascent cups were labeled with an antibody directed against FITC (blue). SWAP70 was immunolabeled (magenta). The movie shows a 3D-rotation of a single SWAP70-positive nascent phagosomal cup. Scale bar, approximately 2 μm.
Time-lapse epi-fluorescence imaging of dendritic cells transiently expressing SWAP70-GFP (green) and RAB5A-RFP (magenta) and pulsed with IgG-opsonized zymosan. Images were acquired every 15 sec. BF: bright field. Scale bar, 5 μm.
Time-lapse epi-fluorescence imaging of dendritic cells transiently expressing the GFP-tagged PH-domain of TAPP2 (PLEKHA2) (binds to PI(3,4)P2; green) and SWAP70-mCherry (magenta) and pulsed with IgG-opsonized zymosan. Images were acquired every 15 sec. BF: bright field. Scale bar, 5 μm.
Time-lapse epi-fluorescence imaging of dendritic cells transiently expressing the GFP-tagged PH-domain of AKT (binds to PI(3,4,5)P3; green) and SWAP70-mCherry (magenta) and pulsed with IgG-opsonized zymosan. Images were acquired every 15 sec. BF: bright field. Scale bar, 5 μm.
Time-lapse epi-fluorescence imaging of dendritic cells transiently expressing the GFP-tagged PH-domain of PLCδ1 (PLCD1; binds to PI(4,5)P2; green) and SWAP70-mCherry (magenta) and pulsed with IgG-opsonized zymosan. Images were acquired every 15 sec. BF: bright field. Scale bar, 5 μm.
Time-lapse epi-fluorescence imaging of dendritic cells transiently expressing the GFP-tagged PX-domain of NCF4 (p40phox; binds to PI(3)P; green) and SWAP70-mCherry (magenta) and pulsed with IgG-opsonized zymosan. Images were acquired every 15 sec. BF: bright field. Scale bar, 5 μm.
Time-lapse epi-fluorescence imaging of dendritic cells transiently expressing SWAP70-GFP (green) and RAC1-RFP (magenta) and pulsed with IgG-opsonized zymosan. Images were acquired every 15 sec. BF: bright field. Scale bar, 5 μm.
Dendritic cells were pulsed with zymosan (blue; autofluorescent core) and immunostained for SWAP70 (magenta) and RAC1 (green). The movie shows a 3D-rotation of a single SWAP70-positive phagosome. Note the alignment of SWAP70 and RAC1on the surface of the phagosomes. Yellow lines indicate the orientation of SWAP70 and RAC1-positive filamentous structures. Scale bar, approximately 2 μm.
Dendritic cells were pulsed with zymosan (blue; autofluorescent core) and immunostained for RAC1 (magenta) and actin (green). The movie shows a 3D-rotation of a single SWAP70-positive phagosome. Note the alignment of RAC1and F-actin on the surface of the phagosomes. Yellow lines indicate the orientation of RAC1and F-actin-positive filamentous structures. Scale bar, approximately 2 μm.
References
- Allen L.A., Aderem A. Mechanisms of phagocytosis. Curr. Opin. Immunol. 1996;8:36–40. doi: 10.1016/s0952-7915(96)80102-6. [DOI] [PubMed] [Google Scholar]
- Anes E., Kühnel M.P., Bos E., Moniz-Pereira J., Habermann A., Griffiths G. Selected lipids activate phagosome actin assembly and maturation resulting in killing of pathogenic mycobacteria. Nat. Cell Biol. 2003;5:793–802. doi: 10.1038/ncb1036. [DOI] [PubMed] [Google Scholar]
- Bahaie N.S., Hosseinkhani M.R., Ge X.N., Kang B.N., Ha S.G., Blumenthal M.S., Jessberger R., Rao S.P., Sriramarao P. Regulation of eosinophil trafficking by SWAP-70 and its role in allergic airway inflammation. J. Immunol. 2012;188:1479–1490. doi: 10.4049/jimmunol.1102253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranov M.V., Ter Beest M., Reinieren-Beeren I., Cambi A., Figdor C.G., van den Bogaart G. Podosomes of dendritic cells facilitate antigen sampling. J. Cell Sci. 2014;127:1052–1064. doi: 10.1242/jcs.141226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas P.S., Gupta S., Stirzaker R.A., Kumar V., Jessberger R., Lu T.T., Bhagat G., Pernis A.B. Dual regulation of IRF4 function in T and B cells is required for the coordination of T-B cell interactions and the prevention of autoimmunity. J. Exp. Med. 2012;209:581–596. doi: 10.1084/jem.20111195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borggrefe T., Masat L., Wabl M., Riwar B., Cattoretti G., Jessberger R. Cellular, intracellular, and developmental expression patterns of murine SWAP-70. Eur. J. Immunol. 1999;29:1812–1822. doi: 10.1002/(SICI)1521-4141(199906)29:06<1812::AID-IMMU1812>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Bretscher A., Edwards K., Fehon R.G. ERM proteins and merlin: integrators at the cell cortex. Nat. Rev. Mol. Cell Biol. 2002;3:586–599. doi: 10.1038/nrm882. [DOI] [PubMed] [Google Scholar]
- Brooks R., Fuhler G.M., Iyer S., Smith M.J., Park M.Y., Paraiso K.H., Engelman R.W., Kerr W.G. SHIP1 inhibition increases immunoregulatory capacity and triggers apoptosis of hematopoietic cancer cells. J. Immunol. 2010;184:3582–3589. doi: 10.4049/jimmunol.0902844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustelo X.R., Sauzeau V., Berenjeno I.M. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. BioEssays. 2007;29:356–370. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J.G., Cullen P.J. Coincidence detection in phosphoinositide signaling. Trends Cell Biol. 2005;15:540–547. doi: 10.1016/j.tcb.2005.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron E., Hall A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science. 1998;282:1717–1721. doi: 10.1126/science.282.5394.1717. [DOI] [PubMed] [Google Scholar]
- Cernuda-Morollón E., Millán J., Shipman M., Marelli-Berg F.M., Ridley A.J. Rac activation by the T-cell receptor inhibits T cell migration. PLoS ONE. 2010;5:e12393. doi: 10.1371/journal.pone.0012393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón-Martínez C.A., Kiessling N., Winterhoff M., Faix J., Müller-Reichert T., Jessberger R. The switch-associated protein 70 (SWAP-70) bundles actin filaments and contributes to the regulation of F-actin dynamics. J. Biol. Chem. 2013;288:28687–28703. doi: 10.1074/jbc.M113.461277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiyomaru T., Tatarano S., Kawakami K., Enokida H., Yoshino H., Nohata N., Fuse M., Seki N., Nakagawa M. SWAP70, actin-binding protein, function as an oncogene targeting tumor-suppressive miR-145 in prostate cancer. Prostate. 2011;71:1559–1567. doi: 10.1002/pros.21372. [DOI] [PubMed] [Google Scholar]
- Chopin M., Quemeneur L., Ripich T., Jessberger R. SWAP-70 controls formation of the splenic marginal zone through regulating T1B-cell differentiation. Eur. J. Immunol. 2010;40:3544–3556. doi: 10.1002/eji.201040556. [DOI] [PubMed] [Google Scholar]
- de Keijzer S., Meddens M.B., Kilic D., Joosten B., Reinieren-Beeren I., Lidke D.S., Cambi A. Interleukin-4 alters early phagosome phenotype by modulating class I PI3K dependent lipid remodeling and protein recruitment. PLoS ONE. 2011;6:e22328. doi: 10.1371/journal.pone.0022328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries I.J., Eggert A.A., Scharenborg N.M., Vissers J.L., Lesterhuis W.J., Boerman O.C., Punt C.J., Adema G.J., Figdor C.G. Phenotypical and functional characterization of clinical grade dendritic cells. J. Immunother. 2002;25:429–438. doi: 10.1097/00002371-200209000-00007. [DOI] [PubMed] [Google Scholar]
- Dingjan I., Verboogen D.R., Paardekooper L.M., Revelo N.H., Sittig S.P., Visser L.J., Mollard G.F., Henriet S.S., Figdor C.G., Ter Beest M., van den Bogaart G. Lipid peroxidation causes endosomal antigen release for cross-presentation. Sci. Rep. 2016;6:22064. doi: 10.1038/srep22064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobek A., Kralova J., Skopcova T., Kucova M., Novák P., Angelisová P., Otahal P., Alberich-Jorda M., Brdicka T. PSTPIP2, a protein associated with autoinflammatory disease, interacts with inhibitory enzymes SHIP1 and Csk. J. Immunol. 2015;195:3416–3426. doi: 10.4049/jimmunol.1401494. [DOI] [PubMed] [Google Scholar]
- Dwyer J., Azzi S., Leclair H.M., Georges S., Carlotti A., Treps L., Galan-Moya E.M., Alexia C., Dupin N., Bidère N., Gavard J. The guanine exchange factor SWAP70 mediates vGPCR-induced endothelial plasticity. Cell Commun. Signal. 2015;13:11. doi: 10.1186/s12964-015-0090-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdağ E., Tüzün E., Uğurel E., Cavuş F., Sehitoğlu E., Giriş M., Vural B., Eraksoy M., Akman-Demir G. Switch-associated protein 70 antibodies in multiple sclerosis: relationship between increased serum levels and clinical relapse. Inflamm. Res. 2012;61:927–930. doi: 10.1007/s00011-012-0488-9. [DOI] [PubMed] [Google Scholar]
- Ferrari G., Langen H., Naito M., Pieters J. A coat protein on phagosomes involved in the intracellular survival of mycobacteria. Cell. 1999;97:435–447. doi: 10.1016/s0092-8674(00)80754-0. [DOI] [PubMed] [Google Scholar]
- Fleming I.N., Gray A., Downes C.P. Regulation of the Rac1-specific exchange factor Tiam1 involves both phosphoinositide 3-kinase-dependent and -independent components. Biochem. J. 2000;351:173–182. doi: 10.1042/0264-6021:3510173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman S.A., Grinstein S. Phagocytosis: receptors, signal integration, and the cytoskeleton. Immunol. Rev. 2014;262:193–215. doi: 10.1111/imr.12212. [DOI] [PubMed] [Google Scholar]
- Fuhler G.M., Brooks R., Toms B., Iyer S., Gengo E.A., Park M.Y., Gumbleton M., Viernes D.R., Chisholm J.D., Kerr W.G. Therapeutic potential of SH2 domain-containing inositol-5′-phosphatase 1 (SHIP1) and SHIP2 inhibition in cancer. Mol. Med. 2012;18:65–75. doi: 10.2119/molmed.2011.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui Y., Tanaka T., Tachikawa H., Ihara S. SWAP-70 is required for oncogenic transformation by v-Src in mouse embryo fibroblasts. Biochem. Biophys. Res. Commun. 2007;356:512–516. doi: 10.1016/j.bbrc.2007.03.011. [DOI] [PubMed] [Google Scholar]
- Garbe A.I., Roscher A., Schüler C., Lutter A.H., Glösmann M., Bernhardt R., Chopin M., Hempel U., Hofbauer L.C., Rammelt S. Regulation of bone mass and osteoclast function depend on the F-actin modulator SWAP-70. J. Bone Miner. Res. 2012;27:2085–2096. doi: 10.1002/jbmr.1670. [DOI] [PubMed] [Google Scholar]
- Gomez-Cambronero J. Structure analysis between the SWAP-70 RHO-GEF and the newly described PLD2-GEF. Small GTPases. 2012;3:202–208. doi: 10.4161/sgtp.20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodridge H.S., Wolf A.J., Underhill D.M. Beta-glucan recognition by the innate immune system. Immunol. Rev. 2009;230:38–50. doi: 10.1111/j.1600-065X.2009.00793.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodridge H.S., Underhill D.M., Touret N. Mechanisms of Fc receptor and dectin-1 activation for phagocytosis. Traffic. 2012;13:1062–1071. doi: 10.1111/j.1600-0854.2012.01382.x. [DOI] [PubMed] [Google Scholar]
- Götz A., Jessberger R. Dendritic cell podosome dynamics does not depend on the F-actin regulator SWAP-70. PLoS ONE. 2013;8:e60642. doi: 10.1371/journal.pone.0060642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg S., Grinstein S. Phagocytosis and innate immunity. Curr. Opin. Immunol. 2002;14:136–145. doi: 10.1016/s0952-7915(01)00309-0. [DOI] [PubMed] [Google Scholar]
- Greenberg S., el Khoury J., di Virgilio F., Kaplan E.M., Silverstein S.C. Ca(2+)-independent F-actin assembly and disassembly during Fc receptor-mediated phagocytosis in mouse macrophages. J. Cell Biol. 1991;113:757–767. doi: 10.1083/jcb.113.4.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross B., Borggrefe T., Wabl M., Sivalenka R.R., Bennett M., Rossi A.B., Jessberger R. SWAP-70-deficient mast cells are impaired in development and IgE-mediated degranulation. Eur. J. Immunol. 2002;32:1121–1128. doi: 10.1002/1521-4141(200204)32:4<1121::AID-IMMU1121>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Gu H., Botelho R.J., Yu M., Grinstein S., Neel B.G. Critical role for scaffolding adapter Gab2 in Fc gamma R-mediated phagocytosis. J. Cell Biol. 2003;161:1151–1161. doi: 10.1083/jcb.200212158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haugh J.M., Codazzi F., Teruel M., Meyer T. Spatial sensing in fibroblasts mediated by 3′ phosphoinositides. J. Cell Biol. 2000;151:1269–1280. doi: 10.1083/jcb.151.6.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill K., Welch H.C. Purification of P-Rex1 from neutrophils and nucleotide exchange assay. Methods Enzymol. 2006;406:26–41. doi: 10.1016/S0076-6879(06)06003-4. [DOI] [PubMed] [Google Scholar]
- Hilpelä P., Oberbanscheidt P., Hahne P., Hund M., Kalhammer G., Small J.V., Bähler M. SWAP-70 identifies a transitional subset of actin filaments in motile cells. Mol. Biol. Cell. 2003;14:3242–3253. doi: 10.1091/mbc.E03-01-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan A., Yakubchyk Y., Chabot J., Obagi C., Daher E., Maekawa K., Gee S.H. The phosphoinositol 3,4-bisphosphate-binding protein TAPP1 interacts with syntrophins and regulates actin cytoskeletal organization. J. Biol. Chem. 2004;279:53717–53724. doi: 10.1074/jbc.M410654200. [DOI] [PubMed] [Google Scholar]
- Hoppe A.D., Swanson J.A. Cdc42, Rac1, and Rac2 display distinct patterns of activation during phagocytosis. Mol. Biol. Cell. 2004;15:3509–3519. doi: 10.1091/mbc.E03-11-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horan K.A., Watanabe K., Kong A.M., Bailey C.G., Rasko J.E., Sasaki T., Mitchell C.A. Regulation of FcgammaR-stimulated phagocytosis by the 72-kDa inositol polyphosphate 5-phosphatase: SHIP1, but not the 72-kDa 5-phosphatase, regulates complement receptor 3 mediated phagocytosis by differential recruitment of these 5-phosphatases to the phagocytic cup. Blood. 2007;110:4480–4491. doi: 10.1182/blood-2007-02-073874. [DOI] [PubMed] [Google Scholar]
- Huang N.N., Becker S., Boularan C., Kamenyeva O., Vural A., Hwang I.Y., Shi C.S., Kehrl J.H. Canonical and noncanonical g-protein signaling helps coordinate actin dynamics to promote macrophage phagocytosis of zymosan. Mol. Cell. Biol. 2014;34:4186–4199. doi: 10.1128/MCB.00325-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara S., Oka T., Fukui Y. Direct binding of SWAP-70 to non-muscle actin is required for membrane ruffling. J. Cell Sci. 2006;119:500–507. doi: 10.1242/jcs.02767. [DOI] [PubMed] [Google Scholar]
- Jungmichel S., Sylvestersen K.B., Choudhary C., Nguyen S., Mann M., Nielsen M.L. Specificity and commonality of the phosphoinositide-binding proteome analyzed by quantitative mass spectrometry. Cell Rep. 2014;6:578–591. doi: 10.1016/j.celrep.2013.12.038. [DOI] [PubMed] [Google Scholar]
- Kanai F., Liu H., Field S.J., Akbary H., Matsuo T., Brown G.E., Cantley L.C., Yaffe M.B. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat. Cell Biol. 2001;3:675–678. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- Kimbara N., Dohi N., Miyamoto M., Asai S., Okada H., Okada N. Diagnostic surface expression of SWAP-70 on HIV-1 infected T cells. Microbiol. Immunol. 2006;50:235–242. doi: 10.1111/j.1348-0421.2006.tb03790.x. [DOI] [PubMed] [Google Scholar]
- Kjeken R., Egeberg M., Habermann A., Kuehnel M., Peyron P., Floetenmeyer M., Walther P., Jahraus A., Defacque H., Kuznetsov S.A., Griffiths G. Fusion between phagosomes, early and late endosomes: a role for actin in fusion between late, but not early endocytic organelles. Mol. Biol. Cell. 2004;15:345–358. doi: 10.1091/mbc.E03-05-0334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klippel A., Kavanaugh W.M., Pot D., Williams L.T. A specific product of phosphatidylinositol 3-kinase directly activates the protein kinase Akt through its pleckstrin homology domain. Mol. Cell. Biol. 1997;17:338–344. doi: 10.1128/mcb.17.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuiper J.W., Pluk H., Oerlemans F., van Leeuwen F.N., de Lange F., Fransen J., Wieringa B. Creatine kinase-mediated ATP supply fuels actin-based events in phagocytosis. PLoS Biol. 2008;6:e51. doi: 10.1371/journal.pbio.0060051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon M.A., Falasca M., Schlessinger J., Ferguson K. Regulatory recruitment of signalling molecules to the cell membrane by pleckstrinhomology domains. Trends Cell Biol. 1997;7:237–242. doi: 10.1016/S0962-8924(97)01065-9. [DOI] [PubMed] [Google Scholar]
- Levin R., Grinstein S., Schlam D. Phosphoinositides in phagocytosis and macropinocytosis. Biochim. Biophys. Acta. 2015;1851:805–823. doi: 10.1016/j.bbalip.2014.09.005. [DOI] [PubMed] [Google Scholar]
- Li H., Marshall A.J. Phosphatidylinositol (3,4) bisphosphate-specific phosphatases and effector proteins: A distinct branch of PI3K signaling. Cell. Signal. 2015;27:1789–1798. doi: 10.1016/j.cellsig.2015.05.013. [DOI] [PubMed] [Google Scholar]
- Li X., Wang X., Zhang X., Zhao M., Tsang W.L., Zhang Y., Yau R.G., Weisman L.S., Xu H. Genetically encoded fluorescent probe to visualize intracellular phosphatidylinositol 3,5-bisphosphate localization and dynamics. Proc. Natl. Acad. Sci. USA. 2013;110:21165–21170. doi: 10.1073/pnas.1311864110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl D., Griffiths G. Transient assembly of F-actin by phagosomes delays phagosome fusion with lysosomes in cargo-overloaded macrophages. J. Cell Sci. 2009;122:2935–2945. doi: 10.1242/jcs.048355. [DOI] [PubMed] [Google Scholar]
- Maekawa M., Terasaka S., Mochizuki Y., Kawai K., Ikeda Y., Araki N., Skolnik E.Y., Taguchi T., Arai H. Sequential breakdown of 3-phosphorylated phosphoinositides is essential for the completion of macropinocytosis. Proc. Natl. Acad. Sci. USA. 2014;111:E978–E987. doi: 10.1073/pnas.1311029111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall A.J., Krahn A.K., Ma K., Duronio V., Hou S. TAPP1 and TAPP2 are targets of phosphatidylinositol 3-kinase signaling in B cells: sustained plasma membrane recruitment triggered by the B-cell antigen receptor. Mol. Cell. Biol. 2002;22:5479–5491. doi: 10.1128/MCB.22.15.5479-5491.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May R.C., Caron E., Hall A., Machesky L.M. Involvement of the Arp2/3 complex in phagocytosis mediated by FcgammaR or CR3. Nat. Cell Biol. 2000;2:246–248. doi: 10.1038/35008673. [DOI] [PubMed] [Google Scholar]
- Missy K., Van Poucke V., Raynal P., Viala C., Mauco G., Plantavid M., Chap H., Payrastre B. Lipid products of phosphoinositide 3-kinase interact with Rac1 GTPase and stimulate GDP dissociation. J. Biol. Chem. 1998;273:30279–30286. doi: 10.1074/jbc.273.46.30279. [DOI] [PubMed] [Google Scholar]
- Murugan A.K., Ihara S., Tokuda E., Uematsu K., Tsuchida N., Fukui Y. SWAP-70 is important for invasive phenotypes of mouse embryo fibroblasts transformed by v-Src. IUBMB Life. 2008;60:236–240. doi: 10.1002/iub.33. [DOI] [PubMed] [Google Scholar]
- Oberbanscheidt P., Balkow S., Kühnl J., Grabbe S., Bähler M. SWAP-70 associates transiently with macropinosomes. Eur. J. Cell Biol. 2007;86:13–24. doi: 10.1016/j.ejcb.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Ocana-Morgner C., Wahren C., Jessberger R. SWAP-70 regulates RhoA/RhoB-dependent MHCII surface localization in dendritic cells. Blood. 2009;113:1474–1482. doi: 10.1182/blood-2008-04-152587. [DOI] [PubMed] [Google Scholar]
- Ocaña-Morgner C., Reichardt P., Chopin M., Braungart S., Wahren C., Gunzer M., Jessberger R. Sphingosine 1-phosphate-induced motility and endocytosis of dendritic cells is regulated by SWAP-70 through RhoA. J. Immunol. 2011;186:5345–5355. doi: 10.4049/jimmunol.1003461. [DOI] [PubMed] [Google Scholar]
- Ocaña-Morgner C., Götz A., Wahren C., Jessberger R. SWAP-70 restricts spontaneous maturation of dendritic cells. J. Immunol. 2013;190:5545–5558. doi: 10.4049/jimmunol.1203095. [DOI] [PubMed] [Google Scholar]
- Pearce G., Angeli V., Randolph G.J., Junt T., von Andrian U., Schnittler H.J., Jessberger R. Signaling protein SWAP-70 is required for efficient B cell homing to lymphoid organs. Nat. Immunol. 2006;7:827–834. doi: 10.1038/ni1365. [DOI] [PubMed] [Google Scholar]
- Pearce G., Audzevich T., Jessberger R. SYK regulates B-cell migration by phosphorylation of the F-actin interacting protein SWAP-70. Blood. 2011;117:1574–1584. doi: 10.1182/blood-2010-07-295659. [DOI] [PubMed] [Google Scholar]
- Riedl J., Crevenna A.H., Kessenbrock K., Yu J.H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T.A., Werb Z. Lifeact: a versatile marker to visualize F-actin. Nat. Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripich T., Jessberger R. SWAP-70 regulates erythropoiesis by controlling α4 integrin. Haematologica. 2011;96:1743–1752. doi: 10.3324/haematol.2011.050468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohatgi R., Nollau P., Ho H.Y., Kirschner M.W., Mayer B.J. Nck and phosphatidylinositol 4,5-bisphosphate synergistically activate actin polymerization through the N-WASP-Arp2/3 pathway. J. Biol. Chem. 2001;276:26448–26452. doi: 10.1074/jbc.M103856200. [DOI] [PubMed] [Google Scholar]
- Rosen S.A., Gaffney P.R., Spiess B., Gould I.R. Understanding the relative affinity and specificity of the pleckstrin homology domain of protein kinase B for inositol phosphates. Phys. Chem. Chem. Phys. 2012;14:929–936. doi: 10.1039/c1cp22240f. [DOI] [PubMed] [Google Scholar]
- Rossman K.L., Der C.J., Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat. Rev. Mol. Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Schlam D., Bagshaw R.D., Freeman S.A., Collins R.F., Pawson T., Fairn G.D., Grinstein S. Phosphoinositide 3-kinase enables phagocytosis of large particles by terminating actin assembly through Rac/Cdc42 GTPase-activating proteins. Nat. Commun. 2015;6:8623. doi: 10.1038/ncomms9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitter T., Pils S., Sakk V., Frank R., Fischer K.D., Hauck C.R. The granulocyte receptor carcinoembryonic antigen-related cell adhesion molecule 3 (CEACAM3) directly associates with Vav to promote phagocytosis of human pathogens. J. Immunol. 2007;178:3797–3805. doi: 10.4049/jimmunol.178.6.3797. [DOI] [PubMed] [Google Scholar]
- Shinohara M., Terada Y., Iwamatsu A., Shinohara A., Mochizuki N., Higuchi M., Gotoh Y., Ihara S., Nagata S., Itoh H. SWAP-70 is a guanine-nucleotide-exchange factor that mediates signalling of membrane ruffling. Nature. 2002;416:759–763. doi: 10.1038/416759a. [DOI] [PubMed] [Google Scholar]
- Shu C.L., Jing-Yang Lai, Su L.C., Chuu C.P., Fukui Y. SWAP-70: a new type of oncogene. PLoS ONE. 2013;8:e59245. doi: 10.1371/journal.pone.0059245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen A., Lippé R., Christoforidis S., Gaullier J.M., Brech A., Callaghan J., Toh B.H., Murphy C., Zerial M., Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- Sivalenka R.R., Jessberger R. SWAP-70 regulates c-kit-induced mast cell activation, cell-cell adhesion, and migration. Mol. Cell. Biol. 2004;24:10277–10288. doi: 10.1128/MCB.24.23.10277-10288.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivalenka R.R., Sinha M., Jessberger R. SWAP-70 regulates mast cell FcepsilonRI-mediated signaling and anaphylaxis. Eur. J. Immunol. 2008;38:841–854. doi: 10.1002/eji.200737597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer T.P., Ahn S., Meyer T. Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 1998;8:343–346. doi: 10.1016/s0960-9822(98)70135-6. [DOI] [PubMed] [Google Scholar]
- Swanson J.A. Shaping cups into phagosomes and macropinosomes. Nat. Rev. Mol. Cell Biol. 2008;9:639–649. doi: 10.1038/nrm2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terebiznik M.R., Vieira O.V., Marcus S.L., Slade A., Yip C.M., Trimble W.S., Meyer T., Finlay B.B., Grinstein S. Elimination of host cell PtdIns(4,5)P(2) by bacterial SigD promotes membrane fission during invasion by Salmonella. Nat. Cell Biol. 2002;4:766–773. doi: 10.1038/ncb854. [DOI] [PubMed] [Google Scholar]
- Türkoğlu R., Gencer M., Ekmekçi D., Ulusoy C., Erdağ E., Sehitoğlu E., Cavuş F., Haytural H., Küçükerden M., Yalçınkaya N. Switch-associated protein 70 antibodies in multiple sclerosis: possible association with disease progression. Med. Princ. Pract. 2014;23:239–245. doi: 10.1159/000358221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Donk N.W., Lokhorst H.M., Nijhuis E.H., Kamphuis M.M., Bloem A.C. Geranylgeranylated proteins are involved in the regulation of myeloma cell growth. Clin. Cancer Res. 2005;11:429–439. [PubMed] [Google Scholar]
- van den Bogaart G., ter Beest M. Domains of phosphoinositides in the plasma membrane. In: Cambi A., Lidke D.S., editors. Cell Membrane Nanodomains. CRC Press; 2014. pp. 173–198. [Google Scholar]
- Vonderheit A., Helenius A. Rab7 associates with early endosomes to mediate sorting and transport of Semliki forest virus to late endosomes. PLoS Biol. 2005;3:e233. doi: 10.1371/journal.pbio.0030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vural B., Demirkan A., Ugurel E., Kalaylioglu-Wheeler Z., Esen B.A., Gure A.O., Gül A., Ozbek U. Seroreactivity against PTEN-induced putative kinase 1 (PINK1) in Turkish patients with Behçet’s disease. Clin. Exp. Rheumatol. 2009;27(2, Suppl 53):S67–S72. [PubMed] [Google Scholar]
- Wakamatsu I., Ihara S., Fukui Y. Mutational analysis on the function of the SWAP-70 PH domain. Mol. Cell. Biochem. 2006;293:137–145. doi: 10.1007/s11010-006-9236-1. [DOI] [PubMed] [Google Scholar]
- Welliver T.P., Swanson J.A. A growth factor signaling cascade confined to circular ruffles in macrophages. Biol. Open. 2012;1:754–760. doi: 10.1242/bio.20121784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam P.T., Theriot J.A. Repeated cycles of rapid actin assembly and disassembly on epithelial cell phagosomes. Mol. Biol. Cell. 2004;15:5647–5658. doi: 10.1091/mbc.E04-06-0509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K., Imai K., Akamatsu A., Mihashi M., Hayashi N., Shimamoto K., Kawasaki T. SWAP70 functions as a Rac/Rop guanine nucleotide-exchange factor in rice. Plant J. 2012;70:389–397. doi: 10.1111/j.1365-313X.2011.04874.x. [DOI] [PubMed] [Google Scholar]
- Yonemura S., Matsui T., Tsukita S., Tsukita S. Rho-dependent and -independent activation mechanisms of ezrin/radixin/moesin proteins: an essential role for polyphosphoinositides in vivo. J. Cell Sci. 2002;115:2569–2580. doi: 10.1242/jcs.115.12.2569. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time-lapse epi-fluorescence imaging of dendritic cells transiently expressing SWAP70-GFP (green) and LifeAct-RFP (magenta) and pulsed with IgG-opsonized zymosan. Images were acquired every 20 sec. BF: bright field. Scale bar, 5 μm.
The animated reconstruction from Figure 1F shows concentric F-actin rings on the phagosomal surface. Dendritic cells were pulsed with zymosan (blue; autofluorescent core) and immunostained for SWAP70 (magenta). Total membranes were labeled with the fixable endocytic marker mCLING (green; Revelo et al., 2014). The bright mCLING-labeled structures surrounding the phagosome are endo/lysosomal compartments. The movies show 3D-rotations of single SWAP70-positive phagosomes. Note the alignment of SWAP70 in concentric fibers (movies 2–4) and rings (movies 5–6) on the surface of the phagosomes. The ventral membrane is positioned on the surface of the microscope cover glass; the dorsal membrane is facing away from it. Scale bars, approximately 2 μm.
The animated reconstruction from Figure 1F shows concentric F-actin rings on the pagosomal surface. Dendritic cells were pulsed with zymosan (blue; autofluorescent core) and immunostained for SWAP70 (magenta). Total membranes were labeled with the fixable endocytic marker mCLING (green; Revelo et al., 2014). The bright mCLING-labeled structures surrounding the phagosome are endo/lysosomal compartments. The movies show 3D-rotations of single SWAP70-positive phagosomes. Note the alignment of SWAP70 in concentric fibers (movies 2–4) and rings (movies 5–6) on the surface of the phagosomes. The ventral membrane is positioned on the surface of the microscope cover glass; the dorsal membrane is facing away from it. Scale bars, approximately 2 μm.
The animated reconstruction from Figure 1F shows concentric F-actin rings on the phagosomal surface. Dendritic cells were pulsed with zymosan (blue; autofluorescent core) and immunostained for SWAP70 (magenta). Total membranes were labeled with the fixable endocytic marker mCLING (green; Revelo et al., 2014). The bright mCLING-labeled structures surrounding the phagosome are endo/lysosomal compartments. The movies show 3D-rotations of single SWAP70-positive phagosomes. Note the alignment of SWAP70 in concentric fibers (movies 2–4) and rings (movies 5–6) on the surface of the phagosomes. The ventral membrane is positioned on the surface of the microscope cover glass; the dorsal membrane is facing away from it. Scale bars, approximately 2 μm.
The animated reconstruction from Figure 1F shows concentric F-actin rings on the phagosomal surface. Dendritic cells were pulsed with zymosan (blue; autofluorescent core) and immunostained for SWAP70 (magenta). Total membranes were labeled with the fixable endocytic marker mCLING (green; Revelo et al., 2014). The bright mCLING-labeled structures surrounding the phagosome are endo/lysosomal compartments. The movies show 3D-rotations of single SWAP70-positive phagosomes. Note the alignment of SWAP70 in concentric fibers (movies 2–4) and rings (movies 5–6) on the surface of the phagosomes. The ventral membrane is positioned on the surface of the microscope cover glass; the dorsal membrane is facing away from it. Scale bars, approximately 2 μm.
The animated reconstruction from Figure 1F shows concentric F-actin rings on the phagosomal surface. Dendritic cells were pulsed with zymosan (blue; autofluorescent core) and immunostained for SWAP70 (magenta). Total membranes were labeled with the fixable endocytic marker mCLING (green; Revelo et al., 2014). The bright mCLING-labeled structures surrounding the phagosome are endo/lysosomal compartments. The movies show 3D-rotations of single SWAP70-positive phagosomes. Note the alignment of SWAP70 in concentric fibers (movies 2–4) and rings (movies 5–6) on the surface of the phagosomes. The ventral membrane is positioned on the surface of the microscope cover glass; the dorsal membrane is facing away from it. Scale bars, approximately 2 μm.
Dendritic cells were pulsed with zymosan (blue; autofluorescent core) and immunostained for SWAP70 (magenta) a1nd actin (green). The movie shows a 3D-rotation of a single SWAP70-positive phagosome. Note the alignment of SWAP70 and F-actin on the surface of the phagosomes. Yellow lines indicate the orientation of F-actin and SWAP70-positive filamentous structures. Scale bar, approximately 2 μm.
3D-confocal stack of a nascent phagosome. FITC-labeled zymosan (green) was phagocytosed by dendritic cells. Uninternalized zymosan and nascent cups were labeled with an antibody directed against FITC (blue). SWAP70 was immunolabeled (magenta). The movie shows a 3D-rotation of a single SWAP70-positive nascent phagosomal cup. Scale bar, approximately 2 μm.
Time-lapse epi-fluorescence imaging of dendritic cells transiently expressing SWAP70-GFP (green) and RAB5A-RFP (magenta) and pulsed with IgG-opsonized zymosan. Images were acquired every 15 sec. BF: bright field. Scale bar, 5 μm.
Time-lapse epi-fluorescence imaging of dendritic cells transiently expressing the GFP-tagged PH-domain of TAPP2 (PLEKHA2) (binds to PI(3,4)P2; green) and SWAP70-mCherry (magenta) and pulsed with IgG-opsonized zymosan. Images were acquired every 15 sec. BF: bright field. Scale bar, 5 μm.
Time-lapse epi-fluorescence imaging of dendritic cells transiently expressing the GFP-tagged PH-domain of AKT (binds to PI(3,4,5)P3; green) and SWAP70-mCherry (magenta) and pulsed with IgG-opsonized zymosan. Images were acquired every 15 sec. BF: bright field. Scale bar, 5 μm.
Time-lapse epi-fluorescence imaging of dendritic cells transiently expressing the GFP-tagged PH-domain of PLCδ1 (PLCD1; binds to PI(4,5)P2; green) and SWAP70-mCherry (magenta) and pulsed with IgG-opsonized zymosan. Images were acquired every 15 sec. BF: bright field. Scale bar, 5 μm.
Time-lapse epi-fluorescence imaging of dendritic cells transiently expressing the GFP-tagged PX-domain of NCF4 (p40phox; binds to PI(3)P; green) and SWAP70-mCherry (magenta) and pulsed with IgG-opsonized zymosan. Images were acquired every 15 sec. BF: bright field. Scale bar, 5 μm.
Time-lapse epi-fluorescence imaging of dendritic cells transiently expressing SWAP70-GFP (green) and RAC1-RFP (magenta) and pulsed with IgG-opsonized zymosan. Images were acquired every 15 sec. BF: bright field. Scale bar, 5 μm.
Dendritic cells were pulsed with zymosan (blue; autofluorescent core) and immunostained for SWAP70 (magenta) and RAC1 (green). The movie shows a 3D-rotation of a single SWAP70-positive phagosome. Note the alignment of SWAP70 and RAC1on the surface of the phagosomes. Yellow lines indicate the orientation of SWAP70 and RAC1-positive filamentous structures. Scale bar, approximately 2 μm.
Dendritic cells were pulsed with zymosan (blue; autofluorescent core) and immunostained for RAC1 (magenta) and actin (green). The movie shows a 3D-rotation of a single SWAP70-positive phagosome. Note the alignment of RAC1and F-actin on the surface of the phagosomes. Yellow lines indicate the orientation of RAC1and F-actin-positive filamentous structures. Scale bar, approximately 2 μm.