Abstract
L27 is a protein-binding domain that can assemble essential proteins for signaling and cell polarity into complexes by interacting in a heterodimeric manner. One of these protein complexes is the PATJ/PALS1/Crumbs tripartite complex, which is crucial for the establishment and maintenance of cell polarity. To reveal the structural basis underlining the obligate heterodimerization, we have determined the crystal structure of the PALS1-L27N/PATJ-L27 heterodimer complex. Each L27 domain is composed of three helices. The two L27 domains heterodimerize by building a compact structure consisting of a four-helix bundle formed by the first two helices of each L27 domain and one coiled-coil formed by the third helix of each domain. The large hydrophobic packing interactions contributed by all the helices of both L27 domains predominantly drive the heterodimer formation, which is likely to be a general feature of L27 domains. Combined with mutational studies, we can begin to understand the structural basis for the specificity of L27 binding pairs. Our results provide unique insights into L27 domain heterodimer complex, which is critical for cell polarization.
Keywords: heterodimer, L27, PALS1, PATJ, structure
Introduction
Recently several protein complexes, for example, the Lin-2/Lin-7/Lin-10 complex (Simske et al, 1996; Borg et al, 1998; Kaech et al, 1998), the PATJ/PALS1/Crumbs complex (Knust et al, 1993; Bhat et al, 1999; Roh et al, 2002, 2003; Straight et al, 2004), and the Cdc42/Par3/Par6/aPKC complex (Joberty et al, 2000; Lin et al, 2000; Wodarz et al, 2000; Petronczki and Knoblich, 2001), have been identified as important factors in protein targeting and cell polarity. The evolutionarily conserved tripartite complexes of PATJ/PALS1/Crumbs in mammals and PATJ/Stardust/Crumbs in Drosophila are crucial for determining cell polarity (Knust et al, 1993; Bachmann et al, 2001; Hong et al, 2001; Lemmers et al, 2002; Roh et al, 2002, 2003; Straight et al, 2004). The Lin-2/Lin-7/Lin-10 proteins control the targeting of the Caenorhabditis elegans EGF receptor LET-23 to the basolateral surface, and their mammalian counterparts have been identified in the brain as possible trafficking proteins (Simske et al, 1996; Borg et al, 1998; Butz et al, 1998; Kaech et al, 1998). The structural basis of the protein–protein interactions that contribute to these complexes is poorly understood at this time.
In the Lin-2/Lin-7/Lin-10 complex and their mammalian homologs, the Lin-2 and Lin-7 proteins interact through a homologous region on both proteins designated the L27 domain after Lin-2/Lin-7. This domain exists once in Lin-7 and twice in Lin-2 (Doerks et al, 2000). The two tandem L27 domains in Lin-2 are called L27N and L27C domains. The L27 domain of Lin-7 binds to the L27C domain of Lin-2. Further studies identified multiple Lin-7 binding partners, including PALS1, PALS2, Dlg2 and Dlg3, that are similar to Lin-2 and interact with Lin-7 with like heterotypical interactions through similar domains (Butz et al, 1998; Kamberov et al, 2000). In addition, the L27N domain of mammalian Lin-2 (also known as CASK) was found to bind to a homologous region at the extreme N-terminus of SAP97 (Lee et al, 2002). To date, L27 domains have not been found to form homo-oligomers.
In the complex composed of PATJ, PALS1, and Crumbs, PALS1, which like Lin-2 contains two tandem L27 modules, interacts with PATJ through its L27N domain. PATJ is composed of a single N-terminal L27 domain (also known as the MRE domain) (Roh et al, 2002; Roh and Margolis, 2003) followed by 10 PDZ domains. It is the L27 domain of PATJ that mediates the interaction with PALS1. Crumbs, a transmembrane protein, binds to the PDZ domain of PALS1 with its cytoplasmic tail (Bachmann et al, 2001; Hong et al, 2001; Roh et al, 2002).
The L27 domains in the different proteins exhibit high diversity in terms of location, copy number, and primary sequence (Figure 1). The binding preferences are also very diversified (Table I), as supported by the finding that both the L27N and L27C domains of Dlg2 and Dlg3 were required to bind to SAP97 (Karnak et al, 2002). The primary role of L27 domains is to form and maintain multiple protein complexes.
Figure 1.
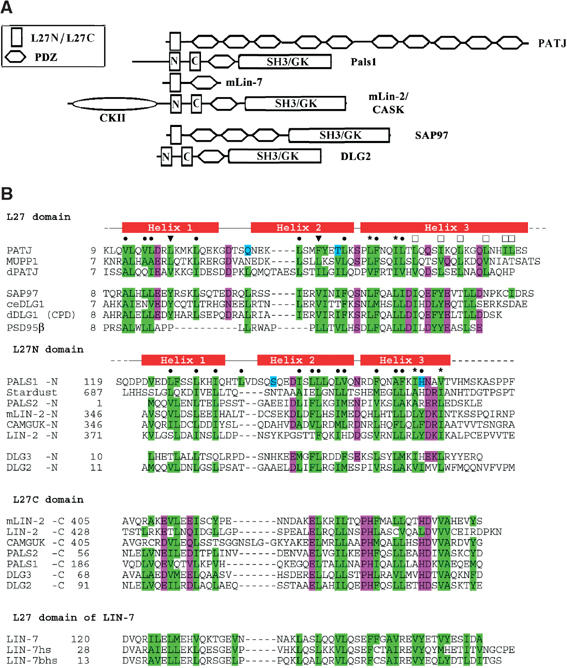
Domain structure of L27-containing proteins and sequence alignment of L27 domains. (A) The L27 domain is present in multidomain proteins such as the MAGUK proteins that contain additional protein–protein interaction modules such as the PDZ, SH3, and GK domains. (B) Structure-based sequence alignment of L27 domains. Residues involved in hydrophobic packing interactions are in a green background; other residues packing inside are in cyan; additionally conserved or highly similar residues are in magenta. Residues packing within the heterodimer interface are denoted by • (or ▾ for residues that were mutated). Residues packing in the interface between two heterodimers are labeled by □, while those residues packing into both interfaces by *.
Table 1.
Different binding types of L27 domains
| Domain 1 | Domain 2 | Reference | ||
|---|---|---|---|---|
| Type-1 binding | ||||
| PATJ | L27 | PALS1 | L27N | Roh et al (2002) |
| MUPP1 | L27 | PALS1 | L27N | Roh et al (2002) |
| DPATJ | L27 | Stardust | L27 | Roh et al (2002) |
| SAP97 | L27 | mLIN-2/CASK | L27N | Lee et al (2002) |
| PSD95β | L27 | mLIN-2/CASK | L27N | Chetkovich et al (2002) |
| PSD95β | L27 | Hrs | Coiled-coil region | Chetkovich et al (2002) |
| dDLG1-CPD | L27 | CAMGUK | L27N | Lee et al (2002) |
| dDLG1-CPD | L27 | mLIN-2 | L27N | Lee et al (2002) |
| Type-2 binding | ||||
| ceLIN-7 | L27 | ceLIN-2 | L27C | Kaech et al (1998) and Harris et al (2002) |
| mLIN-2 | L27C | Kaech et al (1998) and Harris et al (2002) | ||
| mLIN-7 | L27 | mLIN-2 | L27C | Kamberov et al (2000) |
| PALS1 | L27C | Kamberov et al (2000) | ||
| PALS2 | L27C | Kamberov et al (2000) | ||
| DLG3 | L27C | Butz et al (1998) and Kamberov et al (2000) | ||
| DLG2 | L27C | Butz et al (1998) and Kamberov et al (2000) | ||
| Type-3 binding | ||||
| SAP97 | L27 | DLG3 | Both L27N and L27C | Karnak et al (2002) |
| SAP97 | L27 | DLG2 | Both L27N and L27C | D Karnak and B Margolis (unpublished) |
| dDLG |
L27 |
DLG3 |
Both L27N and L27C |
D Karnak and B Margolis (unpublished) |
| d: Drosophila; m: mammals; ce: C. elegans. | ||||
Study of individual L27 domains revealed largely unfolded domains that require the formation of obligate heterodimers to achieve well-folded structures (Harris et al, 2002). However, how the hetero-L27 domains interact with each other and how discrimination between the different L27 domains is achieved is still unknown. We selected the L27 domain of PATJ (L27PATJ) and the L27N domain of PALS1 (L27PALS1N) as a model binding pair to study the structural basis of the heterotypical binding. We used a coexpression system to obtain the two domains and crystallized the complex. The 2.1 Å resolution crystal structure of the L27PATJ/L27PALS1N heterodimer reveals the general features of L27 domains and establishes a heterodimerization mechanism driven by hydrophobic interactions. Understanding the structural requirements that allow these proteins to form stable interactions through L27 modules could provide a means to control the formation and function of these complexes.
Results
Overall structure
The structure of the L27PATJ/L27PALS1N heterodimer complex was solved by single-wavelength anomalous dispersion (SAD) method from a selenomethionine-containing protein crystal and refined to 2.1 Å with Rfactor=24.2%/Rfree=26.2% and excellent geometry (Table II). The three-dimensional structure of one asymmetric unit in the crystal of the L27PATJ/L27PALS1N complex is illustrated in Figure 2A. The model does not contain the last 10 residues of the L27PALS1N domain or any residues from either the His tag fused to the N-terminus of L27PALS1N or the S tag fused to the C-terminus of L27PATJ.
Table 2.
Summary of data collection, phasing, and refinement statistics
| Native | Se-Met | |
|---|---|---|
| Data collection (APS-SerCAT) | ||
| Wavelength (Å) | 1.0722 | 0.97932 (Se peak) |
| Resolution limit (Å) | 2.1 | 3.5 |
| No. of measured reflections (last shella) | 97 523 (12874) | 71 697 (5795) |
| No. of unique reflections (last shell) | 25 594 (3464) | 11 212 (900) |
| Redundancy (last shell) | 3.8 (3.7) | 6.4 (6.43) |
| Completeness (last shell) (%) | 94.3 (99.1) | 99.6 (100) |
| I/sigma (last shell) | 6.95 (2.74) | 21.65 (17.0) |
| Rsym (last shell) (%)b | 11.6 (55.0) | 6.5 (8.7) |
| Space group | R32 | R32 |
| Unit cell dimensions (Å) | a=111.06 | a=110.84 |
| c=193.69 | c=194.5 | |
| No. of complexes in asymmetric unitc | 2 | 2 |
| SAD phasing | ||
| Phasing resolution (Å) | 3.5 | |
| Overall figure of meritd | 0.33/0.81 | |
| Refinement | ||
| Resolution range (Å) | 2.1 | |
| Rwork/Rfree (%) | 24.2 (26.2) | |
| No. of reflections for refinement | 24 357 (89.7%) | |
| No. of reflections for testing set | 1236 (4.6%) | |
| No. of protein residues/non-H atoms | 217/1777 | |
| No. of water molecules | 98 | |
| R.m.s.d. bond lengths (Å) | 0.0057 | |
| R.m.s.d. bond angles (deg) | 0.9184 | |
| R.m.s.d. improper angles (deg) | 0.6712 | |
| R.m.s.d. dihedral angles (deg) | 15.995 | |
| Residues in favored Ramachandran region | 98% | |
| Residues in additionally allowed region | 2% | |
| Residues in generously or disallowed region | 0 | |
| Estimated coordinate error (Å) | 0.29 | |
| Average B-factor for | ||
| Main-chain atoms | 49.9 | |
| Side-chain atoms | 54.8 | |
| Water molecules | 70.9 | |
| All atoms | 53.8 | |
| B value r.m.s.d. for | ||
| Bonded main-chain atoms | 1.689 | |
| Bonded side-chain atoms | 2.875 | |
| Angle main-chain atoms | 2.587 | |
| Angle side-chain atoms |
4.472 |
|
| a Last shell is 2.2–2.1 Å for native data and 3.6–3.5 Å for Se-Met SAD data; all reflections used. | ||
| b Rsym=∑∣I−〈I〉∣/∑I. | ||
| c Each complex has two molecules with one from PALS1-L27N and the other from PATJ-L27. | ||
| d Overall figure of merit after SOLVE/Resolve at the same resolution. | ||
Figure 2.
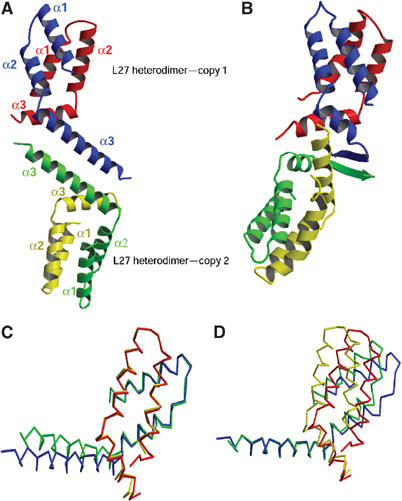
Structure of the L27PALS1N/L27PATJ heterodimer. (A) Overall structure of the two PALS1–PATJ L27 domain complexes present in the crystallographic asymmetric unit. Red/yellow: PALS1; blue/green: PATJ. (B) Ribbon diagram of the SAP97 L27 and the mLin2 L27N complex solved by NMR (Feng et al, 2004). Based on sequence alignment (Figure 1B) and domain classification, the SAP97 L27 domain is colored in blue and green, analogous to the PATJ L27 domain, whereas mLin2 is colored in red and yellow, analogous to the PALS1 L27N domain. Note the similarity in the heterodimer formation to that of the L27PALS1N/L27PATJ heterodimer, and the striking difference in the interface between the two heterodimers. (C) Overlay of the two heterodimers in the asymmetric unit based on Cα atoms belonging to Helix 1 and Helix 2 of the PATJ L27 domain. Red/yellow: PALS1; blue/green: PATJ. (D) Overlay of the two heterodimers in the asymmetric unit based on Cα atoms from Helix 3 of the PATJ L27 domain. All structural figures were generated with MOLSCRIPT (Kraulis, 1991) and RASTER3D (Merrit and Murphy, 1994).
The purified protein solution used for crystallization consisted of approximately equal amounts of the two L27 domains, as determined by SDS–PAGE and immunoblotting (data not shown). The components of the complex in our structure also have a 1:1 ratio and form heterodimers as the proto unit, as suggested previously (Doerks et al, 2000; Harris et al, 2002). The asymmetric unit is comprised of two copies of the heterodimer formed between the L27PATJ domain and the L27PALS1N domain. Each L27 domain of both PATJ and PALS1 is composed of three helices. Helix 1 and Helix 2 of each L27 domain are connected by a loop of several residues and form an antiparallel coiled-coil. Only one residue connects Helix 2 and Helix 3; this residue makes a kink between Helix 2 and Helix 3 resulting in a rotation of Helix 3 relative to Helix 2. Two notable differences are evident between Helix 3 of the two L27 domains. First is the length of this helix, which makes six turns in L27PATJ compared to only three turns in L27PALS1N. Second, the angle formed between Helix 2 and Helix 3 is different. As a result of these differences, L27PALS1N Helix 3 is situated at one end of the antiparallel coiled-coil formed by Helix 1 and Helix 2, while Helix 3 of the L27PATJ domain extends further away from its coiled-coil domain. Helix 3 of L27PATJ in each heterodimer extrudes further outside, forming a long antiparallel coiled-coil with its counterpart from another heterodimer in the asymmetric unit.
In each heterodimer, the antiparallel coiled-coils of L27PALS1N and L27PATJ pack against each other with an angle of about 45° to form a four-helical bundle. In addition, Helix 3 of L27PALS1N packs against the first part of Helix 3 of L27PATJ again with an angle of about 45° forming a short coiled-coil. The coiled-coil formed by Helix 3 of both L27PATJ and L27PALS1N covers one end of the four-helical bundle, forming a compact structure. Each of the helices in the heterodimer can interact with multiple other helices with different hydrophobic sides (i.e. within and between L27 domains). Although the four-helical bundle formed by two coiled-coils is a common folding motif that has been seen in many other structures (Gruber and Lupas, 2003), the motif composed of a four-helical bundle plus another coiled-coil packing to one end of the bundle seems to be unique. A search using the Dali Server (http://www.ebi.ac.uk) did not find homologous folding motifs of reasonable similarity to the L27 heterodimer structure, except for the recently published NMR structure (Feng et al, 2004) of the SAP97 L27/mLin2 L27N complex (Figure 2B), which became available during the revision of this manuscript.
Difference between the two heterodimers in the asymmetric unit
Comparison of the two copies of the heterodimers that make up the asymmetric unit shows that the two heterodimer complexes are very similar to each other in overall shape, but also reveals clear differences between the two in the orientation of the third helix relative to the four-helical bundle. Using 31 Cα atoms from Helix 1 and Helix 2 of L27PATJ to align the two heterodimers results in a root mean square deviation (r.m.s.d.) of only 0.51 Å. Helix 1 and Helix 2 of the two L27PATJ domains overlay very well to each other, while the long Helix 3 of L27PATJ in the two heterodimers make an angle of about 15° relative to each other (Figure 2C). Interestingly, although no atoms were chosen from L27PALS1N to calculate the superimposition, the two L27PALS1N domains also overlay very well throughout most of the three-helical structure with the exception of the latter half of the third helix, which shows minor differences. On the other hand, using 11 Cα atoms of Helix 3 of L27PATJ to superimpose the two heterodimers onto each other (r.m.s.d. 0.35 Å), as shown in Figure 2D, the four-helical bundle of the two heterodimers shows clear conformational changes as a rigid body. However, the conformations of the third helix in the two L27PALS1N domains are still quite close to each other. Thus we conclude that while both the four-helical bundle and the coiled-coil formed by the third helices of L27PALS1N and L27PATJ are quite rigid, the orientation between them has some flexibility.
Comparison between the PALS1 L27N and the PATJ L27 domains
The L27 domain is a widely used motif in different proteins and is reported to form heterodimers with specific partners (Figure 1A and Table I). According to a structure-based sequence alignment (Figure 1B), the L27 domains share similar overall structure. However, differences exist to make individual L27 domains correctly select the cognate interaction partners avoiding incorrect or self-association. Comparison between L27PATJ and L27PALS1N in the same heterodimer shows a similar topology but clear differences in overall shape (Figure 3A). One difference is the length and orientation of the third helix. L27PATJ has a much longer Helix 3 and a more open conformation in terms of the relative orientation of Helix 3 to the first two helices. When all four L27 domains from the asymmetric unit are overlaid (Figure 3C), we find that the L27PATJ domains adopt a more extended conformation (with some variation between the two copies in the asymmetric unit) while the L27PALS1N domains adopt a more compact conformation. This again suggests that Helix 3 of L27 domains has a relatively flexible orientation. Other differences between L27PALS1N and L27PATJ are the shape of the loop that connects Helix 1 and Helix 2 and the length of Helix 1 and Helix 2. These overall differences, plus the differences in side chains of the residues, could enable similar L27 domains to discriminate different L27 domains and select for the correct binding partner.
Figure 3.
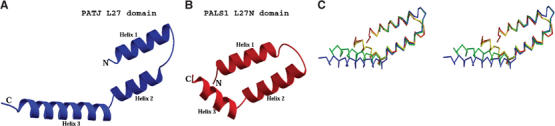
Comparison of the L27PALS1N and L27PATJ domains. Ribbon diagram of the PATJ L27 domain (A) and of the PALS1 L27 domain (B). (C) Stereoview of a Cα trace of the overlaid L27PALS1N and L27PATJ domains. Red/yellow: PALS1; blue/green: PATJ. (The red and blue belong to one heterodimer in the asymmetric unit, and yellow and green belong to the other copy.)
L27PALS1N and L27PATJ form heterodimers predominantly by hydrophobic interactions
In the heterodimer formed by L27PALS1N and L27PATJ, extensive packing interactions between hydrophobic residues from both polypeptides can be found. Almost all of the hydrophobic residues are buried either in intra- or inter-polypeptide surfaces; among the residues buried, most of them pack inside the heterodimer between L27PALS1N and L27PATJ, forming a large hydrophobic interface (Figure 4). Residues from all the six helices from both L27 domains contribute to the hydrophobic interactions that stabilize the heterodimer. The residues are well packed against each other, forming a very compact structure. Most of the hydrophobic residues seem to have a dual function, contributing both to the intramolecular coiled-coil of each protein (via interaction between the respective Helix 1 and Helix 2 of each domain) and to the intermolecular interface between the two L27 domains. Only a few buried residues are not involved in the stabilization of the heterodimer, and are either found in the coiled-coils formed between Helix 1 and Helix 2 of each domain or at the latter half of Helix 3 of L27PATJ, which is involved in the packing between two heterodimers in the asymmetric unit. This will be discussed later.
Figure 4.
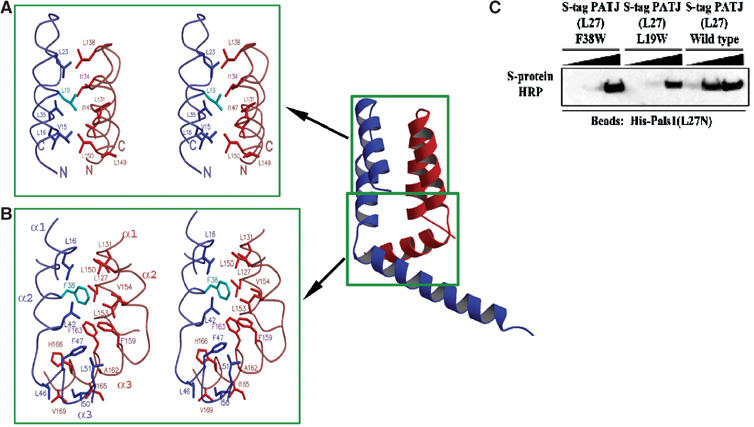
The PALS1–PATJ heterodimer is stabilized by hydrophobic interactions. (A) Stereoview of the hydrophobic packing interactions between the PATJ L27 domain (blue/cyan) and the PALS1 L27N domain (red) focusing on the section of the four-helical bundle closest to the loop that connects the first and second helices of each L27 domain. Mutated residues in PATJ are colored in cyan. (B) Similar to (A), but now the focus is on the interactions in the hydrophobic core formed at the interface of all six helices from both L27 domains. (C) Substituting for a tryptophan at position 19 or 38 of L27PATJ decreases binding to L27PALS1N. Equal amounts of His-L27PALS1N were immobilized on Ni-NTA agarose beads. The lysate from cells expressing wild-type S-tag-L27PATJ or the indicated mutated proteins was added to these beads (5, 50, or 500 μl diluted to 500 μl) and incubated for 2 h at 4°C. Beads were then washed three times in HNTG. Sample buffer was then added and precipitates were subjected to SDS–PAGE and transferred to nitrocellulose. S-tagged proteins were visualized via immunoblotting with S-protein HRP.
In addition to the hydrophobic interactions within the four-helix bundle formed by the first two helices of each domain, there is a large hydrophobic core in the heterodimer formed at the interface between the four-helical bundle and the coiled-coil consisting of the third helix of each L27 domains. Residues located in Helix 3, such as Phe159 and Phe163 of L27PALS1N, and Phe47 and Phe38 from L27PATJ, in addition to numerous other hydrophobic residues such as Leu16, Leu42, and Leu51 from L27PATJ and Leu127, Leu150, Leu127, Leu153, and Val154 from L27PALS1N come together, making extensive and continuous hydrophobic packing interactions (Figure 4B).
The hydrophobic surface area buried by the interactions between L27PALS1N and L27PATJ is approximately 2015 Å2 calculated with CNS (Brünger et al, 1998), accounting for about 51% of the total surface area of L27PALS1N alone and about 39% of the total surface area of L27PATJ alone. Such a large hydrophobic interaction holds the two L27 domains together very tightly, as also suggested by the fact that we co-purified the two using the His6 tag fused only to the PALS1 L27. The large buried hydrophobic surface also suggests that the L27 domains have to come together to form obligate dimers or maybe higher order structures to obtain a well-folded structure. This was suggested by our experiments in which either L27 domain expressed alone was insoluble or highly aggregated (data not shown).
In addition to the hydrophobic interactions in the heterodimer, we also identified several hydrogen bonds and salt bridges that are formed between the side chains of L27PALS1N and L27PATJ. Asp28, Lys26, Gln31, and Gln55 of L27PATJ form hydrogen bonds with His133, Thr137, Ser130, and Asp158 of L27PALS1N, respectively. Lys34, Arg18, and Lys58 of L27PATJ form salt bridges with Asp116, Asp140/Asp146, and Asp158 of L27PALS1N, respectively. These bonds are on the surface of the heterodimer and probably play a minor role in the formation of the heterodimer since most of these residues are not well conserved. However, they could help individual L27 domains achieve the required specificity and affinity.
L27 domains from different proteins exhibit large diversity in sequences and binding preferences as shown in Figure 1 and Table I; yet, they all have a similar array of hydrophobic residues (Figure 1B). How do they identify the correct partner among the numerous L27 domains in the cell? It is known that subtle difference in the array as well as the type and number of buried hydrophobic residues could lead to significant differences in the length and shape of the coiled-coils formed (Gruber and Lupas, 2003). The resulting, albeit small difference in the binding surfaces can be exploited to achieve the required specificities and affinities. To test this, we mutated singly Leu19 and Phe38 of L27PATJ to a tryptophan, which would presumably cause steric hindrance in the interface between the two L27PALS1N and L27PATJ in the heterodimer (Figure 4A and B). As expected, the binding affinity of these mutants to L27PALS1N was greatly reduced (Figure 4C).
Interactions between the two heterodimers in the asymmetric unit
In our structure there are two L27PALS1N/L27PATJ heterodimers in the asymmetric unit (Figure 2A). However, it has been reported by Harris et al (2002) that the L27 domains form a simple heterodimer with 1:1 stoichiometry in solution, not 2:2 tetramer. This caused us to examine the relevance of the interaction between the two heterodimers in the structure. In the asymmetric unit, Helix 3 of two L27PATJ domains forms a long antiparallel coiled-coil. Seven hydrophobic residues (excluding Leu68 of L27PATJ, which is from the cloning vector) from each L27PATJ participate in this coiled-coil (Figure 5A and B). Notably, four of the seven hydrophobic residues are conserved in PATJ-like Type-1 L27/MRE domains according to our structured-based sequence alignment (Figure 1B). No relevant interactions exist between PALS1 of one heterodimer and PATJ from the second heterodimer in the asymmetric unit. There are no hydrogen bonds or salt bridges found between the two heterodimers in the crystal asymmetric unit. In total, a significant surface area (1469 Å2) is buried between the two heterodimers in the asymmetric unit.
Figure 5.
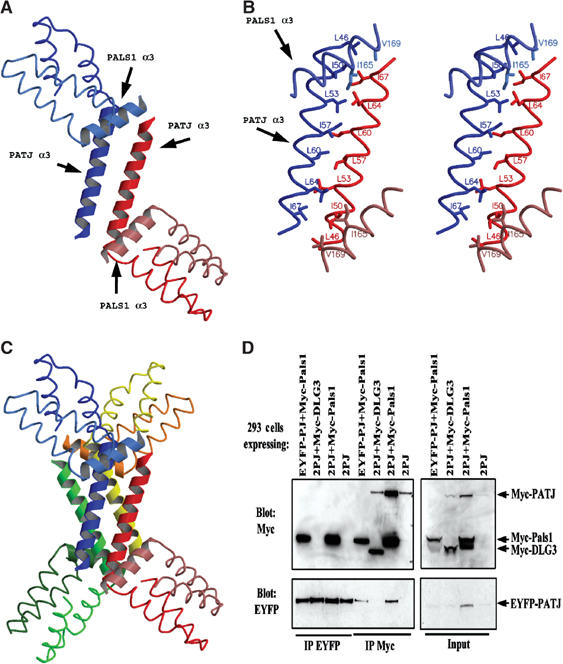
Analysis of possible higher oligomeric states formed by L27 domains. (A) The third helix of each PATJ L27 domain in the asymmetric unit interacts with each other through a long coiled-coil. One heterodimer is colored in red and the other in blue. Note, the coloring scheme in this figure is different from that used in the previous figures. (B) Stereoview showing the detailed interactions between the two L27PATJ domains in the asymmetric unit. (C) The four-helical bundle formed between two asymmetric units. (D) The L27PATJ domain does not dimerize in the presence or absence of Myc-L27PALS1N. Lysates from 293 cells expressing the indicated proteins were subjected to immunoprecipitations with the indicated antibodies (2PJ=Myc-PATJ+EYFP-PATJ). The resultant immunoprecipitates were resolved by SDS–PAGE and transferred to nitrocellulose. Membranes were first immunoblotted with anti-Myc antibodies, stripped, and then immunoblotted with anti-EYFP antibodies. Input represents 5% of the material used in the immunoprecipitations.
Further examination of the packing sites in the crystal revealed a large four-helical bundle formed between two asymmetric units (Figure 5C). This provides the largest crystal-packing surface among all the contact sites. This four-helical bundle is composed solely of Helix 3 from four L27PATJ domains, which belong to two asymmetric units, that is, the long coiled-coil formed between the third helix of each L27PATJ from one asymmetric packs against another from a second asymmetric unit in a back-to-back manner. However, no hydrophobic residues, except Leu60 of each L27PATJ, which points to the center of the four-helix bundle, are involved in the interface between the two long coiled-coils. This helical bundle is maintained by numerous hydrogen bonds between the two coiled-coils and each helix from one coiled-coil interacts with the two helices in the second coiled-coil.
The above crystal-packing analysis supports a model in which two heterodimers form a dimer-of-dimers via hydrophobic residues contained in Helix 3 of PATJ. The larger complex composed of four heterodimers is less likely to be physiologically relevant. To investigate the possibility of a higher oligomeric complex, we utilized two different tags on L27PATJ and tested for their interaction in the presence or absence of L27PALS1N. As shown in Figure 5D, no such interaction could be detected. In this experiment, Myc-PATJ and EYFP PATJ were coexpressed but did not co-immunoprecipitate. In contrast, Myc-PALS1 co-immunoprecipitated with EYFP-PATJ. Thus, in this experiment we could detect heterodimerization between PALS1 and PATJ but not the homointeraction between two PATJ molecules.
To obtain more conclusive evidence for the nature of higher order complex formation, we performed analytical ultracentrifugation sedimentation equilibrium analysis. Multiple models were attempted to fit the experimental data, including a single ideal species (variance: 1.239 × 10−4), a two-component, noninteracting species model (variance: 0.206 × 10−4), and a reversibly associating monomer–dimer system (variance: 0.289 × 10−4). The excellent variance results for both the two-component and monomer–dimer models on the one hand, and the poor variance of the single ideal species model indicates the presence of multiple components. Because of the additional fitting parameters and only a marginally better variance in the two-component model compared to the monomer–dimer model, we feel that the monomer–dimer equilibrium model best describes the data. Monte Carlo analysis of the monomer–dimer fit resulted in a molecular weight of 15 492 kDa and a dissociation constant of 6.99 μM. The determined monomer molecular weight is in good agreement with the theoretical molecular weight of 17.5 kDa determined for the heterodimer. Plots of the residuals and overlays for the monomer–dimer fit, and of the relative concentration distribution of monomer and dimer at different total concentrations are shown in Supplementary Figures 1 and 2.
Discussion
Establishment and maintenance of cell polarity are very important for cell differentiation, development, and cell function. The mammalian PALS1 and its homolog Drosophila Stardust protein are crucial for the establishment of polarized epithelia (Bachmann et al, 2001; Hong et al, 2001; Straight et al, 2004). PALS1 forms a complex with two other proteins, the PDZ domain protein PATJ and the apical transmembrane protein Crumbs (Bachmann et al, 2001; Hong et al, 2001; Roh et al, 2002; Makarova et al, 2003). In the tripartite complex, PALS1 functions as an adaptor, recruits Crumbs and PATJ by using its PDZ domain to bind the cytoplasmic tail of Crumbs and its L27N domain to bind the L27/MRE domain of PATJ (Lemmers et al, 2002; Roh et al, 2002). L27 domains have been established as a protein-binding module that brings multiple proteins into complexes, for example, the Lin-2/Lin-7/Lin-10 complex in C. elegans, by forming heterodimers with each other (Doerks et al, 2000; Harris et al, 2002). Visualization of the interaction between PALS1 and PATJ through the L27 domains thus will provide clear understanding of the nature of binding and could suggest possible ways to further investigate the functions of individual proteins in this complex as well as the relationship between them in the context of cell polarity. Moreover, it could also provide us with the important recognition rules that govern the interaction between the large numbers of the different L27 domains in the cell (Doerks et al, 2000).
Based on the nature of proteins containing the L27 domains and also a sequence alignment, we divided the L27 domains into four categories as shown in Figure 1B. Proteins like PATJ or SAP97 that possess only a single L27 domain at the extreme N-terminus comprise the first group. The second and third categories belong to MAGUK proteins such as Lin-2 or PALS1 that have tandem L27 domains, designated L27N and L27C, respectively (Doerks et al, 2000). The fourth group of L27 domains comprises the L27 domain in the Lin-7 protein and its homologs. In each category, the distribution of the hydrophobic residues is highly similar indicating a comparable three-dimensional structure. In other words, we predict that the structures of L27 domains within a category will be more similar to each other than to those L27 domains of different categories.
When analyzing the complexes formed by L27/L27 domain interactions rather than the individual domains, it is useful to segregate the complexes into three types according to their binding preferences. Type-1 L27 heterodimers are formed by L27 domains from categories I and II, excluding Dlg2 and Dlg3. Examples would be the complexes formed by PATJ and PALS1, or SAP97 and Lin-2. It is the N-terminal module of category II proteins that takes part in Type-1 interactions. In contrast, in Type-2 binding, it is the C-terminal L27 module (category III domains) that participates in binding L27 domains of category IV. An example of a Type-2 complex is Lin-7/Lin-2. The third type of binding, Type-3, requires the participation of both L27N and L27C for complex formation. Here examples include the interaction of SAP97 with both of the L27 domains of Dlg2 or Dlg3.
Based on the sequence alignment, our structure of the PALS1–PATJ L27 complex suggests a general mechanism for how L27 domains bind each other. The L27 domains all have a similar array of hydrophobic residues. Each L27 domain should consist of three helices, the first two of which form an antiparallel coiled-coil. Two L27 domains come together to form a four-helical bundle with the antiparallel coiled-coils formed by the first two helices. The third helix of each domain forms another coiled-coil packing at one end of the four-helix bundle, creating a large hydrophobic interface. The hydrophobic interactions are the major force that drives heterodimer formation. This mechanism also explains the linked folding and binding for the L27 domains as previously suggested by Harris et al (2002). An exception to this general mechanism is that both the L27N domain and L27C domain are required for Dlg3 and Dlg2 to bind to the L27/MRE domain of SAP97 (Karnak et al, 2002). It is possible that the three L27 domains could form a six-helical bundle or that each of the L27N or L27C domains contributes one helix and thus forms a similar four-helical bundle with the SAP97 L27 domain. This will require further investigation.
L27 domain heterodimerization requires a matched surface since displacement of one residue in the hydrophobic core with a larger or smaller residue could lead to either steric hindrance or less compact packing lowering binding affinities. For example, SAP97 and its homologs all have a Tyr18, while the corresponding residue in PATJ is Leu19. Our structure shows that a tyrosine is too large to be accommodated in PALS1, and this could account for the lack of observed binding between SAP97 and PALS1. Also as expected, mutation of Leu19 to tryptophan significantly decreased the binding between PALS1 and PATJ (Figure 4C). This could provide a novel mechanism used by evolution to develop a vast number of specific binding pairs with limited mutation of the primary sequence.
Additionally, hydrogen bonds and salt bridges formed between binding pairs could provide one more way of adjusting binding affinity and specificity. For example, the single mutation D225N in PALS1 L27C domain abrogates binding between PALS1 and mLin-7 (Kamberov et al, 2000). Although these interactions are not the major force in dimer formation of our structure, they could provide subtle differences between different L27 domains and provide additional regulatory mechanisms by modulating binding affinities.
The small L27 domains interact with other L27 domains to organize large protein complexes (Roh et al, 2002). It will be interesting to determine if L27 domain proteins can form higher order complexes using the heterodimer as a proto unit. The interactions between two heterodimers that compose the asymmetric unit suggest how a dimer-of-heterodimers can be formed (Figure 5A and B). Higher order oligomerization could also be formed as suggested by the crystal packing (Figure 5C). However, co-immunoprecipitation assays using mammalian cell lysates failed to detect a higher order unit than the basic heterodimer (Figure 5D). By analytical ultracentrifugation, an equilibrium between the basic heterodimer unit and a dimer-of-heterodimers with a kDa of 7 μM was observed. This suggests that the formation of the dimer-of-heterodimers is protein concentration dependent, and could explain the fact that we failed to detect an interaction between heterodimers in the co-immunoprecipitation assay. In addition, the detergent required for cell lysis may have disrupted the interaction during the co-immunoprecipitation experiments. In contrast, the high protein concentration used in the crystallization of the L27 domains could act to promote the formation of the dimer-of-heterodimers. It should be noted that high local concentrations of PATJ may be achieved at condensed tight junctions and thus the dimer-of-heterodimers may contribute to the avidity of tight junction protein interactions.
During the preparation of this manuscript, the NMR structure of the L27 heterodimer formed by SAP97 and Lin-2 was reported (Feng et al, 2004). Despite the overall similarity in the overall topology in the heterodimers, there are several notable differences between our PALS1–PATJ complex and the SAP97–Lin-2 complex (Figure 2A and B). At the level of individual domains, there are clear differences in the length and orientation of helices. As a result, the superposition of the PATJ L27 domain and the SAP97 L27 domain results in a significant r.m.s.d. of 4.7 Å for 41 Cα atoms. Overlay of PALS1 L27N and mLin-2 L27N domains is not meaningful due to substantial structural differences, such as the curvature of Helix 1 in mLin-2 L27N that is not observed in its counterpart in PALS1. In addition, while we observe a single long Helix 3 in L27PATJ, in the structure by Feng et al the corresponding helix in the SAP97 L27 domain is much shorter and is followed by a β-strand. A further important difference between the NMR and X-ray structures is the variability of the angles between Helix 2 and Helix 3 only observed in our structure. This apparent flexibility between these helices is notable when comparing the two L27PATJ domains in our asymmetric unit (Figure 2C). Since in the NMR structure the two heterodimers were treated as identical molecules, this phenomenon was not observed.
The most striking difference between the two L27 complex structures is observed in the interface between two heterodimers. Whereas in the L27PATJ/L27PALS1N complex the formation of a dimer-of-heterodimers is solely mediated by hydrophobic residues in Helix 3 of two PATJ L27 domains (Figure 5A and B), in the SAP97/mLin-2 L27 complex the dimer-of-heterodimer interaction is mediated by four helices from two heterodimers, with each L27 contributing its third helix; additionally, no direct interactions were observed between the third helices of the two SAP97 L27 or the mLin-2 L27 domains (Feng et al, 2004). The differences between our results and those by Feng et al can be due to the construct used in the NMR study. While we coexpressed each L27 domain as individual polypeptides, Feng et al used a construct in which the two L27 domains were tethered into a single chain. This construct could have resulted in the destruction of the latter half of Helix 3 of the SAP97 L27 domain and the artificial formation of the β-strand in its place. This could also be the cause for the different relative orientation between the two heterodimers in our crystal structure and their NMR structure, as shown in Figure 2A and B.
A second important difference between our results and those reported by Feng et al concerns the stability of the dimer-of-heterodimers. Feng et al (2004), based on their structure and analytical ultracentrifugation data, conclude that the formation of the dimer-of-heterodimers is ‘structurally indispensable for L27 domain-mediated protein assembly'. However, our results question this generalization. Analytical ultracentrifugation data indicate an equilibrium between individual heterodimers and a dimer-of-heterodimers. These data support a model in which the dimer-of-heterodimers can form but is only of moderate-to-weak stability. The discrepancy between our data and that by Feng et al can be explained by the much higher concentrations used in their ultracentrifugation experiments in comparison to those we used. It is possible that the use of the tethered construct by Feng et al also contributed to the stable dimer-of-heterodimers they observed.
In conclusion, our 2.1 Å crystal structure of the PATJ L27 domain and PALS1 L27N domain complex provides a clear and precise model for one of the L27 domain binding pairs. The demonstration of the general rules for binding and special residues for specificity could help greatly in investigation of the functions of the PALS1/PATJ/Crumbs tripartite complex in cell polarity.
Materials and methods
Protein expression and purification
PCR products coding for the L27N domain of mouse PALS1 (amino acids 123–180) and the L27 domain of human PATJ (amino acids 9–67) were subcloned into pSJ7(NusA) and pACYC-Duet-1 (Novagen) by standard methods. The L27 domain of either PALS1 or PATJ was not soluble when expressed individually in bacteria. When expressed as NusA fusion proteins, both L27PALS1N and L27PATJ were soluble; however, the fusion proteins readily precipitated upon removal of the NusA tag (data not shown). Therefore, a coexpression vector was constructed using pACYCDuet-1. The proteins were coexpressed as two separate polypeptides with a His6 tag fused at the N-terminus of L27PALS1N and an S tag (15 residues) fused at the C-terminus of L27PATJ. BL21 (DE3) cells harboring the resulting vector were grown at 37°C and induced with IPTG for 6 h. The soluble coexpressed proteins were purified to high purity by taking advantage of the His tag fused to L27PALS1N domain employing an Ni-NTA agarose column. Further purification was performed with a Superdex-75 gel filtration column. The purity of the protein complex was verified, and existence of both polypeptides at approximately equal amount was confirmed by SAS–PAGE and immunoblotting with antibodies against the His and S tags, respectively. The selenomethionine-incorporated protein complex was expressed following the methionine pathway inhibition procedures (Doublie, 1997) and purified as the native proteins except that 5 mM DTT was added to protect selenomethionine after the Ni-NTA chromatography. For all the proteins purified, no attempt was made to remove the fusion tags.
Crystallization and data collection
The protein solution was kept in 25 mM Tris–HCl (pH 7.5) and 100 mM NaCl at 5.5 mg/ml at −80°C before crystallization. The detergent CHAPS was added to the protein solution at a final concentration of 8 mM before crystallization. Crystals were grown by mixing 1 μl of protein solution and 1 μl of reservoir solution containing 2.5 M ammonium sulfate (pH 5.6) and 10% glycerol and equilibrated against the 1 ml reservoir at 20°C by the hanging-drop method. Without CHAPS, the crystals were highly twinned. Crystals were frozen in liquid nitrogen with cryo-oil (Hampton Research) as cryoprotectant. X-ray data were collected at the Advanced Photon Source, Argonne National Laboratories, using the ID beamline of SER-CAT. A single-wavelength data set to 2.6 Å was collected at the peak of absorption for selenium from a selenomethionine crystal and a native data set to 2.1 Å. The data sets were processed with XDS (Kabsch, 1993). The crystals belong to the space group R32 with four molecules (two heterodimer complexes) per asymmetric unit. Data collection and processing statistics are provided in Table II.
Crystallographic structure solution and refinement
The structure solution was obtained by using the data set collected from a single selenomethionine derivative crystal using the SAD method with SOLVE/RESOLVE package (Terwilliger and Berendzen, 1999). The highest resolution of initial phasing was good to 3.5 Å. The map showed very clear electron density for multiple α-helices packing against each other. Phasing at or extending the phase with density modification to higher resolution gave worse maps. A model for the main-chain atoms was built with ARP (Perrakis et al, 1999) by combining the phase at 3.5 Å and the 2.6 Å data set. Further model building was carried out with O (Jones et al, 1991) and refinement with CNS (Brünger et al, 1998). Correct tracing was facilitated by the positions of the selenium sites in the L27PATJ domain found with SOLVE and the predicted similar coiled-coil α-helical structure between the L27PATJ domain and L27PALS1N domain according to the sequence alignment. The partially refined model from selenomethionine protein was then used to refine against the 2.1 Å native data set. Several cycles of torsion angle dynamics, least squares minimization, individual B-factor refinement alternated with manual rebuilding, and addition of water molecules yielded an Rfactor=24.2% and Rfree=26.2%. The final model contains two copies of the L27PATJ/L27PALS1N heterodimer complexes and 98 water molecules. In one copy, the L27PATJ domain contains residues 12–67 and two additional residues at the C-terminus from the cloning vector and the L27PALS1N domain contains residues 120–170 and three additional residues at the N-terminus from the vector. In the second copy, the L27PATJ domain contains residues 10–67 and two additional residues at the C-terminus from the vector and the L27PALS1N domain contains residues 123–170. All the other residues, including the His6 and S tags had no observable electron density. PROCHECK (Laskowski et al, 1993) revealed that 98% residues are within the favored and 2% are in additionally allowed regions. Statistics of the structure determination and refinement are presented in Table II.
Protein binding assay
Construction of plasmids coding for Myc-PATJ, EYFP-PATJ, Myc-PALS1, and Myc-DLG3 has been reported elsewhere (Borg et al, 1998; Kamberov et al, 2000; Karnak et al, 2002; Roh et al, 2002). Plasmids were transformed into BL21DE3 and lysed by the lysozyme method as described previously (Borg et al, 1998). Crude lysate aliquots were collected before pelleting the lysate. Pellet proteins were solubilized in sample buffer and boiled for 15 min prior to SDS–PAGE. Hexahistidine-tagged proteins were purified on Ni-NTA agarose beads from lysate supplemented with imidazole at a final concentration of 50 mM due to excessive background binding at lower imidazole concentrations. Beads were washed three times in buffer containing 10 mM potassium monophosphate, pH 7.8, 300 mM sodium chloride, 50 mM imidazole, 8% glycerol, and 0.2% Triton X-100. S-tagged proteins were purified on S-protein agarose beads and washed three times in HNTG (50 mM HEPES, pH 7.5, 150 mM NaCl, 10% glycerol, and 0.1% Triton X-100). Following SDS–PAGE, proteins were either transferred to nitrocellulose or visualized using Simply BlueTM Safestain (Invitrogen). His-tagged proteins were visualized by immunoblotting with monoclonal anti-His tag antibodies (Novagen). S-tagged proteins were visualized with S-protein HRP (Novagen).
HEK293 cells were transfected with these plasmids using Fugene 6 transfection reagent (Roche) according to the manufacturer's protocols. Cells were lysed as previously described using 1% Triton X-100-based lysis buffer (Borg et al, 1998). Myc-tagged proteins were immunoprecipitated with mouse monoclonal anti-Myc(9E10) and immunoblotted with mouse monoclonal anti-Myc(4A6). EYFP-tagged proteins were immunoprecipitated and immunoblotted with pan-XFP full-length A.v polyclonal antibody (BD Living Colors, catalog number 8372-2, Clontech). Where indicated, nitrocellulose membranes were stripped in buffer contained 2% SDS and 20 mM Tris (pH 6.7) for 30 min at 55°C. After extensive washing in Tris-buffered saline (TBS) containing 0.1% Triton X-100, and regular TBS, membranes were reprobed.
Analytical ultracentrifugation
All sedimentation equilibrium experiments were performed with a Beckman Optima XL-A at the Keck Biophysics Facility at Northwestern University. Equilibrium and Monte Carlo analyses were performed with UltraScan version 6.2 (http://www.ultrascan.uthscsa.edu). All samples were analyzed in a buffer containing 20 mM potassium phosphate buffer (pH 7.2) and 100 mM NaCl. Sedimentation equilibrium experiments were performed at 20°C and five speeds (16, 23.2, 30.5, 37.7, and 50 krpm). Multiple loading concentrations ranging between 0.3 and 0.7 OD were measured at the given wavelength, data exceeding 0.9 OD were excluded from the fit. Using the globally fitted extinction profile, the extinction coefficients were found to be E280=1280 OD M−1 cm−1, E230=53 058.6 OD M−1 cm−1, and E220=158 243 OD M−1 cm−1. The extinction values determined in this fashion were then used to convert optical densities to molar concentration units at the measured wavelengths. Data in the concentration range between 0 and 16 μM were examined. Data were fitted to multiple models. The most appropriate model was chosen based on visual inspection of the residual run patterns, and based on the best statistics.
Supplementary Material
Supplementary Figures
Acknowledgments
This study was supported by National Institute of Diabetes and Digestive and Kidney grants DK061397 to YL and AL and DK58208 to BM. BM is an investigator of the Howard Hughes Medical Institute. Data were collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences, under contract no. W-31-109-Eng-38. Model and structure factors have been deposited with PDB code 1VF6. The development of the UltraScan software is supported by the National Science Foundation through grant DBI-9974819 to BD. We thankfully acknowledge the use of instruments in the Keck Biophysics Facility at Northwestern University (http://www.biochem.northwestern.edu/Keck/keckmain.html).
References
- Bachmann A, Schneider M, Theilenberg E, Grawe F, Knust E (2001) Drosophila Stardust is a partner of Crumbs in the control of epithelial cell polarity. Nature 414: 638–643 [DOI] [PubMed] [Google Scholar]
- Bhat MA, Izaddoost S, Lu Y, Cho KO, Choi KW, Bellen HJ (1999) Discs Lost, a novel multi-PDZ domain protein, establishes and maintains epithelial polarity. Cell 96: 833–845 [DOI] [PubMed] [Google Scholar]
- Borg JP, Straight SW, Kaech SM, de Taddeo-Borg M, Kroon DE, Karnak D, Turner RS, Kim SK, Margolis B (1998) Identification of an evolutionarily conserved heterotrimeric protein complex involved in protein targeting. J Biol Chem 273: 31633–31636 [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang J-S, Kuszewskli J, Nilges N, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography and NMR system (CNS): a new software system for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Butz S, Okamoto M, Sudhof TC (1998) A tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Cell 94: 773–782 [DOI] [PubMed] [Google Scholar]
- Chetkovich DM, Bunn RC, Kuo SH, Kawasaki Y, Kohwi M, Bredt DS (2002) Postsynaptic targeting of alternative postsynaptic density-95 isoforms by distinct mechanisms. J Neurosci 22: 6415–6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerks T, Bork P, Kamberov E, Makarova O, Muecke S, Margolis B (2000) L27, a novel heterodimerization domain in receptor targeting proteins Lin-2 and Lin-7. Trends Biochem Sci 25: 317–318 [DOI] [PubMed] [Google Scholar]
- Doublie S (1997) Preparation of selenomethionyl proteins for phase determination. In Methods in Enzymology, Carter CW, Sweet RM (eds) Vol. 276, pp 461–472. San Diego: Academic Press [PubMed] [Google Scholar]
- Feng W, Long JF, Fan JS, Suetake T, Zhang M (2004) The tetrameric L27 domain complex as an organization platform for supramolecular assemblies. Nat Struct Mol Biol 11: 475–480 [DOI] [PubMed] [Google Scholar]
- Gruber M, Lupas AN (2003) Historical review: another 50th anniversary—new periodicities in coiled coils. Trends Biochem Sci 28: 679–685 [DOI] [PubMed] [Google Scholar]
- Harris BZ, Venkatasubrahmanyam S, Lim WA (2002) Coordinated folding and association of the LIN-2, -7 (L27) domain. An obligate heterodimerization involved in assembly of signaling and cell polarity complexes. J Biol Chem 277: 34902–34908 [DOI] [PubMed] [Google Scholar]
- Hong Y, Stronach B, Perrimon N, Jan LY, Jan YN (2001) Drosophila Stardust interacts with Crumbs to control polarity of epithelia but not neuroblasts. Nature 414: 634–638 [DOI] [PubMed] [Google Scholar]
- Joberty G, Petersen C, Gao L, Macara IG (2000) The cell-polarity protein Par6 links Par3 and atypical protein kinase C to Cdc42. Nat Cell Biol 2: 531–539 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zhou J-Y, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Crystallogr 24: 795–800 [Google Scholar]
- Kaech SM, Whitfield CW, Kim SK (1998) The LIN-2/LIN-7/LIN-10 complex mediates basolateral membrane localization of the C. elegans EGF receptor LET-23 in vulval epithelial cells. Cell 94: 761–771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamberov E, Makarova O, Roh M, Liu A, Karnak D, Straight S, Margolis B (2000) Molecular cloning and characterization of Pals, proteins associated with mLin-7. J Biol Chem 275: 11425–11431 [DOI] [PubMed] [Google Scholar]
- Karnak D, Lee S, Margolis B (2002) Identification of multiple binding partners for the amino-terminal domain of synapse-associated protein 97. J Biol Chem 277: 46730–46735 [DOI] [PubMed] [Google Scholar]
- Knust E, Tepass U, Wodarz A (1993) Crumbs and stardust, two genes of Drosophila required for the development of epithelial cell polarity. Dev Suppl 261–268 [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Lee S, Fan S, Makarova O, Straight S, Margolis B (2002) A novel and conserved protein–protein interaction domain of mammalian Lin-2/CASK binds and recruits SAP97 to the lateral surface of epithelia. Mol Cell Biol 22: 1778–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmers C, Medina E, Delgrossi MH, Michel D, Arsanto JP, Le Bivic A (2002) hINADl/PATJ, a homolog of discs lost, interacts with crumbs and localizes to tight junctions in human epithelial cells. J Biol Chem 277: 25408–25415 [DOI] [PubMed] [Google Scholar]
- Lin D, Edwards AS, Fawcett JP, Mbamalu G, Scott JD, Pawson T (2000) A mammalian PAR-3–PAR-6 complex implicated in Cdc42/Rac1 and aPKC signalling and cell polarity. Nat Cell Biol 2: 540–547 [DOI] [PubMed] [Google Scholar]
- Makarova O, Roh MH, Liu CJ, Laurinec S, Margolis B (2003) Mammalian Crumbs3 is a small transmembrane protein linked to protein associated with Lin-7 (Pals1). Gene 302: 21–29 [DOI] [PubMed] [Google Scholar]
- Merrit EA, Murphy MEP (1994) Raster3D Version 2.0—a program for photorealistic molecular graphics. Acta Crystallogr D 50: 869–873 [DOI] [PubMed] [Google Scholar]
- Perrakis A, Morris R, Lamzin VS (1999) Automated protein model building combined with iterative structure refinement. Nat Struct Biol 6: 458–463 [DOI] [PubMed] [Google Scholar]
- Petronczki M, Knoblich JA (2001) DmPAR-6 directs epithelial polarity and asymmetric cell division of neuroblasts in Drosophila. Nat Cell Biol 3: 43–49 [DOI] [PubMed] [Google Scholar]
- Roh MH, Fan S, Liu CJ, Margolis B (2003) The Crumbs3–Pals1 complex participates in the establishment of polarity in mammalian epithelial cells. J Cell Sci 116: 2895–2906 [DOI] [PubMed] [Google Scholar]
- Roh MH, Makarova O, Liu CJ, Shin K, Lee S, Laurinec S, Goyal M, Wiggins R, Margolis B (2002) The Maguk protein, Pals1, functions as an adapter, linking mammalian homologues of Crumbs and Discs Lost. J Cell Biol 157: 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh MH, Margolis B (2003) Composition and function of PDZ protein complexes during cell polarization. Am J Physiol Renal Physiol 285: F377–F387 [DOI] [PubMed] [Google Scholar]
- Simske JS, Kaech SM, Harp SA, Kim SK (1996) LET-23 receptor localization by the cell junction protein LIN-7 during C. elegans vulval induction. Cell 85: 195–204 [DOI] [PubMed] [Google Scholar]
- Straight SW, Shin K, Fogg VC, Fan S, Liu CJ, Roh M, Margolis B (2004) Loss of PALS1 expression leads to tight junction and polarity defects. Mol Biol Cell 15: 1981–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J (1999) Automated structure solution for MIR and MAD. Acta Crystallogr D 55: 849–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodarz A, Ramrath A, Grimm A, Knust E (2000) Drosophila atypical protein kinase C associates with Bazooka and controls polarity of epithelia and neuroblasts. J Cell Biol 150: 1361–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures


