Abstract
In Drosophila, dosage compensation is controlled by the male-specific lethal (MSL) complex consisting of at least five proteins and two noncoding RNAs, roX1 and roX2. The roX RNAs function in targeting MSL complex to the X chromosome, and roX transgenes can nucleate spreading of the MSL complex into flanking chromatin when inserted on an autosome. An MSL-binding site (DHS, DNaseI hypersensitive site) has been identified in each roX gene. Here, we investigate the functions of the DHS using transgenic deletion analyses and reporter assays. We find that MSL interaction with the DHS counteracts constitutive repression at roX1, resulting in male-specific expression of roX1 RNA. Surprisingly, the DHS is not required for initiation of cis spreading of MSL complex, instead local transcription of roX RNAs correlates with extensive spreading.
Keywords: dosage compensation, noncoding RNA, transcriptional regulation
Introduction
Transcription in eukaryotes utilizes at least two levels of regulation: gene-specific control that operates locally on individual genes and global modulation of larger domains by chromatin composition or remodeling. Dosage compensation is an example of interplay between these two regulatory mechanisms that has evolved to make X-linked gene expression equivalent in males with one X chromosome and females with two. In Drosophila, dosage compensation is achieved by increasing the transcription of most X-linked genes two-fold in males (Lucchesi, 1998; Meller and Kuroda, 2002). This requires at least five proteins: MLE (maleless), MSL1, MSL2 and MSL3 (male-specific lethal 1, 2 and 3, respectively), and MOF (males absent on the first), and one of two roX (RNA on X) RNAs. MLE and MOF have enzymatic activities that are essential for dosage compensation: MLE is a DExH RNA helicase (Kuroda et al, 1991; Lee et al, 1997) and MOF is a MYST family histone acetyltransferase (Hilfiker et al, 1997; Akhtar and Becker, 2000; Smith et al, 2000). JIL-1, a histone H3 kinase, also associates with the MSL proteins (Jin et al, 2000). The MSL proteins, JIL-1 and roX RNAs bind in a precise pattern along the length of the male X chromosome, resulting in enrichment of chromatin modifications associated with hypertranscription, such as histone H4 acetylated at lysine 16 and H3 phosphorylated at serine 10 (Turner et al, 1992; Wang et al, 2001).
The two noncoding RNAs, roX1 and roX2, are functionally redundant (Meller and Rattner, 2002) even though they have very little sequence homology and are distinct in size (3.7 kb for roX1 RNA versus 0.5–1.2 kb for roX2 RNA) (Amrein and Axel, 1997; Smith et al, 2000). Deletion of either roX gene has no effect on males. Missing both of them, however, results in male lethality (Meller et al, 1997; Meller and Rattner, 2002). The MSL-binding pattern on the X chromosome is drastically disrupted in these roX1roX2 double mutant males, suggesting that roX RNAs are important for correctly targeting MSL complex to the X chromosome. Both roX genes are located on X and overlap two of ∼35 chromatin entry sites (CESs), which are proposed to be high-affinity sites for MSL complexes due to their ability to recruit partial MSL complex in some msl mutant backgrounds (Palmer et al, 1994; Lyman et al, 1997; Gu et al, 2000). Remarkably, when either roX gene is moved to an autosome as a transgene, it can recruit the MSL complex to the insertion site, from which the complex can extensively spread into flanking autosomal DNA (Kelley et al, 1999). This spreading from roX transgenes is most prominently seen if the endogenous roX genes on the X chromosome are deleted (Park et al, 2002). Recent data have shown that spreading can also occur on the X chromosome and suggested that roX genes may function as the major nucleation sites for MSL complex spreading on the X chromosome (Oh et al, 2003). However, spreading in cis from roX genes or the ∼35 CESs cannot be the only mechanism for MSL targeting to the X chromosome, as X to autosome transpositions that lack a mapped CES can still attract MSL complexes (Oh et al, 2004). DNaseI hypersensitivity and transgenic deletion mapping have identified an ∼200 bp MSL-binding site in each roX gene, designated here as DHS (DNaseI hypersensitive site), initially proposed to be the site from which MSL complexes can spread (Kageyama et al, 2001). Sequence alignments reveal short stretches of evolutionarily conserved consensus elements in both DHSs and mutagenesis data have suggested that they are essential for MSL binding (Park et al, 2003).
roX RNAs are male specific (Amrein and Axel, 1997; Meller et al, 1997). Female flies carrying a roX transgene driven by a constitutive promoter fail to accumulate roX RNA unless the complete MSL complex is also ectopically expressed, indicating that the male-specific expression of roX RNA is at least partially caused by the MSL-dependent stabilization of roX RNAs (Meller et al, 2000). Little is known about the regulation of roX genes at the transcriptional level. A recent model proposes that the rate of roX RNA transcription needs to match the rate of MSL complex assembly to regulate the distribution of complexes on the X chromosome (Oh et al, 2003), implying that roX RNA transcription should be finely regulated. The finding that the DHSs in roX genes attract MSL complex raises the question: can MSL complex regulate roX transcription through interaction with the DHS? In this case, the end result would be much greater than two-fold regulation since roX RNAs are male specific.
In the present paper, we dissect the functions of the roX-DHS and study the regulation of roX RNA transcription. We find evidence that the roX-DHS regulates male-specific roX RNA expression by enhancing transcription in males through interaction with the MSL complex, and by repressing transcription in females utilizing unknown repressors. Surprisingly, the roX-DHS is not essential for spreading of the MSL complex; instead, a low level of local roX RNA transcription is sufficient for the ability to spread extensively in cis from roX genes. We propose a model for regulation of roX sex specificity during development in which roX RNA transcription is locked into the male mode by the proper assembly of the MSL complex on the male X chromosome.
Results
DNase I hypersensitive sites in roX genes positively regulate roX RNA levels in males
The roX1-DHS is located near the middle of the roX1 transcription unit, while the roX2-DHS is located downstream of the major roX2 3′ end. Both DHSs are less than 300 bp and share short stretches of conserved sequences. Although transcribed, the DHS sequences are not required for roX RNA stability or function (Park et al, 2003; Stuckenholz et al, 2003), allowing us to make DHS deletions in genomic roX constructs to determine their roles as DNA elements.
GMroX1 and GMroX2 are transgenes that contain full-length roX1 or roX2 genes and flanking sequences (Figure 1A). Both transgenes express roX RNAs at a level comparable to the endogenous roX genes (Figure 1B–D) and rescue roX1roX2 double mutant males (Table I). We deleted ∼300 bp encompassing the DHS from each GMroX transgene to make GMroX1-ΔDHS and GMroX2-ΔDHS (Figure 1A). By performing Northern analyses in either a roX1− or a roX2− background, we found that the level of roX RNA expressed from GMroX1-ΔDHS or GMroX2-ΔDHS was dramatically decreased in most transgenic lines. The average expression in GMroX1-ΔDHS lines was 16.5% of wild type, with a range of 1–70% (Figure 1B), while roX2 expression was reduced to 5% of wild type, with a range of 0.1–20%, and was barely detectable by Northern analysis (Figure 1C). Complementation experiments showed a marked decrease in rescue of roX1roX2 double mutants by GMroX-ΔDHS transgenes, and the rescue frequency correlated with roX RNA levels (Table I legend).
Figure 1.

DHS positively regulates roX RNA levels in males. (A) Structures of the transgenes. GMroX1 and GMroX2 contain full-length roX genomic sequences along with partial segments from flanking genes as indicated by an arrow. In both ΔDHS transgenes, ∼300 bp encompassing the DHS are deleted. In GMroX2-DHS-mut, three blocks of consensus sequences are mutagenized, as indicated by ‘***'. (B–D) Northern blots from adult males to compare RNA level among GMroX, GMroX-ΔDHS and GMroX2-DHS-mut. All transgenic lines carry either a mutant allele (roX1ex6) at their endogenous roX1 locus (B), or are deleted for endogenous roX2 (C, D). Hybridization for rp49 is the loading control in all Northern blots. (B) roX1 Northern. Lane 1: wild-type males; lane 2: GMroX1-67B; lanes 3–12: different GMroX1-ΔDHS lines with the site of insertion indicated. Quantification of roX1 RNA by a phosphoimager is shown as relative to the wild-type level (designated as 1.0), after normalization to rp49 levels. The roX1-ΔDHS transcript is ∼300 nt shorter than wild-type roX1 RNA. (C) roX2 Northern. Lane 1: wild-type males; lanes 2 and 3: GMroX2-86F and 97F, respectively; lanes 4–10: different roX2-ΔDHS transgenic lines with the site of insertion and RNA quantification indicated. (D) roX2 Northern. Lanes 1 and 2: GMroX2-86F and 97F, respectively; lanes 3–9: GMroX2-DHS-mut transgenic lines.
Table 1.
Rescue of roX1roX2 mutants
| Transgene | No. of lines testeda | Rescue range (%)b | Average of rescue frequency (%)c |
|---|---|---|---|
| GMroX1 | 5 | 60–92 | 86 |
| GMroX1-ΔDHS | 12 | 1–57 | 26 |
| GMroX2 | 4 | 65–85 | 82 |
| GMroX2-ΔDHS | 9 | 0.2–15 | 3.5 |
|
GMroX2-DHS-mut |
6 |
0.1–5 |
1.2 |
| yw roX1ex6 Df(1)roX252 [w+4Δ4.3] virgins were crossed with males carrying each transgene. The rescue frequency was calculated as the percentage of expected by comparing the number of male and female progeny. | |||
| a The number of independent lines that were tested for each transgene. | |||
| b Numbers summarize the range of rescue frequency, from the lowest to the highest. For GMroX1-DHS, the lines expressing more roX1 RNA generally had higher rescue. For GMroX2-ΔDHS, three lines (39D, 23B and 49A) showed >10% rescue, while all the other lines showed <3% rescue. | |||
| c Numbers represent the average rescue frequency for each transgene. | |||
roX1-DHS and roX2-DHS share evolutionarily conserved sequences that are essential for attracting MSL complex (Park et al, 2003). To test if these sequences are important for roX RNA expression, we mutagenized the three most conserved elements in roX2-DHS to make the GMroX2-DHS-mut transgene (Figure 1A; see Materials and methods). Consistent with the ΔDHS data, mutagenesis of the DHS in roX2 also abolished roX2 expression in most transgenic lines (Figure 1D).
DHS-dependent MSL binding is not essential for MSL spreading from roX genes
MSL complex not only binds to roX genes but also can spread in cis from these loci up to ∼1 Mbp into flanking sequences. Based on this observation, our lab previously proposed that MSL complexes spread from ∼35 CESs to paint the entire 21 Mbp of X euchromatin (Kelley et al, 1999). The discovery of each DHS as a sequence-specific MSL-binding site within the roX1 or roX2 genes prompted us to test if deletion of the DHS would abolish spreading from GMroX1 and GMroX2 transgenes.
We assayed ΔDHS transgenes for spreading in roX1roX2 mutant males, in which extensive spreading from wild-type GMroX1 or GMroX2 is very consistent due to lack of competition from endogenous roX genes (Figure 2A and E, and Table II; Park et al, 2002). To our surprise, three out of nine GMroX2-ΔDHS and seven out of 12 GMroX1-ΔDHS lines showed variable but extensive MSL spreading in the absence of the DHS (Figure 2B–D and F–H, and Table II). Unlike wild-type GMroX transgenes, spreading was mosaic, seen in some nuclei but not others. In fact, many nuclei showed no MSL association with the transgene, demonstrating that no robust MSL-binding sites remain in these constructs. Transcription of roX RNAs is thought to be required for extensive spreading (Park et al, 2002), and we found that whether a transgenic line showed spreading correlated with its roX RNA level seen by Northern analysis (Figure 1C and Table II legend). These results indicate that cis spreading of the MSL complex from roX transgenes does not require high-affinity DNA sequences for MSL binding within the transgenes themselves. It is important to note, however, that an isolated multimer of roX1-DHS occasionally nucleates limited spreading of the MSL complex (Kageyama et al, 2001). We propose that although DHS is not essential for spreading, it may facilitate the process by recruiting a high concentration of MSL complexes to a specific location (see Discussion).
Figure 2.

DHS is not essential for spreading of the MSL complex from roX loci. Polytene chromosome immunostaining by anti-MSL1 antibody (red). DNA is stained with DAPI (blue). GMroX1 (A) and GMroX2 (E) transgenes provide nucleation sites for extensive spreading (arrowhead) of the MSL complex in ∼100% of nuclei in roX1roX2 mutants. (B) GMroX1-ΔDHS-68A, no binding and spreading at the transgene (arrow). (C) The same transgenic line as in (B) but a different nucleus showing extensive spreading (arrowhead). (D) GMroX1-ΔDHS-25B with no binding and spreading detected (arrow). (F) GMroX2-ΔDHS-50A with no binding and spreading at the transgene (arrow). (G) GMroX2-ΔDHS-23B shows extensive spreading in some nuclei (arrowhead). (H) GMroX2-DHS-mut-70B with limited spreading (arrowhead).
Table 2.
MSL spreading at transgene loci in roX1roX2 mutants
| Transgene | No. of lines testeda | No. of lines showing spreading | % of nuclei showing spreadingb |
|---|---|---|---|
| GMroX1 | 5 | 5 | ∼95 |
| GMroX2 | 6 | 6 | ∼98 |
| GMroX1-ΔDHS | 12 | 7 | 5–82 |
| GMroX2-ΔDHS | 9 | 3 | 3–23 |
|
GMroX2-DHS-mut |
6 |
4 |
7–55 |
| a The number of independent lines that were tested for spreading for each transgene. | |||
| b For GMroX1 and GMroX2, every line showed spreading. The numbers represent the average percentage of nuclei showing spreading for each of these transgenes. For the mutant transgenes, spreading was only seen in a few lines and the percentage of nuclei showing spreading varied from line to line. The number indicates the range of this variation. For GMroX1-ΔDHS, the seven lines with spreading are 72D, 62A, 80A, 68A, 27E, 92A and 93D. The three GMroX2-ΔDHS lines that showed spreading are 39D, 23B and 49A. In GMroX2-DHS-mut flies, the spreading was more frequently seen than in GMroX2-ΔDHS flies, but was usually very limited (2–3 bands). A total of 120–250 nuclei were counted for each line. | |||
roX1 reporter constructs recapitulate DHS-mediated sex-specific regulation
Our analysis of GMroX-ΔDHS transgenes suggested that MSL binding at the DHS upregulates expression of roX RNAs. To study regulation of the roX1 promoter directly, we employed a reporter assay. We identified three distinct 5′ ends of roX1 RNA (see Supplementary information) and therefore chose a 750 bp region that encompassed all three for promoter analysis (Figure 3A). The ProX1 construct was made by inserting this segment upstream of a promoterless eGFP reporter. A second construct, ProX1-DHS, was made by adding the roX1-DHS downstream of the SV40 polyA site of the eGFP reporter (Figure 3A). To minimize possible position effects, both constructs were made in vectors that contain gypsy insulators flanking the reporter cassette (Barolo et al, 2000). eGFP fluorescence was not detected in ProX1 or ProX1-DHS transgenic flies, but this was not unexpected, as our constructs were predicted to produce roX1-eGFP fusion transcripts that contained nonproductive ATG codons from the roX1 5′ region. Therefore, we directly measured eGFP RNA levels by Northern analysis and real-time RT–PCR, normalized to rp49 or PKA mRNA levels. As shown in Figure 3B, no transcription was detected from promoterless eGFP (lanes 2 and 3). In the absence of the DHS, we detected transcription from ProX1 in both sexes, with males showing a slightly higher level of transcription than females (∼2-fold) (Figure 3B, lanes 4–7). When the DHS was added downstream of the 3′ end of eGFP, reporter transcription increased in males (Figure 3B, lanes 8 and 10 versus lanes 4 and 6), but also decreased in females (Figure 3B, lanes 9 and 11 versus lanes 5 and 7), resulting in male-specific expression. Quantitative real-time RT–PCR showed that by adding the DHS, reporter transcription increased 2- to 2.5-fold in males and decreased ∼3-fold in females (Figure 3D). We tested four independent insertion sites for each reporter and got consistent results with P-values <0.05, as evaluated by the Student's t-test.
Figure 3.

DHS directly regulates roX1 promoter activity. (A) Reporter constructs. ProX1 contains 750 bp sequences (gray) from the roX1-5′ region inserted upstream of a promoterless eGFP reporter (green). ProX1-DHS is the same as ProX1 except that the DHS (pink) is added downstream of the SV40 polyA site at the 3′ end of eGFP. The blue bar under the reporter construct represents the probe used in Northern experiments and the red bar indicates the amplified region in real-time PCR. The 750 bp roX1-5′ region contains three roX1 5′ ends mapped by RNase protection assay (see Supplementary information), indicated by arrows, two of which correspond to the start sites of roX1 cDNAs c3 and c20, respectively. (B) Northern blot of reporter RNA from transgenic males (m) or females (f). Lane 1: Hsp70-eGFP; lanes 2 and 3: promoterless eGFP; lanes 4 and 5: ProX1-1; lanes 6 and 7: ProX1-5; lanes 8 and 9: ProX1-DHS-1; lanes 10 and 11: ProX1-DHS-4. Note: The reporters express a roX1-eGFP fusion RNA that is slightly larger (see Supplementary information) than eGFP mRNA in lane 1. (C) Northern blot of reporter RNA from males (m) or females (f), in the presence (+) or absence (−) of an Hsp83-MSL2 transgene. Lanes 1–3: ProX1-4; lanes 4–6: ProX1-DHS-1; lanes 7–9: ProX1-DHS-5; hybridization for rp49 acts as the loading control. (D) Quantitative real-time RT–PCR of ProX1 and ProX1-DHS males (M), females (F) and females with ectopic expression of MSL2 (F+MSL2). For each sample, eGFP transcription has been normalized to the RNA level of the internal control gene pka. Relative eGFP transcription is presented as a ratio to the RNA level of the calibrator, which is the sample with the lowest expression level of eGFP. Here, ProX1-DHS-1 has the lowest expression and is chosen as the calibrator (designated as 1). For each sample, data represent the mean±s.d. from three independent experiments.
DHS-mediated regulation requires MSL proteins
MSL complex is assembled only in males, due to translational repression of the key subunit, MSL2, by Sex Lethal protein in females (Bashaw and Baker, 1997; Kelley et al, 1997). When MSL2 is ectopically expressed in females, functional MSL complex is formed in these animals (Kelley et al, 1995). To determine if the difference in reporter expression between males and females was due to the male-specific MSL complex, we asked whether MSL2 expression in ProX1 and ProX1-DHS females would alter their reporter transcription. Northern analysis and quantitative RT–PCR showed no effect on ProX1 females by ectopic expression of MSL2 protein (Figure 3C and D); therefore, the different level of basal expression between ProX1 males and females is not MSL dependent. In contrast, a robust increase of transcription in ProX1-DHS females was seen in the presence of MSL complexes (Figure 3C and D), suggesting that MSL complexes act through the DHS to mediate positive regulation of roX1 RNA expression. Although the DHS normally lies within the roX1 transcription unit, it is not required for roX1 RNA function or stability (Stuckenholz et al, 2003). Here, the DHS functions downstream of the transcription unit in the ProX1-DHS construct and is not incorporated into the resulting roX1-eGFP reporter RNA (see Supplementary information), strongly suggesting that it functions as a DNA element and therefore is unlikely to influence RNA stability. Our results favor a model in which MSL complexes positively regulate roX1 transcription by interacting with the DHS DNA element.
MSL complex consists of at least five proteins, MSL1, MSL2, MSL3, MLE and MOF, and two roX RNAs. MSL1 and MSL2 proteins are proposed to form the ‘core' of the complex, which remains at ∼35 CESs in the absence of other MSL proteins (Lyman et al, 1997; Copps et al, 1998). MSL3, MLE and MOF, on the other hand, are not essential for binding to most CESs. To test which MSL proteins are required for roX1 transcription, we assayed the ProX1-DHS reporter in Hsp83-MSL2 females mutated for msl1, msl3, mle or mof. Quantitative real-time RT–PCR assays showed that reporter transcription was dramatically decreased in msl1, mle and msl3 mutants (Figure 4A). msl1 and mle had the most severe effect in that reporter transcription was decreased to wild-type female level. We tested two different msl1 alleles with similar results: msl1L60 is a 2 kb deletion removing most of the coding region (Chang and Kuroda, 1998); msl1L183 is a point mutation that disrupts MSL1 function without changing the protein level (R Kelley, personal communication). In contrast, high-level transcription is maintained in mof1 mutants (Figure 4A) carrying a point mutation that disrupts the acetyltransferase function of MOF (Hilfiker et al, 1997). These results suggest that roX1 transcription requires MSL1, 2, 3 and MLE, but not the function of MOF. However, recent observations have suggested that mof1 retains sufficient enzymatic activity to acetylate histone H4 at CESs (Sass et al, 2003), raising the possibility that residual enzymatic activity may contribute to roX1 transcription.
Figure 4.
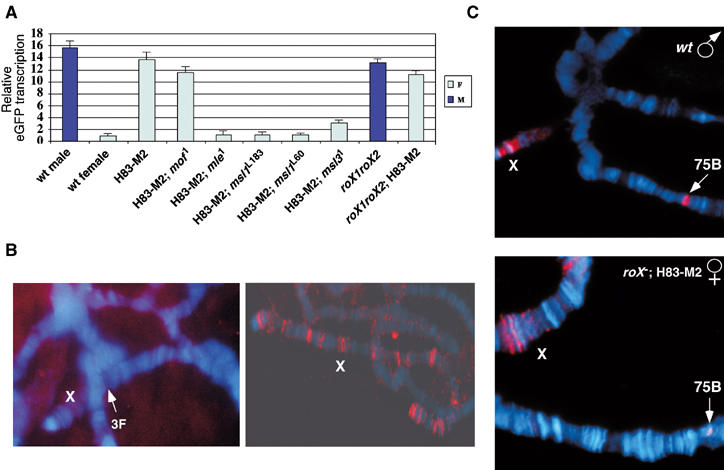
DHS-mediated regulation requires MSL proteins but not roX RNA. (A) Quantitative real-time RT–PCR of ProX1-DHS males (M) or females (F) in wild-type or different msl or roX1roX2 mutant backgrounds (as indicated at the bottom). Results are represented as in Figure 3D, with the transcription level of wild-type female designated as 1. (B) Left panel: in situ hybridization of roX1 RNA (red) in msl3 [Hsp83-MSL2] females. No roX1 RNA was detected on X. We also failed to detect transcription at the endogenous roX1 locus (3F), as indicated by the arrow. Right panel: in the presence of an Hsp83-roX1 transgene on an autosome, roX1 RNA was detected at many CESs on the X chromosome. (C) Polytene chromosomes from ProX1-DHS-1 stained with anti-MSL1 antibody (red). The top panel shows the male chromosomes in a wild-type background; the bottom panel shows the chromosomes from females with ectopic MSL2 protein but mutant for roX1 and roX2. The arrows indicate the transgene loci. X: X chromosome. DNA was stained with DAPI (blue) in (B, C).
In a previous study, females expressing MSL2 but mutant for msl3 failed to incorporate roX1 RNA at the ∼35 binding sites for partial MSL complexes, although roX2 RNA was consistently detected (Meller et al, 2000). The interpretation at the time was that roX2 RNA might be assembled into MSL complex earlier than roX1 RNA, and that incorporation of roX1 RNA required MSL3. However, the results of our studies raised the strong possibility that the roX RNAs have the same ability to assemble into incomplete complex, but that roX1 transcription is repressed when lacking MSL3. To test this idea, we performed in situ hybridization for roX1 RNA in msl3 [Hsp83-MSL2] females with or without an Hsp83-roX1cDNA transgene on an autosome. We found that roX1 RNA was not detected on female X chromosomes when lacking MSL3 (Figure 4B, left panel). However, when roX1 RNA was expressed constitutively from the Hsp83 promoter, it was detected at many CESs (Figure 4B, right panel). This strongly suggests that endogenous roX1 transcription is indeed repressed in the absence of MSL3 and this deficiency in roX1 expression can be overcome by an actively transcribed roX1 transgene.
roX RNAs are not required for positive regulation of ProX1-DHS
We next tested whether roX RNAs autoregulate roX1 transcription. Our previous study showed that MSL complex was not detectable at the DHS in the absence of roX RNAs (Park et al, 2003), suggesting that we would see a decrease of ProX1-DHS reporter transcription in roX1roX2 double mutant males. In contrast, we found that the ProX1-DHS reporter still retained a high level of transcription in roX− males (Figure 4A). To verify this result, we tested females that ectopically express MSL complex but are mutant for roX genes. Consistent with the result in males, ProX1-DHS reporter transcription remained high in females expressing a full set of MSL proteins but no roX RNAs (Figure 4A). To re-examine if a weak MSL–DHS interaction could occur in the absence of roX RNA, we performed chromosome immunostaining in these female third-instar larvae, which are healthy and have much better chromosomal morphology than roX1roX2 mutant males assayed in our earlier work (Park et al, 2003). Weak MSL-binding signals at the ProX1-DHS transgene insertion sites were detected in some nuclei (Figure 4C), suggesting that MSL complex without RNA components can interact with DHS in a weak and/or transient way. Surprisingly, roX RNAs stabilize this interaction, apparently without having a strong effect on roX1 transcription.
MSL proteins counteract constitutive repression at the roX1 promoter
Our reporter assay showed that in the presence of the DHS, transcription from the roX1 promoter was repressed in females compared to basal transcription in the absence of the DHS (Figure 3B and D), raising the question of how roX1 transcription is regulated in females. roX1 RNA is expressed in both males and females during early embryonic development, but it disappears in females and remains in males as the MSL complex becomes established (Meller et al, 1997). This suggests that early roX1 expression is MSL independent. Consistent with this, our results indicate that the isolated roX1 promoter has constitutive basal activity in both sexes. The lack of roX1 expression in females at later stages could be explained by unknown repressor proteins that may interact with DHS and be normally counteracted by MSL complexes in males. To determine whether repression is truly female specific or it can happen in males that lack MSL complexes, we analyzed msl mutant males.
All MSL proteins are required for male viability, but some msl mutant males survive to the late third-instar larval stage, making it possible to measure their RNA levels. We have shown that MSL complex increases roX1 reporter transcription about two-fold in males (Figure 3B and C). If there were no repression in males, we would expect to see the transcription decreased by half in msl mutant males. However, as shown by quantitative real-time RT–PCR, we found that in mle1/mleγ38 male larvae, ProX1-DHS reporter transcription is significantly decreased (more than 10-fold on average) compared to their mle1/CyOy+ brothers (Figure 5A), suggesting that repression can occur in males as well as in females. msl31/msl3mak1 males also have more than a four-fold decrease of transcription, although not as low as mle males (Figure 5B). In contrast, ProX1 reporter transcription is not affected in either msl mutant male (Figure 5A and B), confirming that the decrease of transcription is caused by the DHS and not other sequences. Taken together, our results from msl mutant males suggest that roX1 transcription is regulated by both positive and negative factors, through interaction with a shared cis element, the DHS.
Figure 5.
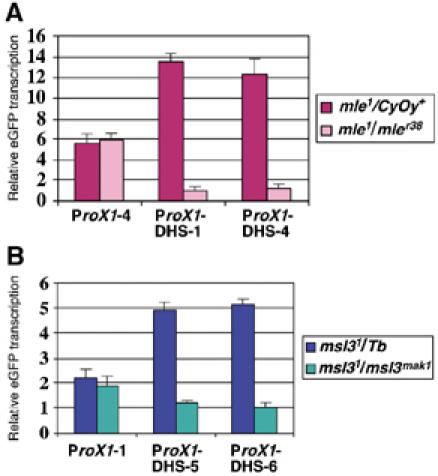
The roX1 promoter is repressed in msl mutant males. Real-time RT–PCR of ProX1 or ProX1-DHS reporter transcription from mle (A) and msl3 (B) mutant male larvae. Results are represented as in Figures 3 and 4, with the transcription level of ProX1-DHS-1 and ProX1-DHS-6 designated as 1 in (A) and (B), respectively.
Discussion
roX RNAs are male specific and play essential functions in dosage compensation. They require MSL complex for RNA stabilization, which contributes at least partially to their male-specific expression pattern (Meller et al, 2000). In this paper, we analyzed regulation of roX RNA transcription by assessing the function of DHS, the primary MSL-binding sites in roX genes. Using transgenic deletion analyses and reporter assays, we found that roX1 transcription is directly regulated by DHS. Genetic data demonstrated that this regulation requires MSL1, 2, 3 and MLE, but not roX RNAs. We further verified this regulation at the endogenous roX1 locus. We propose that the DHS regulates roX1 transcription in both sexes. It acts as a derepressor/enhancer in males to attract MSL complex to activate roX1 transcription, and as a silencer in females to downregulate roX1 RNA transcription. At this point, we have not identified potential repressors. The fact that repression can happen in both sexes in the absence of MSL complex suggests that it may represent the default state. Interestingly, a recent study has shown that MSL complex can overcome silencing of mini-white-containing transgenes inserted into various silent chromatin environments, allowing male-specific activation of mini-white through binding to an adjacent roX1 segment (Kelley and Kuroda, 2003). The underlying mechanism of this antagonism of repression may be similar to what we propose here for the regulation of roX1 transcription.
The finding that the roX1 promoter has MSL-independent basal activity is consistent with previous results that roX1 RNA is expressed in both sexes at the early embryonic stage before MSL complexes are established (Meller et al, 1997). Surprisingly, we found that transcription from the roX1 promoter was only increased two-fold by MSL–DHS interaction, suggesting that MSL complex may use the same strategy to upregulate roX genes as to modulate chromosome-wide transcription. However, we cannot exclude the possibility that, at a functional roX1 locus, MSL complex may act more robustly. For example, nascent roX1 RNA may continually attract MSL proteins to assemble locally to provide a strong positive feedback loop on local transcription. This may explain why we see a large difference in roX1 RNA levels between GMroX1 and GMroX1-ΔDHS flies. That the DHS can regulate roX expression and is required for genomic transgene function does not rule out additional levels of regulation at the endogenous roX1 locus on the X chromosome. Rattner and Meller (2004) have recently proposed that MSL2 alone can upregulate transcription of roX1 in females and that this is DHS independent. The magnitude of this regulation by MSL2 and its site and mechanism of action remain to be determined.
As shown in Figure 6, we propose a model in which transcription of roX1 RNA is developmentally regulated by antagonism between repression and activation. In the early embryonic stages, before MSL complex is established, roX1 RNA is transcribed in both sexes from an MSL-independent basal promoter. Later, in females unknown repressors downregulate roX1 transcription by interacting with DHS, while in males roX1 transcription is maintained and enhanced as MSL complex becomes established. This could occur by successful competition of MSL complexes with repressors for binding to the DHS. Alternatively, both MSL complex and repressors could bind to DHS simultaneously, with the outcome being a finely tuned regulation of roX1 RNA transcription. A question raised by this model is why roX1 transcription needs to be repressed in the absence of MSL complex. One possibility is that roX1 transcription is harmful for female development and thus has to be shut down. Although females carrying a roX1 cDNA transgene driven by a constitutive promoter have no obvious phenotype (Meller et al, 2000), this could be a form of redundant control to ensure fidelity of dosage compensation. Another possibility is that repression is a default mechanism, which is primarily utilized in males to adjust roX1 transcription to an appropriate level. This adjustment might be achieved by maintaining a balance between enhancement and repression.
Figure 6.
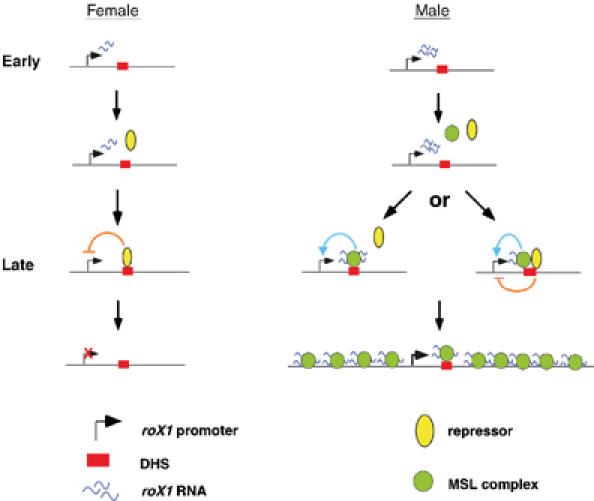
Model for regulation of roX1 transcription. See text for details.
roX loci overlap two of ∼35 CESs, which were proposed to be nucleation sites for MSL spreading. The identification of DHS as the primary MSL-binding site prompted a hypothesis that DHS carries the CES function of roX loci. Surprisingly, we found that roX genes can still nucleate spreading in the absence of DHS, as long as transcription occurred at these loci. Perhaps initial RNA binding can lead to a histone-binding/modification cycle in the absence of a specific DNA interaction, as proposed for HP1 spreading to form heterochromatin (Bannister et al, 2001; Lachner et al, 2001).
Finally, although our data suggest that DHS does not function as a spreading initiation site, it may facilitate the efficient spreading from roX loci due to its high binding affinity for the MSL complex. This may explain why we only saw spreading in some nuclei in the GMroX-ΔDHS stocks. The local concentration of the MSL complex is suggested to be important for cis spreading (Oh et al, 2003). Nascent roX RNA may be the most important attractant to concentrate the MSL complex at the site of assembly (Park et al, 2002), but DHS may also help recruit and maintain this local MSL pool. The previous observation that a multimer of roX1-DHS can occasionally nucleate limited spreading (Kageyama et al, 2001) may reflect this function.
Materials and methods
Transgene construction and transformation
GMroX1-ΔDHS was constructed by two-step subcloning. The sequences upstream of DHS (2.0 kb) were amplified from pCaSpeR-GMroX1 using primers ΔDHS-3 (5′-CGGAATTCAAGTTCACCAGCTC-3′) and ΔDHS-10 (5′-AAAAAGCGGCGCC TTCTCGAAACGCAAG-3′), sequenced and subcloned into EcoRI/NotI-digested pCaSpeR3. The sequences downstream to DHS (2.6 kb) were amplified by primers ΔDHS-9 (5′-AGCAAGGCCTTTCCTATCGAACTG-3′) and ΔDHS-11 (5′-AAAGCGGCCG CTCTGGAAAGACC-3′), sequenced and then subcloned into the NotI site in the former construct to make the final pCaSpeR-GMroX1-ΔDHS transgene construct. To construct GMroX2-ΔDHS, an XbaI–SpeI GMroX2 fragment linked to a PCR fragment (forward primer 5′-ACTAGTTATGACAAATAAAGAC-3′ and reverse primer 5′-GATATCGCA GATTGAAGAATTGAAG-3′) containing the sequences 3′ of DHS was subcloned into pCaSpeR3. To make GMroX2-DHS-mut, PCR-mediated mutagenesis was performed as described previously to mutagenize blocks 1, 2 and 4 (Park et al, 2003). The mutant roX2-DHS was confirmed by sequencing and then subcloned into GMroX2-ΔDHS at the SpeI site. ProX1 was made by subcloning a 750 bp EcoRI fragment containing the roX1 5′ region amplified from pCaSpeR-GMroX1 (forward primer 5′-TGTAGTTGGCTGTAAA TACG-3′ and reverse primer 5′-TACATCTTGCCAGAGATTTCG-3′) into pGreen-Pelican (Barolo et al, 2000). ProX1-DHS was made by subcloning DHS into the SpeI site of ProX1.
Transgenic flies were made by P element-mediated transformation (Spradling and Rubin, 1982).
Northern analysis
Fly total RNA was isolated using the TRIzol Reagent (Invitrogen), and 20 μg of RNA was used per lane. Hybridization was performed as described (Church and Gilbert, 1984), using PCR fragments amplified from roX1 c20 or pCaSpeR-GMroX2 as the probes. eGFP probe was prepared by PCR amplification of eGFP coding region (forward primer 5′-ATGGTGA GCAAGGGCGAGGAG-3′ and reverse primer 5′-CTTGTACAGCTCGTCCATGCC-3′). Quantification of Northern blots was performed using a Phosphoimager (Molecular Dynamics).
Polytene chromosome immunostaining and in situ hybridization
Preparation of polytene chromosomes and immunostaining using anti-MSL1 antibody were as previously described (Kelley et al, 1999).
In situ hybridization was performed as described previously (Meller et al, 2000) except that hybridization was at 46°C rather than 42°C. A digoxygenin-labeled antisense RNA from the 3′ part of roX1 (1.8 kb EcoR1/BglII fragment) was used as the probe.
Quantitative real-time RT–PCR
DNaseI-treated total RNA from ProX1 or ProX1-DHS flies was subjected to first-strand cDNA synthesis using SuperScriptII reverse transcriptase (Invitrogen) and primed by oligo dT. Real-time PCR was performed using the ABI PRISM 7000 Sequence Detection System (Applied Biosystems). The pka gene was used as the internal reference for normalizing variance in the quality of RNA and the amount of input cDNA. The primer sequences for eGFP were as follows: forward 5′-AGCACGACTTCTTCAAGTCCG-3′ and reverse 5′-GTGTCGCCCTCGAACTTCAC-3′. The primer sequences for pka were as follows: forward 5′-TTCTCGGAGCCGCACTCGCGCTTCTAC-3′ and reverse 5′-CAATCAGCAGATTCTC CGGCT-3′. PCR amplification was performed in triplicate in a 25 μl final volume containing 1 × SYBR green PCR buffer, 5.5 mM MgCl2, 200 μM dNTP, 200 nM of each primer, 0.6 U platinum Taq DNA polymerase (Invitrogen) and 1 μl of cDNA. The PCR protocol used an initial denaturing step at 94°C for 10 min followed by 40 cycles of 94°C for 30 s, 52°C for 30 s and 72°C for 1 min. Dissociation curve analysis was run at the end of 40 cycles to verify PCR product identity. Relative quantification of eGFP transcription was determined by comparative CT method based on the manufacturer's instructions (ABI Prism 7700 sequence detection system User Bulletin #2, Applied Biosystems). Standard curve for each set of primers was constructed using a serial dilution of cDNA to verify equal amplification efficiency of the two systems.
Fly stocks and genetic crosses
GMroX1-ΔDHS, GMroX2-ΔDHS or GMroX2-DHS-mut males were crossed with yw roX1ex6, w Df(1)roX252; [w+ 4Δ4.3] or yw roX1ex6Df(1)roX252 [w+ 4Δ4.3] females. The resulting male progeny carrying an autosomal transgene and mutant alleles of roX1, roX2 or both were collected for Northern analyses or chromosomal immunostaining.
To generate msl1L183, msl1L60, mle or msl3 mutant females with ectopic MSL2 expression, [Hsp83-MSL2] females homozygous for each mutant allele were crossed to [ProX1] or [ProX1-DHS] males heterozygous for msl1L183, msl1L60, mle or msl3. The resulting homozygous mutant females were identified by the absence of CyO or Tb phenotype. To generate mof1 mutant females, w cv mof1; [Hsp83-MSL2] females were crossed to w cv mof1; CyO [mof+]18H1/[ProX1] or [ProX1-DHS] and mof1 female adults were selected by the absence of CyO. To generate roX1roX2 females with ectopic MSL2 expression, yw roX1ex6Df(1)roX252 [w+ 4Δ4.3]; msl31[Hsp83-MSL2] females were crossed with yw roX1ex6Df(1)roX252 [w+ 4Δ4.3]; +/CyO[GMroX1-39A]; [ProX1-DHS] males. The resulting yw roX1ex6Df(1)roX252 [w+ 4Δ4.3]; +/+; msl31[Hsp83-MSL2]/[ProX1-DHS] females were identified by the absence of CyO. mle1/mleγ38 male larvae were generated by crossing yw; pr mle1; [ProX1] or [ProX1-DHS] females with yw; mleγ38/CyOy+ males. For msl3 mutant males, yw; [ProX1] or [ProX1-DHS]; msl31 females were crossed to yw; msl3mak1red e/TM6 Tb males and the resulting non-Tb male larvae were selected. yw; msl31 [Hsp83-MSL2]/msl31 [H83-roX1cDNA20] females were generated by crossing yw; msl31 [H83-roX1cDNA20] females with yw; msl31 [Hsp83-MSL2]/TM6 Tb males and non-Tb females were selected in the next generation.
Supplementary Material
Supplementary information
Acknowledgments
We are grateful to Drs R Kelley and V Meller, and members of the Kuroda lab for helpful discussions and critical reading of the manuscript. We thank R Kelley for fly stocks, R Richman for MSL antibodies, and X Chu and H Kennedy for technical assistance. This work was supported by the National Institutes of Health (GM45744) and the Howard Hughes Medical Institute. MIK is an HHMI Investigator.
References
- Akhtar A, Becker PB (2000) Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell 5: 367–375 [DOI] [PubMed] [Google Scholar]
- Amrein H, Axel R (1997) Genes expressed in neurons of adult male Drosophila. Cell 88: 459–469 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 [DOI] [PubMed] [Google Scholar]
- Barolo S, Carver LA, Posakony JW (2000) GFP and beta-galactosidase transformation vectors for promoter/enhancer analysis in Drosophila. Biotechniques 29: 726–732 [DOI] [PubMed] [Google Scholar]
- Bashaw GJ, Baker BS (1997) The regulation of the Drosophila msl-2 gene reveals a function for Sex-lethal in translational control. Cell 89: 789–798 [DOI] [PubMed] [Google Scholar]
- Chang KA, Kuroda MI (1998) Modulation of MSL1 abundance in female Drosophila contributes to the sex specificity of dosage compensation. Genetics 150: 699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church GM, Gilbert W (1984) Genomic sequencing. Proc Natl Acad Sci USA 81: 1991–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copps K, Richman RR, Lyman LM, Chang KA, Rampersad-Ammons J, Kuroda MI (1998) Complex formation by the Drosophila MSL proteins: role of the MSL2 RING finger in protein complex assembly. EMBO J 17: 5409–5417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Wei X, Pannuti A, Lucchesi JC (2000) Targeting the chromatin-remodeling MSL complex of Drosophila to its sites of action on the X chromosome requires both acetyl transferase and ATPase activities. EMBO J 19: 5202–5211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilfiker A, Hilfiker-Kleiner D, Pannuti A, Lucchesi JC (1997) mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. EMBO J 16: 2054–2060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Wang Y, Johansen J, Johansen KM (2000) JIL-1, a chromosomal kinase implicated in regulation of chromatin structure, associates with the male specific lethal (MSL) dosage compensation complex. J Cell Biol 149: 1005–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama Y, Mengus G, Gilfillan G, Kennedy HG, Stuckenholz C, Kelley RL, Becker PB, Kuroda MI (2001) Association and spreading of the Drosophila dosage compensation complex from a discrete roX1 chromatin entry site. EMBO J 20: 2236–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Kuroda MI (2003) The Drosophila roX1 RNA gene can overcome silent chromatin by recruiting the male-specific lethal dosage compensation complex. Genetics 164: 565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI (1999) Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell 98: 513–522 [DOI] [PubMed] [Google Scholar]
- Kelley RL, Solovyeva I, Lyman LM, Richman R, Solovyev V, Kuroda MI (1995) Expression of Msl-2 causes assembly of dosage compensation regulators on the X chromosomes and female lethality in Drosophila. Cell 81: 867–877 [DOI] [PubMed] [Google Scholar]
- Kelley RL, Wang J, Bell L, Kuroda MI (1997) Sex lethal controls dosage compensation in Drosophila by a nonsplicing mechanism. Nature 387: 195–199 [DOI] [PubMed] [Google Scholar]
- Kuroda MI, Kernan MJ, Kreber R, Ganetzky B, Baker BS (1991) The maleless protein associates with the X chromosome to regulate dosage compensation in Drosophila. Cell 66: 935–947 [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116–120 [DOI] [PubMed] [Google Scholar]
- Lee C-G, Chang KA, Kuroda MI, Hurwitz J (1997) The NTPase/helicase activities of Drosophila Maleless, an essential factor in dosage compensation. EMBO J 16: 2671–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucchesi JC (1998) Dosage compensation in flies and worms: the ups and downs of X-chromosome regulation. Curr Opin Genet Dev 8: 179–184 [DOI] [PubMed] [Google Scholar]
- Lyman LM, Copps K, Rastelli L, Kelley RL, Kuroda MI (1997) Drosophila male-specific lethal-2 protein: structure/function analysis and dependence on MSL-1 for chromosome association. Genetics 147: 1743–1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller VH, Gordadze PR, Park Y, Chu X, Stuckenholz C, Kelley RL, Kuroda MI (2000) Ordered assembly of roX RNAs into MSL complexes on the dosage compensated X chromosome in Drosophila. Curr Biol 10: 136–143 [DOI] [PubMed] [Google Scholar]
- Meller VH, Kuroda MI (2002) Sex and the single X chromosome. Adv Genet 46: 1–24 [DOI] [PubMed] [Google Scholar]
- Meller VH, Rattner BP (2002) The roX RNAs encode redundant male-specific lethal transcripts required for targeting of the MSL complex. EMBO J 21: 1084–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meller VH, Wu KH, Roman G, Kuroda MI, Davis RL (1997) roX1 RNA paints the X chromosome of male Drosophila and is regulated by the dosage compensation system. Cell 88: 445–457 [DOI] [PubMed] [Google Scholar]
- Oh H, Bone JR, Kuroda MI (2004) Multiple classes of MSL binding sites target dosage compensation to the X chromosome of Drosophila. Curr Biol 14: 481–487 [DOI] [PubMed] [Google Scholar]
- Oh H, Park Y, Kuroda MI (2003) Local spreading of MSL complexes from roX genes on the Drosophila X chromosome. Genes Dev 17: 1334–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer MJ, Richman R, Richter L, Kuroda MI (1994) Sex-specific regulation of the male-specific lethal-1 dosage compensation gene in Drosophila. Genes Dev 8: 698–706 [DOI] [PubMed] [Google Scholar]
- Park Y, Kelley RL, Oh H, Kuroda MI, Meller VH (2002) Extent of chromatin spreading determined by roX RNA recruitment of MSL proteins. Science 298: 1620–1623 [DOI] [PubMed] [Google Scholar]
- Park Y, Megnus G, Bai X, Kageyama Y, Meller VH, Becker PB, Kuroda MI (2003) Sequence-specific targeting of Drosophila roX genes by the MSL dosage compensation complex. Mol Cell 11: 977–986 [DOI] [PubMed] [Google Scholar]
- Rattner BP, Meller VH (2004) Drosophila Male-Specific Lethal 2 protein controls sex-specific expression of the roX genes. Genetics 166: 1825–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sass GL, Pannuti A, Lucchesi JC (2003) Male-specific lethal complex of Drosophila targets activated regions of the X chromosome for chromatin remodeling. Proc Natl Acad Sci USA 100: 8287–8291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ER, Pannuti A, Gu W, Steurnagel A, Cook RG, Allis CD, Lucchesi JC (2000) The Drosophila MSL complex acetylates histone H4 at lysine 16, a chromatin modification linked to dosage compensation. Mol Cell Biol 20: 312–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC, Rubin GM (1982) Transposition of cloned P-elements into Drosophila germ line chromosomes. Science 218: 341–347 [DOI] [PubMed] [Google Scholar]
- Stuckenholz C, Meller VH, Kuroda MI (2003) Functional redundancy within roX1, a noncoding RNA involved in dosage compensation in Drosophila melanogaster. Genetics 164: 1003–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner BM, Birley AJ, Lavender J (1992) Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell 69: 375–384 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang W, Jin Y, Johansen J, Johansen KM (2001) The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell 105: 433–443 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information


