Abstract
During latency, Kaposi's sarcoma-associated herpesvirus (KSHV) is thought to replicate once and to be partitioned in synchrony with the cell cycle of the host. In this replication cycle, the KSHV terminal repeat (TR) sequence functions as a replication origin, assisted by the latency-associated nuclear antigen (LANA). Thus, TR seems to function as a cis element for the replication and partitioning of the KSHV genome. Viral replication and partitioning are also likely to require cellular factors that interact with TR in either a LANA-dependent or -independent manner. Here, we sought to identify factors that associate with TR by using a TR DNA column and found that poly(ADP-ribose) polymerase 1 (PARP1) and known replication factors, including ORC2, CDC6, and Mcm7, bound to TR. PARP1 bound directly to a specific region within TR independent of LANA, and LANA was poly(ADP-ribosyl)ated by PARP1. Drugs such as hydroxyurea and niacinamide, which raise or lower PARP activity, respectively, affected the virus copy number in infected cells. Thus, the poly(ADP-ribosyl)ation status of LANA appears to affect the replication and/or maintenance of the viral genome. Drugs that specifically up-regulate PARP activity may lead to the disappearance of latent KSHV.
Kaposi's sarcoma-associated herpesvirus (KSHV) or human herpesvirus 8 (HHV8) was discovered in Kaposi's sarcoma lesions (12) and is strongly associated with multicentric Castleman's disease and primary effusion lymphomas, which are predominantly found in AIDS patients (9). KSHV belongs to the gammaherpesviruses, and most of the virus is in the latent phase in these diseases (13).
Epstein-Barr virus (EBV), another human gammaherpesvirus, also establishes latency and is associated with epithelial and many kinds of lymphoid tumors (28). Several genes with potential oncogenic activity, latency-associated nuclear antigen (LANA), K-cyclin (ORF72), vFLIP (ORF71), K15, and LANA2 (vIRF3) of KSHV (17, 20, 46, 55) and EBNA1, LMP1, LMP2, and EBER of EBV (40), are expressed during latency, and the presence of the virus seems to be a minimum requirement for cancer formation (10) and to be tightly linked to the growth activity of the virus-positive B-cell lymphoma lines. Therefore, even in the context of serious disease, control of the latent infection itself could be an important strategy in treatment of these cancers.
In latent replication, the KSHV genome is thought to replicate once per cell cycle, in synchrony with the host replication machinery, as reported for EBV (1, 27, 54). Recent studies of KSHV latent replication and maintenance have revealed that (i) the terminal repeat sequence functions as a replication origin (3, 4) and (ii) LANA binds to a specific sequence within the terminal repeat (TR) and is an essential viral gene product for latent viral replication (3, 4, 27). These two facts are important for understanding how the KSHV genome replicates in the latent phase. In fact, the two viral components TR and LANA appeared to be sufficient to execute viral replication in a transient transfection assay; the presence of several copies of TR and LANA could maintain the viral genome in cells for a few months (3).
During latency, the relationship between TR and LANA is similar to that of oriP and EBNA1 in EBV. oriP is a cis-acting element containing a dyad symmetry region and a family of repeat sequences. There is little similarity among the nucleotide sequences of TR and oriP and the primary amino acid sequences of LANA and EBNA1. However, the functional domains of LANA and EBNA1 have some similarities and can be divided into three basic regions: a chromosome-binding site and nuclear localization signal at the N-terminal end, a repeating amino acid region in the central portion, which is a DE repeat in LANA and a GA repeat in EBNA1, and DNA-binding and dimerization domains in the C-terminal region. These defined regions may reflect functional and conformational similarities of the proteins (5, 6, 24).
Although TR and LANA are the most important components for the latent replication of KSHV, the origin itself within TR has not yet been precisely determined (27), and in contrast to simian virus 40 large T antigen, LANA does not seem to have helicase activity, which is one of the essential activities required for DNA replication. Therefore, it appears that other factors must associate with TR, either dependent on or independent of LANA, and we sought to identify such factors. Using a TR DNA column with whole-cell extract from BC3 (a KSHV-infected cell line) and BJAB (a KSHV-negative, EBV-negative cell line), we found that poly(ADP-ribose) polymerase 1 (PARP1; EC 2.4.2.30), replication factors such as origin recognition complexes (ORCs) and minichromosome maintenance factors (MCMs), heterochromatin factors such as HP1 alpha and MeCP2, and methylated histone H3 bound to TR under stringent conditions. The binding of most of the factors was dependent on LANA, but PARP1 could bind TR directly, at a specific region of TR. Furthermore, we found that LANA was a target of PARP1 and was poly(ADP-ribosyl)ated by it. The effects of hydroxyurea (HU), 3-aminobenzamide (3-AB), and niacinamide (NA) suggested that PARP's activity could affect KSHV viral replication and/or maintenance. These findings further our understanding of the mechanism for viral DNA maintenance in latently infected cells and suggest that PARP-targeting drugs may be exploited to eliminate live KSHV.
MATERIALS AND METHODS
Cells.
BC3, a KSHV-positive and EBV-negative primary effusion lymphoma cell line (11), was maintained in RPMI 1640 medium containing 20% heat-inactivated fetal bovine serum, 100 IU of penicillin G/ml, and 10 μg of streptomycin/ml in a 5% CO2 atmosphere. BJAB, a KSHV-negative and EBV-negative Burkitt lymphoma cell line, was grown under the same conditions, except with 10% fetal bovine serum.
Plasmids.
p009 was generated by deleting the BglII fragment from a cosmid clone Z6 (44). The resultant plasmid contained eight copies of the TR. The NotI fragment of TR was directly inserted into pBluescript II (pBSII) (Stratagene), resulting in pBSII TR (NotI), and PCR-amplified TR was inserted into the XbaI site of the vector, resulting in pBSII TR (XbaI). Primers with an XbaI site at the end, 5′-AATCTAGACGTGAACACCCCGCGCCCCGCGC-3′ and 5′-AATCTAGATAGTGTCCAGGGCTCCACGTAGC-3′, were designed for both ends of TR based on the GenBank accession no. U75699 sequence and used to construct the plasmid. pBSII TR (XbaI) was digested with NotI and XbaI, and the resultant fragments were subcloned into the NotI-XbaI site of pBSII, resulting in pBSII TR 1 (nucleotides 1 to 387) and 2 (nucleotides 388 to 801), respectively. A deletion mutant was generated from pBSII TR 2 (nucleotides 388 to 801) with an Exonuclease III Kit (Takara), resulting in pBSII TR 2-1 (nucleotides 388 to 695). The pBSII TR 2-1 plasmid was digested with NotI and EcoRV, and the fragment of nucleotides 388 to 695 was gel purified and then digested with Sau3AI. The generated fragments, nucleotides 388 to 548 and 549 to 801, were respectively inserted into the NotI-BamHI and BamHI-EcoRV sites of pBSII. These two plasmids were termed pBSII TR 2-1-1 (nucleotides 388 to 548) and pBSII TR 2-1-2 (nucleotides 549 to 695). The nucleotide numbers are based on the GenBank accession no. U75699 sequence.
The KSHV oriLyt common sequence (nucleotides 23137 to 24270) (K-oriLyt CS) (2) was amplified by PCR with 5′-AACTCGAGCTAATTTGCATGCGCTAATCCT-3′ and 5′-AACTCGAGCCAAGAGATCCGTCCCACGTG-3′ as the primers and the genomic DNA from BC3 cells as the template. The amplified fragment was digested with XhoI and inserted into the XhoI site of pBSII, resulting in pBSII K-oriLyt CS.
TR column preparation.
p009 was amplified by routine molecular biology techniques. The entire 801-bp TR fragment was purified after digesting the vector with a NotI restriction enzyme (Roche) and separating the fragments on a 1% agarose gel by using a DNA fragment purification kit (Wizard PCR and DNA purification system; Promega). About 1 mg of the purified fragment was incubated with CNBr-activated Sepharose (Amersham Bioscience) (∼1-ml bed volume) according to the manufacturer's protocol. The unbound reactors were inactivated, the Sepharose was extensively washed with the binding buffer [HEPES-HCl (pH 7.9), 1 mM MgCl2, 1 mM dithiothreitol (DTT), 150 mM NaCl, 0.1% bovine serum albumin (BSA), 0.4 mg of sonicated salmon sperm DNA (sssDNA)/ml, 10 μg of poly(dI-dC) (Amersham Bioscience)/ml, 25% glycerol], and the Sepharose was packed into an Econocolumn (Bio-Rad), resulting in TR(+)Sepharose. CNBr-activated Sepharose without the DNA fragment was generated in the same way, to exclude proteins binding nonspecifically to Sepharose, and termed TR(−)Sepharose.
The K-oriLyt CS mentioned above was purified and bound to CNBr-activated Sepharose as described above for TR, resulting in K-oriLyt CS Sepharose.
Lysate preparation, column binding, and fractionation.
Exponentially growing BC3 and BJAB cells (∼3 × 108 cells, each) were harvested by spinning them down and washing them twice with phosphate-buffered saline (PBS) without calcium and magnesium. The cell pellet was then suspended in whole-cell extraction buffer (HEPES-HCl [pH 7.9], 1 mM MgCl2, 1 mM DTT, 0.45 M NaCl, 25% glycerol), and the suspension was mixed with 50% (vol/vol) 0.3-mm-diameter glass beads and lysed in a cell destruction machine (Multibeads Shocker; Yasui Kikai Co. Ltd.) by subjecting them to 30 s of mincing at 30-s intervals at 4°C for 10 min. The extracted material was separated by centrifugation at 10, 000 × g for 30 min at 4°C. The soluble fraction was adjusted to 150 mM NaCl by adding two volumes of NaCl-free whole-cell extraction buffer stepwise. The lysate was adjusted to 0.1% BSA, 0.4 mg of sssDNA/ml, and 10 μg of poly(dI-dC)/ml. The columns were equilibrated with a binding buffer [HEPES-HCl (pH 7.9), 1 mM MgCl2, 1 mM DTT, 150 mM NaCl, 25% glycerol, 0.4 mg of sssDNA/ml, 10 μg poly(dI-dC)/ml, and 0.1% BSA]. The material was passed through the TR(−)Sepharose and then loaded onto the TR(+)Sepharose column. The material that passed through the TR(+)Sepharose fraction was stored as the unbound fraction. After washing the column with the binding buffer, the bound material was eluted with the same buffer containing increasing concentrations of KCl from 0.2 to 1.0 M, with a 0.1 M step, and finally with 2.5 M KCl (1 ml/fraction).
To determine whether the binding to TR was specific, we obtained the protein binding profiles by using K-oriLyt CS Sepharose with the same procedure.
MALDI-TOF (MS) analyses.
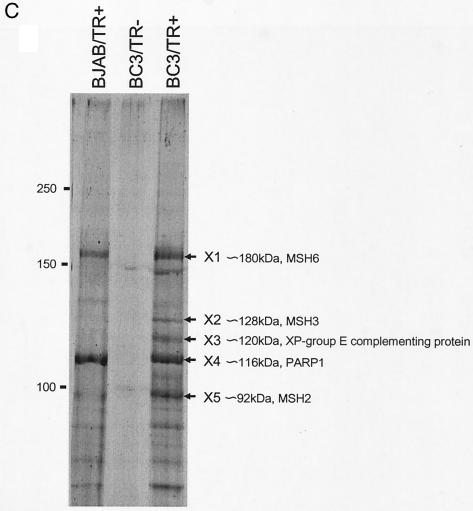
Ten microliters of each fraction from the DNA column was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and stained with Sypro-Ruby (Bio-Rad). The staining pattern was visualized and analyzed by the FX laser-scanning system (Bio-Rad) and Quantity One software (Bio-Rad). Bands in a typically stained pattern around 105 to 150 kDa were cut out of the gel and subjected to matrix-assisted laser desorption ionization-time of flight (mass spectrometry) [MALDI-TOF (MS)] analysis (Apro Science). In this step, five gel slices termed X1 to X5 were analyzed and identified as Mut S homologue 2 (MSH2), PARP1, XP-group E complementing protein, MSH3, and MSH6, respectively (see Results) (Fig. 1B). Confirmation of these proteins by Western blotting was dependent on the availability of antibodies, and of these, PARP1 and MSH6 were confirmed to bind with TR in a TR-specific manner (see below and Results).
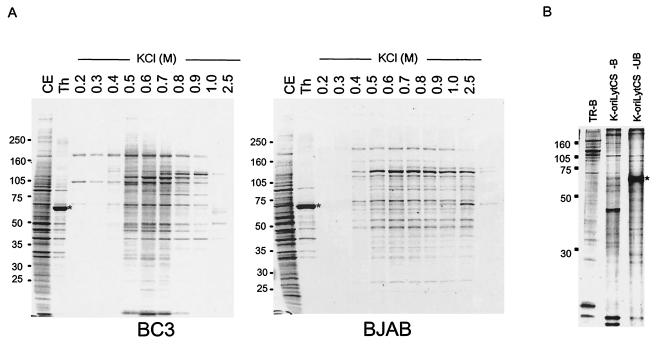
FIG. 1.
Elution profiles from TR(+)Sepharose and K-oriLytCS Sepharose. (A) Elution profiles of BC3 (left) and BJAB (right) cells from TR(+) Sepharose. CE and Th are the profiles of the cell extract (0.1%) and pass-through fractions (0.01%) run on SDS-4 to 20% PAGE, respectively. The bound fraction was eluted with the KCl concentration shown above each lane (0.2 to 2.5 M). The asterisk (*) indicates the position of BSA (0.1%) added to the cell extract just before passing it through the column. (B) Comparison of the elution profiles of the TR(+) Sepharose-bound (TR-B), K-oriLytCS-bound (K-oriLyt CS-B), and non-K-oriLytCS-bound (K-oriLyt CS-UB) fractions. The bound profiles were obtained by eluting all of the bound proteins with 2.5 M KCl. An aliquot (0.1%) of each fraction was separated by SDS-PAGE and visualized with Sypro-Ruby. (C) Genes identified by MS. Typically, the Sypro-Ruby-stained bands from the fractions (0.6 M KCl) of either BC3 or BJAB cell extracts that bound TR(+)Sepharose (BC3/TR+ and BJAB/TR+, respectively) and BC3 extract that bound TR(−)Sepharose (BC3/TR−) were designated X1 to X5 and were analyzed by MALDI-TOF (MS). The identifications of gene products are shown to the right of each band.
Western blot analyses.
A 5-μl aliquot of each fraction was separated by SDS-PAGE and then subjected to Western blot analysis. In this experiment, antibodies against LANA (AB gene), ORC2 (MBL), MCMs (MBL), CDC6 (Sigma), PARP1 (MBL), MSH3 (BD Bioscience), MSH6 (BD Bioscience), PCNA (BD Bioscience), and poly(ADP-ribose) polymer (PAR polymer) (Trevigen) were purchased. The concentrations of each first antibody and the appropriate secondary antibodies conjugated with horseradish peroxidase (Amersham Bioscience) were based on the manufacturer's information. The signal was detected with a commercial reagent (Super Signal West Pico; Pierce) on X-ray film (Rx-U; Fuji Film).
Indirect immunofluorescence assay (IFA).
BC3 cells were harvested and washed twice with PBS. An aliquot was resuspended in PBS, placed on a glass slide, and air dried. The cells were fixed in 4% paraformaldehyde overnight at 4°C. The fixed cells were soaked in distilled water, 70% ethanol, 80% ethanol, 100% ethanol, and again in 100% ethanol, for 5 min each, and air dried again. Cells treated with several drugs such as TPA (12-O-tetradecanoylphorbol 13-acetate [Sigma], 25 ng/ml), HU (50 μM), and NA (10 mM) were prepared in the same way. The cells were then incubated with the first antibody at the concentration recommended by the manufacturer. The anti-PARP1 antibody was a mouse monoclonal antibody (MBL), and the anti-LANA antibody was a rabbit polyclonal antibody (a gift from T. Sata) (31). A mouse monoclonal anti-RTA antibody was used to detect RTA expression (15). The secondary antibodies were goat anti-mouse immunoglobulin G (IgG) antibodies conjugated with Alexa 546 (Molecular Probes-Invitrogen) and goat anti-rabbit IgG antibodies conjugated with Alexa 488 (Molecular Probes-Invitrogen). In the case of replication and transcription activator (RTA), goat anti-mouse IgG antibodies conjugated with Alexa 488 (Molecular Probes-Invitrogen) were used as the secondary antibody. After each incubation with the first and second antibodies, the cells were washed with PBS containing 0.1% Tween 20 (Sigma) (PBS-T) three times. The cells were counterstained with PBS-T containing 0.1 μg of 4′,6′-diamidino-2-phenylindole, dilactate (DAPI) (Molecular Probes)/ml and finally mounted in 90% glycerol-PBS. The signal was observed by confocal fluorescence microscopy (Zeiss 510 META).
Binding assay of TR with PARP1 by ELISA.
The entire TR and TR fragments 1 (nucleotides 1 to 386), 2 (nucleotides 387 to 801), and 2-1 (nucleotides 388 to 695) were amplified by PCR with T7 and reverse primers biotinylated at the 5′ ends. The multiple-cloning site of pBSII was amplified by using the same primers in the same way without biotinylated primers. The amplified fragments were purified with a DNA fragment purification kit (Wizard PCR and DNA purification system; Promega). One microgram of an anti-PARP1 antibody (MBL) was fixed on each well of an enzyme-linked immunosorbent assay (ELISA) plate. The wells were blocked with 1% bovine serum albumin (Nakalai Tesque) in PBS for 1 h at 37°C followed by overnight incubation at 4°C. The wells were washed with PBS-T extensively and then incubated with whole-cell extract adjusted to 150 mM NaCl overnight at 4°C. The wells were washed extensively with PBS-T, 1 pmol of the biotinylated fragments in binding buffer [HEPES-HCl (pH 7.9), 2 mM MgCl2, 0.5 mM DTT, 100 mM KCl, 10 μg of poly(dI-dC)/ml, and 0.01 pmol of nonbiotinylated multiple-cloning site/reaction] was added, and the plates were incubated for 30 min at room temperature (RT). The wells were washed and incubated with streptavidin conjugated with alkaline phosphatase (streptavidin-AP) (Amersham Bioscience) in PBS-T for 30 min at RT, and then the bound streptavidin-AP was detected colorimetrically with p-nitrophenyl phosphate (pNpp) tablets (Bio-Rad) and a diethanolamine buffer according to the manufacturer's protocol (Bio-Rad). The color intensity was measured with an absorbance meter (Benchmark; Bio-Rad) at 405 nm.
Electrophoretic mobility shift assay (EMSA).
The following oligonucleotides were synthesized for probes: 5′-GCGCTTGCTTTCGTTTTCTCCCGCG-3′ and 3′-AACGAAAGCAAAAGAGGGCGCCGGG-5′ (nucleotides 412 to 440), 5′-GCGGCCCCCCGGGCGCGAGCCGCGCGGCGG-3′ and 3′-GGGGGGCCCGCGCTCGGCGCGCCGCCGCCG-5′ (nucleotides 434 to 467), 5′-GGCGCCTCCCGCCCGGGCAT-3′ and 3′-GGAGGGCGGGCCCGTACCCCGGCG-5′ (nucleotides 597 to 624), and 5′-CCCGGGCAGCGAGGGAAGGGG-3′ and 3′-CCGTCGCTCCCTTCCCCCGCGG-5′ (nucleotides 684 to 709). The nucleotide numbers in parenthesis refer to the KSHV terminal sequence (GenBank accession no. U75699). These sense and antisense oligonucleotides were annealed, and 0.1 pmol of the olilgonucleotides was fill-in labeled with [α-32P]dCTP, dATP, dGTP, dTTP, and Klenow fragment (New England Biolabs). The labeled oligonucleotides were filtered through a Sephadex G25 spin column (Amersham Bioscience), followed by ethanol precipitation. The total incorporation of [α-32P]dCTP was measured, and approximately 10,000 cpm of probe was used per reaction. Human PARP1 (0.1 μg; Trevigen) was incubated with each oligonucleotide probe for 30 min at RT in a 20-μl reaction mixture containing 20 mM HEPES-HCl (pH 7.9), 1 mM MgCl2, 0.5 mM DTT, 100 mM KCl, 10 μg of poly(dI-dC)/ml, and 4% Ficoll and analyzed as described previously (45). For competition analyses, a 100-fold excess of annealed cold (nonlabeled) oligonucleotides were included in each reaction mixture. Two micrograms of either polyclonal rabbit anti-PARP1 antibodies (Upstate) or normal rabbit IgG was added for the supershift analyses.
Poly(ADP-ribosyl)ation assay of LANA.
BC3 cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 20 mM EDTA), and then the LANA protein was immunoprecipitated from the extract with a rat monoclonal anti-LANA antibody (AB gene). As a negative control, normal rat IgG was used for the immunoprecipitation. After washing with RIPA buffer three times and Tris-buffered saline (Tris-HCl [pH 8.0], 150 mM NaCl), the material that was immunoprecipitated with the anti-LANA antibody or the normal rat IgG was subjected to a poly(ADP-ribosyl)ation assay. The assay was performed in 50 μM biotin-NAD, 10 μM NAD, and 0.1 μg of human PARP (Trevigen) in 100 μl of reaction buffer (50 mM Tris-HCl [pH 8.0], 0.5 mM DTT, 4 mM MgCl2) at 25°C for 1 h. After adding 20% trichloroacetic acid to stop the reaction and precipitate the protein, the samples were spun. The pellet was washed with ethanol, then washed once with diethyl ether, and dried. The sample was then solubilized in RIPA buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 20 mM EDTA [pH 7.5]) and then subjected to Western blot analysis, where the signal was detected with streptavidin conjugated with horseradish peroxidase (streptavidin-HRP; Amersham Bioscience) and SuperSignal West Pico (Pierce).
HU, NA, and 3-AB treatment of infected cells.
The BC3 cells were treated with HU (50 μM), NA (10 mM), and 3-AB (3 mM) for 5 days. The medium was changed every 2 days to a fresh one containing the drugs. The cells were harvested by centrifugation 1, 3, and 5 days after the drug treatment, and the live cell number was counted by the trypan blue exclusion method. Genomic DNA was extracted on each test day (Wizard SV genomic DNA purification system; Promega). DNA (∼1 ng/reaction) was subjected to real-time PCR with a Roche Light Cycler. The primers based on the K-oriLyt CS sequence (5′-AGGACACCGGTCCCATTTCCCACG-3′ and 5′-CAGCTGCGCTAGGATTAAAGG-3′) were used to measure the KSHV copy number, and the primers based on the human beta-globin gene (5′-ATGGTGCACCTGACTCCTGAGGAG-3′ for the sense strand and 5′-CACCCTGAAGTTCTCAGGATCCAC-3′ for the antisense strand) were used to determine the amount of genomic DNA for precise normalization.
RESULTS
TR binds to distinct factors.
Although details of the latent origin of KSHV have been unclear, studies have shown that TR provides a replication origin in the KSHV latent infection and functions like an autonomous replicating sequence cooperatively with LANA, another essential factor for replication in viral latency (3, 4, 27). LANA is a TR-binding protein that lacks helicase or any other enzymatic activity for DNA replication. If TR and LANA are sufficient viral proteins for viral replication, they must recruit other cellular factors that are also required for viral DNA replication. In the EBV replication system, the primary targets for this are cellular ORCs, CDC6, and MCMs when the viral replication is dependent on the host cell cycle (14, 19, 48, 49). However, the primary nucleotide and amino acid sequences of TR and LANA are completely different from those of EBV oriP and EBNA1, respectively, and it would be interesting to determine whether the same cellular replication machinery components are also associated with TR and/or LANA. Thus, to find factors that bind TR, we generated a TR column conjugated with CNBr-activated Sepharose (Amersham Bioscience) and then sought to identify the factors that bound to it. This kind of analysis, however, frequently includes nonspecific DNA-binding proteins. Therefore, we first passed the lysates through TR(−)Sepharose and added a relatively high concentration of BSA (0.1%), sssDNA (0.4 mg/ml), and poly(dI-dC) (10 μg/ml) to the samples and elution buffers. The profile of proteins eluted from the TR(+)Sepharose column with increasing KCl concentration was quite different between the BC3 (a KSHV-infected cell line) and BJAB (a KSHV-negative, EBV-negative cell line) cell extracts (Fig. 1A), and the profile was completely different from that of K-oriLyt CS Sepharose (Fig. 1B). Thus, TR seemed to bind to specific proteins, some of which were specific for KSHV-infected cells.
SyproRuby-stained proteins at around 105 to 160 kDa were found in sufficient amounts for MALDI-TOF (MS) analysis, and gel slices designated X1 to X5 from the material from the BC3 cell extracts that bound TR(+)Sepharose were subjected to the analysis (Fig. 1C). They were identified as MSH6, MSH3, XP-group E complementing protein, PARP1, and MSH2 (Fig. 1C), respectively. MSH6 and PARP1 seemed to bind TR directly (Fig. 2). This binding was TR-specific, given that we could not detect them on K-oriLyt CS. The identities of MSH3, MSH2, and XP-group E complementing protein have not been confirmed because of the unavailability of antibodies against them.
FIG. 2.
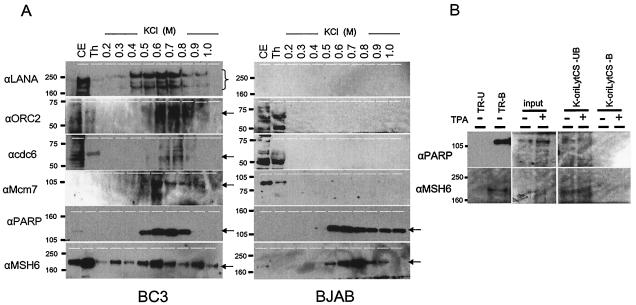
Western blot analysis of the TR(+)Sepharose bound and unbound fractions of BC3 and BJAB cell extracts. (A) Comparison of factors bound to the TR(+)Sepharose column. An aliquot of cell extract (CE; 0.1%), pass-through (Th; 0.01%), and KCl-eluted fractions of BC3 and BJAB cells were separated by SDS-4 to 20% PAGE, blotted on a polyvinylidene difluoride membrane, and analyzed with the commercially available antibodies shown to the left of the panel (α, anti). Typically detected bands are indicated by a bracket for LANA and arrows for ORC2, CDC6, Mcm7, and MSH6. (B) Comparison of bound and unbound fractions to TR(+)Sepharose and K-oriLyt CS Sepharose. K-oriLyt CS was also tested for TPA-uninduced or -induced materials. Binding of PARP1 and MSH6 to TR and K-oriLyt CS was examined. +, present; −, absent.
Cellular replication machinery factors interact with TR.
As mentioned above, if TR and LANA are sufficient for the viral replication in latency, cellular replication factors are likely to associate with them. Some of the TR-bound proteins were not well distinguished or in sufficient amounts in the SyproRuby-stained gels for MALDI-TOF (MS) analysis. Therefore, we tried to detect likely candidates by Western blot analysis with commercially available antibodies. In this analysis, ORC2, CDC6, Mcm7, and LANA were detected on the blot. This binding appeared to be dependent on the association between LANA and TR, since they were not detected in the BJAB cell extracts (Fig. 2A, right). These findings suggested that LANA could recruit cellular DNA replication machinery factors to the TR to initiate viral DNA replication in synchrony with the cellular DNA replication cycle.
PARP1 binds TR.
MALDI-TOF (MS) analysis revealed that several MSH, MSH2, MSH3, and MSH6, XP-group E complementing protein, and PARP1 bound to the TR, as mentioned above. These are all DNA repair-related factors. These proteins may bind DNA nonspecifically; however, even if they bind TR specifically, the reason for their association is unclear at present.
PARP1 seemed to bind TR directly, independent of the presence of KSHV gene products, but this binding was slightly different from that of PARP1 in the BJAB cell extract, as confirmed by Western blot analysis (see below). The binding of PARP1 to TR was neither LANA dependent nor infected cell dependent (Fig. 2A). Nevertheless, the elution profile was close to that of LANA in BC3 cells but was different in BJAB cell extracts, suggesting that the binding of LANA to TR may affect that of PARP1. This binding appeared to be TR specific, given that PARP1 did not bind with K-oriLyt CS (Fig. 2B). Thus, we concluded that PARP1 bound to the KSHV sequence of replication origin, TR, in a sequence-specific manner.
PARP is an abundant enzyme in cells that catalyzes the poly(ADP-ribosyl)ation of proteins and modulates protein function (7). PARP1 binds to specific DNA sequences and affects transcriptional activity in some cases (29, 33, 41). Moreover, it has been reported to bind the EBV oriP region and modulate its function (18). Therefore, we next examined the possible role of PARP1 in KSHV replication and maintenance. It would be especially meaningful if PARP1 had a common function in the replication and maintenance in latency of different gammaherpesviruses, despite the lack of similarity in the primary sequences of the oriP of EBV and TR and of EBNA1 and LANA.
PARP1 colocalizes with LANA in infected cells.
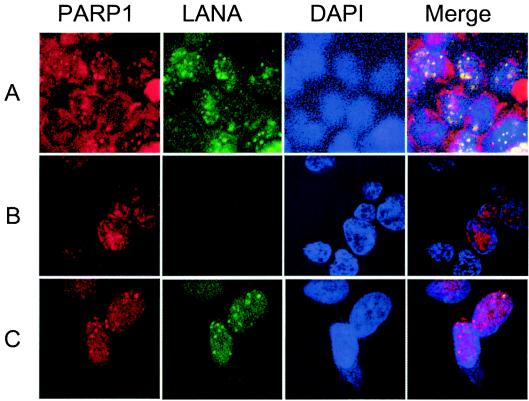
LANA shows a typical dot pattern in the nuclei of infected cells and in uninfected cells transfected with a TR-containing plasmid (reference 3 and our personal communication) by IFA, where the dots are coincident with the presence of the viral genome. Therefore, if PARP1 bound with TR, it would show a similar dot pattern. We found PARP1 (Alexa 546, red) was colocalized with LANA (Alexa 488, green) in the nuclei of infected cells (BC3) (Fig. 3A) and in uninfected cells stably transfected with a LANA expression vector and TR-containing plasmid (BAC TR2) (S. Sakakibara, personal communication) (Fig. 3C), although the colocalization was not complete. On the other hand, PARP1 was typically seen in the perinuclear zone and as small dot-like structures in a weak, diffuse pattern in the nuclei of uninfected cells (BJAB) (Fig. 3B). These staining patterns are reasonable, since PARP1 is thought to be a nuclear protein, although a recent report showed it to be localized to the centrosome and involved in centrosome function (30). These data further confirmed that PARP1 bound TR in the nuclei of infected cells.
FIG. 3.
PARP1 is colocalized with LANA. Anti-PARP1 (Alexa 546, red), anti-LANA (Alexa 488, green), and DAPI (blue) were used to stain nuclei, and merged images are also shown for each case. All of the data were obtained with a Zeiss 510 laser confocal fluorescent microscope. BC3 cells (A), BJAB cells (B), and 293 cells (C) expressing LANA and containing multiple copies of BAC TR-2, a TR-containing bacterial artificial chromosome construct.
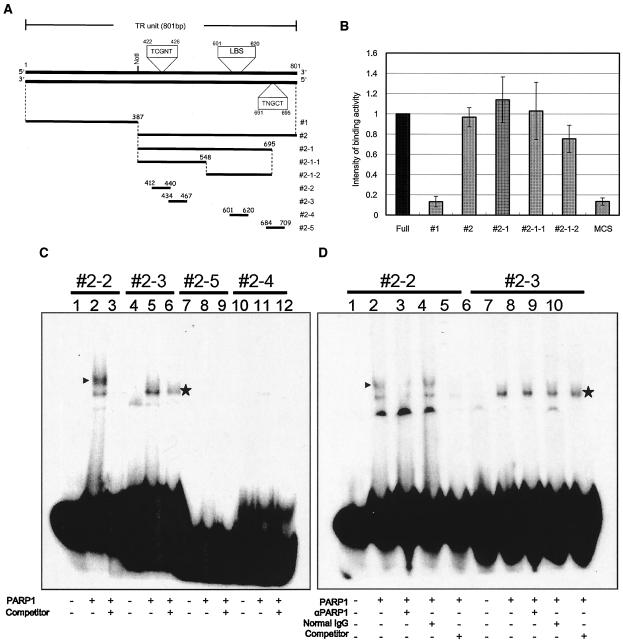
PARP1 binds to a specific region within TR.
PARP1 bound to TR but not to K-oriLyt CS (Fig. 2B), suggesting that PARP1 recognizes a specific nucleotide sequence within TR. The entire TR is 801 bp long, which was too long for analysis by EMSA. To shorten the fragment, several deletion mutants were constructed. The TR was first divided into two parts, 1 (nucleotides 1 to 387) and 2 (nucleotides 388 to 801). The full TR and its parts were amplified with biotinylated primers based on the vector pBSII by PCR. PARP1 in the whole-cell extract was captured by an anti-PARP1 antibody (MBL) and immobilized on an ELISA plate (Pierce). The biotinylated TR or its fragments were incubated with the captured PARP1 in a binding buffer, and then the protein and bound DNA fragment were incubated with streptavidin conjugated with AP (Fig. 4A). The bound fragments were detected by their colorimetric intensities with pNpp (Bio-Rad) as a substrate with a Bio-Rad ELISA meter at 405 nm. In this experiment, fragment 2 showed stronger binding activity than fragment 1 (Fig. 4A and B). We next prepared three fragments: 2-1 (nucleotides 388 to 695), 2-1-1 (nucleotides 388 to 548), and 2-1-2 (nucleotides 549 to 695). Both fragments 2-1-1 and 2-1-2 contained a reported PARP1-binding sequence (TCGNT) (41) and seemed able to bind PARP1, with fragment 2-1-2 showing a weaker binding activity than fragment 2-1-1. This finding suggested that TCGNT may be a target sequence for PARP1. Next, fragments 2-1-1 and 2-1-2 were further divided into smaller pieces to determine and confirm the binding region more precisely. In this experiment, the fragments were prepared as synthetic oligonucleotides, and each oligonucleotide, sense and antisense, was annealed and labeled at the 3′ end with [α-32P]dCTP and Klenow fragment by a fill-in reaction and analyzed by EMSA with purified human PARP1 (Trevigen). As shown in Fig. 4C, a region of fragment 2-2 seemed to bind specifically with PARP1, but the others did not. The faster-migrating band (Fig. 4C and D) seen with the fragment 2-3 probe seemed to be nonspecific because this shifted band was less competed with the competitor and actually was not competed in the same kind of experiment shown in Fig. 4D (lane 10). Further analysis with an anti-PARP1 antibody in an EMSA revealed that the shifted complex in fragment 2-2 was specific for PARP1 (Fig. 4D), since the shifted band disappeared with the addition of the anti-PARP1 antibody. This was probably because the antibody was generated against the DNA binding region of PARP1, and the antibody inhibited the binding of PARP1 to the DNA. Therefore, region 2-2 was found to be the most important for PARP1 binding. Region 2-5 did not show binding with PARP1 in the EMSA, even though it contained a similar sequence and showed moderate binding to PARP1 in the binding ELISA. The difference between the ELISA and EMSA results may be due to experimental stringency and a possible effect of the flanking sequence on the binding activity.
FIG. 4.
PARP1-binding assay. (A) Schematic representation of the TR fragments used for PARP1-binding experiments with ELISA and EMSA. Constructs 1, 2, 2-1, 2-1-1, and 2-1-2 were used in ELISAs, and constructs 2-2 to 2-5 were used in EMSAs. The position of the LANA-binding sequence (LBS) and the suspected PARP1-binding sequences (5′-TCGNT-3′) reported by Nirodi et al. (41) are also shown. (B) PARP1 binding by ELISA. The bound biotinylated fragment was detected colorimetrically by using a streptavidin-biotin-AP complex and pNpp as a substrate. The optical density at 405 nm was measured, and the arbitrary scale of binding intensity for each fragment is shown, where the intensity for the full-length TR was set at 1. (C) EMSA. Each fragment was fill-in labeled with Klenow fragment and [α-32P]dCTP. Lanes 1, 4, 7, and 10, probe only; lanes 2, 5, 8, and 11, probes were incubated with 100 ng of human PARP1 (Trevigen); lanes 3, 6, 9, and 12, each probe was incubated with 100 ng of human PARP1 and a 100-fold excess of each nonlabeled probe. Specifically shifted bands are indicated by arrowheads. (D) EMSA with specific antibodies against PARP1. In this analysis, constructs 2-2 and 2-3 were analyzed. Lanes 1 and 6, probes only; lanes 2 and 7, probes were incubated with 100 ng of human PARP1; lanes 3 and 8, probes were incubated with rabbit polyclonal anti-PARP1 (αPARP1) antibodies (2 μg) and 100 ng of human PARP1; lanes 4 and 9, probes were incubated with normal rabbit IgG (2 μg) and 100 ng of human PARP1; lanes 5 and 10, probes were incubated with 100 ng of human PARP1 and a 100-fold excess of nonlabeled probes. The band that specifically shifted and disappeared with the addition of antibody is indicated by an arrowhead. Asterisks denote nonspecific binding. +, present; −, absent.
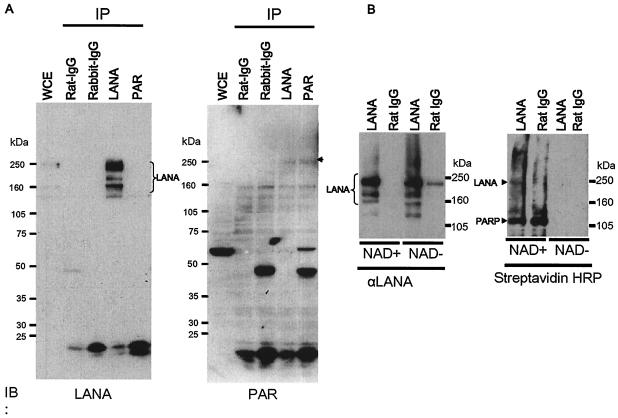
LANA is a target of poly(ADP-ribosyl)ation.
As mentioned above, PARP1 bound to TR and colocalized with LANA in the nucleus, suggesting that LANA may be a target of poly(ADP-ribosyl)ation, which affects the function of proteins, usually by disabling them. We thought that a physical interaction between LANA and PARP1 may be important, as suggested by their similar elution profiles from the TR(+)Sepharose column. Therefore, we tried simply to immunoprecipitate LANA with an anti-LANA antibody to see if PARP1 was coimmunopurified with this method. LANA was strongly visualized in Western blot analysis. In contrast, PARP1 was not detected well under this condition. This finding suggested that LANA did not simply form a stable complex with PARP1 through a protein-protein interaction. Nevertheless, it was still possible that LANA was a target of poly(ADP-ribosyl)ation, since LANA and PARP1 could be very close to each other when bound on the TR, and p53, which is a target of poly(ADP-ribosyl)ation, has not been reported to form a complex with PARP1 (30). Furthermore, PARP1 poly(ADP-ribosyl)ates EBNA1 without any evidence of a physical interaction between them (18). When LANA was immunoprecipitated, an anti-PAR antibody reacted with some protein at the same mobility as LANA (Fig. 5A). On the other hand, when poly(ADP-ribosyl)ated protein was immunoprecipitated with the anti-PAR antibody, LANA was hard to detect, probably because only a very small portion of the LANA molecules was poly(ADP-ribosyl)ated under ordinary culture conditions or because poly(ADP-ribosyl)ated LANA reacted only weakly with the anti-LANA antibody (Fig. 5A). Next, we immunoprecipitated LANA from BC3 cells with a rat anti-LANA antibody and subjected it to a poly(ADP-ribosyl)ation assay with biotinylated NAD. Streptavidin-HRP was used to visualize its incorporation on the blot. As shown in Fig. 5B, the immunoprecipitated LANA was detected with streptavidin-HRP and thus was poly(ADP-ribosyl)ated. LANA was immunoprecipitated well in the concomitant experiment without biotinylated NAD, but streptavidin-HRP did not react with it at all. Thus, we concluded that LANA was poly(ADP-ribosyl)ated.
FIG. 5.
Poly(ADP-ribosyl)ation assay of LANA. (A) Immunoprecipitation followed by Western blot analysis. Whole-cell extracts (WCE) from BC3 cells were immunoprecipitated (IP) with the designated antibodies (2 μg of each) and immunoblotted (IB) with a rat monoclonal anti-LANA antibody (left) or rabbit polyclonal anti-PAR antibodies (right). Multiple forms of LANA are indicated. The arrowhead shows poly(ADP-ribosyl)ated LANA detected by the PAR antibodies. (B) Biotinylated NAD incorporation assay. LANA was immunoprecipitated with either a rat monoclonal anti-LANA (αLANA) antibody (2 μg) or normal rat IgG (2 μg). The immunoprecipitates were incubated with biotinylated NAD and 100 ng of purified human PARP1 (Trevigen) as described in Materials and Methods. The samples were then fractionated by SDS-PAGE and blotted with either a rat monoclonal anti-LANA antibody (2 μg) (left) or streptavidin-HRP (right). The immunoprecipitated LANA (left) and NAD-incorporated LANA and PARP1 are indicated by arrowheads. +, present; −, absent.
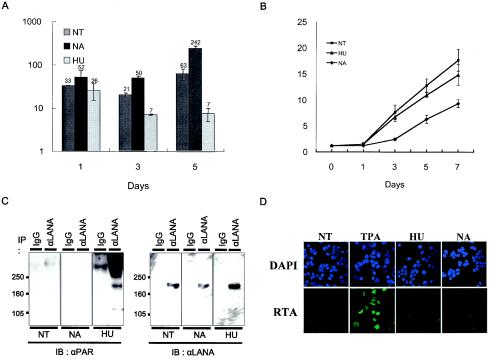
PARP activity modulates virus copy number in infected cells.
The finding that LANA was poly(ADP-ribosyl)ated led us to hypothesize that such modification may affect the function of LANA, thereby regulating the replication and maintenance of the viral genome. To address whether the virus copy number was affected by PARP activity, the infected (BC3) cells were treated with HU, NA, and 3-AB, and the viral genome copy number was measured by real-time PCR. Among these compounds, HU raises and NA and 3-AB decrease PARP activity (8). When the cells were treated with HU, the virus copy number decreased gradually from 30 copies/pg of DNA to 7 copies/pg of DNA. On the other hand, when cells were treated with NA and 3-AB (data not shown), the copy number increased to approximately 240 copies/pg of DNA (Fig. 6A). This finding suggests that PARP activity is relevant to the virus copy number. In this assay, there was the possibility that drug treatment led to the reactivation of virus replication especially in cases of NA and 3-AB treatment. To test this possibility, we assessed the viral lytic gene expression with IFA (Fig. 6D) and Western blot analyses (data not shown). We checked lytic gene expression 5 and 7 days after drug treatment with IFA but could not detect it. The results indicated that no viral lytic gene expression was induced by treating cells with these drugs. However, cell growth was inhibited to some extent with NA (Fig. 6B). The decrease in PARP activity may reduce cellular DNA replication and/or partitioning but not viral replication, resulting in an increase in virus copy number per cell, or alternatively, the massive increase in virus copy number by the drugs may have had a negative effect on cell growth. The decrease in the virus copy number with HU was linked with the increase in poly(ADP-ribosyl)ation of LANA (Fig. 6C) and caused a slight decrease in cell growth (Fig. 6B). This result suggests that the more active PARP is, the smaller the virus copy number is. The presence of KSHV in the cell may have some advantage for cell growth, but the copy number may be an important factor.
FIG. 6.
Viral genome copy number and cell viability in BC3 cells treated with drugs affecting PARP activity. BC3 cells were untreated (NT) or treated with NA or HU as described in Materials and Methods and tested for viral genome copy number by real-time PCR (A) and for cell viability by the trypan blue method (B). (A) Viral genome copy number in drug-treated cells. The viral genome copy number was estimated by amplifying the K-oriLyt CS region and normalized to the β-globin gene content by real-time PCR on the designated day. The number of copies is shown on a logarithmic scale as viral copy number per picogram of DNA. The value above the bar shows a mean value for each case. (B) Cell viability of dru-treated cells. The viable cells were counted on the designated day as described. (C) HU treatment leads to the increase in poly(ADP-ribosyl)ation of LANA. BC3 cells were treated with NT (nontreatment), TPA (25 ng/ml; Sigma), HU (50 μM), or NA (10 mM), and 3 days after treatment, cells were harvested and solubilized with RIPA buffer and the soluble fraction was immunoprecipitated (IP) with either a preimmune rat IgG or a rat monoclonal anti-LANA antibody. The immunoprecipitated fraction was fractionated by SDS-PAGE and immunoblotted (IB) with either rabbit polyclonal anti-PAR (αPAR) antibodies or a rat monoclonal anti-LANA (αLANA) antibody. (D) No lytic gene is expressed by drug treatment. BC3 cells were fixed with 4% paraformaldehyde-PBS 1 day after treatment with NT (nontreatment), TPA (25 ng/ml; Sigma), HU (50 μM), or NA (10 mM). Then the cells were incubated with a mouse monoclonal anti-RTA antibody, followed by goat anti-mouse IgG conjugated with Alexa 488 (Molecular Probes-Invitrogen) (green signal). The signal was detected with a confocal fluorescence microscope (Zeiss 510) (original magnification, ×600). The nucleus was counterstained with DAPI.
DISCUSSION
The TR sequence of the KSHV genome is an essential cis element for viral replication in latency. The TR functions like an autonomous replicating sequence, and LANA binds to a specific sequence within the TR and supports viral replication (4, 27, 37, 38). LANA also interacts with many cellular factors, such as RING3 (39), pRb (43), p53 (22, 32), HP1 (37), CREB-binding protein (36), mSin3 (35), MeCP2 (32), DEK (34), histone H1 (16), ORCs (38), GSK-3β (23), and Suv39H1 (personal communication). Most of these factors are involved in transcription, chromatin structure, and replication, and some or all of them may be required for viral maintenance as episomes and govern the transcription program in latency. Consistent with this idea, LANA is known to be an essential factor for maintaining the episomal form of the viral genome.
Previous studies indicate that TR and LANA are necessary and sufficient for viral replication and maintenance (3, 27). The details about this mechanism, however, have been unclear. Unlike the simian virus 40 large T antigen, LANA does not have any enzymatic activity; therefore, during latency, the replication machinery must be recruited to the TR in synchrony with the cell cycle. Although LANA has been shown to interact directly with ORCs 1 to 5 by the glutathione S-transferase pull-down method (38), the details about these interactions and their purpose remain unclear. Here we sought to identify cellular factors that associate with TR by generating a TR column and analyzing the factors binding to it. We found that cellular replication machinery components such as CDC6 and MCM7, as well as ORC2, bound to TR in a LANA-dependent manner, which was confirmed by Western blot analysis. These factors are likely to support the viral replication that is dependent on the cell cycle, in cooperation with LANA. ORCs do not appear to recognize any specific nucleotide sequences (52), and thus, it is probably very important that a DNA-binding protein, such as LANA in this case, lies on the DNA to recruit such DNA replication machinery factors, although the details of this process remain to be elucidated. When we discuss their dependence on LANA, we should be more careful, since we used different cell lines, BC3 (KSHV positive) and BJAB (KSHV negative), whose genetic backgrounds are not totally identical, though both are thought to originate from a B cell lineage. Thus, not only some viral factors but also some cell-specific factors may be required for interaction with LANA.
We also found that some DNA repair components, such as PARP1, MSH2, MSH3, and MSH6, which were identified by MALDI-TOF (MS), interacted with TR. Among these molecules, PARP1 and MSH6 were confirmed to be TR specific and did not bind K-oriLyt CS. PARP1 bound TR independent of LANA, and the other factors may form a complex with PARP1 (42).
In this report, we focused on PARP1 because it is reported to be a multifunctional protein and was recently found to act as a component of the centromere and the centrosome (21, 30, 47), which suggests that it may be a key factor for chromatin structure (51), replicated genome segregation, and partitioning (50), although it is best known as a target substrate for the caspases in apoptosis (53). In KSHV, a recent report showed that PARP1 interacts with RTA, an immediate-early protein that is a key factor for the induction of lytic replication (25).
We found that PARP1 bound TR independent of LANA, since the PARP1 in the BJAB lysate also bound TR. Although PARP1 is known to bind double-stranded DNA nonspecifically, under the conditions we used, its binding was specific for TR, given that PARP1 did not bind to K-oriLyt CS. Two 5′-TCGNT-3′ motifs, which are present in the regulatory region of the CXCL1 (41) gene, are also in TR. Furthermore, PARP1 is reported to be located in the centromere and the neocentromere and to bind to a 5′-GTGAAAAAG-3′ motif in the human centromeric α-satellite DNA and a tandemly repeated AT28 sequence in a cloned region of neocentromere DNA (21). These sequences are completely different from each other, yet PARP1 was shown to bind to each of them in a specific manner.
Thus, the PARP1-binding sequence may be quite degenerate. We found that one of the two 5′-TCGNT-3′ motifs in the TR may have a higher specificity for PARP1 than the other. This probably means that PARP1's binding to DNA is affected by other factors, such as the flanking sequences and/or the presence of binding proteins such as LANA. In addition, the colocalization of PARP1 and LANA in the infected cells further supported the idea that they interact in vivo and suggested that LANA may be a target of poly(ADP-ribosyl)ation; however, their physical interaction was not proven, as is also the case for PARP1 and p53 (30). An incorporation assay using biotinylated NAD revealed that LANA was a target of PARP1, and it is likely that the function of LANA is modulated by this modification. HU treatment, which increases PARP1 activity, led to a decrease in the viral genome copy number per cell without affecting cell growth, suggesting that the viral replication and partitioning functions are dependent on the ribosylation status of LANA, although the details remain to be elucidated. On the other hand, NA or 3-AB treatment, which causes a decrease in PARP activity, led to an increase in the viral genome copy number. Thus, the PARP1 activity modulated the viral copy number per cell, and therefore, the level of poly(ADP-ribosyl)ation activity may be critical for viral genome replication and maintenance. Since PARP1 is involved in the function and structure of centromeres and centrosomes (21, 30, 51), it is reasonable to propose that the replication and partitioning of the viral genome in latency are affected by PARP1.
PARP1 has also been reported to be involved in EBV genome replication and maintenance (18). PARP1 binds to the EBV oriP dyad symmetry region in a sequence-specific manner, given that it did not bind to the Zta responsive element. Another similar enzyme, tankylase, which is a telomere-associated PARP (7), also binds to EBV oriP in an EBNA1-dependent manner. EBNA1 is poly(ADP-ribosyl)ated, and drugs affecting PARP activity modulate the EBV copy number (18).
Poly(ADP-ribosyl)ation is involved in a variety of physiological and pathophysiological phenomena, such as DNA base excision repair, DNA damage signaling, regulation of genomic instability, and regulation of transcription and proteasomal function (7). PARP1 is a key enzyme and an important factor regulating the host genome structure, replication, and partitioning (33) (6, 26). Its main activity is the poly(ADP-ribosyl)ation of proteins, with the poly(ADP-ribosyl)ated proteins usually losing their activity (29). Although different from the ubiquitin system, poly(ADP-ribosyl)ation may be another protein disabling system. Viruses with an extrachromosomal genome, such as the gammaherpesviruses KSHV and EBV, must utilize cellular components in combination with viral gene products to continue to reside in the host cells. Among these, PARP1 may be used by gammaherpesviruses to modulate this process, and it serves as a potential target for drugs aimed at reducing the levels of these viruses in cells.
REFERENCES
- 1.Adams, A. 1987. Replication of latent Epstein-Barr virus genomes in Raji cells. J. Virol. 61:1743-1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.AuCoin, D. P., K. S. Colletti, Y. Xu, S. A. Cei, and G. S. Pari. 2002. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains two functional lytic origins of DNA replication. J. Virol. 76:7890-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 4.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochkarev, A., J. A. Barwell, R. A. Pfuetzner, E. Bochkareva, L. Frappier, and A. M. Edwards. 1996. Crystal structure of the DNA-binding domain of the Epstein-Barr virus origin-binding protein, EBNA1, bound to DNA. Cell 84:791-800. [DOI] [PubMed] [Google Scholar]
- 6.Bochkarev, A., E. Bochkareva, L. Frappier, and A. M. Edwards. 1998. The 2.2 A structure of a permanganate-sensitive DNA site bound by the Epstein-Barr virus origin binding protein, EBNA1. J. Mol. Biol. 284:1273-1278. [DOI] [PubMed] [Google Scholar]
- 7.Burkle, A. 2001. Physiology and pathophysiology of poly(ADP-ribosyl)ation. Bioessays 23:795-806. [DOI] [PubMed] [Google Scholar]
- 8.Burkle, A. 2001. Poly(APD-ribosyl)ation, a DNA damage-driven protein modification and regulator of genomic instability. Cancer Lett. 163:1-5. [DOI] [PubMed] [Google Scholar]
- 9.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 10.Cesarman, E., and D. M. Knowles. 1997. Kaposi's sarcoma-associated herpesvirus: a lymphotropic human herpesvirus associated with Kaposi's sarcoma, primary effusion lymphoma, and multicentric Castleman's disease. Semin. Diagn. Pathol. 14:54-66. [PubMed] [Google Scholar]
- 11.Cesarman, E., P. S. Moore, P. H. Rao, G. Inghirami, D. M. Knowles, and Y. Chang. 1995. In vitro establishment and characterization of two acquired immunodeficiency syndrome-related lymphoma cell lines (BC-1 and BC-2) containing Kaposi's sarcoma-associated herpesvirus-like (KSHV) DNA sequences. Blood 86:2708-2714. [PubMed] [Google Scholar]
- 12.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 13.Chang, Y., and P. S. Moore. 1996. Kaposi's sarcoma (KS)-associated herpesvirus and its role in KS. Infect. Agents Dis. 5:215-222. [PubMed] [Google Scholar]
- 14.Chaudhuri, B., H. Xu, I. Todorov, A. Dutta, and J. L. Yates. 2001. Human DNA replication initiation factors, ORC and MCM, associate with oriP of Epstein-Barr virus. Proc. Natl. Acad. Sci. USA 98:10085-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, J., K. Ueda, S. Sakakibara, T. Okuno, C. Parravicini, M. Corbellino, and K. Yamanishi. 2001. Activation of latent Kaposi's sarcoma-associated herpesvirus by demethylation of the promoter of the lytic transactivator. Proc. Natl. Acad. Sci. USA 98:4119-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotter, M. A., II, and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 17.Damania, B., and J. U. Jung. 2001. Comparative analysis of the transforming mechanisms of Epstein-Barr virus, Kaposi's sarcoma-associated herpesvirus, and Herpesvirus saimiri. Adv. Cancer Res. 80:51-82. [DOI] [PubMed] [Google Scholar]
- 18.Deng, Z., L. Lezina, C. J. Chen, S. Shtivelband, W. So, and P. M. Lieberman. 2002. Telomeric proteins regulate episomal maintenance of Epstein-Barr virus origin of plasmid replication. Mol. Cell 9:493-503. [DOI] [PubMed] [Google Scholar]
- 19.Dhar, S. K., K. Yoshida, Y. Machida, P. Khaira, B. Chaudhuri, J. A. Wohlschlegel, M. Leffak, J. Yates, and A. Dutta. 2001. Replication from oriP of Epstein-Barr virus requires human ORC and is inhibited by geminin. Cell 106:287-296. [DOI] [PubMed] [Google Scholar]
- 20.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Earle, E., A. Saxena, A. MacDonald, D. F. Hudson, L. G. Shaffer, R. Saffery, M. R. Cancilla, S. M. Cutts, E. Howman, and K. H. Choo. 2000. Poly(ADP-ribose) polymerase at active centromeres and neocentromeres at metaphase. Hum. Mol. Genet. 9:187-194. [DOI] [PubMed] [Google Scholar]
- 22.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 23.Fujimuro, M., F. Y. Wu, C. ApRhys, H. Kajumbula, D. B. Young, G. S. Hayward, and S. D. Hayward. 2003. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 9:300-306. [DOI] [PubMed] [Google Scholar]
- 24.Grundhoff, A., and D. Ganem. 2003. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus permits replication of terminal repeat-containing plasmids. J. Virol. 77:2779-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gwack, Y., H. Nakamura, S. H. Lee, J. Souvlis, J. T. Yustein, S. Gygi, H. J. Kung, and J. U. Jung. 2003. Poly(ADP-ribose) polymerase 1 and Ste20-like kinase hKFC act as transcriptional repressors for gamma-2 herpesvirus lytic replication. Mol. Cell. Biol. 23:8282-8294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassa, P. O., and M. O. Hottiger. 2002. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cell. Mol. Life Sci. 59:1534-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu, J., A. C. Garber, and R. Renne. 2002. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 76:11677-11687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imai, S., S. Koizumi, M. Sugiura, M. Tokunaga, Y. Uemura, N. Yamamoto, S. Tanaka, E. Sato, and T. Osato. 1994. Gastric carcinoma: monoclonal epithelial malignant cells expressing Epstein-Barr virus latent infection protein. Proc. Natl. Acad. Sci. USA 91:9131-9135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kameoka, M., K. Ota, T. Tetsuka, Y. Tanaka, A. Itaya, T. Okamoto, and K. Yoshihara. 2000. Evidence for regulation of NF-kappaB by poly(ADP-ribose) polymerase. Biochem. J. 346:641-649. [PMC free article] [PubMed] [Google Scholar]
- 30.Kanai, M., W. M. Tong, E. Sugihara, Z. Q. Wang, K. Fukasawa, and M. Miwa. 2003. Involvement of poly(ADP-ribose) polymerase 1 and poly(ADP-ribosyl)ation in regulation of centrosome function. Mol. Cell. Biol. 23:2451-2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katano, H., T. Iwasaki, N. Baba, M. Terai, S. Mori, A. Iwamoto, T. Kurata, and T. Sata. 2000. Identification of antigenic proteins encoded by human herpesvirus 8 and seroprevalence in the general population and among patients with and without Kaposi's sarcoma. J. Virol. 74:3478-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Katano, H., Y. Sato, and T. Sata. 2001. Expression of p53 and human herpesvirus-8 (HHV-8)-encoded latency-associated nuclear antigen with inhibition of apoptosis in HHV-8-associated malignancies. Cancer 92:3076-3084. [DOI] [PubMed] [Google Scholar]
- 33.Kraus, W. L., and J. T. Lis. 2003. PARP goes transcription. Cell 113:677-683. [DOI] [PubMed] [Google Scholar]
- 34.Krithivas, A., M. Fujimuro, M. Weidner, D. B. Young, and S. D. Hayward. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596-11604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krithivas, A., D. B. Young, G. Liao, D. Greene, and S. D. Hayward. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 74:9637-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim, C., Y. Gwack, S. Hwang, S. Kim, and J. Choe. 2001. The transcriptional activity of cAMP response element-binding protein-binding protein is modulated by the latency associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Biol. Chem. 276:31016-31022. [DOI] [PubMed] [Google Scholar]
- 37.Lim, C., D. Lee, T. Seo, C. Choi, and J. Choe. 2003. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus functionally interacts with heterochromatin protein 1. J. Biol. Chem. 278:7397-7405. [DOI] [PubMed] [Google Scholar]
- 38.Lim, C., H. Sohn, D. Lee, Y. Gwack, and J. Choe. 2002. Functional dissection of latency-associated nuclear antigen 1 of Kaposi's sarcoma-associated herpesvirus involved in latent DNA replication and transcription of terminal repeats of the viral genome. J. Virol. 76:10320-10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mattsson, K., C. Kiss, G. M. Platt, G. R. Simpson, E. Kashuba, G. Klein, T. F. Schulz, and L. Szekely. 2002. Latent nuclear antigen of Kaposi's sarcoma herpesvirus/human herpesvirus-8 induces and relocates RING3 to nuclear heterochromatin regions. J. Gen. Virol. 83:179-188. [DOI] [PubMed] [Google Scholar]
- 40.Middleton, T., T. A. Gahn, J. M. Martin, and B. Sugden. 1991. Immortalizing genes of Epstein-Barr virus. Adv. Virus Res. 40:19-55. [DOI] [PubMed] [Google Scholar]
- 41.Nirodi, C., S. NagDas, S. P. Gygi, G. Olson, R. Aebersold, and A. Richmond. 2001. A role for poly(ADP-ribose) polymerase in the transcriptional regulation of the melanoma growth stimulatory activity (CXCL1) gene expression. J. Biol. Chem. 276:9366-9374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pleschke, J. M., H. E. Kleczkowska, M. Strohm, and F. R. Althaus. 2000. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 275:40974-40980. [DOI] [PubMed] [Google Scholar]
- 43.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 44.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sakakibara, S., K. Ueda, J. Chen, T. Okuno, and K. Yamanishi. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sarid, R., S. J. Olsen, and P. S. Moore. 1999. Kaposi's sarcoma-associated herpesvirus: epidemiology, virology, and molecular biology. Adv. Virus Res. 52:139-232. [DOI] [PubMed] [Google Scholar]
- 47.Saxena, A., R. Saffery, L. H. Wong, P. Kalitsis, and K. H. Choo. 2002. Centromere proteins Cenpa, Cenpb, and Bub3 interact with poly(ADP-ribose) polymerase-1 protein and are poly(ADP-ribosyl)ated. J. Biol. Chem. 277:26921-26926. [DOI] [PubMed] [Google Scholar]
- 48.Schepers, A., M. Ritzi, K. Bousset, E. Kremmer, J. L. Yates, J. Harwood, J. F. Diffley, and W. Hammerschmidt. 2001. Human origin recognition complex binds to the region of the latent origin of DNA replication of Epstein-Barr virus. EMBO J. 20:4588-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugden, B. 2002. In the beginning: a viral origin exploits the cell. Trends Biochem. Sci. 27:1-3. [DOI] [PubMed] [Google Scholar]
- 50.Tong, W. M., U. Cortes, M. P. Hande, H. Ohgaki, L. R. Cavalli, P. M. Lansdorp, B. R. Haddad, and Z. Q. Wang. 2002. Synergistic role of Ku80 and poly(ADP-ribose) polymerase in suppressing chromosomal aberrations and liver cancer formation. Cancer Res. 62:6990-6996. [PubMed] [Google Scholar]
- 51.Tulin, A., D. Stewart, and A. C. Spradling. 2002. The Drosophila heterochromatic gene encoding poly(ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes Dev. 16:2108-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vashee, S., C. Cvetic, W. Lu, P. Simancek, T. J. Kelly, and J. C. Walter. 2003. Sequence-independent DNA binding and replication initiation by the human origin recognition complex. Genes Dev. 17:1894-1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang, Z. Q., L. Stingl, C. Morrison, M. Jantsch, M. Los, K. Schulze-Osthoff, and E. F. Wagner. 1997. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 11:2347-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yates, J. L., and N. Guan. 1991. Epstein-Barr virus-derived plasmids replicate only once per cell cycle and are not amplified after entry into cells. J. Virol. 65:483-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong, W., H. Wang, B. Herndier, and D. Ganem. 1996. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc. Natl. Acad. Sci. USA 93:6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]