Abstract
Gene expression of the nonsegmented negative-strand RNA viruses is determined by the position of each gene relative to that the single 3′ promoter. The general order of genes among all of the viruses of the order Mononegavirales is highly conserved. In previous work we generated recombinant viruses in which the order of the three central genes of the prototypical rhabdovirus, vesicular stomatitis virus, was rearranged to all six possible permutations. While some of these viruses replicated less well than the wild type when assayed by single-step growth analyses in BSC-1 cells, others replicated as well or slightly better. In the work reported here, we used competition assays to compare the fitness of the viruses with alternative gene orders to that of the wild-type (wt) virus. We found that the relative fitness of these recombinant viruses depended on the multiplicity of infection (MOI) but not on the population size. However, during competitions at low MOI, when complementation cannot compensate for the defects of the populations with rearranged genomes, the virus with the wt gene order was always the most fit.
The viruses of the order Mononegavirales have nonsegmented negative-sense RNA genomes which contain from 5 to 10 genes. Expression of the genes is controlled primarily at the level of transcription by the order of genes on the genome relative to the position of the single 3′ promoter. Transcription is obligatorily sequential, and due to attenuation at each intergenic junction there is a progressive decrease in the transcription of genes that are farther from the promoter (1, 2, 14). The order of genes in the viruses of the order Mononegavirales is highly conserved, and it has been suggested that control of gene expression by gene position relative to the promoter may account for this conservation (19). Proteins required in large amounts such as the nucleocapsid protein, N, the phosphoprotein, P, and the matrix protein, M, are encoded at promoter proximal positions, while those needed in catalytic amounts such as the large subunit of the RNA-dependent RNA polymerase, L, are encoded promoter distally. The genes encoding the viral surface glycoprotein(s) are usually located between the M protein gene and the L protein gene and their exact order differs among the different virus families.
Vesicular stomatitis virus (VSV), the prototype of the rhabdovirus family, contains five genes in its negative-stranded RNA genome in the order 3′-N, P, M, G, L-5′. VSV is the model system in which many of the investigations of the mechanisms controlling gene expression and the basic molecular biology of the negative-stranded RNA viruses were carried out (4, 5). It is also a system in which many of the important principles of evolutionary genetics as well as the distinctive features of RNA virus evolution have been tested (6, 8, 17).
In previous work Wertz et al. investigated the importance of gene order to the biology of VSV by recovering viruses from cDNA in which the nucleocapsid (N) gene had been moved from its promoter proximal position to second, third, or fourth in the gene order without introducing any other changes into the recovered viral genomes (25). The translocation of the N gene to successive positions away from the promoter resulted in a stepwise reduction in N mRNA and protein synthesis and directly demonstrated that the position of the gene determined its level of expression. The N protein is needed in stoichiometric amounts to support genome encapsidation and replication. The movement of the N gene away from the promoter and the stepwise reduction in N protein synthesis also resulted in a stepwise reduction in the ability of these viruses to replicate in cell culture and in their lethality for animals (10, 25). In other work Ball et al. manipulated an infectious cDNA clone of VSV to rearrange the positions of the three central P, M, and G genes to all six possible gene orders while leaving the positions of the N and L genes unchanged and introducing no other sequence changes (3). Infectious viruses were recovered that had the three central genes rearranged to all six possible orders, and gene expression was found to be in concordance with position relative to that of the 3′ promoter. Some of these viruses with rearranged genomes showed a decreased ability to replicate in BSC-1 cells, whereas others replicated as well as or slightly better than virus having the wild-type (wt) gene order. These data suggested that despite the highly conserved gene arrangements of viruses of the order Mononegavirales, certain gene rearrangements were not lethal or even necessarily detrimental as measured by single-step growth analyses.
Growth curves can be a useful tool to characterize viral strains, but they may have limitations. For instance, very small differences in titer or time of release may remain unnoticed but have an important contribution to the success or failure of a virus when it replicates in the presence of competitors. To carry out a finer study of the replicative ability of viruses that have rearranged genomes, we have used the competition assays developed by John Holland and coworkers (13) to evaluate the relative levels of fitness of viruses with rearranged genomes in comparison to those having the wt gene order. These studies are based on the use of a genetically marked surrogate wt as a standard competitor during infection. In the work reported here fitness assays have allowed us to detect fitness defects in all viruses with rearranged genomes that have been tested. These differences occur in a manner dependent on the multiplicity of infection (MOI).
MATERIALS AND METHODS
Cells and virus strains.
Four types of host cells were employed: Vero cells, BSC40 cells, BHK-21 clone 13 cells (BHK-cl.13) (15, 21), and BHK-21 cells from John Holland's laboratory (BHK-JH). Cell lines and methods for single-step growth analysis of the rearranged viruses in BHK-21.c13, BSC40, and Vero cells were as described previously (3, 9, 10). BHK-JH cells were grown in minimal essential medium (MEM) with 7% bovine calf serum and 0.06% proteose peptone #3 (Gibco). All titrations were done in T-25 flasks, with approximately 2 × 106 cells per flask. The hybridoma that produces I1 monoclonal antibody (MAb) was kindly provided by Douglas Lyles (16). Large stocks of I1-MAb were prepared as previously described (7, 13), aliquoted, and frozen until needed.
The wt cDNA (gene order NPMGL) is the sequence of Indiana serotype, San Juan strain, except for the G open reading frame, which was cloned from cDNA of the Orsay strain (26). Several recombinant viruses with different gene orders, including strains GMP (NGMPL), MGP (NMGPL), GPM (NGPML), MPG (NMPGL), and G1N2 (GNPML), were constructed from this background. The generation and characterization of these viruses has been described in detail elsewhere (3, 9).
Fitness assays.
Relative fitness was determined by direct competition between a genetically marked surrogate wt strain (designated strain RU; see Results and Discussion) and each of the recombinant VSV strains. Fitness assays were done as previously described (13). Mixtures of test and competitor virus at an approximate ratio between 10:1 and 20:1 (total inoculum size, 2 × 105 PFU) were used to infect a fresh monolayer of BHK-JH cells. Virus mixtures were allowed to replicate for 24 h. The yield was used, upon appropriate dilution, for a second competition passage under the same conditions. Original mixtures and viral yields after each passage were titrated in triplicate in the presence and absence of I1 MAb to quantify changes in the test strain/strain RU ratios. These changes were normalized by the ratio of the original mixture and log transformed, and the fit of the regression was used to calculate a slope which represents the fitness value (13). Competition passages were done in T-25 flasks with 2 × 106 to 2.5 × 106 cells unless otherwise indicated. Regular fitness assays, which are typically done at an MOI of 0.1 with 2 × 105 PFU, were slightly modified to carry out competition passages at different MOI. The sizes of the initial inoculum were 2 × 103, 2 × 106, and 2 × 107 for competitions done at MOI of 0.001, 1, and 10, respectively. When the MOI was 1 or higher, a single competition passage was done to avoid potential accumulation of defective interfering particles (12). When the MOI was 0.1 or lower, fitness assays were continued for between one and five competition passages. Each fitness value was calculated from at least two determinations.
Growth curves.
Burst sizes in Vero cells, BSC40 cells, and BHK-cl.13 cells were done at an MOI of 3 following standard procedures (3, 9). To determine growth curves in BHK-JH cells, infections were done in T-75 flasks with approximately 6 to 7 × 106 cells at an MOI of 0.001 in MEM with 7% fetal bovine serum. Adsorption and entry were allowed by 10 min of incubation at room temperature followed by 40 min of incubation at 37°C. The inoculum was removed, and 15 ml of fresh MEM was added. Each growth curve was done at least three times. Samples were taken from 0 to 50 h, appropriately diluted, and titrated on semiconfluent BHK-JH cell monolayers in T-25 flasks. Burst sizes in BHK-JH cells were calculated from infections at an initial MOI of 0.1.
RESULTS AND DISCUSSION
VSV strains with rearranged genomes can behave as wt strains in growth curve experiments.
We compared levels of viral production during infection of several cell types. These included BHK-cl.13, BHK-JH, BSC40, and Vero cells. We measured burst size of strains carrying the wt gene order (NPMGL) and strains with several rearrangements, including GMP (NGMPL), MGP (NMGPL), GPM (NGPML), MPG (NMPGL), and G1N2 (GNPML). In these assays, replication of all these viruses in all four cell lines was less than twofold different from that of the wt except for viruses G1N2 and, in one cell line, GPM (Table 1 and data not shown). For BHK-21 host cells, the burst sizes at 30 h postinfection were as follows: in BHK-cl.13 cells, 1,115 PFU/cell for strain PMG (wt) and 285 PFU/cell (0.35 of the burst size of strain PMG [wt]) for strain G1N2; in BHK-JH cells, 6,400 PFU/cell for strain PMG (wt) and 740 PFU/cell (0.12 of the burst size of strain PMG [wt]) for strain G1N2.
TABLE 1.
Burst size of VSV strains with rearrangements of the three internal genes in different host cell types at 30 H post infection
| Strain | Burst size in PFU/cell (ratio)a
|
|||
|---|---|---|---|---|
| BHK-cl.13 | BHK-JH | BSC40 | Vero | |
| GMP | 3,957 (1.9) | 5,800 (0.91) | 361 (1.4) | 1,130 (2) |
| MGP | 4,347 (2.1) | 6,200 (0.97) | 522 (2) | 1,435 (2.5) |
| PMG (wt) | 2,087 (1.0) | 6,400 (1.0) | 265 (1.0) | 565 (1.0) |
| GPM | 1,304 (0.62) | 6,000 (0.94) | 100 (0.38) | 313 (0.6) |
| MPG | 2,348 (1.1) | 5,900 (0.92) | 213 (0.8) | 565 (1.0) |
The ratios of the burst size to that of PMG (wt) are shown in parentheses.
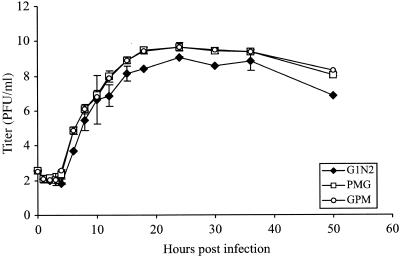
Next we measured growth curves in BHK-JH cells to test whether there were differences in the kinetics of progeny release. The data for three strains, the wt strain as the control and strains GPM and G1N2, which had burst sizes whose values were among the lowest (see above), are shown (Table 1). Figure 1 shows the results of growth curve experiments carried out with BHK-JH cells at an MOI of 0.001 PFU/cell. Strain G1N2 showed differences in the kinetics of virus production compared to the wt strain, but the growth curve of strain GPM was virtually indistinguishable from that of the wt. The results obtained with G1N2 are consistent with those previously observed during the characterization of curves with BHK-cl.13 cells at an MOI of 3 PFU/cell (9). Since the growth analyses described above showed no clear disadvantage in replication as assayed by examination of the growth curves of the viruses with the central genes rearranged, we next used fitness assays to examine relative replicative abilities at a higher level of resolution.
FIG. 1.
Growth curves of recombinant VSV strains. Flasks were infected at an MOI of 0.001 to 0.005. Results show the averages of three independent experiments carried out with strains PMG (wt), GPM, and G1N2. Error bars represent the standard errors when they are larger than the symbol.
Generation of strain RU, a genetically marked surrogate wt strain.
To compare the relative levels of replicative ability (i.e., fitness) of the rearranged viruses by competition assays, we needed a neutral genetic marker that would allow distinction between the two competitors. We chose to use a MAb-resistant mutation (MARM) as a marker, since it had been successfully used in experiments with the Mudd-Summers strain of VSV-Indiana (reviewed in reference 17). To generate a MARM surrogate wt virus, a stock of undiluted wt virus (strain PMG) (250 μl) was mixed with 250 μl of I1 MAb (16) and incubated at 37°C for 1 h. The mixture was plated on a fresh monolayer in a T-25 flask with 2 × 106 cells and incubated for 10 min at room temperature and for 40 min at 37°C. The inoculum was washed three times with MEM supplemented with 7% fetal bovine serum and overlaid with 5 ml of fresh medium-0.3% agar. Infected cells were then incubated at 37°C for 24 h. Several plaques of normal size were picked and amplified once in T-150 flasks with 1.2 × 107 to 1.5 × 107 BHK-JH cells at an MOI of 0.001. The resistance of the clones to I1 MAb was tested by plaque assay in the presence and absence of I1 MAb in the agar overlay. Sensitive clones were discarded, and the resistant clones were further characterized by competition against the parental wt strain (strain PMG). The competitions were initially done on T-25 flasks at an MOI of 0.1 and carried out for five to six passages, as described in Material and Methods. One of the MARM clones, which was designated RU, remained at a stable frequency during the first competition assay (Fig. 2), and it was chosen for further characterization. We carried out additional fitness determinations at an MOI of 0.1 and also replicas of fitness assays at MOI of 0.001, 1, and 10. The average relative fitness resulting from the complete data set was 1.00 ± 0.02, and strain RU was chosen as the genetically marked surrogate wt to carry out fitness analyses of the rearranged strains.
FIG. 2.
Fitness competitions between strains PMG (wt) and RU at an MOI of 0.001. The triplicate ratio measurements at each competition passage of a randomly chosen assay are presented. The solid line corresponds to the fit of the regression. The broken line indicates no changes in the relative ratios of strains PMG and RU.
Fitness assays reveal differences in the replicative abilities of viruses with genome rearrangements.
To perform a more refined characterization of the replicative ability of the recombinant virus strains, we carried out fitness assays between each rearranged strain and the genetically marked MARM surrogate wt (strain RU) in BHK-JH cells. Figure 3 shows representative fitness assays of all the rearranged viruses in competition with RU at an MOI of 0.001. All the rearranged viruses exhibited a significant handicap in fitness compared to the wt strain in spite of the fact that in burst size and growth curve analyses, only strain G1N2 showed disadvantages compared to the wt strain.
FIG. 3.
Representative fitness assays of strains with rearranged genomes at an MOI of 0.001. Fitness assays are presented in two panels for clarity; note the difference in the scale of the y axes. Each point represents the average of the triplicate determination of ratios at each passage. Panel A shows competitions between strains RU and GMP (solid diamonds), RU and GPM (solid circles), and RU and G1N2 (open circles). Panel B shows competitions between strain RU and MGP (open squares) and RU and MPG (solid squares).
The low-fitness phenotype exhibited by viruses with rearranged genomes shows that the wt gene arrangement in viruses of the order Mononegavirales exhibited the highest fitness in the environment in which fitness was tested. These results emphasize the need to carry out competition assays when determining subtle effects of genetic changes on a particular phenotype. While differences in growth curves may be informative, these data show that growth curves which may appear indistinguishable from a wt standard can conceal significant differences in fitness.
The fitness of rearranged genomes varies in a MOI-dependent manner.
In a second set of experiments we carried out competition experiments with BHK-JH cells under standard conditions (13), that is, using an MOI of 0.1 during competitions. These determinations consistently rendered slightly higher relative fitness values than those obtained with an MOI of 0.001 (Fig. 4), suggesting MOI-dependent selection during competition. We then analyzed fitness at MOI of 1 and 10 and observed a gradual approximation of all fitness values towards neutrality (1.0) (Fig. 4).
FIG. 4.
Fitness dependence on MOI during competition between strain RU and the rearranged strains. The MOI in these experiments was increased by increasing the total population size of the inoculum. Error bars show the standard error of each data point. The figure shows the results for strains GMP (solid diamonds), MGP (solid squares), GPM (solid triangles), MPG (solid circles), and G1N2 (open circles).
The differences described above could be due to changes in the MOI during competition, but an alternative explanation was that the results were an effect of the inoculum size. Competitions at an MOI of 10 were done with 10,000-fold more virus than those at an MOI of 0.001, so the population size of the inoculum could have played a significant role in the outcome of the competition by allowing sampling of more beneficial mutations for either competitor. To distinguish between MOI-dependent and population size-dependent selection we carried out additional experiments with strain G1N2, which showed replicative disadvantages both in growth curves and fitness assays at low MOI. These new competitions were done at MOI of 0.1 and 1 but used monolayers with 24 × 106 BHK-JH cells grown in T-300 flasks. Table 2 shows that the fitness values were not significantly different for different population sizes as long as the MOI was kept constant. However, using a specific population size did not result in identical fitness values when the MOI was changed (Table 2). These results have been confirmed with MARM N, another low-fitness VSV strain (18, 27), demonstrating that MOI-dependent selection has a strong effect during competitions. MOI-dependent selection has been previously reported for other viruses, including foot-and-mouth disease virus (20) and phage φ-6 (22-24), that had been selected for improved replication at high MOI. In contrast with the work done with foot-and-mouth disease virus and φ-6, none of the VSV populations had been previously selected for increased fitness at high MOI.
TABLE 2.
Relative fitness values of GIN2 at various MOI and population sizesa
| Population size (PFU) | MOI (PFU/cell)b
|
P | ||
|---|---|---|---|---|
| 0.1 | 1 | 10 | ||
| 2 × 105 | 0.17 ± 0.01 | |||
| 2 × 106 | 0.163 ± 0.002 | 0.44 ± 0.05 | 0.0052 | |
| 2 × 107 | 0.5 ± 0.1 | 0.98 ± 0.03 | 0.01 | |
| P | 0.53 | 0.62 | ||
P values represent the probability of fitness values being the same (unpaired t test).
Number of PFU of competition mixture/cell.
A third potential cause for fitness differences at different MOI is the extent of replication. At high MOI a single round of infection takes place, while at low MOI viral progeny results from at least two rounds of infection. The additional round(s) of infection may help a better competitor express its phenotype. While this possibility cannot be formally ruled out with the present data, it is unlikely that the different number of rounds of infection had a major impact in the results. Work done with MARM N, a low-fitness mutant of VSV Mudd-Summers strain, showed that the fitness phenotype during low-MOI, 24-h infections (as done for this report) corresponds to a composite between the fitness for a single round of replication at low MOI and the fitness for a single round of replication at high MOI (27). The burst size of strain PMG (wt) in BHK-JH cells is close to 6,000 to 7,000 PFU/cell. In a monolayer of 2.5 × 106 cells infected at an MOI of 0.1 with a Poisson distribution, the population of the progeny after the first round of infection is well over 109. Since there are fewer than 106 uninfected cells remaining, the MOI for the second round of infection is very high, and this second round has little to contribute to fitness differences due to complementation. Even for the least fit virus, G1N2, the burst size is enough to result in a second, very high MOI round of infection. Differences in the extent of replication could also arise if the number of rounds of replication that take place at high or low MOI within an infected cell were different. With the present data we cannot conclude which is the case.
The disadvantage of strains with rearranged genomes is due to unbalanced mRNA production and the consequent bias in protein concentration. At low MOI the levels of coinfection are low, and each competitor replicates inside the infected cell with the amount of protein dictated by the expression profile of its own genome. In contrast, at high MOI the probability of coinfection increases. Once inside the cell, and in the presumed absence of compartmentalization, all the templates share the same protein concentration and therefore have the same effective fitness. It is interesting that complementation caused effective fitness improvement of deleterious mutants with defects in cis-acting signals.
Our results fit well with this interpretation. The most noteworthy effect is seen at an MOI of 1. Values at MOI of 0.001 and 0.1 are relatively similar, because the level of coinfection is very small in both cases (in spite of a 100-fold difference in MOI values). At an MOI of 10, on the other hand, all the cells that are infected by RU are also infected by the rearranged competitor. According to the Poisson distribution at an MOI of 1, a cell can be uninfected, doubly infected, or singly infected, a situation qualitatively different from that occurring at an MOI only 10-fold higher or 10-fold lower. The fitness values achieved at an MOI of 10, reaching neutrality or slightly below neutrality (Fig. 4), are consistent with a model of unrestricted complementation. If there were limitations in the level of coinfection or in the availability of viral products inside the coinfected cell, one would expect less-efficient complementation and only a partial restoration of fitness.
It is worth noting that this is not a situation in which strain RU rescues the rearranged strain. Coinfection of strain RU and a rearranged genome results in the averaging of fitness for both competitors. For instance, the burst size of strain G1N2 is significantly lower than that of the wt strain (or strain RU) (see above). The competition experiments were done with a 10- to 20-fold excess of strain G1N2. According to the Poisson distribution, an MOI of 9 for strain G1N2 translates into infection of virtually every cell in the monolayer, and therefore strain RU will always replicate in the presence of G1N2. If RU were rescuing G1N2, one would expect the burst size of the progeny to reach values close to wt values. Instead, in BHK-JH cells the burst size after a 24-h competition passage (900 ± 50 PFU/cell) was similar to that of strain G1N2 alone (2,100 ± 900 PFU/cell) and much lower than that of the wt strain (7,500 ± 630 PFU/cell). Thus, apparent neutrality of strain G1N2 during competitions at high MOI resulted from suboptimal strain RU (wt) replication in cells with imbalanced protein concentration and not from any marked improvement of G1N2 performance. These results, and our interpretation, are also in agreement with the theoretical predictions of Frank's model in that selective differences should disappear with increasing MOI due to complementation (11).
Coinfection is probably common during virus spread within the infected host. After the initial phases of infection in a host, viral load increases and high viremia promotes multiple infections. These indeed must take place, as suggested by the exceedingly frequent isolation of recombinants and reassortants, which can only be generated during coinfection, in members of a variety of viral families, including diverse examples such as poliovirus, dengue fever virus, influenza virus, and human immunodeficiency virus type 1 (reviewed in reference 6).
Conclusions.
We have demonstrated that mutant viruses with indistinguishable growth curves nevertheless can have different fitness levels. Fitness assays have higher sensitivity and are shown here to be able to discriminate differences not revealed in single-step growth analyses. Thus, fitness assays may be necessary to fully examine the potential effects of a mutation on a particular phenotype. The analyses of the group of viruses with rearranged genomes presented in this report support the idea that the gene order in wt VSV is superior to other potential combinations. Finally, the fate of a deleterious genome rearrangement is strongly dependent on the probability of complementation during coinfection with other competitors. Under conditions of high MOI, which probably reflect the conditions under which the intrahost spread of infection occurs, inferior genomes may extend their survival despite—and, indeed, because of—the presence of fitter competitors. Complementation should thus be recognized as an important determinant of the fate of selective mutations in viruses.
Acknowledgments
We are grateful to K. Masters and I. Keren for assistance and to Susi Remold for helpful comments. I1 hybridoma cells were the kind gift of Douglas Lyles.
The work was supported by National Institute of Allergy and Infectious Diseases (National Institutes of Health) grants R01-AI45686 to I.S.N., AI 18270 to L.A.B., and R37-AI12464 to G.W.W.
REFERENCES
- 1.Abraham, G., and A. K. Banerjee. 1976. Sequential transcription of the genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 73:1504-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball, A. L., and C. N. White. 1976. Order of transcription of genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 76:44-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball, L., C. R. Pringle, B. Flanagan, V. P. Perepelitsa, and G. W. Wertz. 1999. Phenotypic consequences of rearranging the P, M, and G genes of vesicular stomatitis virus. J. Virol. 73:4705-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr, J. N., S. P. J. Whelan, and G. W. Wertz. 2002. Transcriptional control of the RNA-dependent RNA polymerase of vesicular stomatitis virus. Biochim. Biophys. Acta 1577:323-353. [DOI] [PubMed] [Google Scholar]
- 5.De, B. P., T. Das, and A. K. Banerjee. 1997. Role of cellular kinases in the gene expression of nonsegmented negative strand RNA viruses. Biol. Chem. 378:489-493. [PubMed] [Google Scholar]
- 6.Domingo, E., C. Biebricher, M. Eigen, and J. J. Holland. 2001. Quasispecies and RNA virus evolution: principles and consequences. Landes Bioscience, Georgetown, Tex.
- 7.Duarte, E. A., I. S. Novella, S. Ledesma, D. K. Clarke, A. Moya, S. F. Elena, E. Domingo, and J. J. Holland. 1994. Subclonal components of consensus fitness in an RNA virus clone. J. Virol. 68:4295-4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elena, S. F., and R. E. Lenski. 2003. Evolution experiments with microorganisms: the dynamics and genetic basis of adaptation. Nat. Rev. Genet. 4:457-469. [DOI] [PubMed] [Google Scholar]
- 9.Flanagan, E. B., L. A. Ball, and G. W. Wertz. 2000. Moving the glycoprotein gene of vesicular stomatitis virus to promoter-proximal positions accelerates and enhances the protective immune response. J. Virol. 74:7895-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flanagan, E. B., J. M. Zamparo, L. Ball, L. L. Rodriguez, and G. W. Wertz. 2001. Rearrangement of the genes of vesicular stomatitis virus eliminates clinical disease in the natural host: new strategy for vaccine development. J. Virol. 75:6107-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank, S. A. 2001. Multiplicity of infection and the evolution of hybrid incompatibility in segmented viruses. Heredity 87:522-529. [DOI] [PubMed] [Google Scholar]
- 12.Holland, J. J. 1991. Defective viral genomes, p. 151-165. In B. Fields and D. Knipe (ed.), Fundamental virology. Raven Press, New York, N.Y.
- 13.Holland, J. J., J. C. de la Torre, D. C. Clarke, and E. Duarte. 1991. Quantitation of relative fitness and great adaptability of clonal populations of RNA viruses. J. Virol. 65:2960-2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iverson, L. E., and J. K. Rose. 1981. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell 23:477-484. [DOI] [PubMed] [Google Scholar]
- 15.Kaariainen, L., and P. J. Gomatos. 1969. A kinetic analysis of the synthesis in BHK-21 cells of RNA specific for Semliki forest virus. J. Gen. Virol. 5:251-265. [DOI] [PubMed] [Google Scholar]
- 16.Lefrancois, L., and D. S. Lyles. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. 1. Analysis of neutralizing epitopes with monoclonal antibodies. Virology 121:157-167. [PubMed] [Google Scholar]
- 17.Novella, I. S. 2003. Contributions of vesicular stomatitis virus to the understanding of RNA virus evolution. Curr. Opin. Microbiol. 6:399-405. [DOI] [PubMed] [Google Scholar]
- 18.Novella, I. S., D. D. Reissig, and C. O. Wilke. 2004. Density-dependent selection in vesicular stomatitis virus. J. Virol. 78:5799-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pringle, C. R., and A. J. Easton. 1997. Monopartite negative-strand RNA genomes. Semin. Virol. 8:49-57. [Google Scholar]
- 20.Sevilla, N., C. M. Ruiz-Jarabo, G. Gomez-Mariano, E. Baranowski, and E. Domingo. 1998. An RNA virus can adapt to the multiplicity of infection. J. Gen. Virol. 79:2971-2980. [DOI] [PubMed] [Google Scholar]
- 21.Stoker, M. 1962. Characteristics of normal and transformed clones arising from BHK21 cells exposed to polyoma virus. Virology 16:649-651. [DOI] [PubMed] [Google Scholar]
- 22.Turner, P. E., and L. Chao. 2002. Escape from prisoner's dilemma in RNA phage phi-6. Am. Nat. 161:497-505. [DOI] [PubMed] [Google Scholar]
- 23.Turner, P. E., and L. Chao. 1999. Prisoner's dilemma in RNA phage phi-6. Nature 398:441-443. [DOI] [PubMed] [Google Scholar]
- 24.Turner, P. E., and L. Chao. 1998. Sex and the evolution of intra-host competition in RNA virus phi-6. Genetics 150:523-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wertz, G. W., V. P. Perepelitsa, and L. A. Ball. 1998. Gene rearrangement attenuates expression and lethality of a nonsegmented negative strand RNA virus. Proc. Natl. Acad. Sci. USA 95:3501-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whelan, S. P. J., L. A. Ball, J. N. Barr, and C. W. Wertz. 1995. Efficient recovery of infectious vesicular stomatitis virus entirely from cDNA clones. Proc. Natl. Acad. Sci. USA 92:8388-8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilke, C. O., D. Reissig, and I. S. Novella. 2004. Replication at periodically changing multiplicity of infection promotes stable coexistence of competing viral populations. Evolution 58:900-905. [DOI] [PubMed] [Google Scholar]