Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV) encodes proteins required for RNA transcription and genome replication as large polyproteins that are proteolytically processed by virus-encoded proteinases to produce mature replicase proteins. In this report, we generated antibodies against SARS-CoV predicted replicase protein and used the antibodies to identify and characterize 12 of the 16 predicted mature replicase proteins (nsp1, nsp2, nsp3, nsp4, nsp5, nsp8, nsp9, nsp12, nsp13, nsp14, nsp15, and nsp16) in SARS-CoV-infected Vero cells. Immunoblot analysis of infected-cell lysates identified proteins of the predicted sizes. Immunofluorescence microscopy detected similar patterns of punctate perinuclear and distributed cytoplasmic foci with all replicase antibodies and as early as 6 h postinfection. Dual-labeling studies demonstrated colocalization of replicase protein nsp8 with nsp2 and nsp3 in cytoplasmic complexes and also with LC3, a protein marker for autophagic vacuoles. Antibodies directed against mouse hepatitis virus (MHV) virions and against the putative RNA-dependent RNA polymerase (Pol) detected SARS-CoV nucleocapsid and nsp12 (Pol), respectively, in SARS-CoV-infected Vero cells. These results confirm the predicted protein processing pattern for mature SARS-CoV replicase proteins, demonstrate localization of replicase proteins to cytoplasmic complexes containing markers for autophagosome membranes, and suggest conservation of protein epitopes in the replicase and nucleocapsid of SARS-CoV and the group II coronavirus, MHV. Further, the results demonstrate the ability of replicase antibodies to detect SARS-CoV-infected cells as early as 6 h postinfection and thus represent important tools for studies of SARS-CoV replication, inhibition, and diagnosis.
The etiologic agent of severe acute respiratory syndrome (SARS) has been shown to be a new human coronavirus (SARS-CoV) (18, 21, 23, 25). The significant morbidity and mortality, and potential for reemergence, make SARS-CoV a continued worldwide public health threat. Genome sequences of SARS-CoV isolates have provided important information regarding the organization of the genome and its relationship to other coronaviruses (21, 25). The significant conservation of deduced amino acid sequence and predicted protein domains in the replicase polyprotein across multiple coronaviruses indicates that these proteins likely play important, conserved roles in replication (29, 32).
Coronavirus replication in cells is initiated by translation of the two overlapping open reading frames (ORF1a and ORF1b) of the 5′ replicase gene to yield two polyproteins, pp1a and pp1ab (see Fig. 1) (25, 32). Based on studies of other coronaviruses, co- and posttranslational proteolytic processing of the nascent SARS-CoV replicase polyproteins is predicted to be mediated by two virus-encoded proteinases, a 3C-like proteinase (3CLpro) and a papain-like proteinase (PLP) (12, 29, 32). The proteolytic precursors and mature replicase proteins likely mediate the processes of replication complex formation, subgenomic mRNA transcription, and genome replication. Comparison of known polyprotein cleavage sites of other coronaviruses with the deduced amino acid (39) sequence of the SARS-CoV replicase polyprotein has resulted in prediction of 16 mature replicase nonstructural proteins (nsps) (32). However, detection of these mature products in SARS-CoV-infected cells has not been reported.
FIG. 1.
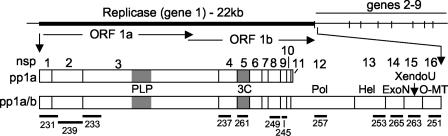
Predicted SARS-CoV replicase gene organization, mature proteins, and antibodies. The location of the SARS-CoV replicase gene in the genome is depicted, with ORF1a and ORF1b shown. The protein domains of polyprotein 1a (pp1a) and the frameshift-fusion polyprotein 1ab (pp1a/b) are shown. Protein domains are indicated by vertical bars, by nonstructural protein number (nsp), and by activities or putative or known functions. The processing of the replicase polyprotein is mediated by two proteinases, the PLP and the 3CL-pro. Black rectangles below proteins indicate regions cloned and expressed as proteins for induction of antibodies (numbers 231 to 265).
The number of mature proteins produced from coronavirus replicase polyproteins suggests a level of complexity and possible virus-encoded functions greater than that of any other known positive-strand RNA virus family. Enzymatic activities have been demonstrated for coronavirus proteinases and the RNA helicase and have been predicted for an RNA-dependent RNA polymerase (Pol) and several RNA processing enzymes (19, 27, 39) (see Fig. 1). Recent bioinformatics analyses of the SARS-CoV replicase gene also have predicted functions for mature replicase proteins in RNA processing (29). The development of reverse genetic systems for SARS-CoV and other coronaviruses should make it possible to experimentally determine the role of replicase proteins in replication and pathogenesis (9, 15, 31, 36-38).
In addition to their functions in RNA synthesis, new roles for replicase proteins as potential determinants of pathogenicity and tropism have been identified. Studies with the JHM and A59 strains of mouse hepatitis virus (MHV), which differ in their tropism and pathogenesis, have suggested that the replicase gene may contribute to tropism and pathogenicity (22). The pathology associated with SARS-CoV infection in humans differs from that seen with other human coronavirus infections. Specifically, SARS-CoV causes an atypical pneumonia, and the virus has been detected in multiple organ systems (3, 23), whereas the human coronaviruses 229E and OC43 cause common colds and are found primarily in the pharynx, although neuroinvasion has been reported (1, 10). The role that SARS-CoV replicase proteins have in the broader tropism and increased pathogenicity of SARS-CoV remains to be determined.
In this study, we generated antibodies against predicted SARS-CoV mature replicase proteins and used the antibodies to identify and characterize replicase proteins in SARS-CoV-infected Vero cells by immunofluorescence and immunoblot assays. In immunoblot experiments, antibodies against nsp1, nsp2, nsp3, nsp4, nsp5, nsp8, nsp9, nsp12, nsp13, nsp14, nsp15, and nsp16 detected specific proteins with masses consistent with bioinformatics predictions. Immunofluorescence labeling for multiple replicase proteins demonstrated punctate cytoplasmic complexes by 6 h postinfection (p.i.). Dual-labeling experiments showed colocalization of replicase proteins in cytoplasmic complexes with each other and with light chain 3 (LC3), a protein marker of autophagosome membranes (24). Finally, antibodies against mouse hepatitis virus Pol (nsp12) detected SARS-CoV nsp12 in infected Vero cells, demonstrating epitope conservation between MHV Pol and SARS-CoV nsp12. These results confirm the expression and processing of SARS-CoV replicase proteins and demonstrate their colocalization to cytoplasmic complexes that contain markers for autophagy or autophagy-like processes. Together, these results suggest significant conservation of replicase protein expression and replication complex formation and form the basis for genetic and biochemical studies of SARS-CoV replicase proteins during replication and pathogenesis.
MATERIALS AND METHODS
Cells and antibodies.
Vero-E6 cells were used in all assays in this report (30). Cells were passaged in OptiPRO serum-free medium supplemented with 4 mM glutamine (Gibco). The MHV-Pol and MHV antibodies have been described previously (8, 11). For generation of antibodies against putative SARS-CoV replicase proteins, the SARS-CoV genome was used as a template for reverse transcription-PCR amplification of targeted areas, cloning into pET-23 protein expression vectors, and purification as previously described (4, 8, 11, 20) (Promega). Proteins generated for antibody production were the following: nsp1, nsp2, nsp3 nsp4, nsp5, nsp8, nsp9, nsp12, nsp13, nsp14, nsp15, and nsp16. The specific nucleotides and amino acids cloned are shown in Table 1. New Zealand White rabbits were immunized with the purified proteins for antibody production (Cocalico, Inc.). After initial inoculation, rabbits were boosted at day 14 and day 21, and test bleeds were performed. Rabbits were boosted again at day 49, and sera from production bleeds on day 56 were used in the experiments in this report. For all studies, preimmune sera from the same rabbits were matched with immune sera. For dual-labeling studies, the same antigens and immunization schedule were used to raise antibodies in guinea pigs. In this study, guinea pig anti-nsp8 was used for dual-labeling experiments.
TABLE 1.
SARS-CoV replicase antibodies
| Proteina | Predicted amino acids | Cloned nucleotides | Amino acids | Antiserum (rabbit) | Useb | Predicted/observed mass (kDa) |
|---|---|---|---|---|---|---|
| nsp1 | M1-180G | 265-945 | 1-227 | VU231 | IB/IF | 20/20 |
| nsp2 | A181-818G | 946-2718 | 228-818 | VU239 | IB/IF | 70/70 |
| nsp3 (PLP) | A819-2740G | 2719-3726 | 819-1154 | VU233 | IB/IF | 213/213 |
| nsp4 | K2741-3240Q | 8572-9235 | 2770-2990 | VU237 | IB/IF | 56/38 |
| nsp5 (3CLpro) | S3241-3546Q | 9986-10590 | 3241-3442 | VU261 | IB | 33/30 |
| nsp8 | A3920-4117Q | 12022-12615 | 3920-4117 | VU249 | IB/IF | 22/22 |
| nsp9 | N4118-4230Q | 12616-12954 | 4118-4230 | VU245 | IB/IF | 12/12 |
| nsp12 (PoI) | S4370-5301Q | 14334-15266 | 4690-5001 | VU257 | IB | 106/106 |
| nsp13 (Hel) | A5302-5902Q | 17053-17969 | 5597-5902 | VU253 | IB/IF | 67/67 |
| nsp14 (ExoN) | A5903-6429Q | 18600-19444 | 6112-6393 | VU265 | IB | 60/60 |
| nsp15 (XendoU) | S6430-6775Q | 19746-20425 | 6494-6720 | VU263 | IB | 38/38 |
| nsp16 (2′-O-MT) | A6776-7073N | 20674-21483 | 6804-7073 | VU251 | IB | 33/33 |
Hel, helicase; ExoN, exonuclease of DEDD superfamily; XendoU, poly(U)-specific endoribonuclease; 2′-O-MT, 2′-O-methyltransferase.
IB, immunoblot; IF, immunofluorescence.
Primers.
Primers used for SARS-CoV replicase protein reverse transcription-PCR were as follows (underlined sequence represents restriction endonuclease site): Nsp1 (left, 5′ CATGCCATGGAGAGCCTTGTT 3′ [NcoI]; right, 5′ TTGCTCGAGACCTCTCTTCGACTC 3′ [XhoI]), Nsp2 (left, 5′ TTGGATCCATGGTCTACTGCTGCCGT 3′ [BamHI]; right, 5′ CGCAAGCTTACCCCCTTTTAAGCG 3′ [HindIII]), Nsp3 (left, 5′ CGCCCATGGCACCAATTAAAGGT 3′ [NcoI]; right, 5′ CGCCTCGAGTGCAATATAAACCTG 3′ [XhoI]), Nsp4 (left, 5′ CGCCCATGGTACATACATTGTCA 3′ [NcoI]; right, 5′ CTTCTCGAGAGCTCTGTAATGCTC 3′ [XhoI]), Nsp5 (left, 5′ CGCCCATGGGTGGTTTTAGGAAAA 3′ [NcoI]; right, 5′ CGCCTCGAGTAATGTTATGGTTGT 3′ [XhoI]), Nsp8 (left, 5′ CGCCCATGGCTATTGCTTCAGAA 3′ [NcoI]; right, 5′ CCTCTCGAGCTGTAGTTTAACAGC 3′ [XhoI]), Nsp9 (left, 5′ GCGCCATGGATAATGAACTGAGTCCA 3′ [NcoI]; right, 5′ CTTCTCGAGCTGAAGACGTACTGT 3′ [XhoI]), Nsp12 (left, 5′ CCTCCATGGTTCCACCTACAAGTT 3′ [NcoI]; right, 3′ CGCCTCGAGTATCCTAAGCATGTT 3′ [XhoI]), Nsp13 (left, 5′ CATCCATGGCTCTCTATTACCCA 3′ [NcoI]; right, 5′ CCTCTCGAGTTGTAATGTAGCCAC 3′ [XhoI]); Nsp14 (left, 5′ CCACCATGGACAAACGTGCAACT 3′ [NcoI]; right, 5′ CCTCTCGAGTCGGTACTCATTTGC 3′ [XhoI]), Nsp15 (left, 5′ CTTCCATGGTGCCAGAGATTAAG 3′ [NcoI]; right, 5′ CGCAAGCTTCACAGAACACACACA 3′ [HindIII]), and Nsp16 (left, 5′ CGCCCATGGATTATGGTGAAAATGC 3′ [NcoI]; right, 5′ CGCCTCGAGGTTGTTAACAAGAAT 3′ [XhoI]).
Immunoblot.
SARS-CoV infected-cell lysates were generated by infecting Vero cell monolayers with SARS-CoV (Urbani strain) at a multiplicity of infection (MOI) of one 50% tissue culture infective dose (TCID50) per cell for 24 h and then lysing cells in NP-40 lysis buffer. Lysates were heated to 65°C for 30 min, and sodium dodecyl sulfate (SDS) was added to a final concentration of 2%. Inactivation of virus was confirmed by testing of dilutions of lysates by TCID50. For immunoblot analysis, 20 μl of lysate (equivalent of 2 × 105 cells) was electrophoresed on 4 to 20% gradient SDS-polyacrylamide gel electrophoresis (PAGE) gels (Bio-Rad). Proteins were transferred to nitrocellulose (Protran; Schleicher & Schuell) according to the manufacturer's recommendations. Membranes were incubated in blocking buffer (5% nonfat dried milk in TBS-Tween [50 mM Tris, pH 7.6, 150 mM NaCl]) for 1 h at room temperature (RT). Antibodies were diluted 1:500 in blocking buffer and incubated with the membrane for 1 h at RT. Membranes were washed three times for 15 min at RT with TBS-Tween. Goat anti-rabbit horseradish peroxidase-conjugated antibodies (Promega) were diluted 1:2,000 in blocking buffer and incubated with the membrane for 1 h at RT. Membranes were washed three times for 15 min at RT with TBS-Tween. Antibody label was detected by chemiluminescence using Lumi-Light (Roche) as per the manufacturer's instructions.
Immunofluorescence.
Replicate Vero cell monolayers on glass coverslips were infected with SARS-CoV at an MOI of one TCID50/cell. Inoculum was removed at 1 h p.i. and replaced with fresh OptiPRO serum-free medium. At the indicated times p.i., cells were fixed and permeablized by addition of −20°C 100% methanol. Virus inactivation was confirmed by incubation of coverslips in medium on monolayers of fresh Vero cells. For immunofluorescence, cells were rehydrated in phosphate-buffered saline (PBS) containing 5% bovine serum albumin (BSA). The primary antibody was diluted 1:500 (except MHV Pol [VU145, used at 1:1,000]) in PBS-2% goat serum-0.05% NP-40-1% BSA. Antibodies were allowed to adsorb for 1 h at RT and then washed two times with PBS-0.05% NP-40-1% BSA. Cells were then incubated for 1 h at RT with a 1:1,000 dilution of anti-rabbit secondary antibodies conjugated to Alexa 488 (Molecular Probes, Eugene, Ore.). Cells were washed twice with PBS-0.05% NP-40-1% BSA and then once with PBS prior to being rinsed in deionized H2O and mounted onto glass slides with Aquapolymount (Polysciences, Inc.). Images were acquired on a Zeiss 510 LSM microscope using a ×40, oil immersion lens and a 488-nm krypton-argon laser for excitation. All immunofluorescence infection and labeling experiments were performed at least three times.
SARS-CoV growth in Vero Cells.
In two independent experiments, plates of Vero cells were infected at an MOI of 1 TCID50/cell. The inoculum was allowed to adsorb to cells for 1 h at 37°C. Inoculum was removed, and cells were washed three times with prewarmed medium. Medium samples were obtained at 0, 1, 3, 6, 12, and 24 h p.i. Samples were serially diluted, and titers were determined in quadruplicate on Vero cell monolayers by TCID50.
Comparison of SARS-CoV and MHV replicase protein.
Deduced amino acid sequences from the replicase genes of SARS-CoV and MHV were compared using pairwise alignment in MacVector (Accelrys) with a Blosum 30 matrix, an open gap penalty of 10, and an extend gap penalty of 0.1 in ClustalW alignment. Sequences of predicted mature proteins were based on published bioinformatics analysis (29, 32).
RESULTS
Generation of antibodies to predicted mature SARS-CoV replicase proteins.
Comparison of the SARS-CoV deduced replicase polyprotein with known coronavirus cleavage sites, processing steps, and mature replicase proteins has led to predicted models of polyprotein processing (Fig. 1) (29). These putative cleavage sites were used as a template for cloning and expression of the predicted mature SARS-CoV replicase proteins to generate specific antibodies in rabbits (Table 1). Published bioinformatic analyses predicted the first polyprotein cleavage site (CS1) at 180G↓A181 (32). However, a 3-amino-acid (aa) sequence identical to MHV CS1 (227RG↓V228) suggested a possible additional or alternative cleaveage site. Thus, the recombinant expressed protein for generation of anti-nsp1 antibody incorporated aa 1 to 228.
The antigenic regions expressed for generation of antibodies to the remaining replicase proteins were based on predicted cleavage sites (29, 32). When possible, the entire predicted mature protein of interest was expressed as an antigen for antibody production. For proteins with a predicted molecular mass of >30 kDa or for proteins whose full-length expression in Escherichia coli was toxic, fragments containing predicted antigenic regions were generated. In this report we have studied antibodies against nsp1, -2, -3, -4, -5, -8, -9, -12, -13, -14, -15, and -16. Proteins and antibodies against nsp6, -7, and -10 have been generated but not yet tested (data not shown).
Detection of ORF1a mature protein products by immunoblotting and immunofluorescence.
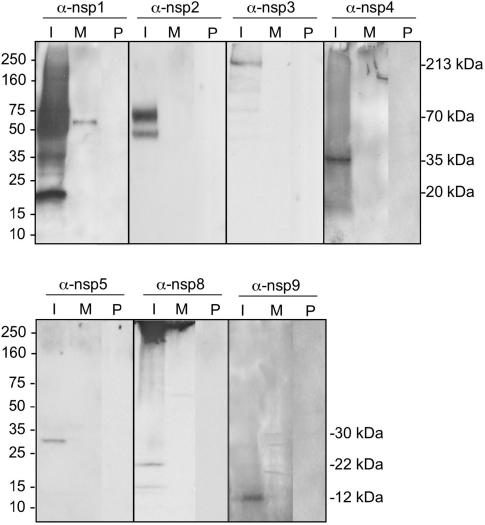
To determine the identity and intracellular localization of ORF1a-encoded mature replicase proteins, antibodies against nsp1, nsp2, nsp3, nsp4, nsp5, nsp8, and nsp9 were used in immunoblot and immunofluorescence experiments. When lysates from SARS-CoV-infected Vero cells at 24 h p.i. were probed by immunoblotting, the anti-nsp1 antibody detected a 20-kDa protein in infected cells (Fig. 2). This is identical to the predicted mass for the nsp1 protein cleaved at 180G↓A181, whereas cleavage at 227RG↓V228 would predict a protein of 25 kDa. The anti-nsp1 serum also detected a discrete band of 35 kDa, as well as a more diffuse signal between 50 and 160 kDa. The identity of these additional bands is unknown, but they may represent oligomers of nsp1. Proteins were not detected in the mock-infected cells or in infected cells probed with preimmune serum from the same animal, indicating that the additional bands detected in infected-cell lysates were not nonspecifically detected viral or cellular proteins.
FIG. 2.
Identification of ORF1a SARS-CoV replicase proteins by immunoblotting. Lysates of SARS-CoV-infected (I) and mock-infected (M) Vero cells were separated by SDS-PAGE, transferred to nitrocellulose, and probed with the anti-nsp1 (α-nsp1), -nsp2, -nsp3, -nsp4, -nsp5, -nsp8 or -nsp9 antibody. Additionally, preimmune (P) sera from the same rabbits were used to immunoblot infected-cell lysates. Marker proteins are to the left of the blots, and observed masses of specific products are to the right of the blots.
The anti-nsp2 antibody detected a 70-kDa protein, consistent with the predicted size of nsp2 cleaved at 180G↓A181 and 818G↓A819. A protein with a molecular mass of ∼48 kDa was also detected from infected-cell lysates, although with less intensity than the 70-kDa product. Whether this represents a processing or degradation product of nsp2 could not be resolved by immunoblotting (Fig. 2). The anti-nsp3 antibody detected a protein of 213 kDa from infected-cell lysates, consistent with the predicted size of a protein spanning 819A to G2740. No immunoreactivity was detected from mock-infected lysates or from infected lysates probed with preimmune serum, demonstrating the specificity of the immune serum (Fig. 2).
The anti-nsp4 antibody detected a protein with an observed mass of 35 kDa, compared with a predicted size of 55 kDa based on a putative cleavage at 2740G↓K2741 and 3240Q↓S3241 (Fig. 2). nsp4 predominantly contains hydrophobic amino acids, similar to the MHV MP1 protein, which, despite a predicted mass of 56 kDa, has an observed molecular mass of 44 kDa (17). The anti-nsp5 antibody, directed against the 3CLpro of SARS-CoV, detected a 30-kDa protein in infected-cell lysates but did not detect any proteins in mock-infected cells or in infected cells probed with preimmune serum (Fig. 2). The anti-nsp8 antibody predominantly detected a 22-kDa protein but also detected a protein of ∼15 kDa from infected lysates. nsp8 has been predicted to be cleaved from the replicase polyprotein at 3919Q↓A3920 and 4117Q↓N4118 and have a mass of 22 kDa. The anti-nsp9 antibody detected a protein with mass of 12 kDa, consistent with a protein spanning 4118N to Q4230 (Fig. 2). The MHV proteins analogous to nsp8 and nsp9 (p22 and p12) are detected in an intermediate precursor with a mass of 150 kDa (26). Neither the anti-nsp8 nor anti-nsp9 antibodies detected a precursor of this size by immunoblotting, suggesting either that the majority of the nsp8 and nsp9 proteins in a SARS-CoV-infected cell are cleaved and the analogous precursor was not present or that precursors could not be recognized by these antibodies in immunoblotting.
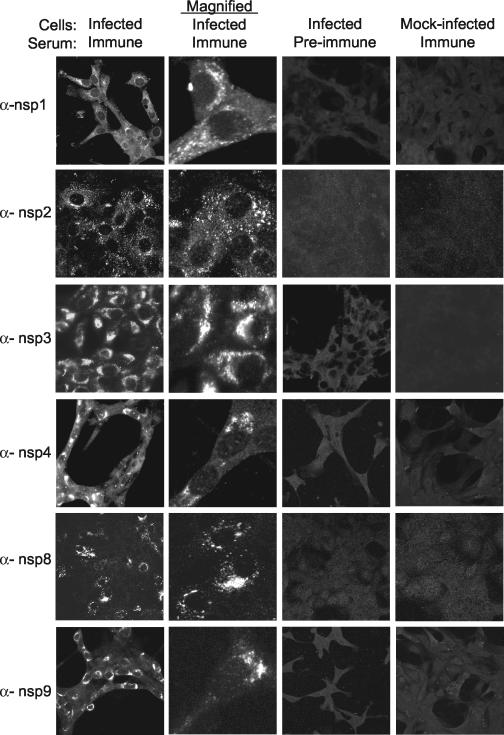
To determine the intracellular localization of SARS-CoV ORF1a replicase proteins, the anti-nsp1, anti-nsp2, anti-nsp3, anti-nsp4, anti-nsp8, and anti-nsp9 antibodies were used in immunofluorescence assays (Fig. 3). Vero cells cultured on glass coverslips were mock infected or infected with SARS-CoV at an MOI of one TCID50/cell for 12 h, followed by fixation with −20°C methanol and processing for immunofluorescence. When infected cells were stained with preimmune sera and mock-infected cells were stained with immune sera, low-level, diffuse, background fluorescence was observed in all cells (Fig. 3, columns 3 and 4). In contrast, when SARS-CoV-infected Vero cells were stained with immune sera against ORF1a proteins, discrete cytoplasmic foci of bright fluorescence were observed (Fig. 3, column 1). The cytoplasmic foci of fluorescence were both perinuclear and distributed throughout the cytoplasm. When infected-cell monolayers were imaged at higher magnification, the cytoplasmic foci resolved into discrete, small structures characteristic of endoplasmic reticulum (ER) or small vesicles (Fig. 3, column 2). With the anti-nsp4 antibody, fluorescence signal also was detected in the nucleoli of infected cells in addition to the cytoplasmic foci. Such nucleolar localization has not been observed for the orthologous MHV protein (MP1). However, anti-nsp4 did detect only a single protein by immunoblotting, and the nucleolar fluorescence was observed only in infected cells with immune serum. Nevertheless, it will be important to use multiple antibodies from different species and different detection approaches before the localization of nsp4 to nucleoli can be concluded.
FIG. 3.
Localization of SARS-CoV ORF1a replicase proteins in Vero cells. Vero cells on glass coverslips were infected with SARS-CoV or were mock infected for 12 h prior to fixation with methanol. Cells were incubated with the indicated antibodies (e.g., anti-nsp1 [α-nsp1]) and a secondary Alexa 488 and imaged by indirect fluorescent-antibody assay, using a Zeiss 510 confocal microscope as described in Materials and Methods. Images in the second column were obtained at higher magnification to show single-cell details of fluorescence labeling.
Replicase proteins can be detected early in infection and prior to increases in virus yield.
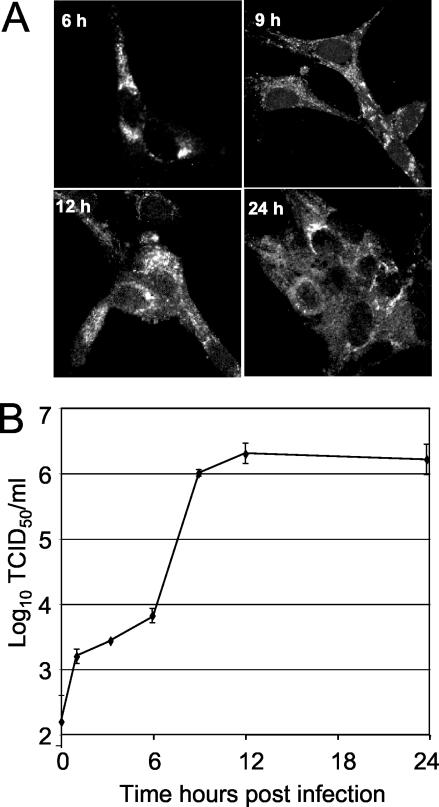
To determine the timing of replicase protein expression, SARS-CoV-infected Vero cells on replicate coverslips were fixed at 3, 6, 9, 12, and 24 h p.i. and stained with anti-nsp1 antibody (Fig. 4A). No specific nsp1 fluorescence was detected at 3 h p.i., but at 6, 9, 12, and 24 h p.i., perinuclear and cytoplasmic foci were readily detected. At 6 h p.i., nearly all Vero cells stained for nsp1, demonstrating the synthesis and targeting of nsp1 early in infection. By 12 and 24 h p.i., more intense, widespread, and in some cases diffuse fluorescence was observed, suggesting a possible change in the distribution or accumulation of nsp1 or degradation of specific foci at later times of infection.
FIG. 4.
(A) Time course of nsp1 detection. Vero cells were infected with SARS-CoV, fixed at 3, 6, 9, 12, and 24 h p.i., and labeled with anti-nsp1 antibodies. No specific signal was observed at 3 h p.i. (not shown). (B) Single-cycle growth of SARS-CoV growth in Vero cells. Vero cell monolayers were infected with SARS-CoV at an MOI of one TCID50/cell, and samples of medium were obtained at times indicated postinfection and assayed by TCID50. Error bars indicate standard deviations of four measurements in each of two independent experiments.
To determine the relationship of the timing of replicase protein detection to that of virus release, a growth assay was performed (Fig. 4B). When Vero cells were infected at an MOI of 1.0 TCID50 per cell and analyzed for supernatant virus at different times postinfection, the maximal rates of virus production were from 9 to 12 h p.i. This exponential release of virus therefore occurred 3 to 6 h after the first detection of nsp1 by immunofluorescence. Interestingly, this experiment also demonstrated that with standard Vero cells, the single-cycle growth of SARS-CoV was very similar to that of MHV in culture, and the results were in apparent contrast to those in the study by Yount et al., showing exponential virus production and peak yields between 24 and 48 h p.i (37), using the same virus strain (Urbani) and cells. However, in the study of Yount et al., an MOI of 0.1 PFU per cell was used for infection, and the results therefore likely reflect multiple rounds of infection. In any case, the comparison of immunofluorescence detection and virus growth in our study strongly suggests a SARS-CoV growth cycle similar to that of MHV and shows that replicase proteins can be readily detected before increases in virus yield.
Identification of ORF1b replicase proteins by immunoblotting and immunofluorescence.
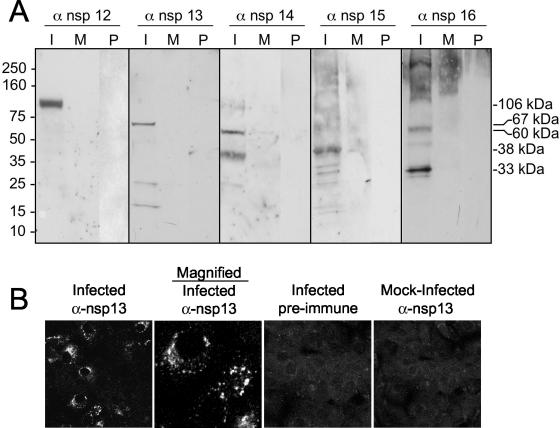
Having shown that ORF1a proteins were detectable by immunoblot and immunofluorescence analysis, we next generated antibodies against the putative mature ORF1b proteins, nsp12, nsp13, nsp14, nsp15, and nsp16. Expression of ORF1b proteins requires a −1 ribosomal frameshift at the 5′ end of ORF1b. This frameshift has been predicted to be ∼30% efficient for other coronaviruses (6, 7). To determine the expression pattern of the ORF1b proteins, we performed immunoblot and immunofluorescence experiments in SARS-CoV-infected Vero cells. The anti-nsp12 antibody, directed against the putative RNA-dependent RNA Pol, detected a single protein with an observed mass of 106 kDa, identical to the predicted mass of a protein cleaved at 4369Q↓S4370 and 5301Q↓A5302 (Fig. 5A). The anti-nsp13 antibody, directed against the RNA helicase, predominantly detected a protein with an observed mass of 67 kDa, consistent with cleavage at 5301Q↓A5302 and 5902Q↓A5903, but to a lesser extent also detected proteins of 22 and 15 kDa (Fig. 5A). The nsp14 protein has been reported to have distant homology with a cellular 3′-to-5′ exonuclease of the DEDD superfamily (29). The anti-nsp14 antibody detected a protein of 60 kDa, identical to the size predicted by analysis of the deduced amino acid sequence. A protein band of 38 kDa was also detected from infected-cell lysates with anti-nsp14 antisera, but neither the 38-kDa nor 60-kDa proteins were detected in mock-infected cells or in infected cells probed with preimmune serum (Fig. 5A). The anti-nsp15 antibody detected a 38-kDa protein from infected-cell lysates and, to a lesser extent, several proteins of lower molecular mass. The nsp15 protein is predicted to have a mass of 38 kDa and has been reported to have domains of a poly(U)-specific endoribonuclease (29) (Fig. 5A). The anti-nsp16 antibody strongly detected a 33-kDa protein in infected cells as well as some proteins of larger molecular masses (approximately 49 kDa, 60 kDa, and several proteins of >110 kDa) (Fig. 5A). Whether any of these proteins represent potential nsp16 precursors could not be determined by immunoblot analysis.
FIG. 5.
Identification of ORF1b SARS-CoV replicase proteins by immunoblotting and immunofluorescence. (A) Lysates of SARS-CoV-infected (I) and mock-infected (M) cells were separated by SDS-PAGE, transferred to nitrocellulose, and probed with the anti-nsp12 (α-nsp12), -nsp13, -nsp14, -nsp15, or -nsp16 antibody. Additionally, preimmune (P) sera were used to probe blots. Mass markers are to the left of the blots, and observed masses of specific proteins are to the right of the blots. (B) Vero cells were infected with SARS-CoV or were mock infected for 12 h prior to fixation with methanol. Cells were labeled with the anti-nsp13 antibody and imaged as described in Materials and Methods. The second column is the image obtained at increased magnification to show detail of the fluorescence image.
The anti-nsp12, -13, -14, -15, and -16 antibodies were next tested in immunofluorescence experiments to determine the subcellular localization of the proteins expressed from ORF1b. In these initial studies, only the nsp13 antibody stained infected cells. At 12 h p.i., nsp13 was detected exclusively in the cytoplasm in bright foci that were predominantly perinuclear but also had foci throughout the cytoplasm, similar to the localizations demonstrated for the ORF1a-expressed proteins. Fluorescence was not detected in mock-infected cells or in infected cells probed with preimmune serum, demonstrating the specificity of the antibody in this assay (Fig. 5B). The remaining ORF1b protein antibodies (anti-nsp12, anti-nsp14, anti-nsp15, and anti-nsp16) did not strongly stain infected Vero cells for immunofluorescence; however, they were detected equivalently to nsp13 in immunoblot analysis, suggesting that protein abundance is not the reason for the lack of detection.
SARS-CoV replicase proteins colocalize with each other and with a marker for autophagic membranes.
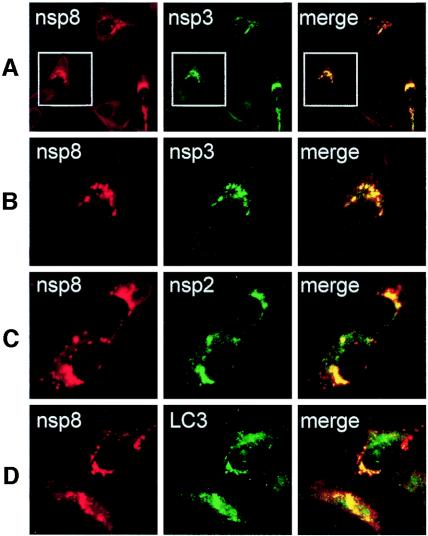
All SARS-CoV anti-replicase antibodies detected similar punctate cytoplasmic complexes by immunofluorescence analysis. In MHV-infected cells, all tested replicase proteins have been shown to colocalize to cytoplasmic replication complexes that are also sites of viral RNA synthesis and that are established on double-membrane vesicles likely derived by induction and usurpation of pathways of cellular autophagy (8, 16, 24, 34). Recent ultrastructural studies of SARS-CoV-infected cells have shown the presence of double-membrane vesicles as well (14). To determine if SARS-CoV replicase proteins colocalized with each other, guinea pig anti-nsp8 antibodies were used in dual-labeling experiments with anti-nsp2 and anti-nsp3 (Fig. 6). Dual labeling with anti-nsp8 and anti-nsp2 in SARS-CoV-infected Vero cells at 6 h p.i. demonstrated readily detectable punctate foci with both antibodies. The signals showed colocalization of nsp8 and nsp2 in all cells where specific signal was detected (Fig. 6C). Similarly, nsp8 and nsp3 were detected in punctate foci with colocalization of signal (Fig. 6A and B). These results corroborated the early detection of replicase proteins observed with nsp1 and demonstrated that the patterns seen with different replicase antibodies actually represented the same complexes. The results suggest that like MHV, the SARS-CoV replicase complexes all localize to cytoplasmic complexes that are sites for viral RNA synthesis.
FIG. 6.
Colocalization of replicase proteins and cellular proteins in SARS-CoV-infected Vero Cells. Vero cells on glass coverslips were infected with SARS-CoV (Urbani strain) at an MOI of 1.0 TCID50 per cell. At 6 h p.i., cells were fixed with 100% methanol and processed for dual labeling using guinea pig anti-nsp8 (red, Alexa 546 in all panels) and rabbit anti-nsp3, anti-nsp 2, or anti-LC3 (green, Alexa 488). (A) Colocalization of nsp8 (red) and nsp3 (green). (B) Magnification of panel A cells (white outline) to show single-cell detail. (C) Colocalization of nsp8 (red) and nsp2 (green). (D) Colocalization of nsp8 (red) and LC3 (green). See the text for details. The merged column shows colocalization of red and green signal as yellow pixels. Images were obtained on a Zeiss LSM 510 confocal microscope.
When SARS-CoV-infected Vero cells were dually labeled for nsp8 and the protein marker for autophagic vacuoles, microtubule-associated protein LC3 (kindly provided by Tamotsu Yoshimori and Noboru Mizushima) (24), LC3 was identified both in large discrete foci and in smaller and distributed foci, consistent with an ER distribution and similar to the pattern seen in MHV-infected DBT cells. In mock-infected Vero cells, only the small distributed foci were observed (data not shown). Dual labeling with anti-nsp8 demonstrated nsp8 in large discrete foci that colocalized with LC3, while the more distributed small foci of LC3 did not colocalize with nsp8 (Fig. 6D). This pattern indicates that nsp8 and likely other replicase proteins localize to a subset of LC3-positive membranes, possibly derived from ER by autophagy or autophagy-like processes.
Conservation of replicase protein epitopes between SARS-CoV and MHV.
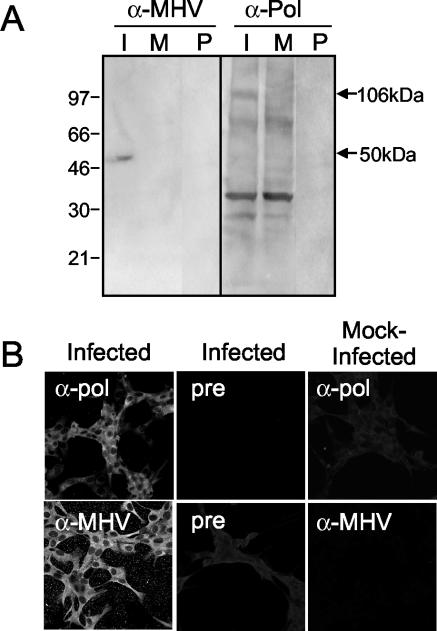
The predicted organization and mature products of the SARS-CoV replicase polyproteins resembles that of the group 2 coronaviruses, such as MHV (29). The deduced amino acid sequences of mature replicase proteins of SARS-CoV and MHV were compared using a Clustal-W analysis (MacVector). Overall, ORF1a had 48% identical or similar residues, while ORF1b had 78% identical or similar residues. Increasing similarity was predicted near the carboxy terminus of ORF1a and throughout ORF1b. The strongest similarity was found in the RNA-dependent RNA Pol (80%) and RNA helicase (83%). In contrast, comparison of SARS-CoV and MHV structural proteins spike, membrane protein, and nucleocapsid predicted 32, 19, and 37% similarity. To determine if predicted similarity between replicase proteins of SARS-CoV and MHV proteins was reflected in their antigenic specificity, antibodies generated against the putative MHV RNA-dependent RNA Pol (ortholog of SARS-CoV nsp12), and against MHV virions were used in immunoblot and immunofluorescence assays in SARS-CoV-infected Vero cells. The MHV anti-Pol antibodies detected a protein with an observed mass of 106 kDa in SARS-CoV-infected lysates (Fig. 7A), identical to the predicted mass (106 kDa) of the putative RNA-dependent RNA polymerase (nsp12 Pol). The protein was not detected by preimmune serum or in mock-infected cells by anti-Pol, demonstrating the specificity of the detection. The anti-MHV antibodies detect the MHV structural proteins spike, membrane protein, and nucleocapsid (11). In lysates of SARS-CoV-infected Vero cells, the MHV antiserum detected only a 50-kDa protein, corresponding to the predicted size of the SARS-CoV nucleocapsid protein (Fig. 7A). This result was interesting because SARS-CoV and MHV nucleocapsid proteins share only 37% amino acid identity; however, this protein was not detected when the membrane was probed with preimmune serum or when mock-infected cells were probed with anti-MHV, suggesting that the detected protein was the SARS-CoV nucleocapsid protein (Fig. 7A).
FIG. 7.
MHV antibodies detect SARS-CoV proteins. (A) Identification of replicase proteins by immunoblotting. SARS-CoV-infected (I) and mock-infected (M) cell lysates were separated by SDS-PAGE, transferred to nitrocellulose, and labeled with anti-MHV (α-MHV) or MHV anti-Pol (α-Pol) antibody. Additionally, preimmune (P) sera were used to label blots. Mass markers are to the left, and masses of specific proteins are given to the right of the blots. (B) Antibodies against MHV-Pol and MHV were used to label SARS-CoV-infected or mock-infected Vero cells by immunofluorescence. Infected Vero cells were stained with immune sera. Fluorescence was not detected in mock-infected cells with immune sera or in infected cells stained with preimmune sera.
When SARS-CoV-infected Vero cells on coverslips were stained with the MHV antibodies, no fluorescence was detected in mock-infected cells or with preimmune sera. In contrast, the anti-MHV and MHV anti-Pol antibodies specifically stained SARS-CoV-infected Vero cells in punctate perinuclear and cytoplasmic foci (Fig. 7B). More diffuse fluorescence also was observed throughout the monolayer with the MHV antisera, similar to the pattern of SARS-CoV antibody labeling at 24 h p.i. Together, the immunoblot and immunofluorescence results suggest epitope conservation between the putative Pol and nucleocapsid proteins of SARS-CoV and MHV and suggest that antibodies against replicase proteins of MHV and SARS-CoV may be useful in studies of protein structure and function.
DISCUSSION
The results in this report demonstrate identification of 12 of the 16 predicted mature SARS-CoV replicase proteins by immunofluorescence, immunoblotting, or both, using antibodies directed against both SARS-CoV and MHV replicase proteins. All replicase proteins identified in immunoblot experiments, with the exception of the highly hydrophobic nsp4, had observed masses consistent with the predicted masses of mature proteins cleaved from the polyprotein. The identities of the additional protein bands recognized by several antisera have not been determined, but their detection only from infected-cell lysates probed with immune serum suggests the possibility of intermediate precursors, oligomers, or alternative cleavage events. Metabolic labeling and pulse-chase experiments will be required define precursor/product relationships among the proteins.
The first SARS-CoV polyprotein cleavage site is predicted to be PLP-mediated cleavage of CS1 at the carboxy terminus of nsp1 (29, 32). However, the SARS-CoV nsp1 contains two putative cleavage sites, at 180G↓A181 and at 227G↓V228, yielding predicted proteins of 20 and 25 kDa, respectively. The 180G↓A181 cleavage site was predicted based on its homology to PLP2 cleavage sites in bovine coronavirus, while the 227G↓V228 site matches the CS1 from MHV (32). SARS-CoV appears to encode only one PLP, which has similarity to PLP2 of other coronaviruses, and the protein identified in SARS-CoV-infected cells was observed to be 20 kDa in mass. Thus, the results in this report support the hypothesis that the first cleavage of the replicase polyprotein occurs at 180G↓A181 and provide further evidence that the SARS-CoV-encoded PLP has substrate specificity most closely matching that of PLP2 from other coronaviruses.
The nsp4 protein was predicted to have a mass of 55 kDa but was observed to have a mass of 35 kDa on SDS-polyacrylamide gels. nsp4 is composed almost exclusively of hydrophobic amino acids and thus may migrate aberrantly on SDS-polyacrylamide gels, similar to the MP1 protein of MHV, which has a predicted mass of 56 kDa but an observed mass of 44 kDa on SDS-polyacrylamide gels. Direct sequencing of the termini of nsp4 will be required for identification of the terminal residues and confirmation of the amino- and carboxy-terminal cleavage sites.
The replicase proteins detected in immunofluorescence assays all displayed similar patterns of localization to discrete cytoplasmic and perinuclear complexes, suggesting that these proteins may form multiprotein cytoplasmic complexes. Studies with MHV have demonstrated that all mature replicase proteins tested colocalize in cytoplasmic complexes that are the sites of RNA transcription and replication (2, 4, 8, 11, 16, 28, 34). The high level of conservation between SARS-CoV and MHV in significant stretches of the replicase polyprotein suggests that the organization of the SARS-CoV replication complexes will have a structure similar to those of MHV and should colocalize with newly synthesized viral RNA. The demonstration that SARS-CoV nsp8 colocalized with nsp2 and nsp3 strongly supports this prediction. In addition, almost all nsp8 signal colocalized with the autophagy marker protein, LC3. The labeling pattern of LC3 in Vero cells was very similar to that seen in murine DBT cells, with constitutive ER-like distribution in mock-infected cells (not shown) and the detection of larger, punctate complexes in virus-infected cells (24). nsp8 colocalized to the larger discrete foci of LC3-positive membranes. This result is consistent with the finding of extensive double-membrane vesicles both in SARS-CoV-infected Vero cells in culture and in pathology specimens from patients (14) and suggests that SARS-CoV may share with MHV and possibly other coronaviruses the strategy of induction and use of pathways of cellular autophagy for formation of replication complexes on double-membrane vesicles.
SARS-CoV replicase proteins were detected by 6 h p.i., prior to exponential increases in supernatant virus. MHV replicase protein and virus release have been detected at times very similar to those described here for SARS-CoV (2, 4, 5, 11, 28), but early detection of SARS-CoV-infected cells by numerous replicase antibodies was interesting, since most studies of SARS-CoV replication published to date have focused on measurements of titer of virus from 24 to 48 h p.i. In this study, time points between 3 and 6 h p.i. were not used, so it is possible that replicase proteins could be detected earlier than 6 h p.i. Furthermore, the detection of SARS-CoV nsp12 (Pol) by MHV anti-Pol antibody demonstrates antigenic conservation between Pol proteins of SARS-CoV and MHV. Surprisingly, this was also true for nucleocapsid, despite limited amino acid similarity overall. These results suggest that replicase antibodies may be useful tools both for studies of SARS-CoV replication and pathogenesis and as approaches to early detection of infected cells. Since there are both highly divergent and highly conserved domains of the replicase gene, it may be possible to identify panels of antibodies that are virus specific and others that are group specific or even broadly useful in detecting multiple coronaviruses. The recent identification of additional novel human coronaviruses and the likelihood of identifying more suggest that differentiating SARS-CoV infection from that from other coronaviruses may be important in any future outbreaks of coronavirus-associated illness (13, 33, 35).
Acknowledgments
This work was supported by Public Health Service grant AI50083-S1 (M.R.D.) and Training Grant in Mechanisms of Vascular Disease, NIH 5 T32 HL07751 (E.P.).
We thank Tamotsu Yoshimori and Noboru Mizushima for LC3 antibodies. Experiments were performed in part through the use of the Cell Imaging Core Resource of the Vanderbilt Ingram Cancer Cener (CA68485 and DK20593). We thank Rachel Graham, Sarah Brockway, and Jennifer Sparks for critical reading of the manuscript.
REFERENCES
- 1.Arbour, N., R. Day, J. Newcombe, and P. J. Talbot. 2000. Neuroinvasion by human respiratory coronaviruses. J. Virol. 74:8913-8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bi, W., J. D. Pinon, S. Hughes, P. J. Bonilla, K. V. Holmes, S. R. Weiss, and J. L. Leibowitz. 1998. Localization of mouse hepatitis virus open reading frame 1a derived proteins. J. Neurovirol. 4:594-605. [DOI] [PubMed] [Google Scholar]
- 3.Booth, C. M., L. M. Matukas, G. A. Tomlinson, A. R. Rachlis, D. B. Rose, H. A. Dwosh, S. L. Walmsley, T. Mazzulli, M. Avendano, P. Derkach, I. E. Ephtimios, I. Kitai, B. D. Mederski, S. B. Shadowitz, W. L. Gold, L. A. Hawryluck, E. Rea, J. S. Chenkin, D. W. Cescon, S. M. Poutanen, and A. S. Detsky. 2003. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. JAMA 289:2801-2809. [DOI] [PubMed] [Google Scholar]
- 4.Bost, A. G., R. H. Carnahan, X. T. Lu, and M. R. Denison. 2000. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 74:3379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bost, A. G., E. Prentice, and M. R. Denison. 2001. Mouse hepatitis virus replicase protein complexes are translocated to sites of M protein accumulation in the ERGIC at late times of infection. Virology 285:21-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brierley, I., M. E. G. Boursnell, M. M. Binns, B. Billimoria, V. C. Blok, T. D. K. Brown, and S. C. Inglis. 1987. An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the coronavirus IBV. EMBO J. 6:3779-3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brierley, I., P. Digard, and S. C. Inglis. 1989. Characterization of an efficient coronavirus ribosomal frameshift signal: requirement for an RNA pseudoknot. Cell 57:537-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockway, S. M., C. T. Clay, X. T. Lu, and M. R. Denison. 2003. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 77:10515-10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casais, R., V. Thiel, S. G. Siddell, D. Cavanagh, and P. Britton. 2001. Reverse genetics system for the avian coronavirus infectious bronchitis virus. J. Virol. 75:12359-12369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denison, M. R. 1998. The common cold: rhinoviruses and coronaviruses, p. 253-280. In R. Dolin and P. F. Wright (ed.), Viral infections of the lung. Marcel Dekker, Inc., New York, N.Y.
- 11.Denison, M. R., W. J. Spaan, Y. van der Meer, C. A. Gibson, A. C. Sims, E. Prentice, and X. T. Lu. 1999. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J. Virol. 73:6862-6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denison, M. R., P. W. Zoltick, S. A. Hughes, B. Giangreco, A. L. Olson, S. Perlman, J. L. Leibowitz, and S. R. Weiss. 1992. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology 189:274-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouchier, R. A., N. G. Hartwig, T. M. Bestebroer, B. Niemeyer, J. C. De Jong, J. H. Simon, and A. D. Osterhaus. 2004. A previously undescribed coronavirus associated with respiratory disease in humans. Proc. Natl. Acad. Sci. USA 101:6212-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldsmith, C. S., K. M. Tatti, T. G. Ksiazek, P. E. Rollin, J. A. Comer, W. W. Lee, P. A. Rota, B. Bankamp, W. J. Bellini, and S. R. Zaki. 2004. Ultrastructural characterization of SARS coronavirus. Emerg. Infect. Dis. 10:320-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gonzalez, J. M., F. Almazan, Z. Penzes, E. Calvo, and L. Enjuanes. 2001. Cloning of a transmissible gastroenteritis coronavirus full-length cDNA. Adv. Exp. Med. Biol. 494:533-536. [DOI] [PubMed] [Google Scholar]
- 16.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 76:3697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanjanahaluethai, A., D. Jukneliene, and S. C. Baker. 2003. Identification of the murine coronavirus MP1 cleavage site recognized by papain-like proteinase 2. J. Virol. 77:7376-7382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 19.Lee, H.-J., C.-K. Shieh, A. E. Gorbalenya, E. V. Koonin, N. LaMonica, J. Tuler, A. Bagdzhadhzyan, and M. M. C. Lai. 1991. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology 180:567-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu, X. T., A. C. Sims, and M. R. Denison. 1998. Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro. J. Virol. 72:2265-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marra, M. A., S. J. Jones, C. R. Astell, R. A. Holt, A. Brooks-Wilson, Y. S. Butterfield, J. Khattra, J. K. Asano, S. A. Barber, S. Y. Chan, A. Cloutier, S. M. Coughlin, D. Freeman, N. Girn, O. L. Griffith, S. R. Leach, M. Mayo, H. McDonald, S. B. Montgomery, P. K. Pandoh, A. S. Petrescu, A. G. Robertson, J. E. Schein, A. Siddiqui, D. E. Smailus, J. M. Stott, G. S. Yang, F. Plummer, A. Andonov, H. Artsob, N. Bastien, K. Bernard, T. F. Booth, D. Bowness, M. Czub, M. Drebot, L. Fernando, R. Flick, M. Garbutt, M. Gray, A. Grolla, S. Jones, H. Feldmann, A. Meyers, A. Kabani, Y. Li, S. Normand, U. Stroher, G. A. Tipples, S. Tyler, R. Vogrig, D. Ward, B. Watson, R. C. Brunham, M. Krajden, M. Petric, D. M. Skowronski, C. Upton, and R. L. Roper. 2003. The genome sequence of the SARS-associated coronavirus. Science 300:1399-1404. [DOI] [PubMed] [Google Scholar]
- 22.Navas, S., and S. R. Weiss. 2003. Murine coronavirus-induced hepatitis: JHM genetic background eliminates A59 spike-determined hepatotropism. J. Virol. 77:4972-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2003. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 279:10136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 26.Schiller, J. J., A. Kanjanahaluethai, and S. C. Baker. 1998. Processing of the coronavirus mhv-jhm polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology 242:288-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seybert, A. 2000. The human coronavirus 229E superfamily 1 helicase has RNA and DNA duplex-unwinding activities with 5′-to-3′ polarity. RNA 6:1056-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi, S. T., J. J. Schiller, A. Kanjanahaluethai, S. Baker, J. Oh, and M. M. C. Lai. 1999. Colocalization and membrane association of murine hepatitis virus gene 1 products and de novo-synthesized viral RNA in infected cells. J. Virol. 73:5957-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331:991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subbarao, K., J. McAuliffe, L. Vogel, G. Fahle, S. Fischer, K. Tatti, M. Packard, W. J. Shieh, S. Zaki, and B. Murphy. 2004. Prior infection and passive transfer of neutralizing antibody prevent replication of severe acute respiratory syndrome coronavirus in the respiratory tract of mice. J. Virol. 78:3572-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thiel, V., J. Herold, B. Schelle, and S. G. Siddell. 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 82:1273-1281. [DOI] [PubMed] [Google Scholar]
- 32.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 84:2305-2315. [DOI] [PubMed] [Google Scholar]
- 33.Van Der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-Van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73:7641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Elden, L. J., A. M. van Loon, F. van Alphen, K. A. Hendriksen, A. I. Hoepelman, M. G. van Kraaij, J. J. Oosterheert, P. Schipper, R. Schuurman, and M. Nijhuis. 2004. Frequent detection of human coronaviruses in clinical specimens from patients with respiratory tract infection by use of a novel real-time reverse-transcriptase polymerase chain reaction. J. Infect. Dis. 189:652-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yount, B. 2000. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 74:10600-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yount, B., K. M. Curtis, E. A. Fritz, L. E. Hensley, P. B. Jahrling, E. Prentice, M. R. Denison, T. W. Geisbert, and R. S. Baric. 2003. Reverse genetics with a full-length infectious cDNA of severe acute respiratory syndrome coronavirus. Proc. Natl. Acad. Sci. USA 100:12995-13000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 76:11065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81:853-879. [DOI] [PubMed] [Google Scholar]