Abstract
Two chimpanzees, 1535 and 1536, became persistently infected following inoculation with RNA transcripts from cDNA clones of hepatitis C virus (HCV). Analysis of the HCV genomes from both animals showed an accumulation of amino acid substitutions over time. The appearance of substitutions in the envelope genes was associated with increased antienvelope antibody titers. However, extensive mutations were not incorporated into hypervariable region 1 (HVR1). A comparison of the nonsynonymous substitution rate/synonymous substitution rate was made at various time points to analyze selective pressure. The highest level of selective pressure occurred during the acute phase and decreased as the infection continued. The nonsynonymous substitution rate was initially higher than the synonymous substitution rate but decreased over time from 3.3 × 10−3 (chimpanzee 1535) and 3.2 × 10−3 (chimpanzee 1536) substitutions/site/year at week 26 to 1.4 × 10−3 (chimpanzee 1535) and 1.7 × 10−3 (chimpanzee 1536) at week 216, while the synonymous substitution rate remained steady at ∼1 × 10−3 substitutions/site/year. Analysis of PCR products using single-stranded conformational polymorphism indicated a low level of heterogeneity in the viral genome. The results of these studies confirm that the persistence of infection is not solely due to changes in HVR1 or heterogeneity and that the majority of variants observed in natural infections could not arise simply through mutation during the time period most humans and chimpanzees are observed. These data also indicate that immune pressure and selection continue throughout the chronic phase.
Hepatitis C virus (HCV) was first identified in 1989 (6) and is the major causative agent of parenterally transmitted non-A, non-B hepatitis. In general, chronic infections in chimpanzees exhibit only very mild hepatitis, while a wide spectrum of disease is observed in humans, ranging from nonapparent to mild to severe chronic active hepatitis or end-stage cirrhosis and, potentially, hepatocellular carcinoma (25). However, the more serious forms of chronic liver disease in association with HCV in humans are usually not seen until at least the third decade after infection. Very few chimpanzees have been monitored for that length of time; therefore, similar long-range illnesses in this animal model cannot be excluded.
The mechanisms leading to viral persistence, which is associated with the more severe forms of liver disease, are as yet undefined. Any single HCV isolate exists as a quasispecies with sequence variability throughout the RNA genome (3, 27). This variation could lead to evasion of the host immune response through the selection of neutralizing antibody or cytotoxic T-lymphocyte escape mutants and thereby the establishment of persistent infection. Evidence for both types of escape mutants have been reported in HCV infections (4, 7, 13, 31). Reports have indicated that hypervariable region 1 (HVR1), located in the N terminus of the E2 protein, evolves more rapidly in vivo than the rest of the viral genome (15, 18) and that it plays a major role in the maintenance of persistent infections (reviewed in reference 17). Previous studies have hypothesized that a higher complexity of virus species provide an indicator of progression to chronicity, particularly in HVR1 (8, 24). However, RNA transcribed from an infectious cDNA clone lacking HVR1 caused a persistent infection in a chimpanzee, indicating that this region is not essential for infection or persistence (10).
The quasispecies nature of natural isolates makes it impossible to distinguish true de novo mutations. In this study, we have examined the molecular evolution of HCV over a 4-year period in two chimpanzees infected with a virus consisting of a single sequence as the starting population. The predominant circulating virus at different times after infection was analyzed by direct sequencing of PCR amplicons. We have been able to monitor the true accumulation of mutations in the HCV genome in chimpanzees over time relative to host responses and viral kinetics. We propose that the majority of variant sequences observed during infections with quasispecies isolates, particularly multiple substitutions observed in HVR1, arise primarily through selection of sequence variants already present in the pool of quasispecies rather than by mutation during the time period most humans and chimpanzees have been observed. Our studies indicate that even over several years only single-amino-acid mutations become fixed at distinct sites and that many more years of infection would be required before significant sequence changes would be achieved.
MATERIALS AND METHODS
Chimpanzees.
The housing, maintenance, and care of the chimpanzees used in this study met requirements for the humane use of animals in scientific research as defined by the National Institutes of Health. Chimpanzee 1535 (Ch1535) and chimpanzee 1536 (Ch1536) were inoculated with H77 RNA transcripts by direct intrahepatic injection (14). Serum samples and liver biopsy samples were collected from the animals for RNA extraction and enzyme-linked immunosorbent assay testing.
RNA extractions and real-time RT-PCR.
Total RNA was prepared from 100 μl of a serum sample or a section of a liver biopsy sample with an area of approximately 1 mm3 using TRIzol (Life Technologies, Gaithersburg, Md.) as previously described (16). RNA pellets were resuspended in 10-μl portions of RNasin-dithiothreitol-water (0.2 U of RNasin per μl of water, 10 mM dithiothreitol) (Promega, Madison, Wis.) and stored at −80°C until use. Negative controls, in the form of serum samples from Ch1535 and Ch1536 prior to inoculation or serum samples from uninoculated chimpanzees, were included in all extractions. RNA levels in serum samples were quantified by real-time reverse transcription-PCR (RT-PCR) (22).
RT-PCR.
RT of purified RNA was performed using the first-strand cDNA synthesis kit (Pharmacia, Uppsala, Sweden) according to the manufacturer's instructions, and 10 pg of random hexamers in a 15- or 33-μl reaction mixture volume. Nested PCRs, consisting of 40 cycles each, were performed on RT products using the Expand High Fidelity PCR system (Boehringer Mannheim, Indianapolis, Ind.) as previously described (16). The HCV genome was amplified as overlapping 400- to 1,100-bp fragments. Primer sequences were based on the cDNA consensus clone (designated p90) used for the synthesis of the original RNA transcripts inoculated into Ch1535 and Ch1536.
Sequence analysis of PCR products.
Following PCR, specific fragments were isolated from ethidium bromide-stained gels using Genelute agarose spin columns (Sigma, St. Louis, Mo.) according to the manufacturer's instructions. Sequencing reactions were performed by SeqWright (Houston, Tex.) using HCV-specific primers, and the data were analyzed using the DNAstar (Madison, Wis.) sequence analysis package.
Enzyme-linked immunosorbent assay for E1E2- and HVR1-specific antibodies.
Sera were tested for HCV-specific antibodies using purified E1E2 protein or biotinylated peptides covering amino acids 384 (aa 384) to 410 of the HCV polyprotein as previously described (16). Mean optical density (OD) values were expressed as P/N ratios calculated by dividing the OD at 405 nm for test sera after infection by that obtained for preimmune serum. The cutoff value was taken as P/N = 2.
SSCP analysis of PCR products.
Single-stranded conformational polymorphism (SSCP) analysis was performed using 20 ng (5 μl) of 164- to 176-bp PCR products denatured with 5 μl of 95% formamide. Products were generated by amplification of cDNA obtained by random-primed RT of viral RNA extracted from serum. DNA was resolved on 8% polyacrylamide-1× Tris-borate-EDTA gels (Novex; Invitrogen). Gels were prerun for 30 min at 100 V and 18°C, then loaded, and run for an additional 2 h at 100 V. Bands were visualized by silver staining.
RESULTS
Viral kinetics during persistence.
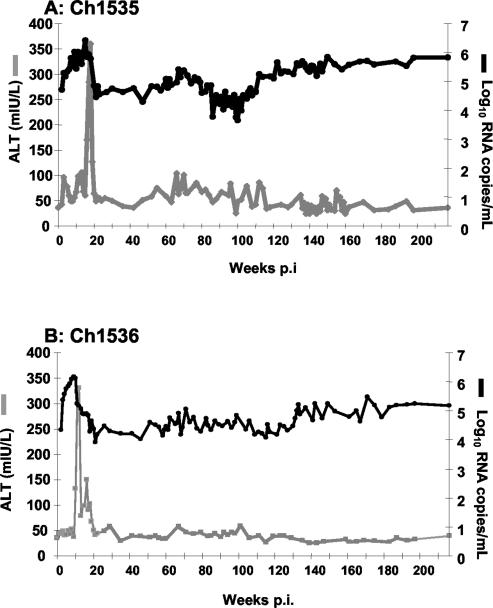
Figure 1A and B show the HCV titer and alanine transaminase (ALT) levels in Ch1535 and Ch1536, respectively, since infection in 1996. There was an exponential increase in viral RNA in both animals following infection, peaking at >106 RNA copies/ml within the first 10 to 12 weeks. ALT elevations, indicative of cellular immune responses in the liver, led to rapid decreases in RNA titers to between 104 and 105 RNA copies/ml; however, both animals developed persistent infections.
FIG. 1.
Clinical and virologic responses up to 216 weeks p.i. in Ch1535 (A) and Ch1536 (B) after inoculation with RNA transcripts representing the HCV infectious cDNA.
Serum RNA titers were measured periodically over a 4-year period. The titers fluctuated during that time in both animals, but the titers in Ch1535 consistently remained approximately 0.5 log10 unit higher than those in Ch1536. Greater fluctuations in viral RNA titers seemed to correlate with greater fluctuations in ALT levels, suggesting that cellular immune responses were active in the liver during these time points. Despite indications of immune responses in both animals throughout the infection, there was no clearance and viral titers remained at or above 104 RNA copies/ml.
HCV genome analysis.
The predominant circulating virus from each animal was analyzed for mutations at 26 (Ch1535) or 22 (Ch1536), 60, 130, and 216 weeks postinfection (p.i.). Analysis of the viral genome sequence isolated from Ch1536 at week 22 indicated no substitutions; therefore, a sample from week 26 was analyzed from this animal. Overlapping fragments were amplified by PCR and directly sequenced. The observed nucleotide and amino acid changes are shown for each animal in Tables 1 and 2.
TABLE 1.
Nucleotide and amino substitutions observed in the predominant circulating virus from serum samples from Ch1535a
| HCV region | NT | NT changeb | AA | AA changeb | Substitutionc at wk:
|
|||
|---|---|---|---|---|---|---|---|---|
| 26 | 60 | 130 | 216 | |||||
| C | 362 | T-C | x | x | x | |||
| E1 | 1199 | T-C | x | x | ||||
| E1 | 1339 | T-C | 333 | V-A | X | |||
| E1d | 1453 | T-C | 371 | V-A | X | X | ||
| E1d | 1458 | G-C | 373 | V-L | X | X | X | |
| E2 | 1512 | A-G | 391 | S-G | X | X | ||
| E2 | 1613 | C-T | x | x | x | |||
| E2 | 1673 | G-T | 444 | Q-H | X | X | ||
| E2 | 1773 | A-G | 478 | S-G | X | X | X | |
| E2 | 1786 | A-T | 482 | E-V | X | |||
| E2 | 1997 | T-C | Mixede | |||||
| E2 | 2351 | A-G | Mixede | |||||
| E2 | 2402 | C-T | x | x | ||||
| E2 | 2499 | G-A | 720 | V-I | X | X | X | |
| p7 | 2603 | C-T | x | x | x | |||
| p7 | 2629 | A-G | 763 | H-R | X | X | ||
| p7 | 2638 | T-C | 766 | V-A | X | X | X | X |
| p7 | 2693 | C-T | x | x | ||||
| NS2 | 2916 | G-A | 859 | V-M | X | X | X | |
| NS2 | 3173 | T-C | x | x | x | |||
| NS2 | 3350 | T-C | x | x | ||||
| NS3 | 3431 | G-T | x | x | ||||
| NS3 | 3632 | C-G | x | x | x | |||
| NS3 | 3749 | C-T | x | x | ||||
| NS3 | 3764 | C-T | x | x | ||||
| NS3d | 4672 | T-A | 1444 | F-S | X | X | ||
| NS3 | 4703 | C-T | x | x | ||||
| NS3 | 4766 | C-T | x | x | ||||
| NS3 | 4938 | G-C | 1533 | A-P | X | X | X | X |
| NS3d | 5244 | G-A | 1635 | V-I | X | X | X | X |
| NS4A | 5440 | T-C | 1700 | V-A | X | X | ||
| NS4B | 5575 | T-C | 1745 | V-A | X | X | ||
| NS4B | 5742 | C-T | x | x | ||||
| NS4B | 5870 | A-G | x | |||||
| NS4B | 5900 | A-G | x | x | ||||
| NS4B | 6083 | A-G | x | x | ||||
| NS4B | 6104 | C-T | x | x | x | |||
| NS5A | 6687 | A-G | Mixede | |||||
| NS5A | 6802 | A-T | 2154 | Y-F | X | |||
| NS5A | 6947 | G-A | x | x | x | |||
| NS5A | 7028 | T-C | x | x | ||||
| NS5B | 7796 | G-T | 2485 | Q-H | X | X | X | |
| NS5B | 7799 | C-A | 2486 | D-E | X | |||
| NS5B | 8243 | G-C | x | |||||
| NS5B | 8552 | C-T | x | x | ||||
| NS5B | 8624 | G-A | x | x | x | |||
| NS5B | 9008 | C-T | x | x | ||||
| NS5B | 9338 | C-T | x | |||||
Nonsynonymous mutations are shown in boldface type. NT, nucleotide; AA, amino acid.
NT and AA changes are shown with the original NT or AA before the hyphen and with the new NT or AA after the hyphen.
Substitutions observed at weeks 26, 60, 130, and 216 p.i. The presence of a synonymous (x) or nonsynonymous (X) substitution is indicated.
Located in previously identified human or chimpanzee T-cell epitopes.
“Mixed” indicates that both the original and the new NT sequences were observed as mixed peaks on the electropherogram.
TABLE 2.
Nucleotide and amino substitutions observed in the predominant circulating virus from serum samples from Ch1536a
| HCV region | NT | NT changeb | AA | AA changeb | Substitutionc at wk:
|
|||
|---|---|---|---|---|---|---|---|---|
| 22 | 60 | 130 | 216 | |||||
| C | 845 | C-T | Mixede | |||||
| E1 | 1043 | C-T | x | |||||
| E1 | 1134 | C-A | 265 | L-I | X | X | X | X |
| E2 | 1540 | T-C | 400 | V-A | X | X | X | |
| E2d | 1548 | C-T | 403 | L-F | Mixede | X | X | |
| E2 | 1673 | G-C | 444 | Q-H | X | |||
| E2d | 1728 | A-C | 463 | T-A | X | X | X | X |
| E2 | 1910 | C-T | Mixede | |||||
| E2 | 1923 | A-G | 528 | S-G | X | X | ||
| E2 | 1941 | A-G | 534 | T-A | X | |||
| E2 | 1997 | T-C | x | |||||
| E2 | 2029 | T-C | 563 | V-A | X | X | ||
| E2d | 2067 | A-G | 576 | N-D | X | |||
| E2 | 2468 | C-T | x | x | x | |||
| p7d | 2689 | T-C | 783 | V-A | X | |||
| p7 | 2718 | G-A | 793 | M-V | X | X | X | |
| NS2 | 2843 | T-C | x | |||||
| NS2 | 2927 | C-T | Mixede | |||||
| NS2 | 2994 | G-A | 885 | V-I | X | |||
| NS2 | 3047 | T-C | x | x | x | |||
| NS3 | 3883 | G-A | 1181 | R-K | X | X | X | X |
| NS3 | 4825 | A-G | 1495 | K-R | Mixede | |||
| NS3 | 4892 | C-T | Mixede | |||||
| NS3 | 5042 | T-C | x | x | x | |||
| NS4A | 5357 | G-T | Mixede | |||||
| NS4A | 5424 | A-G | 1695 | I-V | X | |||
| NS4B | 5984 | T-C | x | |||||
| NS4B | 6245 | T-C | x | |||||
| NS5A | 6484 | G-C | 2048 | G-A | X | |||
| NS5A | 6533 | C-T | x | |||||
| NS5A | 7083 | A-G | 2248 | N-D | X | |||
| NS5A | 7382 | T-C | x | x | x | |||
| NS5A | 7386 | A-G | 2349 | T-A | X | X | X | |
| NS5A | 7575 | G-A | 2412 | A-T | X | X | X | |
| NS5B | 7698 | C-A | 2453 | H-N | X | X | X | X |
| NS5B | 7707 | C-A | 2456 | L-M | X | X | X | |
| NS5B | 7715 | T-C | x | |||||
| NS5B | 8004 | G-A | 2555 | D-N | X | X | X | |
| NS5B | 8576 | A-G | x | |||||
| NS5B | 8846 | T-C | x | |||||
| NS5B | 9135 | C-T | x | x | x | |||
| NS5B | 9193 | G-A | 2951 | R-L | X | |||
| STOP | 9376 | G-A | TGA | TAA | x | x | x | |
Nonsynonymous mutations are shown in boldface type. NT, nucleotide; AA, amino acid.
NT and AA changes are shown with the original NT or AA before the hyphen and with the new NT or AA after the hyphen.
Substitutions observed at weeks 22, 60, 130, and 216 p.i. The presence of a synonymous (x) or nonsynonymous (X) substitution is indicated.
Located in previously identified human or chimpanzee T-cell epitopes.
“Mixed” indicates that both the original and new NT sequences were observed as mixed peaks on the electropherogram.
All but two of the mutations observed at earlier time points were maintained in the viral genomes at subsequent sample dates. This maintenance of the majority of the amino acid substitutions in the circulating virus genome over time confirms that these are not spontaneous PCR errors and that they possibly confer a replicative advantage to the virus in these hosts either through immune escape or adaptation specifically to the chimpanzee. The observation that no mutations were shared between the two animals during the early, presumably adaptive, phase or that mutations did not become dominant until more than 20 weeks into the infection argues more in favor of immune selection or adaptation through regulation of gene expression to maintain persistence.
Positive versus negative or random selection.
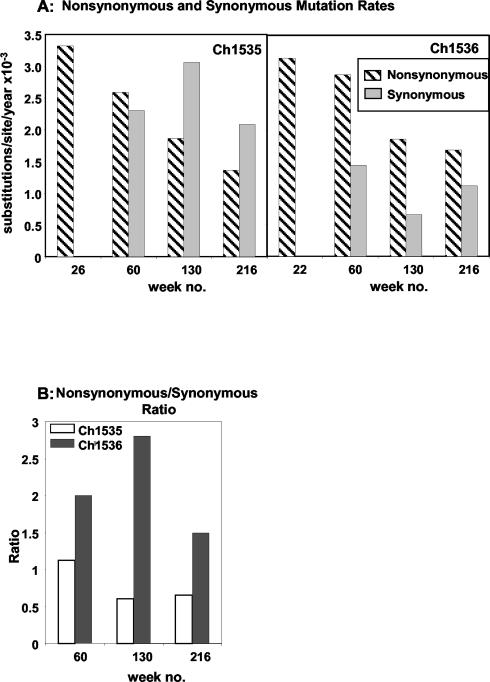
The substitution rates per codon site per year after 60 weeks were calculated as 1.57 × 10−3 (Ch1535) and 1.48 × 10−3 (Ch1536) substitutions/site at the nucleotide level and 2.59 × 10−3 (Ch1535) and 2.88 × 10−3 (Ch1536) substitutions/site at the amino acid level. A method to distinguish positive selection from random accumulation of mutations is to compare the nonsynonymous (protein sequence-altering changes) substitution rate (dN) with the synonymous (silent) substitution rate (dS). These comparisons are shown in Fig. 2A for each animal at each sampling time point. It is significant that for both animals the mutations observed early were 100% nonsynonymous, suggesting that the greatest level of selective pressure occurs during this early phase. When the substitution rates were calculated at the later time points, dN decreased steadily (Fig. 2A) from 3.32 × 10−3 (Ch1535) and 3.14 × 10−3 (Ch1536) substitutions/site/year to 1.36 × 10−3 (Ch1535) and 1.6 × 10−3 (Ch1536) substitutions/site/year. This indicates that fewer amino acid substitutions were acquired and maintained by the virus as the persistent infection progressed, possibly because they no longer confer a replicative advantage due to diminished immune pressure during the chronic phase.
FIG. 2.
(A) Comparison of nonsynonymous and synonymous fixation of mutation rates for Ch1535 and Ch1536 at each sampling time point during the course of the chronic infection. (B) Nonsynonymous substitution rate/synonymous substitution rate ratios for Ch1535 and Ch1536.
The relationship between dN and dS differed somewhat between the animals. The dN/dS ratio for Ch1536 was >1 throughout the study period (Fig. 2B). This would suggest that although the level of selective pressure decreased during chronicity, positive selection was favored. For Ch1535, dN dropped below that for dS from week 130, reflected by a decrease in the ratio to approximately 0.5 (Fig. 2B). This would suggest a significant reduction in positive selective pressure. The dN/dS ratio for weeks 26 and 22 could not be calculated, as no synonymous mutations were detected.
Envelope and HVR1 substitutions.
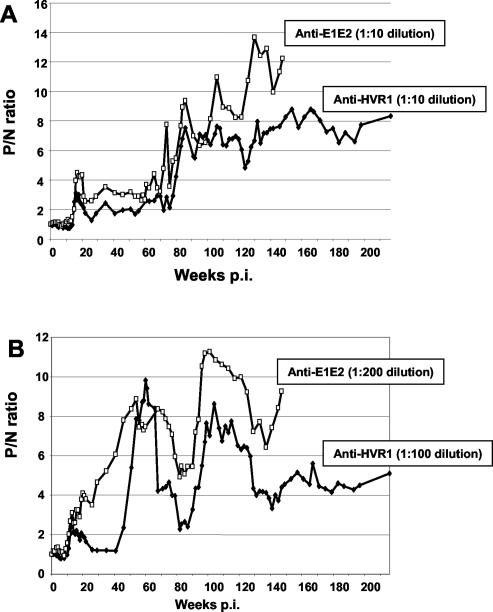
Approximately 50% of the amino acid substitutions were observed in the envelope region; this proportion remained consistent throughout the study period and was higher than for other regions of the genome. The appearance of substitutions seemed to coincide with increased anti-E1E2 antibody titers, suggesting a response to immune pressure. Figure 3 shows antibody responses to the p90 HVR1 peptide and to recombinant E1E2 antigen. The antibody profiles against both antigens are very similar for each animal, with an increase in antibody to HVR1 correlating with an increase to the whole E1E2 protein. Ch1536 developed higher levels of antibody to the envelope region than Ch1535. This increase coincides with a higher amino acid substitution rate in E2 for Ch1536, 6.61 × 10−3 and 4.65 × 10−3 substitutions/site at weeks 130 and 216, respectively, versus 4.41 × 10−3 and 2.66 × 10−3 substitutions/site for the same dates in Ch1535. The concomitant higher antibody level and the dN/dS ratios of 6 and 3.5 at these time points for E2 substitutions in Ch1536 suggest that selective immune pressure took place.
FIG. 3.
Comparison of anti-E1E2 and anti-HVR1 antibody levels in Ch1535 (A) and Ch1536 (B) after inoculation with RNA transcripts representing the HCV infectious cDNA. Responses are represented as P/N ratios with a cutoff value of 2.
Although the envelope appeared to be a site for accumulated mutations, HVR1 did not show extensive sequence diversity following persistent infection, despite increasing antibody to this region. Only one amino acid change, S391G, was observed in virus isolated from Ch1535 at week 130 p.i. In Ch1536, two substitutions were observed at week 60 (V400A) and week 130 (L403F). HVR1 peptides representing the mutated sequences in both animals from aa 391 to 410 were equally well recognized by sera from both chimpanzees taken from early and late time points p.i. (data not shown). These results could indicate that none of these HVR1 substitutions represent escape mutants or that they form part of a conformational epitope.
Genome heterogeneity.
It is possible that although the predominant circulating virus incorporates a low number of stable mutations over time, the overall population of viruses present in serum shows a greater variation.
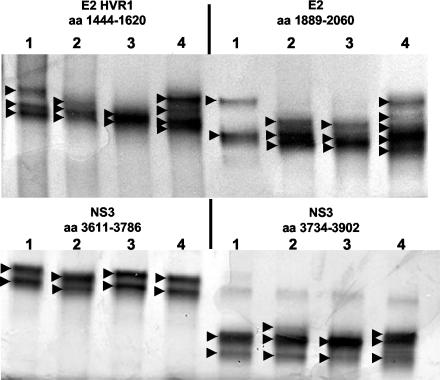
Figure 4 shows the results of SSCP analysis of specific regions from the viral genomes of Ch1535 and Ch1536 compared to the same regions amplified from the p90 cDNA clone and from a 6-week p.i. plasma sample from Ch1397, an animal that received H77 plasma. HVR1 and E2 regions amplified from serum samples from Ch1397 show a more complex band pattern than these same regions from the serum samples of Ch1535 and Ch1536, suggesting a greater diversity of these viral sequences in this animal. The NS3 regions show less complexity for all animals, suggesting that these regions are not areas of extensive heterogeneity even in natural isolates. The different band patterns for Ch1535 and Ch1536 are reflective of different viral sequences in these animals (Tables 1 and 2). Analysis of cloned PCR fragments covering the E2 (including HVR1), NS3, and NS5B regions of virus from Ch1535 and Ch1536 and the H77 inoculum (ranging from 335 to 719 bp in length) also confirmed this reduced heterogeneity in the animals that received the infectious clone (data not shown). Between 80 and 100% of the clones contained the mutations observed from PCR sequencing; the clones that did not carry the mutated nucleotide contained the wild-type sequence. These data confirm a low level of sequence heterogeneity after more than 4 years of HCV persistence in the sera of animals that received clonal infectious RNA.
FIG. 4.
SSCP analysis of HCV regions. The HCV regions were amplified from the p90 cDNA clone (lanes 1), serum from Ch1535 at week 216 (lanes 2), serum from Ch1536 at week 216 (lanes 3), and Ch1397 that received H77 plasma (lanes 4). Arrowheads indicate major bands observed in each sample.
DISCUSSION
In this study, we were able to observe the accumulation of fixed mutations in a viral genome over time. We demonstrated that the viral genomes accumulated mutations over time in all genes except the core region and that the HVR1 region remained stable throughout, incorporating a minimal number of mutations. There appeared to be a slowing of the mutation rate at the amino acid level in both animals, suggesting lower immune pressure, possibly due to weaker T-cell responses relative to those during the acute infection. This observation suggests that persistent nonsynonymous mutations accumulate due to selective pressure rather than simply as a consequence of long-term HCV replication.
Although the virus isolated from the two animals in this study have only one common mutation, several of the amino acid changes are located in previously identified HCV epitopes recognized by human or chimpanzee class I-restricted CD8+ T cells (reviewed in reference 22). There have been several reports in the literature of viral immune escape at the neutralizing antibody and T-cell levels in both chimpanzees and humans (7, 11, 26, 30), giving weight to the argument that these events play a major role in the establishment and maintenance of persistent infections. The observation that higher antienvelope antibody titers in Ch1536 correlated with a greater number of amino acid substitutions in E2 and a higher dN/dS ratio for this gene product suggests that escape from neutralizing antibody occurs during the chronic phase. The low antienvelope antibody levels during the first 20 to 30 weeks of infection suggest that this mechanism occurs more often in the chronic phase than in the acute phase. Antibody to the surface proteins has been shown to have some neutralizing effect in several studies (5, 9, 10, 21), and this hypothesis of escape can now be addressed using the retrovirus pseudoparticle systems recently developed (1, 12). Interestingly, two amino acid changes within the E2 region (aa 403 and 444) occurring late in infection (at week 130) lie within recently identified monoclonal antibody epitopes shown to have potent neutralizing ability using retrovirus pseudotype particles bearing HCV glycoproteins (12). The substitution at residue 444 was the only shared mutation between both animals.
One pattern that is emerging from this and other studies is that HVR1 does not accumulate mutations rapidly in the chimpanzee even with high titers of HVR1 antibody (23). Previous reports that show rapid change of HVR1 over time have used infections that are initiated from quasispecies populations. We consider these to represent selected changes in the dominant sequence from the preexisting population as opposed to de novo mutations. The changes that were incorporated into HVR1 in the virus isolated from Ch1535 and Ch1536 did not appear to represent antibody escape mutants and, given the mutation rates in these two animals, substantially longer periods of time would be required to generate HVR1 regions that are no longer recognized by preexisting antibody. It has been previously suggested that the rate of HCV evolution in chimpanzees and humans is different (23) with weaker positive selection in chimpanzees. This may account for our observations, although other studies have shown higher rates of HCV substitution in chimpanzees (19, 29). These discrepancies using natural isolates in chimpanzees may be due to subtype differences or different levels of complexity. Infections in humans usually result from higher doses of HCV than the <200 infectious doses normally used in chimpanzee studies, which would therefore result in a greater variant population for subsequent selection.
It has been reported that there is a restricted pattern of amino acid replacement within HVR1 (20, 28), with some positions having little or no variation among all genotypes. The substitutions observed in Ch1535 and Ch1536 follow these previously identified patterns of constraint (28). In this study, we also showed very little sequence heterogeneity of the virus after more than 4 years of infection even in HVR1, which would suggest further that there is no causal relationship between HVR1 variation and HCV persistence.
The comparisons between synonymous and nonsynonymous mutations at different stages of disease have shown that the early mutations arising immediately after control of virus replication during the acute phase are 100% nonsynonymous. This result would suggest that the greatest level of immune pressure occurs during this early phase. Analysis of the viral kinetics in these two animals indicates that significant control of viral replication occurs at weeks 10 to 15; this period coincides with ALT elevations and seroconversion, indicators of induction of the adaptive immune response. As the period of persistence increases, the calculated dN/dS ratio becomes <1 for Ch1535 while remaining higher for Ch1536. This again suggests that there is less immune pressure occurring later during the chronic phase of Ch1535. The difference in dN/dS for Ch1535 and Ch1536 could be due to a slightly higher level of replication in Ch1535 (titers are approximately 0.5 log unit higher), which would allow for the introduction of a greater number of random mutations. Alternatively, the replication rate in Ch1536 may be lower due to a relatively stronger immune response, which would in turn result in more nonsynonymous mutations due to selective pressure. This increase in synonymous mutations in Ch1535 was observed at week 130 p.i. after an increase in viral replication of almost 2 log units (Fig. 1A). This event would argue for a reduced or less effective immune response in this animal that allowed higher levels of viral replication and in turn led to a higher incorporation of random mutations.
Previous studies that have examined molecular evolution of HCV have begun with natural isolates consisting of a quasispecies population. Shifts in sequence due to mutations cannot be differentiated from shifts in virus populations. It is possible in any system that amino acid-changing substitutions can occur by chance and confer no selective advantage or disadvantage to the virus. Using infectious clones, the functional significance of mutations can be more accurately studied. Such analyses are currently being addressed using recently developed in vitro systems such as retrovirus pseudotypes (12) or subgenomic HCV replicons (2).
A steady-state level of virus titers in any persistent infection suggests that virus is being produced and cleared at the same rate, suggesting that some immune response is occurring in these chimpanzees during the chronic phase to suppress the virus, albeit less than that occurring during the acute phase. This may be at the T-cell level and/or due to neutralizing antibody production. The substitution data presented in this study support this hypothesis, and exploitation of these immune responses may be the key to combating persistent infections of HCV.
Acknowledgments
We thank Phil Snoy, Kamela Evans-Davis, and Ray Olsen for invaluable veterinary services and Michael Klutch for assistance with automated sequencing.
These studies were supported in part by internal FDA funds, a grant from the National Vaccine Program Office, and a grant from the National Cancer Institute (CA85883).
REFERENCES
- 1.Bartosch, B., J. Dubuisson, and F. L. Cosset. 2003. Infectious hepatitis C virus pseudoparticles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 197:633-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 3.Bukh, J., R. H. Miller, and R. H. Purcell. 1995. Genetic heterogeneity of hepatitis C virus: quasispecies and genotypes. Semin. Liver Dis. 15:41-63. [DOI] [PubMed] [Google Scholar]
- 4.Chang, K. M., B. Rehermann, J. G. McHutchison, C. Pasquinelli, S. Southwood, A. Sette, and F. V. Chisari. 1997. Immunological significance of cytotoxic T lymphocyte epitope variants in patients chronically infected by the hepatitis C virus. J. Clin. Investig. 100:2376-2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choo, Q. L., G. Kuo, R. Ralston, A. Weiner, D. Chien, G. Van Nest, J. Han, K. Berger, K. Thudium, C. Kuo, J. Kansopon, J. McFarland, A. Tabrizi, K. Ching, B. Moss, L. B. Cummins, M. Houghton, and E. Muchmore. 1994. Vaccination of chimpanzees against infection by the hepatitis C virus. Proc. Natl. Acad. Sci. USA 91:1294-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 7.Erickson, A. L., Y. Kimura, S. Igarashi, J. Eichelberger, M. Houghton, J. Sidney, D. McKinney, A. Sette, A. L. Hughes, and C. M. Walker. 2001. The outcome of hepatitis C virus infection is predicted by escape mutations in epitopes targeted by cytotoxic T lymphocytes. Immunity 15:883-895. [DOI] [PubMed] [Google Scholar]
- 8.Farci, P., A. Shimoda, A. Coiana, G. Diaz, G. Peddis, J. C. Melpolder, A. Strazzera, D. Y. Chien, S. J. Munoz, A. Balestrieri, R. H. Purcell, and H. J. Alter. 2000. The outcome of acute hepatitis C predicted by the evolution of the viral quasispecies. Science 288:339-344. [DOI] [PubMed] [Google Scholar]
- 9.Farci, P., A. Shimoda, D. Wong, T. Cabezon, D. De Gioannis, A. Strazzera, Y. Shimizu, M. Shapiro, H. J. Alter, and R. H. Purcell. 1996. Prevention of hepatitis C virus infection in chimpanzees by hyperimmune serum against the hypervariable region 1 of the envelope 2 protein. Proc. Natl. Acad. Sci. USA 93:15394-15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forns, X., R. Thimme, S. Govindarajan, S. U. Emerson, R. H. Purcell, F. V. Chisari, and J. Bukh. 2000. Hepatitis C virus lacking the hypervariable region 1 of the second envelope protein is infectious and causes acute resolving or persistent infection in chimpanzees. Proc. Natl. Acad. Sci. USA 97:13318-13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grakoui, A., N. H. Shoukry, D. J. Woollard, J. H. Han, H. L. Hanson, J. Ghrayeb, K. K. Murthy, C. M. Rice, and C. M. Walker. 2003. HCV persistence and immune evasion in the absence of memory T cell help. Science 302:659-662. [DOI] [PubMed] [Google Scholar]
- 12.Hsu, M., J. Zhang, M. Flint, C. Logvinoff, C. Cheng-Mayer, C. M. Rice, and J. A. McKeating. 2003. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl. Acad. Sci. USA 100:7271-7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaneko, T., T. Moriyama, K. Udaka, K. Hiroishi, H. Kita, H. Okamoto, H. Yagita, K. Okumura, and M. Imawari. 1997. Impaired induction of cytotoxic T lymphocytes by antagonism of a weak agonist borne by a variant hepatitis C virus epitope. Eur. J. Immunol. 27:1782-1787. [DOI] [PubMed] [Google Scholar]
- 14.Kolykhalov, A. A., E. V. Agapov, K. Blight, K. Mihalik, S. M. Feinstone, and C. M. Rice. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570-574. [DOI] [PubMed] [Google Scholar]
- 15.Kurosaki, M., N. Enomoto, F. Marumo, and C. Sato. 1993. Rapid sequence variation of the hypervariable region of hepatitis C virus during the course of chronic infection. Hepatology 18:1293-1299. [PubMed] [Google Scholar]
- 16.Major, M. E., K. Mihalik, J. Fernandez, J. Seidman, D. Kleiner, A. A. Kolykhalov, C. M. Rice, and S. M. Feinstone. 1999. Long-term follow-up of chimpanzees inoculated with the first infectious clone for hepatitis C virus. J. Virol. 73:3317-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Major, M. E., B. Rehermann, and S. M. Feinstone. 2001. Hepatitis C viruses, p. 1127-1161. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 18.Ogata, N., H. J. Alter, R. H. Miller, and R. H. Purcell. 1991. Nucleotide sequence and mutation rate of the H strain of hepatitis C virus. Proc. Natl. Acad. Sci. USA 88:3392-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okada, S. I., Y. Akahane, H. Suzuki, H. Okamoto, and S. Mishiro. 1992. The degree of variability in the amino terminal region of the E2/NS1 protein of hepatitis C virus correlates with responsiveness to interferon therapy in viremic patients. Hepatology 16:619-624. [DOI] [PubMed] [Google Scholar]
- 20.Penin, F., C. Combet, G. Germanidis, P. O. Frainais, G. Deleage, and J. M. Pawlotsky. 2001. Conservation of the conformation and positive charges of hepatitis C virus E2 envelope glycoprotein hypervariable region 1 points to a role in cell attachment. J. Virol. 75:5703-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Puig, M., M. E. Major, K. Mihalik, M. Y. Yu, and S. M. Feinstone. 2004. Immunization of chimpanzees with an envelope protein-based vaccine enhances specific humoral and cellular immune responses that delay hepatitis C virus infection. Vaccine 22:991-1000. [DOI] [PubMed] [Google Scholar]
- 22.Puig, M., K. Mihalik, M. Y. Yu, S. M. Feinstone, and M. E. Major. 2002. Sensitivity and reproducibility of HCV quantitation in chimpanzee sera using TaqMan real-time PCR assay. J. Virol. Methods 105:253-263. [DOI] [PubMed] [Google Scholar]
- 23.Ray, S. C., Q. Mao, R. E. Lanford, S. Bassett, O. Laeyendecker, Y. M. Wang, and D. L. Thomas. 2000. Hypervariable region 1 sequence stability during hepatitis C virus replication in chimpanzees. J. Virol. 74:3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ray, S. C., Y. M. Wang, O. Laeyendecker, J. R. Ticehurst, S. A. Villano, and D. L. Thomas. 1999. Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: hypervariable region 1 as a decoy. J. Virol. 73:2938-2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saito, I., T. Miyamura, A. Ohbayashi, H. Harada, T. Katayama, S. Kikuchi, Y. Watanabe, S. Koi, M. Onji, Y. Ohta, Q. L. Choo, M. Houghton, and G. Kuo. 1990. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 87:6547-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shimizu, Y. K., M. Hijikata, A. Iwamoto, H. J. Alter, R. H. Purcell, and H. Yoshikura. 1994. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J. Virol. 68:1494-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simmonds, P., A. Alberti, H. J. Alter, F. Bonino, D. W. Bradley, C. Brechot, J. T. Brouwer, S. W. Chan, K. Chayama, and D. S. Chen. 1994. A proposed system for the nomenclature of hepatitis C viral genotypes. Hepatology 19:1321-1324. [PubMed] [Google Scholar]
- 28.Smith, D. B. 1999. Evolution of the hypervariable region of hepatitis C virus. J. Viral Hepat. 6(Suppl. 1):41-46. [DOI] [PubMed] [Google Scholar]
- 29.van Doorn, L., I. Capriles, G. Maertens, R. DeLeys, K. Murray, T. Kos, H. Schellekens, and W. Quint. 1995. Sequence evolution of the hypervariable region in the putative envelope region E2/NS1 of hepatitis C virus is correlated with specific humoral immune responses. J. Virol. 69:773-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang, H., T. Bian, S. J. Merrill, and D. D. Eckels. 2002. Sequence variation in the gene encoding the nonstructural 3 protein of hepatitis C virus: evidence for immune selection. J. Mol. Evol. 54:465-473. [DOI] [PubMed] [Google Scholar]
- 31.Wang, H., and D. D. Eckels. 1999. Mutations in immunodominant T cell epitopes derived from the nonstructural 3 protein of hepatitis C virus have the potential for generating escape variants that may have important consequences for T cell recognition. J. Immunol. 162:4177-4183. [PubMed] [Google Scholar]