Abstract
Cellular antiviral responses are mediated partly by the expression of interferon-stimulated genes, triggered by viral genomes, their transcripts and replicative intermediates. Persistent replication of a hepatitis C virus (HCV) replicon suggests that the replicon does not elicit cellular innate antiviral responses. In the present study, we investigated regulatory factors of the interferon-mediated antiviral system in cells expressing an HCV replicon. Luciferase reporter assays revealed that the baseline activity of the interferon-stimulated response element (ISRE) was significantly lower in cells harboring the replicon than in naive cells. Among the proteins involved in the IFN/Jak/STAT pathway and in ISRE activity, the expression level of interferon regulatory factor 1 (IRF-1) was found to be significantly lower in cells harboring the replicon. Transfection of an IRF-1 expression construct into cells harboring the replicon caused an increase of ISRE activity, accompanied by suppression of expression of the HCV replicon. Moreover, in cured Huh7 cells from which the HCV replicon had been eliminated, the expression levels of IRF-1 and ISRE activity also were suppressed, demonstrating that the decrease of IRF-1 is attributable, not to active suppression by the viral proteins, but to adaptation of cells that enables replication of the HCV subgenome. The high permissiveness of the cured cells for the replicon was abolished by transgenic supplementation of IRF-1 expression. Taken together, IRF-1 is one of the key host factors that regulate intracellular HCV replication through modulation of interferon-stimulated-gene-mediated antiviral responses.
Hepatitis C virus (HCV) is one of the most important pathogens causing liver-related morbidity and mortality (3, 10). HCV establishes a persistent infection in the liver, leading to the development of chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma. Interferon (IFN) plays a central role in eliminating HCV, not only following clinical therapeutic application but also as a cellular immune response (30, 39). Cellular innate responses to eliminate viruses are mediated by the IFN-stimulated genes (ISGs), including 2,5-oligoadenylate synthetase (OAS), double-stranded RNA-dependent protein kinase R, and MxA proteins, and by as-yet-uncharacterized genes (35). DNA microarray analyses of liver specimens from a chimpanzee infected with HCV revealed that the expression of various cytokines and chemokines is induced during the course of viral infection and its clearance and that a considerable proportion of the genes are induced by IFN-α or IFN-γ (5).
The control of expression of ISGs is directed by the IFN-stimulated response elements (ISRE) and/or IFN-γ-activated sites (GAS) located in their promoter and/or enhancer regions (30). GAS is the binding site for phosphorylated signal transducer and activator of transcription 1 (STAT1) homodimers called IFN-γ-activated factor (GAF), which in turn is activated through IFN-γ receptor-associated Janus kinase (Jak) (38). Thus, GAS drives the expression of genes induced by IFN-γ (11). On the other hand, ISRE is the binding site for the IFN-stimulated gene factor 3 (ISGF-3) that consists of phosphorylated STAT1 and STAT2, and IFN regulatory factor 9 (IRF-9) (38, 39). ISGF-3 is activated through the binding of IFN-α/β to their receptor (11).
Besides IFN receptor-mediated stimuli, other IRF family members regulate the expression levels of ISGs and IFN-α/β genes through binding to ISRE and a similar DNA sequence, positive regulatory domain (PRD) I, within the IFN-β promoter and PRD-like elements (PRD-LE) within IFN-α promoters (26, 27, 37, 38). These sequences respond differentially to each specific IRF—IRF-1, IRF-3, and IRF-7 in particular (26, 27, 37, 38). IRF-3 and IRF-7 have been identified as direct transducers of virus-mediated signaling and play a critical role in the induction of IFN genes (32, 33). Both IRF-3 and IRF-7, which reside in the cytoplasm, undergo virus-induced phosphorylation and translocate to the nucleus (38). IRF-7 can activate IFN-α and IFN-β genes through binding PRD-LE and PRDI, respectively, whereas IRF-3 predominantly affects the IFN-β gene and some ISGs through binding PRDI and ISRE, respectively (26, 31, 39). IRF-1 was first identified as a regulator of the promoter of the IFN-α/β gene (30). Cellular expression of IRF-1 is induced by various cytokines, such as IFN-α/β/γ, tumor necrosis factor alpha, interleukin-1, interleukin-6, and LIF, and by viral infection (38). The recognition sequence of IRF-1, IRF-E, overlaps with that of ISRE, which binds ISGF-3 (37, 38). IRF-1 appears to function as a regulator of cellular antiviral responses to IFNs by affecting a set of ISGs. Overexpression of IRF-1 induces an antiviral state affecting various viruses, including vesicular stomatitis virus, encephalomyocarditis virus, and Newcastle disease virus (28).
Basic studies of HCV infection and replication were hampered by the lack of efficient cell culture systems. The HCV subgenomic replicon system, developed by Lohmann et al., is an efficient and noncytopathic cellular genomic replication model that has allowed various molecular studies of HCV replication (25). Host cells for the replicon generally are restricted to the human hepatoma cell line, Huh7. Furthermore, for continuous expression of the replicon, certain amino acid substitutions are required for adaptation to the host cells. In contrast, inoculation of these cellular adapted mutant HCV-RNAs into chimpanzee liver failed to induce persistent infection (9). These findings suggest that viral fitness to the host cellular environment is critical for the establishment of continuous replication.
In cells harboring the replicon, the expression of the replicon is abolished by small amounts of exogenous IFN-α/β/γ (6, 13, 16, 36), suggesting intact IFN receptor-mediated cellular antiviral responses. However, in the absence of IFN, persistent and high-level expression of the replicon makes us speculate that intracellular virus-induced antiviral responses are attenuated in the host cells or that those antiviral responses are suppressed by the expression of viral proteins.
In the present study, we investigated the factors associated with cellular antiviral responses and their regulation by IRFs and found that the baseline ISRE activity is decreased in cells expressing the HCV replicon, that the transcriptional decrease of IRF-1 is involved in the downregulation of the ISRE responses and, more importantly, that expression of IRF-1 negatively regulates HCV replication.
MATERIALS AND METHODS
Cell culture.
Human hepatoma Huh7 cells were grown in conventional medium consisting of Dulbecco modified minimal essential medium (Sigma, St. Louis, Mo.) supplemented with 100 IU of penicillin/ml, 100 μg of streptomycin/ml, and 10% fetal bovine serum at 37°C under 5% CO2. G418 (Wako, Osaka, Japan) was added to the culture medium to a final concentration of 200 μg/ml for cells carrying the HCV replicon.
Plasmid constructions.
The plasmid, pHCV1bneo/delS is an HCV subgenomic replicon construct derived from the chimpanzee-infectious clone, HCV-N (GenBank accession no. AF139594, kindly provided by Christoph Seeger) (16). Another HCV-1b replicon construct, pR-J4/S2197P, is constructed from the HC-J4 clone (25a, 40). Both pHCV1bneo/delS and pR-J4/S2197P are bicistronic RNA molecules expressing the neomycin phosphotransferase (NPT) gene and the HCV nonstructural genes spanning from NS3 to NS5B. Plasmid pRep-Feo was derived from pHCV1bneo/delS with replacement of the NPT gene by a fusion of the firefly luciferase (Fluc) and NPT genes (36, 41). pRep-Fluc was constructed from pHCV1bneo/delS by replacing the NPT gene with the firefly luciferase gene for transient-replication assay. A replication-defective replicon construct, pRep-Fluc-NS5Bdel, was constructed from pHC1bneo/delS by introducing a frameshift mutation into NS5B by BclI digestion and served as a replication-negative control.
The full-length human IRF-1 gene was amplified by PCR from human intestine and cloned into pcDNA3 and pcDNA4/TO/myc-His B (Invitrogen, Carlsbad, Calif.) to yield the mammalian expression construct, pcDNA3-IRF-1 and pcDNA4/to/IRF-1-Myc/His. The nucleotide sequence was confirmed. pISRE-TA-Luc (Invitrogen) contained five copies of consensus ISRE motifs upstream of the firefly luciferase gene. pGAS-TA-Luc (Invitrogen) contained two copies of the STAT1 enhancer element upstream of the firefly luciferase gene. pTA-Luc (Invitrogen), which lacks the enhancer element, was used for background determination. pcDNA3.1 (Invitrogen) was used as an empty vector for mock transfection. pRL-CMV (Promega, Madison, Wis.), which expresses the Renilla luciferase protein, was used for correction of transfection efficiency.
In vitro transcription.
The replicon RNA was synthesized in vitro from 1 μg of linearized replicon plasmid by the RiboMax Large Scale RNA Production System (Promega) by using T7 RNA polymerase. After DNase I treatment, the transcribed RNA was purified by the acid guanidinium thiocyanate-phenol-chloroform method with ISOGEN (Nippon Gene, Tokyo, Japan) to remove completely trace amounts of residual template DNA, and resuspended in RNase-free water.
Electroporation and G418 selection to obtain cells harboring the HCV replicon.
Subconfluent Huh7 cells were treated with trypsin, and 5 × 106 cells were resuspended in 500 μl of serum-free Dulbecco modified Eagle medium-F12 medium (Sigma) in the presence of 10 μg of each replicon RNA, transferred to a 4-mm electroporation cuvette (Equi Bio, Middlesex, United Kingdom), and subjected to an electric pulse (1,050 μF and 270 V) by using the EasyJect system (Equi Bio). After electroporation, the cell suspension was diluted with conventional medium and plated in 10-cm-diameter cell culture dishes. After 24 h, G418 was added to a concentration of 200 μg/ml, and the medium was changed twice weekly. At 3 weeks after transfection, G418-resistant colonies appeared, and cell lines harboring continuous replication of the replicon were established. Cell lines Huh7/Rep-N, Huh7/Rep-J4, and Huh7/Rep-Feo were established from HCV1bneo/delS, R-J4/S2197P, and Rep-Feo, respectively.
Establishment of cured Huh7 cells.
Cured Huh7 cells, from which the replicon had been eliminated, were established by treating Huh7/Rep-Feo cells with 100 U of IFN-α/ml for 14 days. Clearance of replicon RNA was confirmed by reverse transcription-PCR (RT-PCR) and by the loss of resistance to G418.
Electroporation and transient-replication assays.
Subconfluent cells were treated with trypsin, and 6 × 106 cells were resuspended in 500 μl of OPTI-MEM in the presence of 5 μg of luciferase-expressing replicon RNA Rep-Fluc and were electroporated as described above. After electroporation, the cell suspension was diluted to 12 ml with conventional medium and seeded onto a 24-well plate. The luciferase activities of the cell lysates were measured by using a single luciferase assay protocol. To correct the efficiency of induction, each value was shown as a relative ratio adjusted by a value at 4 h of electroporation.
Transient transfection.
Transient transfection was performed by using FuGENE-6 transfection reagent (Roche Applied Science, Indianapolis, Ind.) and Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Reporter assays.
To measure the ISRE and GAS activity, cells were seeded at 5 × 104 per well in 24-well plates on the day before transfection. Totals of 400 ng of pISRE-TA-Luc, pGAS-TA-Luc, or pTA-Luc with 1 ng of pRL-CMV were transfected to each well by using 2 μl of Lipofectamine 2000. At 48 h after transfection, dual luciferase assays were performed on the cell lysates. To determine the efficacy of IFN on ISRE activities, the 6-h treatment of IFN-α was made 48 h after the transfection of pISRE-TA-Luc and pRL-CMV. A reporter assay was performed to determine the effect of IRF-1 on ISRE in the cells harboring the replicon. A total of 5 × 104 Huh7/Rep-N cells per well was subcultured onto 24-well plates the day before transfection. A total of 100 ng of pISRE-TA-Luc and various amounts of pcDNA3-IRF-1 with empty vector and 0.1 ng of pRL-CMV, to a total mass of DNA of 400 ng, were transfected by using 2 μl of Lipofectamine 2000. At 48 h after transfection, dual luciferase assays were performed on the cell lysates.
Transfection of pcDNA3-IRF-1 to Huh7/Feo was performed to determine the effect of IRF-1 on the replication of the HCV replicon. A total of 5 × 104 Huh7/Rep-Feo cells per well were subcultured onto 24-well plates, and various amounts of pcDNA3-IRF-1 with empty vector, to a total mass of DNA of 400 ng, were transfected to each well by using 1.2 μl of FuGENE-6. At 48 h after transfection, the cell lysates were collected and single luciferase assays were performed. Luciferase activities were quantified with a luminometer (TD-20/20; Turner Designs, Sunnyvale, Calif.) by using the dual-luciferase reporter assay system (Promega) for dual luciferase assay and the Bright-Glo luciferase assay system (Promega) for single luciferase assay.
RT-PCR and LightCycler-based PCR assay.
Total cellular RNA was extracted from cells by using ISOGEN. Then, 2 μg of total cellular RNA was used to generate cDNA from each sample by using SuperScript II (Invitrogen) reverse transcriptase. The mRNA expression levels were measured with the LightCycler PCR and detection system (Roche). Thermocycling was done in a final volume of 10 μl containing 1 μl of cDNA sample or calibrator, 1.25 mM MgCl2, 0.5 μM concentrations of each primer, and 1 μl of LightCycler FastStart DNA Master SYBR Green 1 mix (Roche). Cycle numbers of the logarithmic linear phase were plotted against the logarithm of the concentration of template DNA. The concentrations of DNA in the samples were calculated by comparing the cycle numbers of the logarithmic linear phase of the samples with the external standards. The results of amplification of the samples were verified on a 1% agarose gel. No nonspecific amplification product was documented. The primers used were as follows: IRF-1 sense (positions 1018 to 1043; 5′-GTAAGGAGGAGCCAGAAATTGACAGC-3′), IRF-1 antisense (positions 1152 to 1175; 5′-CTACGGTGCACAGGGAATGGCCTG-3′), IRF-3 sense (positions 1154 to 1177; 5′-ACGTGCCTCAGGGCCTTGGTAGAA-3′), IRF-3 antisense (positions 1307 to 1330; 5′-TCAGCTCTCCCCAGGGCCCTGGAA-3′), STAT1-α/β sense (positions 2291 to 2320; 5′-ACTGGATATATCAAGACTGAGTTGATTTCT-3′), STAT1-α antisense (positions 2411 to 2440; 5′-GTTCATCATACTGTCGAATTCTACAGAGCC-3′), STAT1-β antisense (positions 2411 to 2440; 5′-GATAGCAATTACAATGGAAAAGTAAAATAC-3′), β-actin sense (positions 1011 to 1040; 5′-ACAATGAAGATCAAGATCATTGCTCCTCCT-3′), and β-actin antisense (positions 1131 to 1160; 5′-TTTGCGGTGGACGATGGAGGGGCCGGACTC-3′).
Preparation of siRNA and transfection.
Sense and antisense strands of small interfering RNA (siRNA) oligonucleotides directed against IRF-1 mRNA were synthesized. The sequences were 5′-CCAAGAACCAGAGAAAAGAdTdT-3′ (sense) and 5′-UCUUUUCUCUGGUUCUUGGdTdT-3′ (antisense). siRNA directed against an unrelated target was used as a negative control.
MTS assays.
To evaluate cell growth and cell viability, dimethylthiazol carboxymethoxyphenyl sulfophenyl tetrazolium (MTS) assays were performed by using the CellTiter 96 AQueous One-Solution cell proliferation assay (Promega).
Preparation of nuclear extracts.
After two washes in ice-cold phosphate-buffered saline, 2 × 107 cells were harvested and centrifuged at 1,000 × g for 5 min in a microcentrifuge. The cell pellet was resuspended in 80 μl of buffer A (10 mM Tris-HCl [pH 7.3], 1.5 mM MgCl2, 10 mM KCl, 1 mM dithiothreitol, 0.4% Nonidet P-40, 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin/ml, 1 μM pepstatin, 50 mM NaF, and 1 mM Na3VO4). After incubation on ice for 5 min, cell lysates were centrifuged, and the nuclear pellet was then resuspended in 75 μl of buffer C (20 mM Tris-HCl [pH 7.3], 1.5 mM MgCl2, 484 mM KCl, 1 mM dithiothreitol, 0.2 mM EDTA, 25% glycerol, 1 mM phenylmethylsulfonyl fluoride, 10 μg of aprotinin/ml, 1 μM pepstatin, 50 mM NaF, and 1 mM Na3VO4), followed by incubation for 30 min at 4°C. Nuclear debris were pelleted with centrifugation at 12,000 × g for 15 min, and the supernatant was used as nuclear extracts (26a).
Western blot analysis.
A total of 25 μg of nuclear extract lysate was electrophoresed through a NuPAGE 10% Bis-Tris gel (Invitrogen) and blotted onto the polyvinylidene difluoride Western blot membranes (Roche). The membranes were incubated with rabbit polyclonal anti-IRF-1 antibody (Santa Cruz Biotechnology, Santa Cruz, Calif.), followed by the addition of peroxidase-labeled anti-rabbit immunoglobulin G (IgG) antibody. Chemiluminescence was detected by using the ECL Western blotting analysis system (Amersham Biosciences, Buckinghamshire, United Kingdom) according to the manufacturer's protocol. The membrane was stripped by washing it with Tris-buffered saline containing 2% sodium dodecyl sulfate and 0.7% 2-mercaptoethanol for 30 min and then incubated with monoclonal anti-NS5A antibody (Biodesign, Saco, Maine), followed by the addition of peroxidase-labeled anti-mouse IgG antibody.
Preparation of a recombinant adenovirus expressing IRF-1.
The PmeI-BamHI-digested fragment of pcDNA4/to/IRF-1-Myc/His containing the IRF-1 gene with a myc/His tag was blunt ended by using a DNA blunting kit (Takara, Otsu, Japan). This fragment was integrated to produce the IRF-1-expressing recombinant adenovirus vector, Ad-IRF-1 by using an adenovirus expression vector kit (Takara). The IRF-1 protein produced by Ad-IRF-1 had a myc/His tag that was used for differentiation from endogenous IRF-1. LacZ-expressing adenovirus, Ad-LacZ, was prepared as a control. The expression of IRF-1 from Ad-IRF-1 was confirmed by Western blot analysis. Cured Huh7 cells were infected with Ad-IRF-1 at multiplicity of infection (MOI) of 3. As a control, pcDNA3-IRF-1 and pcDNA4/to/IRF-1-Myc/His were transfected on cured Huh7 cells by using FuGENE-6. At 48 h after the transfection, nuclear extracts were prepared and Western blot analysis was performed by using anti-IRF-1 antibody and anti-His antibody as previously described.
Statistical analyses.
Statistical analysis was performed by using an unpaired, two-tailed Student t test. P values of <0.05 were considered statistically significant.
RESULTS
ISRE activities in cells harboring the HCV replicon were lower than in naive Huh7.
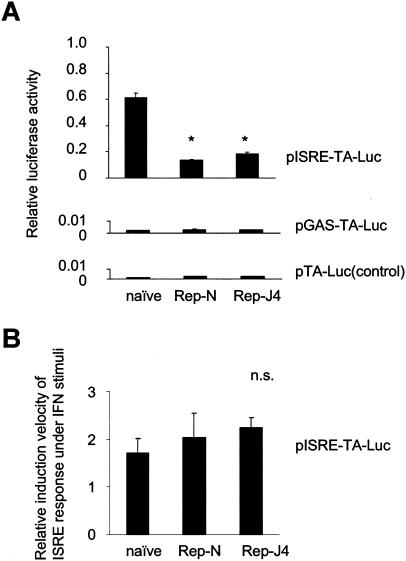
The ISRE and GAS activities of naive Huh7 cells and the Huh7 cell lines harboring the replicon (Huh7/Rep-N and Huh7/Rep-J4) were analyzed by transfection of the reporter constructs, pISRE-TA-Luc and pGAS-TA-Luc (Fig. 1A). The relative ISRE-regulated luciferase activities were significantly lower in the Huh7/Rep-N and Huh7/Rep-J4 cells than in untransfected Huh7 cells (22.6% ± 0.8% and 30.0% ± 1.9% in Huh/Rep-N and Huh7/Rep-J4 relative to naive Huh7 cells, respectively; P < 0.0001). On the other hand, the GAS-luciferase activities were at almost background levels in all cell lines tested. We also examined the ISRE responses to exogenous IFN-α stimuli in cells with or without replicon (Fig. 1B). At 6 h after treatment with 10 U of IFN-α/ml, induction levels of ISRE activity were not significantly different between cell lines (170.1% ± 31.2%, 203.8% ± 51.4%, and 224.3% ± 21.1% in the naive Huh7, Huh7/Rep-N and Huh7-RepJ4, respectively).
FIG. 1.
ISRE activities in Huh7 cells and cells harboring the replicon. (A) The ISRE and GAS activities of naive Huh7 cells and Huh7 cells harboring the replicons (Huh7/Rep-N and Huh7/Rep-J4) were analyzed by reporter assays. These cells were transfected with pISRE-TA-Luc, pGAS-TA-Luc, and pTA-Luc, which lacks the enhancer element, as a control. pRL-CMV was cotransfected to correct the efficiency of transfection, and dual luciferase assays of the cell lysates were performed. The data are means ± the standard deviation (SD). ★, P < 0.0001 relative to naive Huh7 cells. (B) The ISRE responses to exogenous IFN-α stimuli in cells with or without replicon were analyzed by reporter assay. At 48 h after transfection with pISRE-TA-Luc and pRL-CMV, treatment with 10 U of IFN-α/ml was made. At 6 h after IFN-α stimuli, the relative induction levels of ISRE activities over each unstimulated cells were calculated.
IRF-1 expression was decreased in cells expressing the replicon.
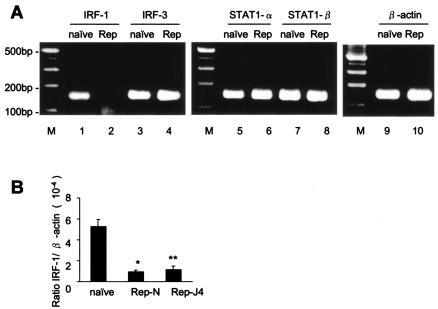
The ISRE activity is regulated by several known cellular factors (30, 38). To examine the levels of cellular factors that bind and regulate ISRE activity, we measured intracellular mRNA expression levels of IRF-1, IRF-3, STAT1-α, and STAT1-β in naive Huh7 and Huh7/Rep cells by RT-PCR analysis (Fig. 2A). The expression level of IRF-1 mRNA in Huh7 cells harboring the replicon was markedly decreased compared to that in naive Huh7. On the other hand, there were no differences in the expression levels of IRF-3, STAT1-α, and STAT1-β between cells with or without the replicon. The levels of IRF-1, IRF-3 and β-actin genes were quantified by using quantitative real-time RT-PCR and normalized by β-actin. The analysis showed that the expression levels of IRF-1 in the two Huh7 cell lines expressing the HCV replicon were significantly lower than in Huh7 cells (17.8% ± 3.2% and 21.9% ± 3.2% relative to naive Huh7 cells [P = 0.0013 and P = 0.0022 in Huh/Rep-N and Huh7/Rep-J4, respectively]; Fig. 2B). Also, in Western blot analysis, IRF-1 protein synthesis was decreased in Huh7 cells expressing replicon compared to naive Huh7 cells (see Fig. 5, lanes 1 to 3). These results suggest that the downregulation of the ISRE activities in cells expressing the replicon is mediated, at least in part, by a decrease in cellular IRF-1 transcription.
FIG. 2.
The expression levels of IRF-1 mRNA were lower in cells harboring the replicon. (A) To determine the levels of cellular factors, we measured intracellular mRNA expression levels of IRF-1, IRF-3, STAT1-α, and STAT1-β in Huh7 and Huh7 cells harboring the replicon by RT-PCR analysis. (B) Quantification of the expression levels of interstitial IRF-1 mRNA was carried out by using real-time RT-PCR. cDNA was transcribed from naive Huh7 cells and cell lines harboring the replicon. Cycle numbers of the logarithmic linear phase were plotted against the logarithm of the concentration of template DNA and are presented as the calculated concentration in units. The data are means ± the SD. ★, P = 0.0013; ★★, P = 0.0022 (relative to naive Huh7 cells).
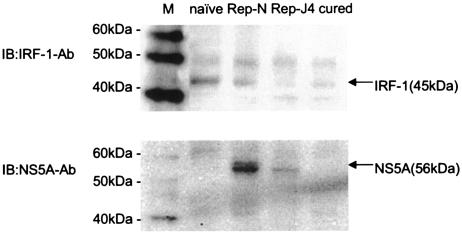
FIG. 5.
Western blot analysis of IRF-1 and NS5A expression in naive Huh7 cells, Huh7/Rep-N and Huh7/Rep-J4 cells, and cured Huh7 cells. Nuclear extracts were isolated from naive Huh7 cells, Huh7 cells harboring the replicons, and cured Huh7 cells. A total of 25 μg of each was separated on a sodium dodecyl sulfate-10% polyacrylamide gel and immunoblotted with anti-IRF-1 antibody. The membrane was reprobed with a rabbit anti-NS5A antibody and analyzed. IB, immunoblotting with the indicated primary antibody.
IRF-1 positively regulated the ISRE activity.
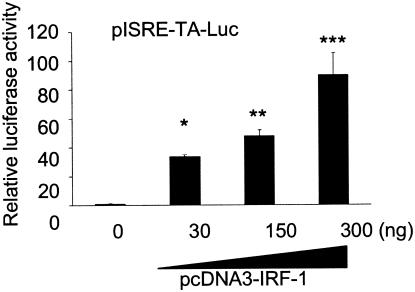
It is known that IRF-1 binds directly not only to IRF-E but also to ISRE and positively regulates the expression of ISGs (27, 37). Therefore, we examined the effects of overexpression and suppression of IRF-1 on ISRE activity in cells expressing the HCV replicon. An IRF-1 expression vector, pcDNA3-IRF-1, was cotransfected with the pISRE-TA-Luc reporter construct into Huh7/Rep-N. After 48 h of transfection, the ISRE-driven luciferase activities were significantly higher, increasing in a dose-dependent manner up to ∼90-fold in cells transfected with pcDNA3-IRF-1 (Fig. 3A).
FIG. 3.
Effect of IRF-1 overexpression and knockdown on the regulation of ISRE activity. The effects of IRF-1 on the regulation of ISRE activity were analyzed in cells expressing the HCV replicon. pRL-CMV (Promega) was cotransfected to correct for the efficiency of induction. Luciferase activities in Huh7/Rep-N cells cotransfected with pISRE-TA-Luc and various amounts of pcDNA3-IRF-1 with empty vector, to a total mass of DNA 400 ng, were assayed 48 h after transfection. The data are means ± the SD. ★, P = 0.004; ★★, P = 0.0023; ★★★, P = 0.0099 (relative to transfection with the empty vector).
Overexpression of IRF-1 suppressed expression of the HCV replicon.
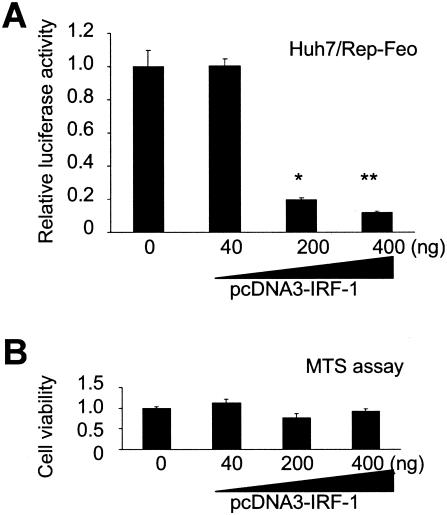
To examine the effect of IRF-1 expression on intracellular HCV replication, pcDNA3-IRF-1 was transfected into Huh7/Rep-Feo cells, in which the replicon expression levels can be monitored readily by luciferase assays. After transfection of pcDNA3-IRF-1 with empty vector, to a total mass of DNA of 400 ng, into Huh7/Rep-Feo cells, the luciferase activities of the Huh7/Rep-Feo decreased significantly in a dose-dependent manner (Fig. 4A). MTS assays of the cells transfected with IRF-1 showed no significant effects on cell growth and viability, demonstrating that the effects of IRF-1 transfection on the expression of the replicon were not due to cytotoxicity (Fig. 4B).
FIG. 4.
Effect of IRF-1 overexpression on the replication of the HCV replicon. The effects of IRF-1 on the cells harboring the HCV replicon were analyzed. (A) Transfection of pcDNA3-IRF-1 into Huh7/Rep-Feo and monitoring of replication levels as luciferase activities in the cell lysates were performed. Empty vector was cotransfected to correct the amounts of transfected vectors to be same quantity, and single luciferase assays of the cell lysates were performed 48 h after transfection. The data are means ± the SD. ★, P = 0.005; ★★, P = 0.0041 (relative to transfection with the empty vector). (B) MTS assays of the cells transfected IRF-1.
IRF-1 remains underexpressed in cured Huh7 cells.
It has been reported that HCV proteins modulate the expression of various IFN-regulated genes (15, 20). To determine whether the decrease of IRF-1 in the cells expressing replicon is due to negative regulatory effects of HCV nonstructural proteins, cured Huh7 cells were used from which the replicon had been removed. Equal amounts of nuclear extract from each were loaded in each lane to perform Western blot analysis of IRF-1 protein. Interestingly, IRF-1 expression in cured Huh7 cells remained decreased to levels similar to the Huh7 cells harboring the replicon (Fig. 5, lanes 2 through 4). Similarly, ISRE activities and expression levels of IRF-1 mRNA in cured Huh7 cells remained lower than in naive Huh7 cells and were similar to those in HCV cells harboring the replicon.
Overexpression of IRF-1 renders cured Huh7 cells nonpermissive for the HCV replicon.
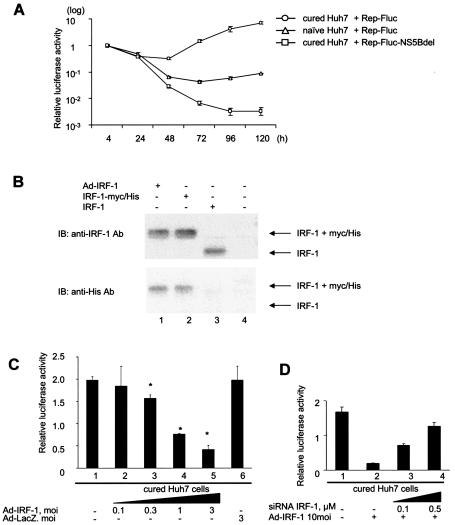
To determine whether the downregulation of IRF-1 enables replication of the HCV replicon, we performed transient-replication assays on the cured Huh7 cells. As reported earlier, the cured Huh7 cells showed increased permissiveness for replication of the HCV replicon (7) (Fig. 6A). The HCV replicon expressing firefly luciferase, Rep-Fluc, was transfected into the cured Huh7 cells and into naive Huh7 cells by electroporation. As a negative control, Rep-Fluc-NS5Bdel also was transfected into the cells. Transfection of the replication-deficient construct, Rep-Fluc-NS5Bdel, showed that the luciferase activity, which was maximal at 4 h, diminished subsequently and reached background levels by 72 h. In contrast, transfection of Rep-Fluc into the cured and naive Huh7 showed that the luciferase activity, once decreased, became elevated again at 48 and 72 h, demonstrating obvious replication. The luciferase activities of the cured Huh7 cells at 48 and 72 h were significantly higher than those of naive Huh7 cells. These results showed that the cured Huh7 cells were highly permissive compared to naive Huh7 cells.
FIG. 6.
Cured Huh7 cells lost their permissiveness for the HCV replicon with overexpression of IRF-1. Transient replication assays were performed on cured Huh7 cells to determine whether the downregulation of IRF-1 enables replication of the HCV replicon. (A) The high permissiveness of cured Huh7 cells is shown. The HCV replicon expressing the firefly luciferase, Rep-Fluc, was transfected into cured Huh7 cells and naive Huh7 cells by electroporation. As a negative control, Rep-Fluc-NS5Bdel also was transfected into the cells. The luciferase activities of the cell lysates were measured. To correct for the efficiency of induction, each value is shown as a ratio relative to the relevant relative light units at 4 h. (B) The IRF-1 protein produced by IRF-1-expressing recombinant adenovirus, Ad-IRF-1, was examined. Cured Huh7 cells were infected at an MOI of 3 with Ad-IRF-1 (lane 1). As a control, pcDNA3-IRF-1 (lane 2) and pcDNA4/to/IRF-1-Myc/His (lane 3) were transfected on cured Huh7 cells by using FuGENE-6. At 48 h after transfection, nuclear extracts were isolated and Western blot analysis was performed with anti-IRF-1 antibody and anti-His antibody, as previously described. (C) To determine the effect of supplementation of IRF-1 on the cured Huh7 cells, various titers of Ad-IRF-1 were used to infect the cured Huh7 cells 3 h before electroporation of Rep-Fluc. The relative values of 72 h over 4 h were calculated. The data are means ± the SD. ★, P < 0.01 (relative to null transfection [lane 1]). (D) To determine whether overexpression of IRF-1 affected the permissiveness of the cured Huh7 cells, a transient assay was performed with an siRNA directed to IRF-1 mRNA. At 3 h before electroporation, the cured Huh7 cells were infected with Ad-IRF-1 at MOI of 10. Rep-Fluc and the siRNA oligonucleotide directed to IRF-1 were transfected by electroporation. The relative values of 72 h over 4 h were calculated.
To examine the effect of IRF-1 supplementation on the cured Huh7 cells, IRF-1-expressing recombinant adenovirus, Ad-IRF-1, was constructed from pcDNA4/to/IRF-1-Myc/His that expresses IRF-1 with the myc-His tag. The expression of IRF-1 in the cured Huh7 cells was examined by Western blot analysis (Fig. 6B). In lysates derived from Ad-IRF-1-infected cells (Fig. 6B, lane 1) and pcDNA4/to/IRF-1-Myc/His-transfected cells (Fig. 6B, lane 2), IRF-1 was detected by both anti-IRF-1 antibody and anti-His antibody. The molecular weights of these IRF-1s are higher thanthat of the IRF-1 derived from pcDNA3-IRF-1 by the addition of myc-His tag (Fig. 6B, lane 3). Various titers of Ad-IRF-1 were used to infect the cured Huh7 cells 3 h before electroporation of Rep-Fluc (Fig. 6C). The relative values of 72 h divided by those for 4 h were calculated. Adenovirus transfection of IRF-1 suppressed the replication of the HCV replicon in the cured Huh7 cells in a dose-dependent manner (Fig. 6C, lanes 2 to 5). Infection of the cured Huh7 cells with Ad-IRF-1 at an MOI of 3 resulted in a loss of permissiveness for the replicon by about 20% (2.0 ± 0.1 and 0.4 ± 0.1, respectively) (Fig. 6C, lanes 1 and 5). To determine whether overexpression of IRF-1 affected the permissiveness of the cured Huh7 cells, a transient assay was performed by using an siRNA directed to IRF-1 mRNA. The cured Huh7 cells were infected with Ad-IRF-1 at an MOI of 10. Rep-Fluc and the siRNA were cotransfected 3 h after infection, and the luciferase activities were measured at 4 and 72 h after transfection. The replication of the HCV replicon was increased by knockdown of IRF-1 in a dose-dependent manner (Fig. 6D).
DISCUSSION
This is the first study of the involvement of IRF-1 in HCV replication. Using Huh7 cell lines harboring an HCV replicon and cells cured of the replicon, we have found that the baseline ISRE activity of cells harboring the HCV replicon is lower than that of naive cells (Fig. 1A). Among the host proteins that bind and regulate ISRE activity, expression of IRF-1 alone was decreased in the cells harboring the replicon (Fig. 2). Because overexpression analyses of IRF-1 have been shown to affect ISRE activity directly (Fig. 3), the transcriptional decrease of IRF-1 in the cells expressing the replicon appears to attenuate ISG responses mediated by the ISRE. Indeed, the overexpression of IRF-1 suppressed replication of the HCV replicon substantially (Fig. 4). One possible explanation was that the decrease in IRF-1 expression was due to active suppression by viral proteins. We found that, although IRF-1 seemed to have been suppressed by the HCV replicon, the cured Huh7 cells, from which the replicon had been eliminated, also showed decreased ISRE activity and a lower expression level of IRF-1 mRNA (Fig. 5). These results suggest that the decrease in IRF-1 found in our present study is not due to active downregulation by expression of viral proteins but possibly to the adaptation of the host cells to increased permissiveness for the replicon through continuous selection of the replicon during culture with G418. Transgenic supplementation of IRF-1 into cured Huh7 cells again abolished their high permissiveness for the replicon (Fig. 6C). Taking all of these findings into consideration, IRF-1 is one of the key cellular factors that modulate levels of ISRE-regulated ISG expression and predominantly affect the intracellular replication of HCV genomic RNA.
Induction of ISGs is mediated through ISRE and/or GAS (11, 39). Our initial results from a reporter assay showed that the ISRE activity in the absence of IFN stimuli was significantly lower in cells harboring the replicon than in naive cells. On the other hand, the activities of GAS were at almost background levels in both naive cells and cells harboring the replicon, suggesting that IFN-γ and the GAF/GAS pathway are much less involved in the cellular antiviral actions in Huh7 cells (Fig. 1). ISRE is bound by ISGF-3 or by IRFs. However, in the absence of IFN stimuli, ISGF-3 is not the main activating factor for ISRE (24). Thus, the IRFs, including IRF-1, IRF-3, and IRF-7, are potential regulators of the baseline ISRE activity. Our present results have shown that IRF-1 was suppressed exclusively in cells harboring the replicon (Fig. 2). These results suggest that the decreased ISRE activity found in the cells harboring the replicon may be attributable to the transcriptional decrease of IRF-1.
The HCV structural and nonstructural proteins have been reported to affect the IFN signaling pathway. Heim et al. reported that expression of the entire HCV polyprotein in Huh7 cells strongly inhibited IFN-α-mediated signal transduction (18). Stable expression of HCV core protein inhibited IFN-induced STAT1 expression through modulation of both GAF and ISGF-3 protein complex formation, but it did not interfere with the activation of the downstream effector genes, IRF-1 and 561, in IFN-treated cells (4). IFN-induced intracellular signaling through the Jak/STAT pathway is impaired in a transgenic mouse that expresses the HCV protein (8). Pflugheber et al. have reported that intracellular expression of NS5A blocks activation of IRF-1 triggered by double-stranded RNA and that those effects play a part in viral persistence (26b). These reports suggested that interference with IFN-induced intracellular signaling by HCV proteins could be the explanation for HCV resistance to IFN treatment.
The expression levels of mRNA are not only controlled by transcriptional regulation but also through posttranscriptional effects such as modulation of mRNA stability. As for IRF-1, several studies have shown that the expression is regulated by transcriptional modulation principally through STATs and NF-κB (17, 29). Our preliminary data of mRNA stability assay by actinomycin D treatment, an inhibitor of transcription elongation, failed to detect a difference in the half-life of IRF-1 mRNA between cell lines with or without the replicon.
We do not have a satisfactory explanation of why IRF-1 was the primary target of clonal knockdown in the cells harboring the replicon. Because IRF-1 has important roles in the antiviral mechanism (14, 21, 30), the lower expression of IRF-1 might be enough to achieve high levels of continuous cellular replication of the HCV replicon. Blight et al. reported that cured Huh7 cells are highly permissive for HCV replication and suggested that the difference in permissiveness of the cured Huh7 cells for HCV replication might be explained by a subpopulation of the parental Huh7 cells that are permissive for replication of the HCV replicon (7). Interestingly, they also have reported that the permissiveness of parental naive Huh7 cells was not altered by treatment with IFN. These results suggest that one of the factors governing permissiveness for the HCV replicon may be induced by the selection of cells with lower levels of IRF-1 expression. Indeed, the overexpression of IRF-1 in the cured Huh7 cells removed the permissiveness for the HCV replicon in parallel with the expression of the transfected IRF-1. It is also possible that a subpopulation of Huh-7 cells, which express lower levels of IRF-1, may subsequently be selected through the antibiotic selection. However, our data have shown that a typical transfection of 5 μg of replicon RNA into 6 × 106 of Huh7 cells yields more than 1,000 G418-resistant colonies and that the numbers of the colonies correlate with the amount of the transfected replicon RNA and not with the numbers of cells seeded. These findings suggest that the decrease in IRF-1 expression in replicon-harboring Huh7 is not due to the selection of cells in which IRF-1 is genetically knocked down.
It has recently been reported that other nonhepatic human cell lines and mouse hepatic cells could also harbor a subgenomic replicon (2, 12, 19, 42). We have performed a preliminary study with an HCV replicon derived from HCV genotype 2a, JFH-1, which replicates in various hepatic and nonhepatic cell lines (12, 19). An assay with human epithelial HeLa cells expressing JFH-1 replicon has shown that the relative ISRE activity was significantly lower in the HeLa/JFH-1 cells than in untransfected HeLa cells (52.4% ± 3.5% in HeLa/JFH-1 cells relative to naive HeLa cells). However, the baseline expression level of IRF-1 mRNA was much lower in HeLa cells with or without replicon than that in Huh7 cells. Although further studies are necessary, our findings suggest that the decreased baseline ISRE activities may at least have contributed to permissiveness and high-level expression of HCV replicon and that the decreased IRF-1 activities may be one mechanism to downregulate ISRE activities in Huh7 cells.
In contrast to the results in cultured cells, the clinical features of IFN, ISGs, and IRF expression are complicated. A study with needle biopsy samples from chronic hepatitis C patients who had not been treated with IFN showed that IFN-γ and IRF-1 mRNA were upregulated and correlated positively with the patients' alanine aminotransferase levels in blood (1). Another group reported that peripheral blood mononuclear cells from patients with hepatitis C showed increased expression of IRF-1 mRNA and higher IRF-1/IRF-2 ratios compared to normal subjects (23). A possible reason for the discrepancy in IRF-1 expression patterns between clinical samples and the replicon cell culture system may be the presence or absence of the immune system. IFN-γ, which is known to be the strongest inducer of IRF-1 (22), is synthesized by natural killer cells, CD4 Th1 cells, and CD8 cytotoxic lymphocytes in response to antigenic stimuli (34). In any case, it has been suggested that IRF-1 is one of the principal host factors involved in cellular responses against viral infection and in clearance of viruses from cells.
In conclusion, our results demonstrate that IRF-1 is an important regulatory factor for baseline HCV replication and has a widely accepted role as a mediator of antiviral action. IRF-1 potentially may be a key molecular target to control HCV replication.
REFERENCES
- 1.Abbate, I., M. Romano, R. Longo, G. Cappiello, O. Lo Iacono, V. Di Marco, C. Paparella, A. Spano, and M. R. Capobianchi. 2003. Endogenous levels of mRNA for IFNs and IFN-related genes in hepatic biopsies of chronic HCV-infected and non-alcoholic steatohepatitis patients. J. Med. Virol. 70:581-587. [DOI] [PubMed] [Google Scholar]
- 2.Ali, S., C. Pellerin, D. Lamarre, and G. Kukolj. 2004. Hepatitis C virus subgenomic replicons in the human embryonic kidney 293 cell line. J. Virol. 78:491-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alter, M. J. 1997. Epidemiology of hepatitis C. Hepatology 26:62S-65S. [DOI] [PubMed]
- 4.Basu, A., K. Meyer, R. B. Ray, and R. Ray. 2001. Hepatitis C virus core protein modulates the interferon-induced transacting factors of Jak/Stat signaling pathway but does not affect the activation of downstream IRF-1 or 561 gene. Virology 288:379-390. [DOI] [PubMed] [Google Scholar]
- 5.Bigger, C. B., K. M. Brasky, and R. E. Lanford. 2001. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J. Virol. 75:7059-7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 7.Blight, K. J., J. A. McKeating, and C. M. Rice. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001-13014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blindenbacher, A., F. H. Duong, L. Hunziker, S. T. Stutvoet, X. Wang, L. Terracciano, D. Moradpour, H. E. Blum, T. Alonzi, M. Tripodi, N. La Monica, and M. H. Heim. 2003. Expression of hepatitis C virus proteins inhibits interferon alpha signaling in the liver of transgenic mice. Gastroenterology 124:1465-1475. [DOI] [PubMed] [Google Scholar]
- 9.Bukh, J., T. Pietschmann, V. Lohmann, N. Krieger, K. Faulk, R. E. Engle, S. Govindarajan, M. Shapiro, M. St Claire, and R. Bartenschlager. 2002. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc. Natl. Acad. Sci. USA 99:14416-14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 11.Darnell, J.E., Jr., I. M. Kerr, and G. R. Stark. 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415-1421. [DOI] [PubMed] [Google Scholar]
- 12.Date T., T. Kato, M. Miyamoto, Z. Zhao, K. Yasui, M. Mizokami, and T. Wakita. 2004. Genotype 2a hepatitis C virus subgenomic replicon can replicate in HepG2 and IMY-N9 cells. J. Biol. Chem. 279:22371-22376. [DOI] [PubMed] [Google Scholar]
- 13.Frese, M., T. Pietschmann, D. Moradpour, O. Haller, and R. Bartenschlager. 2001. Interferon-alpha inhibits hepatitis C virus subgenomic RNA replication by an MxA-independent pathway. J. Gen. Virol. 82:723-733. [DOI] [PubMed] [Google Scholar]
- 14.Frese, M., K. Barth, A. Kaul, V. Lohmann, V. Schwarzle, and R. Bartenschlager. 2003. Hepatitis C virus RNA replication is resistant to tumour necrosis factor-alpha. J. Gen. Virol. 84:1253-1259. [DOI] [PubMed] [Google Scholar]
- 15.Geiss, G. K., V. S. Carter, Y. He, B. K. Kwieciszewski, T. Holzman, M. J. Korth, C. A. Lazaro, N. Fausto, R. E. Bumgarner, and M. G. Katze. 2003. Gene expression profiling of the cellular transcriptional network regulated by alpha/beta interferon and its partial attenuation by the hepatitis c virus nonstructural 5A protein. J. Virol. 77:6367-6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo, J. T., V. V. Bichko, and C. Seeger. 2001. Effect of alpha interferon on the hepatitis C virus replicon. J. Virol. 75:8516-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harada, H., E. Takahashi, S. Itoh, K. Harada, T. A. Hori, and T. Taniguchi. 1994. Structure and regulation of the human interferon regulatory factor 1 (IRF-1) and IRF-2 genes: implications for a gene network in the interferon system. Mol. Cell. Biol. 14:1500-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heim, M. H., D. Moradpour, and H. E. Blum. 1999. Expression of hepatitis C virus proteins inhibits signal transduction through the Jak-STAT pathway. J. Virol. 73:8469-8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kato, T., T. Date, M. Miyamoto, A. Furusaka, K. Tokushige, M. Mizokami, and T. Wakita. 2003. Efficient replication of the genotype 2a hepatitis C virus subgenomic replicon. Gastroenterology 125:1808-1817. [DOI] [PubMed] [Google Scholar]
- 20.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2:675-687. [DOI] [PubMed] [Google Scholar]
- 21.Kimura, T., K. Nakayama, J. Penninger, M. Kitagawa, H. Harada, T. Matsuyama, N. Tanaka, R. Kamijo, J. Vilcek, and T. W. Mak. 1994. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science 264:1921-1924. [DOI] [PubMed] [Google Scholar]
- 22.Kroger, A., M. Koster, K. Schroeder, H. Hauser, and P. P. Mueller. 2002. Activities of IRF-1. J. Interferon Cytokine Res. 22:5-14. [DOI] [PubMed] [Google Scholar]
- 23.Larrea, E., A. Alberdi, Y. Castelruiz, P. Boya, M. P. Civeira, and J. Prieto. 2001. Expression of interferon-alpha subtypes in peripheral mononuclear cells from patients with chronic hepatitis C: a role for interferon-α5. J. Viral. Hepat. 8:103-110. [DOI] [PubMed] [Google Scholar]
- 24.Lau, J. F., J. P. Parisien, and C. M. Horvath. 2000. Interferon regulatory factor subcellular localization is determined by a bipartite nuclear localization signal in the DNA-binding domain and interaction with cytoplasmic retention factors. Proc. Natl. Acad. Sci. USA 97:7278-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 25a.Maekawa, S., N. Enomoto, N. Sakamoto, M. Kurosaki, E. Ueda, T. Kohashi, H. Watanabe, C. H. Chen, T. Yamashiro, Y. Tanabe, N. Kanazawa, M. Nakagawa, C. Sato, and M. Watanabe. Introduction of NS5A mutations enables subgenomic HCV-replicon derived from chimpanzee-infectious HC-J4 isolate to replicate efficiently in Huh-7 cells. J. Viral Hepatitis, in press. [DOI] [PubMed]
- 26.Nakaya, T., M. Sato, N. Hata, M. Asagiri, H. Suemori, S. Noguchi, N. Tanaka, and T. Taniguchi. 2001. Gene induction pathways mediated by distinct IRFs during viral infection. Biochem. Biophys. Res. Commun. 283:1150-1156. [DOI] [PubMed] [Google Scholar]
- 26a.Oshima S., T. Nakamura, S. Namiki, E. Okada, K. Tsuchiya, R. Okamoto, M. Yamazaki, T. Yokota, M. Aida, Y. Yamaguchi, T. Kanai, H. Handa, and M. Watanabe. Interferon regulatory factor 1 (IRF-1) and IRF-2 distinctively up-regulate gene expression and production of interleukin-7 in human intestinal epithelial cells. Mol. Cell. Biol., in press. [DOI] [PMC free article] [PubMed]
- 26b.Pflugheber, J., B. Fredericksen, R. Sumpter, Jr., C. Wang, F. Ware, D. L. Sodora, and M. Gale, Jr. 2002. Regulation of PKR and IRF-1 during hepatitis C virus RNA replication Proc. Natl. Acad. Sci. USA 99:4650-4655. [DOI] [PMC free article] [PubMed]
- 27.Pine, R., T. Decker, D. S. Kessler, D. E. Levy, and J. E. Jr Darnell. 1990. Purification and cloning of interferon-stimulated gene factor 2 (ISGF2): ISGF2 (IRF-1) can bind to the promoters of both beta interferon- and interferon-stimulated genes but is not a primary transcriptional activator of either. Mol. Cell. Biol. 10:2448-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pine, R. 1992. Constitutive expression of an ISGF2/IRF1 transgene leads to interferon-independent activation of interferon-inducible genes and resistance to virus infection. J. Virol. 66:4470-4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pine, R., A. Canova, and C. Schindler. 1994. Tyrosine phosphorylated p91 binds to a single element in the ISGF2/IRF-1 promoter to mediate induction by IFN alpha and IFN gamma and is likely to autoregulate the p91 gene. EMBO. J. 13:158-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Samuel, C. E. 2001. Antiviral actions of interferons. Clin. Microbiol. Rev. 14:778-809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato, M., N. Hata, M. Asagiri, T. Nakaya, T. Taniguchi, and N. Tanaka. 1998. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 441:106-110. [DOI] [PubMed] [Google Scholar]
- 32.Sato, M., H. Suemori, N. Hata, M. Asagiri, K. Ogasawara, K. Nakao, T. Nakaya, M. Katsuki, S. Noguchi, N. Tanaka, and T. Taniguchi. 2000. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity 13:539-548. [DOI] [PubMed] [Google Scholar]
- 33.Sen, G. C. 2001. Viruses and interferons. Annu. Rev. Microbiol. 55:255-281. [DOI] [PubMed] [Google Scholar]
- 34.Shtrichman, R., and C. E. Samuel. 2001. The role of gamma interferon in antimicrobial immunity. Curr. Opin. Microbiol. 4:251-259. [DOI] [PubMed] [Google Scholar]
- 35.Stark, G. R., I. M. Kerr, B. R. Williams, R. H. Silverman, and R. D. Schreiber. 1998. How cells respond to interferons. Annu. Rev. Biochem. 67:227-264. [DOI] [PubMed] [Google Scholar]
- 36.Tanabe, Y., N. Sakamoto, N. Enomoto, M. Kurosaki, E. Ueda, S. Maekawa, T. Yamashiro, M. Nakagawa, C. H. Chen, N. Kanazawa, S. Kakinuma, and M. Watanabe. 2004. Synergistic inhibition of intracellular hepatitis C virus replication by combination of ribavirin and interferon-alpha. J. Infect. Dis. 189:1129-1139. [DOI] [PubMed] [Google Scholar]
- 37.Tanaka, N., T. Kawakami, and T. Taniguchi. 1993. Recognition DNA sequences of interferon regulatory factor 1 (IRF-1) and IRF-2, regulators of cell growth and the interferon system. Mol. Cell. Biol. 13:4531-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taniguchi, T., K. Ogasawara, A. Takaoka, and N. Tanaka. 2001. IRF family of transcription factors as regulators of host defense. Annu. Rev. Immunol. 19:623-655. [DOI] [PubMed] [Google Scholar]
- 39.Taniguchi, T., and A. Takaoka. 2002. The interferon-alpha/beta system in antiviral responses: a multimodal machinery of gene regulation by the IRF family of transcription factors. Curr. Opin. Immunol. 14:111-116. [DOI] [PubMed] [Google Scholar]
- 40.Yanagi, M., M. St Claire, M. Shapiro, S. U. Emerson, R. H. Purcell, and J. Bukh. 1998. Transcripts of a chimeric cDNA clone of hepatitis C virus genotype 1b are infectious in vivo. Virology 244:161-172. [DOI] [PubMed] [Google Scholar]
- 41.Yokota, T., N. Sakamoto, N. Enomoto, Y. Tanabe, M. Miyagishi, S. Maekawa, L. Yi, M. Kurosaki, K. Taira, M. Watanabe, and H. Mizusawa. 2003. Inhibition of intracellular hepatitis C virus replication by synthetic and vector-derived small interfering RNAs. EMBO. Rep. 4:602-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu, Q., J. T. Guo, and C. Seeger. 2003. Replication of hepatitis C virus subgenomes in nonhepatic epithelial and mouse hepatoma cells. J. Virol. 77:9204-9210. [DOI] [PMC free article] [PubMed] [Google Scholar]