Abstract
Herpes simplex virus type 1 (HSV-1) is a large (150-kb) double-stranded DNA virus that forms latent infections in neuronal cells of the human peripheral nervous system. Previous work determined that the HSV-1 genome is found in an ordered nucleosomal structure during latent infection. However, during lytic infection, it was unclear whether viral DNA was in a chromatin state. We examined HSV-1 during lytic infection using micrococcal nuclease digestion and chromatin immunoprecipitation. The HSV-1 genome is at least partially nucleosomal, although apparently not in a regular repeating structure. Analysis of histones associated with HSV-1, within both the promoter and the transcribed regions, revealed covalent amino tail modifications similar to those associated with active host mammalian genes. Certain of the modifications were detected in the temporal order expected of the immediate-early, early, and late gene classes. These data suggest that productive infection may be accompanied by acquisition of a permissive chromatin state.
Herpes simplex virus type 1 (HSV-1) is a double-stranded DNA virus that initially infects epithelial cells. Following the progression of lytic infection, HSV-1 establishes a latent infection of sensory neurons that innervate the site of the primary infection. The pattern of viral gene expression during lytic infection was first defined in cell culture in the early 1970s (12), and this progression has been revealed in elegant molecular detail (45, 46). The immediate-early (IE) genes are activated by the association of VP16, a viral protein that enters the cell as part of the viral tegument, with the cellular proteins host cell factor (HCF) and Oct1 (47). IE genes are classified by their detection, first during infection in the absence of de novo protein synthesis, as well as by their involvement in activating the expression of the early (E) genes. Early genes include those involved in viral DNA replication, such as the thymidine kinase (TK) gene. Expression of the late (L) genes occurs in tandem with viral replication, and most late genes encode structural proteins.
In recent years it was revealed that in eukaryotes, chromatin modulation is a central feature of genomic regulation, including transcription, replication, and DNA repair (31). While the precise mechanisms are not yet elucidated, one consequence of chromatin alteration during transcription is to regulate access to DNA sequences at the promoter and within genes. Since the original description of the nucleosome as a complex formed of 146 bp of DNA sequence wrapped around an octamer of core histones (an H3-H4 histone protein tetramer associating with two H2A-H2B dimers) (17), attention has focused on two general mechanisms to alter chromatin. First, ATP-dependent remodeling enzymes restructure nucleosomes or alter their positions along the DNA (36). Second, covalent modifications occur on the extended amino- and carboxyl-terminal tails of the core histone proteins (3). These modifications include some that are dynamic and reversible, such as lysine acetylation and serine/threonine phosphorylation, and others that are apparently more stable, such as lysine methylation.
The precise modifications and patterns of modifications on the histone tails are currently thought to help dictate specific biological outcomes (14, 43). Histone acetylation is largely involved in transcription activation and includes acetylated lysine 9 (ac-Lys9) and 14 (ac-Lys14) on histone H3 (3, 38). Histone deacetylases (HDACs) remove acetyl groups and hence are generally involved in gene-specific transcriptional repression (20). Acetyltransferases and deacetylases are components of large protein complexes that are targeted to promoters via physical recruitment by DNA-bound activator and repressor proteins (11). Methylation of the lysine residue at position 9 (me-Lys9) on the H3 subunit by histone methyltransferases is associated both with gene-specific repression (33) and with regional genomic silencing (23). One example of this involves the inactivation of the X chromosome in female mammals, whose overall activity is decreased to match the activity of the single male X chromosome (7). In contrast, methylation of lysine 4 (me-Lys4) on H3 is a marker of active chromatin (23). me-Lys4 has a role both in gene-specific activation (18) and in establishing broad euchromatic domains of activity, such as the dosage-compensated single X chromosome in male drosophila, whose overall activity is increased to match that of the two X chromosomes in females (35). Analysis of these modifications in association with a particular DNA sequence can lead to a greater understanding of transcriptional regulation in mammalian genomes and the DNA viruses that infect them.
Using partial micrococcal nuclease (MNase) digestion experiments with mouse trigeminal ganglia, previous work indicated that the HSV-1 genome is predominantly organized as nucleosomes during a latent infection (8). Further, it was found that differentially modified histones may play a regulatory role during HSV-1 latency (19). Previous MNase experiments during lytic infection showed that the majority of the HSV DNA was not in an ordered nucleosomal form (26, 29). We report a reexamination of the association of HSV-1 DNA with histone proteins during the lytic phase of infection. At early times, HSV-1 DNA is associated with histones and appears to be in a partial nucleosomal state. Further, histones that associate with the HSV-1 genome during lytic infection bear covalent modifications representative of transcriptionally active cellular genes.
MATERIALS AND METHODS
Cells and viruses.
Sy5y cells, from a human neuroblastoma cell line, were grown in RPMI 1640 medium supplemented with 10% fetal calf serum and antibiotics. The F strain of HSV-1 was used to infect the cells at a multiplicity of infection of 5.
MNase assay.
Nuclei were isolated from infected and mock-infected Sy5y cells for analysis by MNase digestion. Sy5y cells were resuspended in reticulocyte standard buffer (10 mM Tris-Cl, pH 7.5, 10 mM NaCl, 3 mM MgCl2) with 0.5% NP-40 for cell lysis. The nuclei were then pelleted at 4°C through a 0.33 M sucrose cushion at 500 × g and treated with MNase as described previously (40). Two concentrations of MNase (U.S. Biochemical Corp.) were used, 0.05 U/μl and 5 U/μl, for partial and complete digestion, respectively. MNase digestion proceeded for 5 min at room temperature and was stopped by adding 12 mM EDTA on ice. Then, 100 μg/ml of proteinase K (Fisher Scientific) was added, and cells were incubated overnight at 37°C followed by isolation of DNA by standard procedures. Genomic DNA samples were purified from Sy5y cells with the Wizard genomic DNA purification kit (Promega) and then digested with MNase and processed as described above. Samples were run on a 1.5% agarose gel and then probed with a random primed 32P-labeled (Invitrogen) 12.7-kb PstI-EcoRI fragment from within the joint repeat region of the HSV-1 genome for Southern analysis.
Histone protein isolation.
Histone proteins were isolated from infected and mock-infected Sy5y cells by previously described methods (28). In brief, cells were lysed, and proteins were acid extracted by using 0.2 M hydrochloric acid. Then, samples were dialyzed twice against 0.1 M acetic acid and three times against sterile water. Protein (2 μg for 3me-Lys4 and 50 μg for all others) was loaded onto a sodium dodecyl sulfate-polyacrylamide gel, and proteins were identified by using the following antibodies: anti-histone H3, anti-acetyl-histone H3 (K9), anti-acetyl-histone H3 (K14), anti-dimethyl-histone H3 (K9) from Upstate, and anti-trimethyl-histone H3 (K4) from Abcam. The anti-histone H3 antibody was monoclonal, and all the other antibodies used in these studies were polyclonal antibodies.
Chromatin immunoprecipitation (ChIP) assays.
Cells mock infected and infected with the F strain of HSV-1 were processed as described previously (2). In brief, cells were cross-linked with 1% formaldehyde for 15 min at room temperature. This reaction was stopped by the addition of 0.125 M glycine. Lysed cells were resuspended in IP buffer (20 mM Tris, pH 8.0, 200 mM NaCl, 0.5% Triton X-100, 0.05% deoxycholic acid, 0.5% NP-40, and 1 mM phenylmethylsulfonyl fluoride) and then sonicated. Before IP, one-tenth of the extract was saved for use as the input. The antibodies used for IP are the same as those described above for histone protein isolation.
Quantitative real-time PCR.
An ABI7700 machine with Sequence Detection software was used for quantitative PCR analysis (Applied Biosystems). Primers were designed by using the published HSV-1 sequence and are described in Table 1. SYBR green reagents (Applied Biosystems) were used to test for double-stranded DNA products resulting from the PCRs, and dissociation curve analysis was used to test the specificities of these PCR products. Standard PCR conditions for cycling and dissociation curve analysis as well as data analysis using the relative standard curve method were used as recommended by the manufacturer. Standard curves were made for each of the virus-specific and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) primer sets using known dilutions of viral or cellular DNA. These relative quantities were then normalized to GAPDH. The normalized inputs were compared to normalized immunoprecipitated samples and presented as a ratio. Each sample was done in triplicate, and standard deviations are presented.
TABLE 1.
Primers used for quantitative PCR analysis
| Gene region | Primer sequences (5′ to 3′)b | HSV-1 sequence coordinatesa |
|---|---|---|
| ICP0 promoter | TAACTTATACCCCACGCCTTTC (F) | 124275-124297 |
| TCCGGGTATGGTAATGAGTTTC (R) | 124574-124552 | |
| ICP0 transcript | CCCACTATCAGGTACACCAGCTT (F) | 122849-122871 |
| CTGCGCTGCGACACCTT (R) | 122951-122967 | |
| TK promoter | CTATGATGACACAAACCCCG (F) | 48012-48031 |
| GAGTTTCACGCCACCAAGAT (R) | 47839-47858 | |
| TK 5′ | GCAGCGACCCGCTTAACA (F) | 47879-47896 |
| GAAGAGGTGCGGGAGTTTCAC (R) | 47827-47847 | |
| TK middle | GCGTCGGTCACGGCATAAG (F) | 47393-47411 |
| GGGTGAGATATCGGCCGGG (R) | 47476-47458 | |
| TK 3′ | ACCGTATTGGCAAGCAGCCC (F) | 47066-47085 |
| GCGGCTTGACCTGGCTATGC (R) | 47128-47109 | |
| VP16 promoterc | GCCGCCCCGTACCTCGTGAC (F) | 105301-105320 |
| CAGCCCGCTCCGCTTCTCG (R) | 105508-105526 | |
| VP16 transcript | GAGCAGGCCCTCGATGGTA (F) | 103937-103955 |
| GCGGGAACACGCGTACA (R) | 103987-104003 |
Primer sequences were taken from the HSV-1 sequence available in GenBank (accession no. NC_001806).
F, forward; R, reverse.
The sequence of these primers was generously shared by Steve Triezenberg.
RESULTS
Complete MNase digestion reveals nucleosomes within the viral genome during lytic infection.
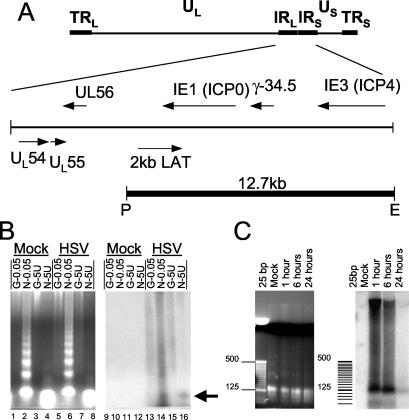
MNase digestion was used to determine whether HSV-1 is associated with nucleosomes and, if so, whether a repeating ordered nucleosomal pattern occurs on HSV-1 DNA. Neuronal Sy5y cells were mock infected or infected with HSV-1 F strain, and nuclei were collected at 24 h postinfection (p.i.) for analysis by MNase digestion. Two differing amounts of MNase were used to generate either partial or complete digestion: the smaller amount generated the classic ladder MNase pattern, and the larger amount revealed DNA of mononucleosome molecular weight. These were visualized with a stained gel (Fig. 1B, left panel), where 0.05 U of MNase produced a DNA ladder (Fig. 1B, lanes 2 and 6), and 5 U yielded a band corresponding to single nucleosome-sized DNA migrating at the bottom of the gel (Fig. 1B, lanes 4 and 8). Note that DNA was detectable only in lanes marked “N,” which were digestions of nuclei (e.g., Fig. 1B, lane 2), and not in those marked “G” (e.g., Fig. 1B, lane 1), which were control genomic DNA purified free of protein prior to digestion.
FIG. 1.
MNase digestion of nuclei isolated from Sy5y cells infected with HSV-1. (A) The HSV-1 genome and a detailed analysis of the unique long (UL) and inverted repeat (IR) region, which encompasses several viral genes. The PstI-EcoRI fragment used as a probe in the Southern analysis is marked as a solid line with ends at P and E. TRL, terminal long repeat; TRS, terminal short repeat; LAT, latency-associated transcript. (B) Mock- and HSV-1-infected samples are labeled as genomic DNA (G) and nuclei (N) that are then digested with either 0.05 or 5 U/μl of MNase. The arrow indicates the location of the mononucleosome band. The left panel shows an ethidium bromide-stained agarose gel. The right panel shows the Southern blot probed with the PstI-EcoRI fragment of HSV-1 as shown above. (C) Mock- and HSV-1-infected nuclei isolated at 1, 6, and 24 h p.i. are digested with 5 U/μl of MNase. The left and right panels show an agarose gel and a Southern blot probed as above, respectively. The samples were loaded at 10 times the quantity shown on the agarose gel for the Southern analysis.
DNA isolated from MNase-treated nuclei was subjected to Southern blotting and hybridized with a DNA probe sequence encompassing most of the joint repeat region of the virus (Fig. 1A). This region of the virus includes ICP0 and ICP4, which are two IE genes, as well as the latency-associated transcript region. Infected samples digested with the smaller amount of enzyme exhibited a smear rather than a discrete ladder (Fig. 1B, lane 14), which is similar to results from previous observations (26, 29). In contrast, digestion with the larger amount of enzyme resulted in a band comigrating with the DNA of approximately mononucleosome molecular weight (Fig. 1B, compare lane 16, arrow, with lane 8). These data suggest that some degree of nucleosomal structure exists within this region of the genome, albeit not in a regular repeating pattern.
We then repeated the experiment and tested earlier time points, during the time IE gene expression is known to increase. The larger amount of MNase (5 U) was used to determine the amount of viral DNA that is associated with the MNase-resistant structure. At the earlier times, 1 and 6 h p.i., there were larger amounts of the DNA digestion product that corresponds to mononucleosome size, 38 and 43% of the total counts, respectively, compared to that detected at 24 h p.i. (8.8%) (Fig. 1C). These data suggest that viral DNA becomes partially assembled into nucleosomes soon after infection.
Overall levels of covalently modified histone H3 in infected cells.
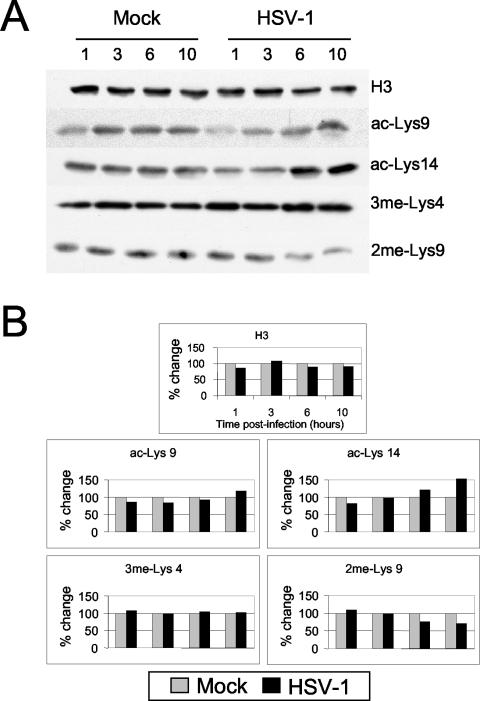
Many viruses have evolved means to alter host gene expression. For example, the virion host shutoff factor in HSV alters host translation (22, 34). Further, in certain conditions, the levels of particular histone modifications are grossly altered, manifesting as increases or decreases of specific modifications in bulk cellular histones. For example, the level of histone H3 Ser-10 phosphorylation in total chromatin is altered during the cell cycle (peaking during mitosis) and when resting cells are mitogenically stimulated (13). We examined cells infected with HSV-1, using Western blot analysis, to determine whether changes occur in the level of histone modifications. Various histone modifications were monitored in cellular histones over the time course of HSV infection and were compared to histones obtained from a mock infection (Fig. 2A). First, the linear range of Western blot detection was determined for each antibody in the set of eight samples (data not shown). The level of histone H3 was fairly constant during the infection (Fig. 2). The level of certain histone modifications reflecting active genes (ac-Lys9 and ac-Lys14) increased 1.5- to 2-fold as the infection progressed, with ac-Lys14 showing the greatest increase (Fig. 2). In contrast, the level of 2me-Lys9, a modification that correlates with inactive genes or with heterochromatic DNA, decreased nearly twofold in infected cells (Fig. 2). These results suggest that the viral infection modestly influences overall levels of specific histone modifications.
FIG. 2.
Western blot analysis of total histone proteins present in Sy5y cells during an HSV-1 infection. (A) Sy5y cells were mock infected or infected with HSV-1 and harvested at the times (in hours) p.i. indicated. Antibodies detecting unmodified histone H3 or the covalently modified amino-terminal tail of histone H3 are indicated. Each histone modification was assayed in at least two independent infections with essentially similar results, and a representative experiment is shown. (B) Quantitation of Western blots presented as the percent change of the HSV-1 samples compared to mock-infected samples (set at 100%) at the same time points.
Histone covalent modifications are associated with promoter regions and increase during lytic HSV-1 infection.
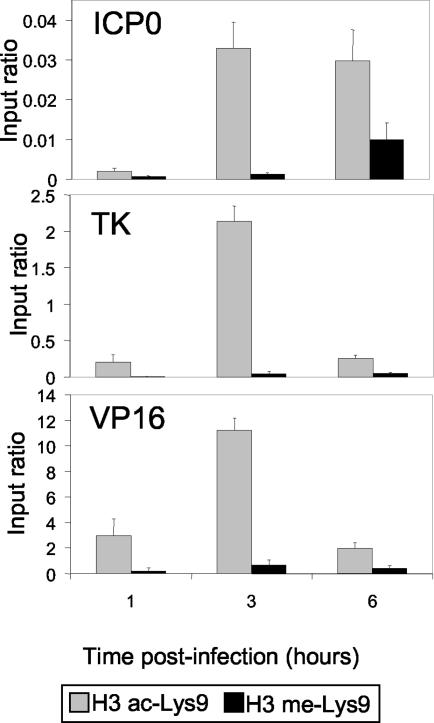
The results described above show that, first, nucleosomes are associated with HSV-1 DNA during infection and, second, there are modest changes in overall cellular levels of histone modifications during infection. Next, using ChIP we investigated whether histones are present and covalently modified at specific HSV-1 genes during the cascade of viral gene expression. Initially we examined histones during the early phase of infection, before and during DNA replication, when gene expression is induced. Samples were collected and cross-linked at 1, 3, and 6 h after infection and processed to yield purified nuclear extract. ChIP assays were performed with antibodies that detect Lys9, either acetylated (representative of gene activity) or methylated (representative of gene inactivity). We used quantitative PCR in real time to examine the immunoprecipitate for promoter DNA corresponding to an IE gene (ICP0), an E gene (TK), and an early/late (E/L) gene (VP16).
Viral DNA associated with histone proteins as early as 1 h p.i. (Fig. 3). All three promoters showed larger relative increases of ac-Lys9, rather than 2me-Lys9, during this time course. The level of acetylation rose significantly at 3 h and then fell by 6 h at the TK and VP16 promoters but remained constant at ICP0. The larger relative increases in ac-Lys9 than in 2me-Lys9 are consistent with the priming of the promoters for transcription.
FIG. 3.
ChIP analysis of histone H3 associated with viral gene promoter regions during HSV-1 lytic infection of Sy5y cells. Nuclei of infected cells were isolated at various times p.i. as indicated. ChIP assays were performed using ac-Lys9 (gray) or 2me-Lys9 (black) antibodies against the amino-terminal tail of histone H3. The values for ChIP samples at the region of interest as determined by quantitative PCR were normalized to GAPDH and then compared to input values. The gene promoters analyzed were ICP0 (upper panel), TK (middle panel), and VP16 (lower panel).
The relative level of ac-Lys9-modified histone H3 was greater for VP16 (as high as 11% of input) and less for TK (∼2% of input). The lowest level was for ICP0, which exhibited a very modest 0.03% of input. This result suggests that the promoters of IE genes may be activated before significant histone association with the HSV-1 genome occurs. This result also suggests that promoter activation of the E and E/L genes may require recruitment of either appropriately modified histones or chromatin-modifying complexes to create the modifications.
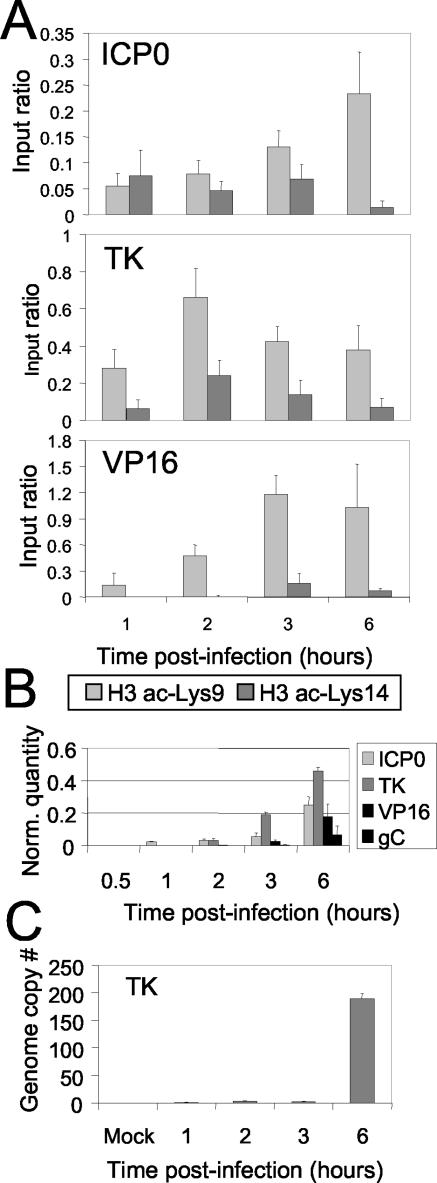
To further test the correlation of gene activity with “active” histone modifications, another ChIP assay was done, comparing levels of ac-Lys9 and ac-Lys14 with those of histone H3 (Fig. 4A; Table 2). Similar to those of ac-Lys9, ac-Lys14 levels also correlated with gene activity. In this experiment one additional time point, 2 h, was analyzed to determine whether the timing of acquisition of the modifications might discriminate different temporal gene classes. Lys-9 acetylation again was apparent for all three genes at 1 h. A peak of modification was detected at 2 h for TK and 3 h for VP16, consistent with the order of their transcription (see below). This correspondence with kinetic class was also apparent for ac-Lys14, where ICP0 (IE) increased at 1 h, TK (E) increased at 2 h, and VP16 (E/L) increased at 3 h. Again, the relative levels were lowest for ICP0, intermediate for TK, and highest for VP16. As a control for the progress of the infection, we examined RNA production for these genes over the time course. ICP0 (IE) was detected at 1 h, TK (E) was detected at 2 h, VP16 (E/L) was detected at 3 h, and gC (L) was detected at 6 h (Fig. 4B). The number of viral genome copies increased dramatically at 6 h (Fig. 4C), after the peak in histone modifications at 2 to 3 h after infection (Fig. 4, compare panels A and C). Thus, there was a strong correlation between the timing of RNA appearance (and not simply the increase in the amount of viral DNA) and the acquisition of “activating” histone modifications for three gene classes.
FIG. 4.
ChIP analysis of histone H3 associated with HSV-1 genes compared to transcription during lytic infection. (A) Methods were the same as those described in the legend to Fig. 3. The results of ChIP analysis using antibodies detecting histone H3 modified at ac-Lys9 (light gray) or ac-Lys14 (dark gray) are shown. Genes were the same as those described in the legend to Fig. 3. (B) Quantitative transcriptional analysis by real-time PCR of the four kinetic classes of HSV-1 genes. The genes analyzed were ICP0 (IE), TK (E), VP16 (E/L), and gC (L), as indicated in the key, and primers for each transcript are shown in Table 1. (C) Genome copy numbers of HSV-1 as quantitated by real-time PCR. Total DNA was isolated and analyzed with a standard curve for copies of the TK gene. PCR primers for TK are designated TK 5′ (Table 1).
TABLE 2.
Input ratios from ChIP experiments shown in Fig. 4
| Gene | Histone modification | Time p.i. (h)
|
|||
|---|---|---|---|---|---|
| 1 | 2 | 3 | 6 | ||
| ICP0 | ac-Lys9 | 0.056 | 0.078 | 0.131 | 0.234 |
| ac-Lys14 | 0.075 | 0.046 | 0.068 | 0.014 | |
| TK | ac-Lys9 | 0.281 | 0.662 | 0.423 | 0.379 |
| ac-Lys14 | 0.062 | 0.240 | 0.138 | 0.071 | |
| VP16 | ac-Lys9 | 0.141 | 0.475 | 1.18 | 1.03 |
| ac-Lys14 | 0 | 0.005 | 0.160 | 0.076 | |
Histone H3 Lys4 methylation preferentially associates with the 5′ end of transcribed regions.
Similar to acetylation of Lys9 and Lys14, methylation of Lys4 on histone H3 has been correlated with gene induction (39). However, there are interesting differences with respect to the location of the modification within chromatin relative to the open reading frame (ORF), as examined in the model eukaryote Saccharomyces cerevisiae. While transcription-linked acetylation is largely confined to the promoter (11, 21), me-Lys4 is associated with the entire ORF (4). Based on these observations, ChIP was performed with me-Lys4 antibody.
Similar to the other “activating” modifications (ac-Lys9 and ac-Lys14), the amount of 3me-Lys4 increased early after infection for ICP0 and then appeared coincidently for TK and VP16 (Fig. 5A). However, the promoter regions of these genes exhibited very low methylation. Instead, the modification was found within the transcribed region for each gene. Interestingly, in contrast to both ac-Lys9 and ac-Lys14, 3me-Lys4 levels were more similar between the three gene classes. In particular, levels of me-Lys4 at ICP0 were half of those at TK.
FIG. 5.
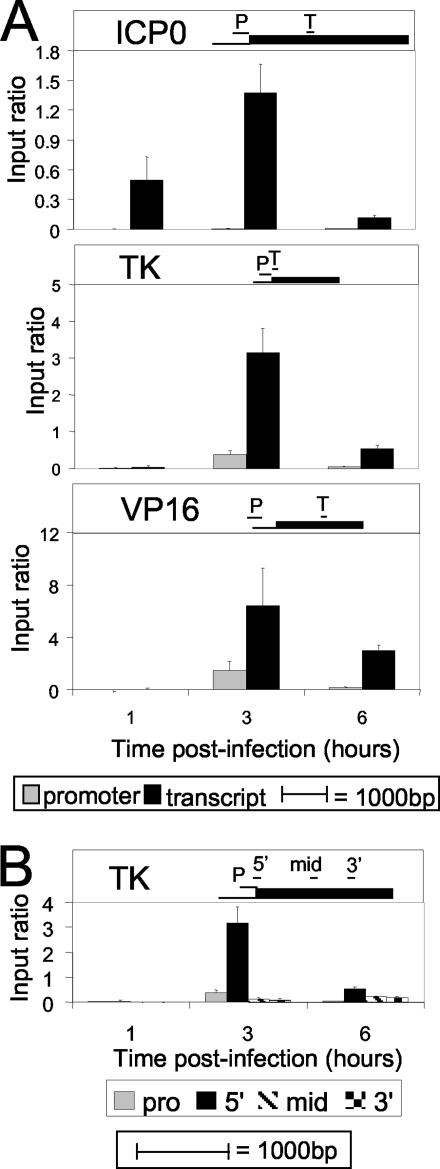
ChIP analysis of histone H3 modified at me-K4 within various viral gene regions during infection. (A) Methods were the same as those described in the legend to Fig. 3. The results of ChIP analysis using antibodies detecting histone H3 modified by 3me-Lys4 within three genes are shown. Viral gene-specific primers were used within the promoter and coding regions of ICP0, TK, and VP16, as indicated. Diagrams showing relative primer locations (to scale) within each gene are shown. (B) ChIP analysis of 3me-K4 within the TK promoter and gene. Methods were the same as those described in the legend to Fig. 3, and the same antibody as that used in panel A was used. Primers to TK were within the promoter and the ORF, as indicted.
It was further observed that trimethylation of Lys-4 on histone H3 is enriched within promoter regions of genes in S. cerevisiae (32). To examine the specificity of the localization of 3me-Lys4 in greater detail, two more regions of the TK transcribed region were tested, i.e., the middle and 3′ end of the transcribed region. Methylation discretely localized within the 5′ end of the gene and was detected at only very low levels within the promoter, middle, or 3′ end of the gene (Fig. 5B).
DISCUSSION
It was previously shown using partial MNase digestion that HSV-1 DNA, when latent in the peripheral nervous system of mice, is present in a nucleosomal structure (8). However, similar experimental approaches led to the conclusion that, during lytic infection, the HSV-1 genome is largely not associated with nucleosomes (26, 29). Nevertheless, many aspects of HSV-1 transcriptional regulation during lytic infection have been shown to be highly similar to that of and to require factors from the mammalian host. For example, HSV-1 genes utilize many host transcriptional activators, such as Oct1, Sp1, and CREB (15, 16, 25, 41), and these activators interact with chromatin-modulating enzymes (1, 5). Taken together, these data led us to reexamine the nucleosomal state of lytically infecting HSV-1 DNA in human neuroblastoma (Sy5y) cells.
HSV-1 DNA is associated with nucleosomes during lytic infection.
In previous similar MNase approaches, a ladder of DNA fragments was not observed within HSV-1 (26, 29). However, we detected a smear of HSV-1 DNA using partial MNase analyis (Fig. 1B). Thus, an alternative conclusion is that nucleosomes are not evenly distributed on the viral genome but occur only in certain regions. Indeed, complete MNase digestion (Fig. 1B and C) yielded a single viral DNA fragment comigrating with mononucleosomes, indicating that the viral genome is associated with nucleosomes during lytic infection. Although we probed with only the joint repeat region of the HSV-1 genome, our further analyses using ChIP (see below) indicate that multiple genes are associated with histones.
Viral gene-associated histones exhibit covalent modifications typical of transcriptional activity.
We initially examined the level of histones bearing specific covalent modifications within bulk histones. Alterations in modification levels were detected upon viral infection and, although modest, trended toward modifications that correlate with gene activity. In particular, the amount of ac-Lys14 increased (Fig. 2), becoming apparent at the time (6 h) of strong HSV-1 gene transcription (Fig. 4B). Although the fraction of total HSV-1 genes is small relative to that of the host genome, this result suggests that HSV-1 could broadly manipulate histone modification levels in order to alter cellular transcription. Based on these results, we speculated that HSV-1 might possess its own histone acetylation or methylation enzymes; however, using computer-based homology searches we have not found enzymatic domains related to either the GNAT or CBP/p300 families of histone acetyltransferases (HATs) or the SET or DOT1 families of histone methyltransferases. Thus, HSV-1 may alter cellular histone levels indirectly by influencing host gene expression (see below for further discussion).
Based on these altered histone modification levels within the host cell, we examined histone modifications at specific HSV-1 genes. Several histone H3 modifications increased during infection, including ac-Lys9 (Fig. 3 and 4), ac-Lys14 (Fig. 4), and 3me-Lys4 (Fig. 5). These results indicate, first, that specific gene regions of the HSV-1 genome become associated with histone proteins during infection and, second, that the modifications showing the greatest increases correlate with gene activity in other eukaryotic systems. By contrast, 2me-Lys9, which is associated with gene repression, did not show similar increases (although the level did increase at the IE gene ICP0 at 6 h; see discussion below). Association with histones occurred rapidly after viral DNA entered the cell, as the ChIP assays revealed signals by 1 h after infection. The number of modification signals continued to increase, and the peak (approximately 2 to 3 h for most assays) may occur before most viral replication, which is strongly detected at approximately 6 h. Indeed, in our assays the peak in ChIP signals occurred well before the peak in viral DNA (Fig. 4C). Thus, it is possible that the portion of newly replicated viral DNA destined to be packaged into virions does not associate with histones, and indeed, encapsidated virus appears not to be nucleosomal (26, 27).
Viral genes posses both promoter and gene-associated patterns of active histone modifications.
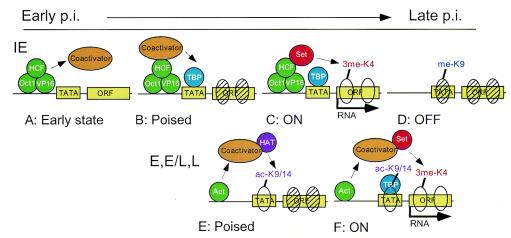
Our results can be interpreted into a model of HSV-1 gene activation in a chromatin context, as shown in Fig. 6. Two classes of modifications associated with gene-specific activation have been distinguished. The first class correlates with transcriptional initiation, typified by histone H3 ac-Lys9 and ac-Lys14. In our assays, the ChIP profile of gene-specific association of these modifications reflected the HSV-1 gene kinetic class. Modifications at ICP0, an IE gene, while very low in number (see below for further discussion), were first detected at 1 h and stayed fairly constant to 3 h. The modifications at TK (E class) peaked at 2 h, and the modifications at VP16 (E/L class) peaked at 3 h (Fig. 4A). Certain HATs that target these residues are known, including CBP/p300 and Gcn5/PCAF. These HATs are recruited (as components of large macromolecular complexes) to promoters via physical association with DNA-bound activators. For example, the VP16 activator, which is part of an Oct1/HCF complex, interacts with both the yeast and human forms of the Gcn5 HAT complex in vitro (44). In fact, the yeast and human Gcn5 coactivator complex contains modules that perform functions other than acetylation, including recruitment of TATA-binding protein (TBP) to the promoter (24). Thus, these data suggest that an early step may be Oct1, HCF, and VP16 recruitment of the coactivator/HAT complexes to activate ICP0 via mechanisms not including histone acetylation (since the ICP0 promoter exhibits very low levels of histone modifications) (Fig. 6A). Once the IE gene activators ICP0 and ICP4 are expressed, they may then recruit coactivator/HAT complexes to the E, E/L, and L gene classes, where histone acetylation at the promoter becomes more prominent (Fig. 6E) (6, 9). Note that the “repressing” modification 2me-Lys9 becomes associated with ICP0 at 6 h, which correlates with evidence that ICP0 expression is decreased at later times (12).
FIG. 6.
Schematic model of transcriptional activation of HSV-1. The upper panel shows an IE gene, such as ICP0, and the lower panel shows an E or E/L gene, such as TK or VP16. Repressed chromatin is represented by hatched ovals (nucleosomes), and permissive chromatin is indicated by open ovals (nucleosomes). TATA-binding protein (TBP) and the TATA box in the proximal promoter are shown. Activators of the IE genes (Oct1, HCF, and VP16) and other activators of the E/L and L genes (Act) are bound to upstream enhancers or promoters. The coactivators are protein complexes that function in recruitment of TBP and histone modifications. HAT represents a histone H3 acetyltransferase, and Set represents a histone H3 methylase. RNA synthesis is indicated by an arrow. Histone-covalent modifications are indicated over the nucleosomes.
The second class of modifications associated with gene activation occurs within gene ORFs and appears to be involved in transcriptional elongation. The best characterized of these is me-Lys4, which correlates with gene induction and occurs most strongly at the 5′ end of ORFs (4, 32, 39). Lys4 methylation appears to be involved in the transition from initiation to elongation of RNA synthesis (10). Consistent with this involvement, we detected the level of 3me-Lys4 rising at the time that RNA synthesis is first detected, at 1 to 2 h for ICP0 (Fig. 6C) and at 3 h for TK and VP16 (Fig. 6F). In addition, we observed higher levels of 3me-Lys4 within ORFs than in promoter regions for ICP0, TK, and VP16. Further, the TK gene showed a striking concentration of 3me-Lys4 at the 5′ end of the ORF compared to the promoter, middle, or 3′ end. The latter relationship had previously been shown only for genes of S. cerevisiae (32), and thus our observations of HSV-1 indicate that the level of 3me-Lys4 is likely to be high at the 5′ ends of mammalian genes as well.
Notably, the three gene classes exhibited more similar levels of 3me-Lys4 within their coding sequences than acetylation within their promoters, including the IE gene, ICP0. These findings suggest that the virus associates with histones by the time ICP0 is ready to be transcribed, and hence 3me-Lys4 on the ICP0 gene may be relevant for IE genes (Fig. 6C).
We do not yet know the portion of infecting HSV-1 that is associated with chromatin nor whether histone modifications are required for transcription of HSV-1. However, our data provide new insights into potential mechanisms of HSV-1 transcriptional regulation during lytic infection. We detected preferential association of activation-relevant histone H3 modifications, including ac-Lys9, ac-Lys14, and 3me-Lys4 at the promoters and ORFs-5′ ends of HSV-1 genes. These observations of 3me-Lys4 may relate to a recently identified histone methyltransferase, Set1, which associates with VP16 and HCF (48). Because lytic HSV-1 was previously believed to be histone free, it was argued that Set1 methylates nonhistone substrates (48); however, our results suggest that Set1 may target histones within the HSV-1 genome.
Potential chromatin-modulating enzymes have previously been shown to be involved in infection with several herpes viruses. The Epstein-Barr virus protein, EBNA3C, may modulate transcription by interacting with a complex of cellular proteins, including a histone acetyltransferase protein, p300, and a protein involved in chromatin remodeling, ProTα (42). During cytomegalovirus infection, histone deacetylases are involved in the repression of IE genes in nonpermissive cells, and in permissive cells, IE promoters are associated with acetylated histones (30). Thus, chromatin remodeling may be involved in regulating infection of different cell types during various stages of viral infection. In addition, the bovine herpesvirus IE protein, bICP0, has been shown to associate with histone deacetylase 1 (HDAC1) and may be involved in stimulating productive infection (49). In the presence of an HSV infection, HDAC1 and HDAC2 have been shown to undergo posttranslational modifications, and HDAC inhibitors accelerate certain aspects of the infection (37). If HDAC enzymatic activity is decreased by these modifications, then this finding may be related to our observation that bulk cellular levels of histone acetylation increase during infection (Fig. 2). Thus, while histones were not shown to be a direct substrate of these enzymes during viral infection, our data suggest that histones are one possible substrate.
In conclusion, we have found that the neurotropic human HSV-1 genome, previously thought to be nucleosome free, is associated with nucleosomes during the lytic stage of viral infection. Our findings suggest that chromatin-modulatory enzymes may play a role in HSV-1 lytic infection and imply that modification of histones in HSV-1 may also be important during the reactivation of the virus from latency (19). Thus, if HSV-1 gene expression is dependent on these histone-modification enzymes, then a new class(es) of viral inhibitors could be developed against these targets. Such inhibitors may provide a means of medical intervention at an early stage of the HSV-1 life cycle.
Acknowledgments
S.L.B. and N.W.F. acknowledge research support from a PO1 grant (NS33768) from NIH. J.R.K. acknowledges support from an NIH training grant “Training in Virology” (T32 AI-07324) at the University of Pennsylvania.
We thank N. C. T. Emre, G. P. Moore, and M. Schwartz for critical reading of the manuscript and T. Block, H.-Y. Su, and members of the Berger and Fraser laboratories for valuable discussions.
REFERENCES
- 1.Asahara, H., B. Santoso, E. Guzman, K. Du, P. A. Cole, I. Davidson, and M. Montminy. 2001. Chromatin-dependent cooperativity between constitutive and inducible activation domains in CREB. Mol. Cell. Biol. 21:7892-7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243-1254. [DOI] [PubMed] [Google Scholar]
- 3.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 4.Bernstein, B. E., E. L. Humphrey, R. L. Erlich, R. Schneider, P. Bouman, J. S. Liu, T. Kouzarides, and S. L. Schreiber. 2002. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc. Natl. Acad. Sci. USA 99:8695-8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boekhoudt, G. H., Z. Guo, G. W. Beresford, and J. M. Boss. 2003. Communication between NF-kappa B and Sp1 controls histone acetylation within the proximal promoter of the monocyte chemoattractant protein 1 gene. J. Immunol. 170:4139-4147. [DOI] [PubMed] [Google Scholar]
- 6.Carrozza, M. J., and N. DeLuca. 1998. The high mobility group protein 1 is a coactivator of herpes simplex virus ICP4 in vitro. J. Virol. 72:6752-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cohen, D. E., and J. T. Lee. 2002. X-chromosome inactivation and the search for chromosome-wide silencers. Curr. Opin. Genet. Dev. 12:219-224. [DOI] [PubMed] [Google Scholar]
- 8.Deshmane, S., and N. W. Fraser. 1989. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J. Virol. 63:943-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grondin, B., and N. DeLuca. 2000. Herpes simplex virus type 1 ICP4 promotes transcription preinitiation complex formation by enhancing the binding of TFIID to DNA. J. Virol. 74:11504-11510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hampsey, M., and D. Reinberg. 2003. Tails of intrigue: phosphorylation of RNA polymerase II mediates histone methylation. Cell 113:429-432. [DOI] [PubMed] [Google Scholar]
- 11.Hassan, A. H., K. E. Neely, M. Vignali, J. C. Reese, and J. L. Workman. 2001. Promoter targeting of chromatin-modifying complexes. Front. Biosci. 6:D1054-D1064. [DOI] [PubMed] [Google Scholar]
- 12.Honess, R. W., and B. Roizman. 1974. Regulation of herpes macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J. Virol. 14:8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu, J. Y., Z. W. Sun, X. Li, M. Reuben, K. Tatchell, D. K. Bishop, J. M. Grushcow, C. J. Brame, J. A. Caldwell, D. F. Hunt, R. Lin, M. M. Smith, and C. D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102:279-291. [DOI] [PubMed] [Google Scholar]
- 14.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 15.Jones, K. A., and R. Tjian. 1985. Sp1 binds to promoter sequences and activates herpes simplex virus ′immediate-early' gene transcription in vitro. Nature 317:179-182. [DOI] [PubMed] [Google Scholar]
- 16.Kenny, J. J., F. C. Krebs, H. T. Hartle, A. E. Gartner, B. Chatton, J. M. Leiden, J. P. Hoeffler, P. C. Weber, and B. Wigdahl. 1994. Identification of a second ATF/CREB-like element in the herpes simplex virus type 1 (HSV-1) latency-associated transcript (LAT) promoter. Virology 200:220-235. [DOI] [PubMed] [Google Scholar]
- 17.Kornberg, R. D. 1974. Chromatin structure: a repeating unit of histones and DNA. Science 184:868-871. [DOI] [PubMed] [Google Scholar]
- 18.Kouzarides, T. 2002. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 12:198-209. [DOI] [PubMed] [Google Scholar]
- 19.Kubat, N. J., R. K. Tran, P. McAnany, and D. C. Bloom. 2004. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J. Virol. 78:1139-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuo, M. H., and C. D. Allis. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20:615-626. [DOI] [PubMed] [Google Scholar]
- 21.Kuo, M. H., J. Zhou, P. Jambeck, M. E. Churchill, and C. D. Allis. 1998. Histone acetyltransferase activity of yeast Gcn5p is required for the activation of target genes in vivo. Genes Dev. 12:627-639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwong, A. D., J. A. Kruper, and N. Frenkel. 1988. Herpes simplex virus virion host shutoff function. J. Virol. 62:912-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lachner, M., and T. Jenuwein. 2002. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 14:286-298. [DOI] [PubMed] [Google Scholar]
- 24.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15:1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leib, D. A., K. C. Nadeau, J. A. Rundle, and P. A. Schaffer. 1991. The promoter of the latency-associated transcripts of herpes simplex type 1 contains a functional cAMP-response element: role of the latency-associated transcripts and cAMP in reactivation of viral latency. Proc. Natl. Acad. Sci. USA 88:48-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leinbach, S. S., and W. C. Summers. 1980. The structure of herpes simplex virus type 1 DNA as probed by micrococcal nuclease digestion. J. Gen. Virol. 51:45-59. [DOI] [PubMed] [Google Scholar]
- 27.Lentine, A. F., and S. L. Bachenheimer. 1990. Intracellular organization of herpes simplex virus type 1 DNA assayed by staphylococcal nuclease sensitivity. Virus Res. 16:275-292. [DOI] [PubMed] [Google Scholar]
- 28.Mizzen, C. A., J. E. Brownell, R. G. Cook, and C. D. Allis. 1999. Histone acetyltransferases: preparation of substrates and assay procedures. Methods Enzymol. 304:675-696. [DOI] [PubMed] [Google Scholar]
- 29.Muggeridge, M. I., and N. W. Fraser. 1986. Chromosomal organization of the herpes simplex virus genome during acute infection of the mouse central nervous system. J. Virol. 59:764-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy, J. C., W. Fischle, E. Verdin, and J. H. Sinclair. 2002. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 21:1112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108:475-487. [DOI] [PubMed] [Google Scholar]
- 32.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen, S. J., R. Schneider, U. M. Bauer, A. J. Bannister, A. Morrison, D. O'Carroll, R. Firestein, M. Cleary, T. Jenuwein, R. E. Herrera, and T. Kouzarides. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561-565. [DOI] [PubMed] [Google Scholar]
- 34.Oroskar, A. A., and G. S. Read. 1989. Control of mRNA stability by the virion host shutoff function of herpes simplex virus. J. Virol. 63:1897-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pannuti, A., and J. C. Lucchesi. 2000. Recycling to remodel: evolution of dosage-compensation complexes. Curr. Opin. Genet. Dev. 10:644-650. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, C. L., and J. L. Workman. 2000. Promoter targeting and chromatin remodeling by the SWI/SNF complex. Curr. Opin. Genet. Dev. 10:187-192. [DOI] [PubMed] [Google Scholar]
- 37.Poon, A. P., Y. Liang, and B. Roizman. 2003. Herpes simplex virus 1 gene expression is accelerated by inhibitors of histone deacetylases in rabbit skin cells infected with a mutant carrying a cDNA copy of the infected-cell protein no. 0. J. Virol. 77:12671-12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice, J. C., and C. D. Allis. 2001. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr. Opin. Cell Biol. 13:263-273. [DOI] [PubMed] [Google Scholar]
- 39.Santos-Rosa, H., R. Schneider, A. J. Bannister, J. Sherriff, B. E. Bernstein, N. C. Emre, S. L. Schreiber, J. Mellor, and T. Kouzarides. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407-411. [DOI] [PubMed] [Google Scholar]
- 40.Spiker, S., M. G. Murray, and W. F. Thompson. 1983. DNase I sensitivity of transcriptionally active genes in intact nuclei and isolated chromatin of plants. Proc. Natl. Acad. Sci. USA 80:815-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stern, S., M. Tanaka, and W. Herr. 1989. The Oct-1 homoeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature 341:624-630. [DOI] [PubMed] [Google Scholar]
- 42.Subramanian, C., S. Hasan, M. Rowe, M. Hottiger, R. Orre, and E. S. Robertson. 2002. Epstein-Barr virus nuclear antigen 3C and prothymosin alpha interact with the p300 transcriptional coactivator at the CH1 and CH3/HAT domains and cooperate in regulation of transcription and histone acetylation. J. Virol. 76:4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 44.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 45.Wagner, E. K., J. F. Guzowski, and J. Singh. 1995. Transcription of the herpes simplex virus genome during productive and latent infection. Prog. Nucleic Acid Res. Mol. Biol. 51:123-165. [DOI] [PubMed] [Google Scholar]
- 46.Weir, J. P. 2001. Regulation of herpes simplex virus gene expression. Gene 271:117-130. [DOI] [PubMed] [Google Scholar]
- 47.Wysocka, J., and W. Herr. 2003. The herpes simplex virus VP16-induced complex: the makings of a regulatory switch. Trends Biochem. Sci. 28:294-304. [DOI] [PubMed] [Google Scholar]
- 48.Wysocka, J., M. P. Myers, C. D. Laherty, R. N. Eisenman, and W. Herr. 2003. Human Sin3 deacetylase and trithorax-related Set1/Ash2 histone H3-K4 methyltransferase are tethered together selectively by the cell-proliferation factor HCF-1. Genes Dev. 17:896-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang, Y., and C. Jones. 2001. The bovine herpesvirus 1 immediate-early protein (bICP0) associates with histone deacetylase 1 to activate transcription. J. Virol. 75:9571-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]