Abstract
African swine fever virus (ASFV), a large icosahedral deoxyvirus, is the causative agent of an economically relevant hemorrhagic disease that affects domestic pigs. The major purpose of the present study was to investigate the nuclear transport activities of the ASFV p37 and p14 proteins, which result from the proteolytic processing of a common precursor. Experiments were performed by using yeast-based nucleocytoplasmic transport assays and by analysis of the subcellular localization of different green fluorescent and Myc fusion proteins in mammalian cells. The results obtained both in yeast and mammalian cells clearly demonstrated that ASFV p14 protein is imported into the nucleus but not exported to the cytoplasm. The ability of p37 protein to be exported from the nucleus to the cytoplasm of both yeast and mammalian cells was also demonstrated, and the results clearly indicate that p37 nuclear export is dependent on the interaction of the protein with the CRM-1 receptor. In addition, p37 was shown to exhibit nuclear import activity in mammalian cells. The p37 protein nuclear import and export abilities described here constitute the first report of a nucleocytoplasmic shuttling protein encoded by the ASFV genome. Overall, the overlapping results obtained for green fluorescent protein fusions and Myc-tagged proteins undoubtedly demonstrate that ASFV p37 and p14 proteins exhibit nucleocytoplasmic transport activities. These findings are significant for understanding the role these proteins play in the replication cycle of ASFV.
African swine fever virus (ASFV), one of the most complex animal viruses, is the only member of the new family Asfarviridae (10). This large enveloped virus is the causative agent of a highly lethal hemorrhagic disease that affects domestic pigs (9, 42).
The ASFV genome is a large linear double-stranded DNA molecule (170 to 190 kbp) that encodes more than 150 polypeptides, including about 50 structural proteins and several enzymes involved in viral DNA replication, viral gene transcription, and protein modification (41, 46).
Although ASFV has been considered for many years to be a virus that replicates exclusively in the cell cytoplasm (9, 42), it has been recently reported that an initial stage of its replication cycle occurs inside the nucleus of infected cells (17, 32). It is therefore reasonable to assume that some of ASFV proteins must be actively transported between the nucleus and cytoplasm of host cells.
The bidirectional transport of macromolecules between the nucleus and cytoplasm is a selective process that occurs exclusively through nuclear pores. The nuclear pore complex is a 125-MDa macromolecular assembly of 50 to 100 polypeptides that produces an aqueous channel with a diameter of 9 nm. The channel allows the passive diffusion of small molecules, including ions, metabolites, and globular proteins of up to ca. 60 kDa. Translocation of macromolecules larger than 60 kDa into and out of the nucleus is an active, energy-dependent process that is mediated by specific sequence motifs: nuclear localization signals (NLSs) and nuclear export signals (NESs) (6, 19, 28).
Classical examples of NLSs are the highly basic motifs found in the simian virus 40 (SV40) large T antigen (22) and in nucleoplasmin (31), although several nonclassical sequences have also been shown to function as NLSs (11, 45).
The best-characterized NESs consist of short peptide sequences of four closely spaced hydrophobic amino acids, such as leucine or isoleucine residues. These leucine-rich NESs were initially identified in the human immunodeficiency virus type 1 (HIV-1) Rev protein (13) and in the protein kinase A inhibitor (43). However, NESs that do not belong to the family of leucine-rich NESs have also been described for various proteins, as exemplified by the atypical NES present in hepatitis D antigen HDAg-L (25).
The direct interaction of proteins bearing leucine-rich NESs with the nuclear export factor CRM-1 (named for chromosome region maintenance 1; also known as exportin-1) is essential for the nuclear export process (15, 16, 29, 39). NESs present in proteins and RanGTP bind directly to CRM-1, and this trimeric complex is then translocated from the nucleus to the cytoplasm through the nuclear pore complex. Although the CRM-1-dependent transport is the best-characterized pathway, CRM-1-independent nuclear export mechanisms have been proposed for various proteins (12, 25, 26).
In the present study we examined the nuclear transport capacity of two ASFV structural proteins—p14 and p37—which are the result of the proteolytic processing of the precursor pp220 that is catalyzed by a viral protease that shares sequence similarity with the proteases of the SUMO-1 family (2, 33). Repression of protease expression inhibits polyprotein processing, leading to the assembly of core-defective, noninfectious viral particles (1).
The initial event of the proteolytic cascade is the separation of the mature protein p150 and the precursor pp90; the cleavage of this precursor results in the mature protein p34 and in preprotein pp55, which finally produces p37 and p14 (37).
Our results clearly demonstrate that ASFV p37 and p14 proteins are involved in nucleocytoplasmic transport. Interestingly, although deriving from a common immediate precursor, these proteins exhibit different transport activities. p14 protein is imported into the nucleus, and p37 protein is both imported into the nucleus and exported from the nucleus to the cell cytoplasm, making it the first nucleocytoplasmic shuttling protein identified in ASFV. Moreover, we were able to determine that p37 protein is exported from the cell nucleus through the CRM-1-dependent nuclear export pathway.
MATERIALS AND METHODS
Plasmid constructs.
cDNAs encoding p14 and p37 were amplified by PCR with ASFV Lisbon 60 strain DNA as a template and with specific primers. The primers introduced BamHI and NsiI recognition sites, respectively, at the 5′ and 3′ ends of the amplified DNA sequences. Each DNA fragment was cloned into the BamHI/PstI restriction sites of either the plasmid for the yeast nuclear import and export assays—pNIA and pNEA (30), respectively—or pCMV-Tag 3C (Stratagene, La Jolla, Calif.), as well as into pEGFP-C1 (Clontech, Palo Alto, Calif.).
pNIAGFP and pNEAGFP were generated by cloning green fluorescent protein (GFP) cDNA downstream of the yeast Gal4p activation domain (Gal4AD) in pNIA or pNEA, as follows. GFP cDNA was amplified by PCR, with pEGFP-C1 as a template and with the primer pair 5′-CGGGATCCCCATGGTGAGCAAGG-3′ and 5′-CGGGATCCACTTGTACAGCTCGTC-3′ that introduced BglII and BamHI recognition sites. The amplified cDNA was then cloned into the BamHI cloning site of pNIA or pNEA.
The primers used to generate pNIAGFPp14, pNEAp14, and Myc-p14 were pri1 (5′-GGATCCTTATGGACGAGGAGAAAACG-3′) and pri2 (5′-ATGCATCTAACCGCCTACCTTTGTA-3′), the primers used to generate pNEAp37 and Myc-p37 were pri3 (5′-GGATCCTTATGGCTGCCCTAACGG-3′) and pri4 (5′-ATGCATCTAGCCTCCCAGTATCATA-3′), the primers used to generate GFPp14 were pri5 (5′-GGATCCATGGACGAGGAGAAAACG-3′) and pri2, and the primers used to generate GFPp37 were pri6 (5′-GGATCCATGGCTGCCCTAACGG-3′) and pri4.
To construct GFPp37NLS, which encodes the classical SV40NLS (PKKKRKV) fused to the C terminus of p37, PCR-based insertion was carried out by using the QuickChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. To perform the insertion, GFPp37 (template) and the primer 5′-CGCGTATGATACTGGGAGGCCCAAAAAAGAAGAGAAAGGTCATGCATTCGACGGTACCGCGGG-3′ and its complementary oligonucleotide were used.
GFPNLS was generated by the same strategy. pEGFP-C1 was used as a template, and the primer 5′-TACAAGTCCGGACTCAGATCTCCAAAAAAGAAGAGAAAGGTCATGCATTCGACGGTACCGCGGG-3′ and its complementary oligonucleotide were used to perform the insertion.
The constructs GFP-GFPp14 and GFP-GFPp37 were generated by PCR amplification with GFPp37 and GFPp14, respectively, as the template. The amplified cDNAs were introduced into BamHI and PstI restriction sites of pEGFP-C1. The primer pair pri7 (5′-GGATCCATGGTGAGCAAGG-3′) and pri2 was used to generate GFP-GFPp14, and the primer pair pri7 and pri4 was used to generate GFP-GFPp37.
The pNEARev (30) and pTat-GFP-NLS (40) plasmids have been described previously.
Proper framing of all DNA constructs was confirmed by DNA sequencing (AGOWA, Berlin, Germany).
Yeast nuclear import and export assays.
The yeast nuclear import and nuclear export assays were performed as described previously by Rhee et al. (30). Briefly, pNIA and pNEA derived constructs were transformed into Saccharomyces cerevisiae strain L40, which contains the two LexA-inducible genes HIS3 and lacZ (20), by the lithium acetate method (21). The transformed yeasts were then plated on selective medium without tryptophan. Since pNIA and pNEA contain the TRP1 gene, only transformed yeasts can grow on this medium.
After growth, a few colonies were spread on minimal medium without tryptophan and in parallel were plated on minimal medium lacking both tryptophan and histidine and supplemented with 10 mM 3-amino-1,2,4-triazole (3AT; Sigma, St. Louis, Mo.), a repressor of yeast endogenous histidine production. For pNEARev, minimal medium lacking both tryptophan and histidine was supplemented with 100 mM 3AT (30). Yeast growth in the absence of histidine was evaluated.
In addition, yeasts plated on tryptophan-deficient medium were transferred to nitrocellulose filters and assayed for β-galactosidase activity (7). Briefly, after disruption of yeast cell membrane by incubation at −70°C for 30 min, the yeast lysate was incubated with the β-galactosidase substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Sigma), and blue color development was evaluated.
For quantitative determination of β-galactosidase activity, an enzymatic assay was performed in liquid cultures (38). After growth in liquid medium without tryptophan, yeast cells were disrupted, and the β-galactosidase chromogenic substrate ONPG (o-nitrophenyl-β-d-galactopyranoside; Sigma) was added in excess. After incubation at 30°C, the reaction was stopped by raising the pH to 11, which inactivates β-galactosidase. The β-galactosidase activity was calculated according to the following equation (27): β-galactosidase units = 1,000 × OD420/t × V × OD600, where OD420 is the optical density at 420 nm of the sample measured after the incubation of yeast cell lysate with ONPG, t is the time of incubation (in minutes) of the yeast cell lysate with ONPG, V is the volume of the sample used in the assay (in milliliters), and OD600 is the optical density at 600 nm of the yeast cell culture at the start of the assay.
Cell culture.
Vero cells were grown and maintained in Dulbecco modified Eagle medium-high glucose (DMEM-HG; Sigma) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (Biochrom KG, Berlin, Germany) and with 100 U of penicillin and 100 μg of streptomycin (Sigma) per ml in a 5% CO2 humidified atmosphere at 37°C.
For the transfection experiments, 7.5 × 104 Vero cells were seeded onto 16-mm glass coverslips on a 12-well plate.
Transfection, leptomycin B (LMB) treatment, and temperature shift assay.
Transfection experiments were performed by using transferrin-associated lipoplexes (35, 36) or Fugene 6 transfection reagent (Roche Molecular Biochemicals, Penzberg, Germany) as follows.
The cationic liposome/transferrin/plasmid DNA complexes containing 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and cholesterol (1:1 molar ratio; Avanti-Polar Lipids, Alabaster, Ala.) were prepared at a cationic lipid/DNA (+/−) charge ratio of 1/1 as described previously (35, 36). For each construct, 3 μg of plasmid DNA per well was used. Vero cells at 50% confluence were incubated with the complexes for 4 h in serum-free medium. After this, the medium was replaced with DMEM-HG containing 10% fetal bovine serum, and the cells were incubated another 24 h to allow gene expression.
Alternatively, Fugene 6 transfection reagent was used, according to the manufacturer's instructions, with 1 μg of plasmid DNA per well.
Where indicated, cells were incubated with 20 ng of LMB (Sigma)/ml in DMEM-HG for 3 h prior to fixation.
The temperature shift assay was performed by incubating transfected cells in DMEM-HG containing cycloheximide (20 mg/ml; Sigma), a protein synthesis inhibitor, for 1 h at 37°C, followed by 3 h of incubation on ice.
Fluorescence microscopy.
For fluorescence analysis of GFP fusion proteins, cells were washed with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 15 min, and rinsed with PBS.
For immunofluorescence analysis of Myc-tagged proteins, cells were washed and fixed with 4% paraformaldehyde as described above. Cells were then permeabilized with 0.2% Triton X-100 for 2 min at room temperature, blocked with 3% bovine serum albumin in PBS at room temperature for 1 h, and incubated for 1 h at room temperature with anti-c-Myc monoclonal antibody (Roche Molecular Biochemicals) diluted 1:100 in blocking solution. After an extensive wash with PBS, cells were incubated with Alexa Fluor 488 goat anti-mouse immunoglobulin G antibody (Molecular Probes Europe BV, Leiden, The Netherlands).
For both direct and indirect fluorescence observations, the coverslips were then inverted and mounted on glass slides with Vectashield mounting medium (Vector Laboratories, Inc., Burlingame, Calif.). The slides were then visualized by using a Bio-Rad MRC 600 fluorescence confocal microscope equipped with an argon/krypton laser.
RESULTS
ASFV p37 protein is exported from the nucleus to the cytoplasm of yeast cells.
To determine whether the ASFV p37 protein undergoes nuclear export, a yeast-based nuclear export assay (30) was used. The rationale for this assay is the expression in yeast cells of a fusion protein encoded by pNEA, comprising mLexA (bacterial LexA modified to inactivate its internal NLS), SV40NLS (SV40 large T-antigen NLS), Gal4AD (yeast Gal4p activation domain), and the protein to be tested, subcloned in-frame downstream of Gal4AD. The expression of the two reporter genes HIS3 and lacZ present in the S. cerevisiae strain L40 under the control of LexA (20) depends on the intracellular localization of the tested fusion protein. If the tested protein is not exported from the nucleus, the fusion product is localized in the yeast cell nucleus due to the presence of SV40NLS. Yeast cells harboring this construct express β-galactosidase and grow in the absence of histidine as a consequence of lacZ and HIS3 gene expression, respectively. On the other hand, if the protein is exported, the fusion product is redirected to the cell cytoplasm, at least partly abolishing the β-galactosidase activity and impeding growth on histidine-deficient medium.
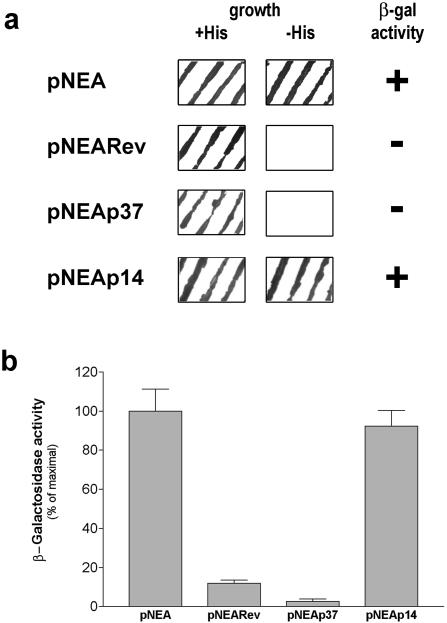
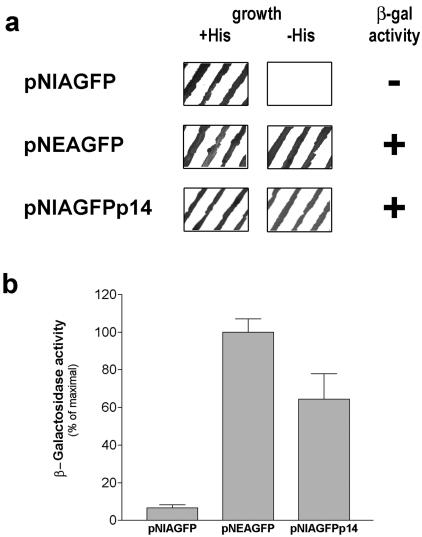
Figure 1a shows that yeast cells expressing the fusion product containing the p37 protein (pNEAp37) did not grow in the absence of histidine, indicating that the reporter HIS3 gene was not activated. Accordingly, this construct did not induce β-galactosidase activity (Fig. 1a), strongly suggesting that p37 is exported from the nucleus to the cytoplasm of yeast cells.
FIG. 1.
Assessment of the ability of ASFV p37 and p14 proteins to be exported from the nucleus to the cytoplasm of yeast cells. (a) Evaluation of yeast cell growth and β-galactosidase activity. The growth of L40 yeasts transformed with the indicated constructs was analyzed both on selective medium without tryptophan (+His) and on medium without tryptophan and histidine (−His) as described in Materials and Methods. The β-galactosidase activity was evaluated by using a colony-lift filter assay. Yeast cells expressing pNEAp37 showed no growth in the absence of histidine and no β-galactosidase activity. Yeast cells expressing pNEAp14 exhibited growth on histidine dropout medium and β-galactosidase activity. (b) Quantitative β-galactosidase activity assay in liquid cultures. After cell growth in minimal medium lacking tryptophan, the β-galactosidase activity was determined as described in Materials and Methods. Yeast cells transformed with pNEAp37 showed residual values of β-galactosidase activity, whereas pNEAp14 induced β-galactosidase activity levels similar to those of the negative control pNEA. The data are expressed as a percentage of maximal enzymatic activity obtained for pNEA alone; standard deviations are shown based on quintuplicates of at least three independent experiments.
In addition, by using the yeast-based nuclear export assay, it was demonstrated that the precursor of both p37 and p14 proteins, the pp55 preprotein, is also exported from the nucleus (data not shown).
Also presented in Fig. 1a are the results obtained for pNEA and pNEARev, which were used as controls for the nuclear export assay. pNEA encodes only the mLexA-SV40NLS-Gal4AD fusion protein that is actively imported into the nucleus, where it leads to maximal expression of the two reporter genes. pNEARev encodes the fusion product containing HIV-1 Rev protein (30), which is known to be exported from the nucleus due to the presence of a leucine-rich NES. When expressed in yeast cells, this fusion protein does not induce either growth on histidine-deficient medium or β-galactosidase activity, and therefore it was used as a positive control for the assay.
Since the filter lift assay only provides a qualitative evaluation of β-galactosidase activity, the liquid culture assay was used to quantify the activity of this enzyme in yeasts expressing the different constructs. Yeast cells harboring pNEAp37 showed, similarly to pNEARev, significantly lower levels of β-galactosidase activity than that observed for the negative control (pNEA) (Fig. 1b), further supporting the ability of p37 protein to undergo nuclear export in yeast cells.
The p37 protein encoded by ASFV is exported from the nucleus of mammalian cells.
To investigate the nuclear export activity of p37 in a more biologically relevant system, studies in ASFV host mammalian cells were performed. To this end, the intracellular localization of GFPp37 fusion protein was determined by confocal fluorescence microscopy in Vero cells.
These experiments showed that GFP protein alone is distributed throughout the cell as a consequence of its low molecular weight, which allows its passive diffusion within the cell (Fig. 2a).
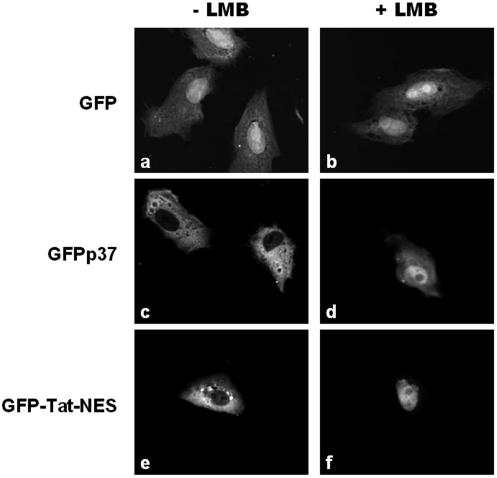
FIG. 2.
Effect of LMB on the nuclear export ability of p37 protein. Subconfluent cultures of Vero cells were transfected with the plasmids encoding GFP, GFPp37, or GFP-Tat-NES. At 24 h after transfection, cells were incubated or not incubated with LMB (20 ng/ml) for 3 h and then fixed, and the subcellular localization of the different proteins was visualized by fluorescence confocal microscopy as described in Materials and Methods. (a and b) GFP is distributed throughout the cell, and its localization is not altered by LMB treatment. (c and d) In untreated cells, GFPp37 fusion protein is localized exclusively in the cytoplasm, whereas in LMB-treated cells GFPp37 accumulates inside the nucleus. (e and f) GFP-Tat-NES fusion protein was used as a control for the inhibitory activity of LMB on the nuclear export mediated by the HIV-1 Rev NES present in that construct.
In contrast, the GFPp37 fusion protein was localized exclusively in the cytoplasm of transfected cells (Fig. 2c). This result is consistent with our observations with the yeast-based nuclear export assay and provides evidence that ASFV p37 protein is exported from the nucleus to the cytoplasm of mammalian cells.
To confirm that the subcellular localization of p37 protein is not a consequence of its fusion to GFP, a new construct of p37 protein tagged with the small epitope Myc was generated and transfected into Vero cells. Similar to what was observed for GFP-p37 (Fig. 2c), Myc-p37 was exclusively localized in the cytoplasm (Fig. 3a), thus demonstrating both that p37 is exported and that GFP has no effect on the subcellular localization of this protein.
FIG. 3.
Effect of LMB on the subcellular localization of Myc-tagged p37 protein. Vero cells were transfected with the expression vector encoding Myc-p37. At 24 h after transfection, cells were either left untreated or treated with 20 ng of LMB/ml for 3 h. Subcellular localization of Myc-p37 was analyzed by immunofluorescence with an antibody to the Myc epitope as described in Materials and Methods. (a) Myc-p37 is localized in the cell cytoplasm. (b) Treatment of cells with LMB induces nuclear accumulation of Myc-p37.
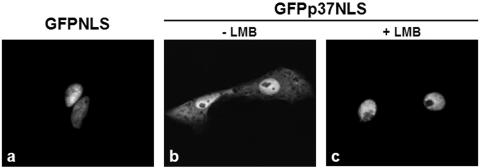
To further confirm the nuclear export ability of p37 and to evaluate the strength of this process, a slightly modified GFP nuclear export assay (14, 39) was performed with a fusion protein that contains the SV40NLS inserted at the C terminus of p37 (GFPp37NLS). This assay explores the competition between the nuclear import promoted by the SV40NLS and the nuclear export capacity of p37 protein.
Vero cells transfected with GFPp37NLS construct displayed fluorescence predominantly in the nucleus, although a significant amount of the signal was detected in the cytoplasm (Fig. 4b), as opposed to what was observed for the control GFPNLS, which is localized exclusively in the cell nucleus (Fig. 4a). This result clearly indicates that the strength of p37 nuclear export is sufficient to partially override the nuclear import mediated by the SV40NLS.
FIG. 4.
Competition assay between the nuclear import mediated by the SV40NLS and the nuclear export capacity of p37 protein. Vero cells were transfected with GFPNLS or GFPp37NLS, and the subcellular localization of these fusion proteins was analyzed by fluorescence microscopy in cells that were treated or not treated with LMB (20 ng/ml, 3 h) as described in Materials and Methods. (a) GFPNLS, a construct that contains the SV40NLS but not a NES, presents an exclusively nuclear localization and is shown for comparison. (b) GFPp37NLS is found preferentially in the nucleus of nontreated cells, but cytoplasmic localization is evident. (c) Treatment of cells with LMB leads to an exclusively nuclear localization of the GFPp37NLS fusion.
Taken together, these results clearly demonstrate that p37 protein is exported from the nucleus to the cytoplasm of mammalian cells.
CRM-1 receptor mediates the nuclear export of ASFV p37 protein.
To investigate the pathway involved in the nuclear export of p37 protein, Vero cells expressing GFPp37 were treated with LMB, a specific inhibitor of the CRM-1-dependent nuclear export pathway (23).
The antibiotic LMB binds directly to a cysteine residue of the CRM-1 receptor critical for the nuclear export (24), thus abolishing its association with NESs and therefore specifically inhibiting the nuclear export of proteins containing these signals.
As described above, cells expressing GFP alone presented an uniform staining in both the nucleus and cytoplasm, whereas in cells expressing GFPp37 the fluorescence is exclusively localized in the cytoplasm (Fig. 2a and c). When cells were treated with LMB, however, the control GFP remained evenly distributed throughout the cell (Fig. 2b), whereas for GFPp37 a dramatic change in its subcellular localization was observed, resulting in its accumulation inside the nucleus of transfected cells (Fig. 2d).
LMB treatment of cells expressing the Myc-p37 protein resulted also in a nuclear accumulation of the protein (Fig. 3b), supporting the involvement of CRM-1 receptor in the nuclear export process of p37.
Cells expressing GFP-Tat-NES, the control for LMB activity, which contains both the NLS of HIV-1 Tat protein and the leucine-rich NES of HIV-1 Rev protein (40), displayed an exclusively cytoplasmic localization (Fig. 2e). In contrast, when the nuclear export driven by the Rev NES was inhibited by LMB, the fusion protein was localized inside the cell nucleus (Fig. 2f). The observation of the same behavior for GFPp37 and Myc-p37 demonstrates that the CRM-1 pathway is responsible for the nuclear export of p37 ASFV protein.
LMB treatment of cells expressing the fusion protein GFPp37NLS abolished the cytoplasmic fluorescence observed in the nontreated cells, leading to an exclusively nuclear localization of the fusion protein (Fig. 4b and c). This finding proves that the cytoplasmic fluorescence observed for nontreated cells was due to a competitive effect of the NES present in p37, further supporting that the nuclear export of p37 protein is dependent on the CRM-1 receptor.
ASFV p37 protein is a nucleocytoplasmic shuttling protein.
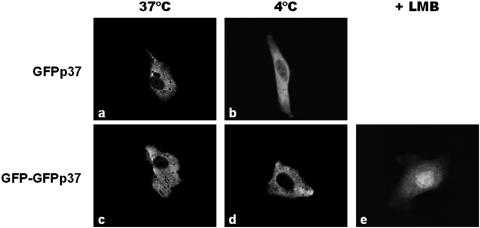
To clarify the process involved in the nuclear accumulation observed for GFPp37 in the presence of LMB, studies of the subcellular localization of GFPp37 at 4°C in the presence of a protein synthesis inhibitor (to avoid de novo GFPp37 synthesis) were performed. At this temperature all active transport processes are blocked, whereas diffusion remains unaffected.
As opposed to what was observed upon incubation of cells expressing GFPp37 at 37°C (Fig. 5a), fluorescence inside the nucleus can be observed upon incubation of cells at 4°C (Fig. 5b), indicating that GFPp37 is able to diffuse between the nucleus and the cytoplasm. Nonetheless, the nuclear fluorescence observed under this condition is significantly lower than in cells treated with LMB at 37°C (Fig. 2d), strongly suggesting that, in addition to diffusion, an active nuclear import process is involved in the nuclear accumulation of GFPp37.
FIG. 5.
Effect of temperature shift and treatment with LMB on the subcellular localization of ASFV p37 fusion proteins. Vero cells were transfected with GFPp37 or GFP-GFPp37. Where indicated, cells were treated with 20 ng of LMB/ml for 3 h. A temperature shift assay was performed by treating the cells with a protein synthesis inhibitor for 1 h, followed by further incubation in the presence of the inhibitor, for 3 h at 37°C (control) or 4°C as described in Materials and Methods. (a and c) At 37°C both GFPp37 and GFP-GFPp37 are localized exclusively in the cytoplasm of transfected cells. (b and d) At 4°C, fluorescence inside the nucleus can be observed for cells expressing GFPp37. In contrast, GFP-GFPp37 remains exclusively in the cytoplasm, indicating that this protein does not diffuse. (e) Upon treatment of cells expressing the nondiffusible GFP-GFPp37 protein with LMB (20 ng/ml, 3 h at 37°C), nuclear accumulation is observed.
Therefore, to investigate the nuclear import ability of p37 protein, an additional construct was generated (GFP-GFPp37) with a molecular weight of 91, clearly greater than the nuclear pore diffusion exclusion limit. Accordingly, experiments performed at 4°C demonstrated that the fusion protein GFP-GFPp37 is not able to diffuse into the nucleus (compare Fig. 5c to d). Nuclear accumulation of the nondiffusible GFP-GFPp37 protein (Fig. 5e) in cells treated with LMB demonstrates that an active process is responsible for the entry of p37 into the nucleus.
Overall, the results obtained clearly demonstrate the nucleocytoplasmic shuttling capacity of p37 protein.
ASFV p14 protein is not exported from the nucleus to the cytoplasm.
In contrast to the results obtained for p37 protein in the yeast nuclear export assay, the fusion product containing p14 protein (pNEAp14) induced yeast cell growth on histidine dropout medium and β-galactosidase activity (Fig. 1a), indicating that p14 protein does not undergo nuclear export.
The levels of β-galactosidase activity quantified by the β-galactosidase liquid culture assay were similar for yeast cells expressing pNEAp14 and pNEA (Fig. 1b), confirming that the fusion protein pNEAp14 is not exported, remaining inside the nucleus where it activates the expression of the lacZ reporter gene.
To extend the present study to mammalian cells, a fusion of p14 and GFP proteins was expressed in Vero cells. Using confocal fluorescence microscopy, GFPp14 fusion protein was found to be localized both in the cytoplasm and nucleus, although exhibiting a more pronounced nuclear localization (Fig. 6a). Treatment of cells expressing GFPp14 with LMB had no effect on the subcellular localization of the fusion protein, as shown in Fig. 6b.
FIG. 6.
Evaluation of the nucleocytoplasmic transport ability of p14 protein in mammalian cells. Vero cells expressing either GFPp14, Myc-p14, or GFP-GFPp14 were analyzed by fluorescence microscopy 24 h posttransfection as described in Materials and Methods. (a and c) GFPp14 and Myc-p14 fusion proteins are predominantly localized in the cell nucleus, although cytoplasmic fluorescence can be detected. (b and d) LMB (20 ng/ml, 3 h) has no effect on the subcellular localization of GFPp14 and of Myc-p14. (e) GFP-GFPp14 fusion protein accumulates inside the nucleus of the transfected cells, although cytoplasmic fluorescence can be observed.
To exclude any effect of GFP on the subcellular localization of p14 protein, a fusion of the epitope Myc with p14 protein was also generated. Similar to what was observed for GFP p14, Myc-tagged p14 protein was localized predominantly in the cell nucleus (Fig. 6c), and its subcellular localization was not affected by LMB treatment (Fig. 6d).
These findings corroborate the results obtained by using the yeast nuclear export assay and, together, demonstrate that ASFV p14 protein is not actively exported from the nucleus to the cytoplasm in either yeast or mammalian cells.
ASFV p14 protein is imported to the nucleus of both yeast and mammalian cells.
Given the predominantly nuclear localization of GFPp14 and Myc-p14 fusion proteins (Fig. 6a and c), a yeast-based nuclear import assay (30) was used as a first approach to determine whether p14 protein enters the nucleus by an active process. This assay is similar to the yeast nuclear export assay described above, except that the protein to be tested was cloned into pNIA, a construct that does not contain the SV40NLS, downstream of Gal4AD. As a consequence, the triple fusion protein comprising mLexA, Gal4AD and the protein being tested is not able to enter the nucleus unless this protein contains an NLS.
If the fusion protein is not imported into the nucleus, it does not activate the expression of the reporter genes lacZ and HIS3, and therefore the yeast cells do not grow on histidine-deficient medium nor present β-galactosidase activity. In contrast, if the tested protein is actively imported, the resulting fusion protein enters the nucleus and, once there, activates the expression of the two reporter genes, which results both in β-galactosidase activity and yeast growth on histidine-deficient medium.
When expressed in yeasts, the pNIA-GFPp14 fusion protein induced β-galactosidase activity and growth on histidine-deficient medium (Fig. 7a). Since this construct, due to the insertion of GFP between Gal4AD and p14, has a molecular weight not compatible with diffusion across the nuclear pore, this result indicates that p14 is actively imported into the nucleus.
FIG. 7.
Evaluation of the nuclear import capacity of ASFV p14 protein in yeasts. (a) Evaluation of yeast cell growth and β-galactosidase activity. The growth of yeast cells transformed with the indicated constructs was evaluated both on minimal medium with histidine but lacking tryptophan (+His) and on minimal medium lacking both tryptophan and histidine (−His) as described in Materials and Methods. A β-galactosidase colony-lift filter assay was performed in yeasts grown on tryptophan-deficient medium. Yeast cells expressing pNIA-GFPp14 show growth on medium without histidine and β-galactosidase activity. (b) The β-galactosidase activity was quantified in liquid cultures after yeast cell growth in minimal medium lacking tryptophan as described in Materials and Methods. The data are expressed as a percentage of maximal enzymatic activity obtained for pNEAGFP; standard deviations are shown based on quintuplicates of at least three independent experiments.
pNIA-GFP and pNEA-GFP are the controls for the nuclear import assay. pNIA-GFP encodes for the fusion protein mLexA-Gal4AD-GFP and was used as negative control because it is not imported into the nucleus, resulting in minimal expression of the two reporter genes (Fig. 7a), whereas pNEA-GFP encodes the fusion protein mLexA-SV40NLS-Gal4AD-GFP which, due to the presence of the SV40NLS, is actively imported into the nucleus, leading to maximal activity of both lacZ and HIS3 genes (Fig. 7a).
The results obtained by using the quantitative β-galactosidase assay show that pNIA-GFPp14 induced levels of β-galactosidase activity comparable to the high levels obtained for pNEA-GFP (Fig. 7b), a finding which confirms that p14 protein is imported into the nucleus of yeasts.
The nuclear import ability of p14 was further confirmed in mammalian cells by determining the subcellular localization of the GFP-GFPp14 fusion protein, by fluorescence confocal microscopy. The fusion protein was localized both in the nucleus and in the cytoplasm, but accumulation inside the cell nucleus was clearly noticeable (Fig. 6e). Given the high molecular weight of the GFP-GFPp14 fusion protein, its accumulation inside the nucleus is clear evidence that p14 protein is imported into the nucleus of mammalian cells.
DISCUSSION
Viral proteins are involved in numerous processes, including the entry, morphogenesis, and egress of viruses, making the detailed characterization of these proteins a key element to better understand the different steps of infection.
The proteins studied here—p37 and p14, together with p34 and p150—are major structural ASFV proteins that are produced by sequential proteolytic processing of pp220 polyprotein precursor, which is a late protein synthesized after DNA replication (37). These four proteins are located at the core shell, which functions as a matrix domain between the DNA-containing nucleoid and the inner envelope. Collectively, they represent ca. 25% of the protein mass of the virus particle and are present in an essentially equimolecular stoichiometry (3, 4).
The nuclear export ability of both p37 and p14 proteins was assessed by using the described yeast-based nuclear export assay. Using this strategy we were able to demonstrate that p37 protein, but not p14 protein, is exported from the nucleus of yeast cells. However, p14 protein was imported into the nucleus, as evaluated by the yeast-based nuclear import assay.
These results were validated in a more relevant biological system by determining the intracellular localization of p37 and p14 proteins fused to GFP or Myc, in mammalian cells, by fluorescence confocal microscopy. In agreement with the results obtained in yeasts, p37 fusion proteins were detected exclusively in the cell cytoplasm, whereas the fusions containing p14 protein were localized both in the nucleus and cytoplasm of mammalian cells, although exhibiting a preferential localization inside the nucleus.
The overlapping results obtained for GFP fusions and Myc- tagged proteins undoubtedly demonstrate that p14 and p37 proteins are responsible for the described nuclear transport abilities.
The nuclear export observed for pp55 preprotein (data not shown), which is the immediate precursor of p14 and p37 proteins, should be therefore attributed to p37 protein.
The nuclear accumulation of p37 fused with GFP or Myc upon LMB treatment demonstrates that the nuclear export exhibited by p37 protein is mediated by the CRM-1 pathway.
The nuclear import ability of p37 protein was evaluated with a fusion protein with a molecular weight above the size limit for diffusion through the nuclear pore complexes. Nuclear accumulation of this nondiffusible protein induced by cell treatment with LMB clearly demonstrates that p37 is also imported into the nucleus.
Although nuclear shuttling proteins have been identified in many other viruses (8, 34, 40, 44), the findings reported here show p37 protein to be the first nucleocytoplasmic shuttling protein encoded by the ASFV genome.
The active transport of p14 protein to the nucleus demonstrated by the yeast-based nuclear import assay was further confirmed in mammalian cells by the determination of the subcellular localization of a p14 fusion protein with a molecular weight above the nuclear pore complex size exclusion limit. In addition to p14 protein (and p37 protein), ASFV I14L protein has been shown to be imported into the cell nucleus (18). Nonetheless, the function of these proteins and the importance of their nuclear import remain to be elucidated.
The generation from a common precursor of two proteins (p14 and p37) exhibiting different nucleocytoplasmic transport activities is, per se, interesting, although the wider biologic and virologic significance of this fact is still unknown.
It has been demonstrated that fusion of SV40NLS with proteins containing NESs that are localized in the cell cytoplasm at the steady state, namely, HIV-1 Vpr (34) and hepatitis B virus X protein (14), results either in an exclusively nuclear localization or in a diffused staining pattern in the cytoplasm and nucleus, respectively. Therefore, our observation of significant cytoplasmic fluorescence in cells transfected with GFPp37NLS is in agreement with the observations for hepatitis B virus X protein (14), clearly indicating that the extent of p37 nuclear export is high, overcoming the potency of the SV40NLS.
To identify possible sequences responsible for the nuclear import of p14 protein and for p37 protein nucleocytoplasmic shuttling, sequence analysis of both proteins was performed. This analysis resulted in the identification of several putative leucine-rich NESs in p37 protein. However, the presence of nonclassical NESs or of a nucleocytoplasmic bidirectional signal in this protein cannot be excluded. Studies are in progress to test the functionality of the identified sequences and to determine the signals responsible for the nuclear import of p14 and p37 proteins.
The lack of homology of ASFV p37 and p14 proteins with other well-characterized proteins in databases makes any prediction of their putative function difficult. Nonetheless, interesting studies on the repression of the expression of their pp220 precursor have demonstrated that pp220 or the products of its proteolytic processing are essential for viral core assembly and envelopment, as well as for the subsequent steps of core formation, including DNA encapsidation and nucleoid maturation (5).
Immunofluorescence (37) and immunoelectron (4) microscopy studies do not report the presence of the products of proteolytic processing of pp220 inside the nucleus of ASFV-infected cells. However, it should be emphasized that these studies were performed at late times of infection (12 to 18 h postinfection), when virus assembly is already occurring.
Based on the existing data, a role for the nucleocytoplasmic shuttling activity of p37 protein and for p14 protein nuclear import at early stages of viral infection cannot be excluded, especially considering the importance that the nucleocytoplasmic transport processes may have at an early stage of infection, when the nuclear phase of ASFV DNA replication is occurring. It is possible that p14 and p37 proteins are involved in the formation of a complex with the viral DNA, facilitating its entry into the nucleus, to initiate DNA replication. In an alternative or complementary scenario, p37 protein could be involved in the export of the viral genome from the nucleus to the cytoplasm of infected cells after the brief nuclear replication phase. The localization of both p14 and p37 proteins in the core shell of the viral particle supports these hypotheses, since this would allow the interaction of these proteins with the viral DNA.
Although additional studies are needed to elucidate the relevance of the nucleocytoplasmic transport activities described here to the function of p14 and p37 proteins, it is reasonable to consider that this capacity must be critical for the role played by these proteins in the replication cycle of the ASFV. Studies are now under way to confirm the nucleocytoplasmic transport activities of p14 and p37 proteins in ASFV-infected cells at early times postinfection and to further elucidate the relevance of this transport to the viral replication cycle.
Acknowledgments
We are grateful to L. Valdeira for kindly providing the ASFV Lisbon 60 strain and G. Pavlakis for providing the pTat-GFP-NES plasmid.
This study was supported by a grant from the Portuguese Foundation for Science and Technology (POCTI/CVT/42700/2001). A. Eulálio and I. Nunes-Correia are recipients of fellowships from the Portuguese Foundation for Science and Technology. The work in the V. Citovsky laboratory was supported by grants from the National Institutes of Health, the National Science Foundation, the U.S. Department of Agriculture, the U.S.-Israel Binational Science Foundation (BSF), and the U.S.-Israel Binational Research and Development Fund (BARD).
REFERENCES
- 1.Alejo, A., G. Andres, and M. L. Salas. 2003. African swine fever virus proteinase is essential for core maturation and infectivity. J. Virol. 77:5571-5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andres, G., A. Alejo, C. Simon-Mateo, and M. L. Salas. 2001. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 276:780-787. [DOI] [PubMed] [Google Scholar]
- 3.Andres, G., A. Alejo, J. Salas, and M. L. Salas. 2002. African swine fever virus polyproteins pp220 and pp62 assemble into the core shell. J. Virol. 76:12473-12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andres, G., C. Simon-Mateo, and E. Vinuela. 1997. Assembly of African swine fever virus: role of polyprotein pp220. J. Virol. 71:2331-2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Andres, G., E. Garcia, M. L. Salas, and J. M. Rodriguez. 2002. Repression of African swine fever virus polyprotein pp220-encoding gene leads to the assembly of icosahedral core-less particles. J. Virol. 76:2654-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bednenko, J., G. Cingolani, and L. Gerace. 2003. Nucleocytoplasmic transport: navigating the channel. Traffic 4:127-135. [DOI] [PubMed] [Google Scholar]
- 7.Breeden, L., and K. Nasmyth. 1985. Regulation of the yeast HO gene. Cold Spring Harbor Symp. Quant. Biol. 50:643-650. [DOI] [PubMed] [Google Scholar]
- 8.Cheng, G., M. E. Brett, and B. He. 2002. Signals that dictate nuclear, nucleolar, and cytoplasmic shuttling of the gamma(1)34.5 protein of herpes simplex virus type 1. J. Virol. 76:9434-9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa, J. V. 1990. Molecular biology of iridoviruses. Kluwer Academic Publishers, Boston, Mass.
- 10.Dixon, L. K., J. V. Costa, J. M. Escribano, D. L. Rock, E. Viñuela, and P. J. Wilkinson. 2000. Virus taxonomy. Seventh report of the International Committee for the Taxonomy of Viruses. Academic Press, Inc., New York, N.Y.
- 11.Fan, X. C., and J. A. Steitz. 1998. HNS, a nuclear-cytoplasmic shuttling sequence in HuR. Proc. Natl. Acad. Sci. USA 95:15293-15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farjot, G., M. Buisson, D. M. Duc, L. Gazzolo, A. Sergeant, and I. Mikaelian. 2000. Epstein-Barr virus EB2 protein exports unspliced RNA via a Crm-1-independent pathway. J. Virol. 74:6068-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fischer, U., J. Huber, W. C. Boelens, I. W. Mattaj, and R. Luhrmann. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82:475-483. [DOI] [PubMed] [Google Scholar]
- 14.Forgues, M., A. J. Marrogi, E. A. Spillare, C. G. Wu, Q. Yang, M. Yoshida, and X. W. Wang. 2001. Interaction of the hepatitis B virus X protein with the Crm1-dependent nuclear export pathway. J. Biol. Chem. 276:22797-22803. [DOI] [PubMed] [Google Scholar]
- 15.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390:308-311. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Beato, R., M. L. Salas, E. Vinuela, and J. Salas. 1992. Role of the host cell nucleus in the replication of African swine fever virus DNA. Virology 188:637-649. [DOI] [PubMed] [Google Scholar]
- 18.Goatley, L. C., M. B. Marron, S. C. Jacobs, J. M. Hammond, J. E. Miskin, C. C. Abrams, G. L. Smith, and L. K. Dixon. 1999. Nuclear and nucleolar localization of an African swine fever virus protein, I14L, that is similar to the herpes simplex virus-encoded virulence factor ICP34.5. J. Gen. Virol. 80(Pt. 3):525-535. [DOI] [PubMed] [Google Scholar]
- 19.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 20.Hollenberg, S. M., R. Sternglanz, P. F. Cheng, and H. Weintraub. 1995. Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol. Cell. Biol. 15:3813-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Kalderon, D., W. D. Richardson, A. F. Markham, and A. E. Smith. 1984. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature 311:33-38. [DOI] [PubMed] [Google Scholar]
- 23.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242:540-547. [DOI] [PubMed] [Google Scholar]
- 24.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96:9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, C. H., S. C. Chang, C. H. Wu, and M. F. Chang. 2001. A novel chromosome region maintenance 1-independent nuclear export signal of the large form of hepatitis delta antigen that is required for the viral assembly. J. Biol. Chem. 276:8142-8148. [DOI] [PubMed] [Google Scholar]
- 26.Lischka, P., O. Rosorius, E. Trommer, and T. Stamminger. 2001. A novel transferable nuclear export signal mediates CRM1-independent nucleocytoplasmic shuttling of the human cytomegalovirus transactivator protein pUL69. EMBO J. 20:7271-7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Nakielny, S., and G. Dreyfuss. 1999. Transport of proteins and RNAs in and out of the nucleus. Cell 99:677-690. [DOI] [PubMed] [Google Scholar]
- 29.Ossareh-Nazari, B., F. Bachelerie, and C. Dargemont. 1997. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 278:141-144. [DOI] [PubMed] [Google Scholar]
- 30.Rhee, Y., F. Gurel, Y. Gafni, C. Dingwall, and V. Citovsky. 2000. A genetic system for detection of protein nuclear import and export. Nat. Biotechnol. 18:433-437. [DOI] [PubMed] [Google Scholar]
- 31.Robbins, J., S. M. Dilworth, R. A. Laskey, and C. Dingwall. 1991. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64:615-623. [DOI] [PubMed] [Google Scholar]
- 32.Rojo, G., R. Garcia-Beato, E. Vinuela, M. L. Salas, and J. Salas. 1999. Replication of African swine fever virus DNA in infected cells. Virology 257:524-536. [DOI] [PubMed] [Google Scholar]
- 33.Rubio, D., A. Alejo, I. Rodriguez, and M. L. Salas. 2003. Polyprotein processing protease of African swine fever virus: purification and biochemical characterization. J. Virol. 77:4444-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherman, M. P., C. M. de Noronha, M. I. Heusch, S. Greene, and W. C. Greene. 2001. Nucleocytoplasmic shuttling by human immunodeficiency virus type 1 Vpr. J. Virol. 75:1522-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simoes, S., V. Slepushkin, P. Pires, R. Gaspar, M. C. P. de Lima, and N. Duzgunes. 1999. Mechanisms of gene transfer mediated by lipoplexes associated with targeting ligands or pH-sensitive peptides. Gene Ther. 6:1798-1807. [DOI] [PubMed] [Google Scholar]
- 36.Simoes, S., V. Slepushkin, R. Gaspar, M. C. P. de Lima, and N. Duzgunes. 1998. Gene delivery by negatively charged ternary complexes of DNA, cationic liposomes and transferrin or fusigenic peptides. Gene Ther. 5:955-964. [DOI] [PubMed] [Google Scholar]
- 37.Simon-Mateo, C., G. Andres, and E. Vinuela. 1993. Polyprotein processing in African swine fever virus: a novel gene expression strategy for a DNA virus. EMBO J. 12:2977-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stachel, S. E., G. An, C. Flores, and E. W. Nester. 1985. A Tn3 lacZ transposon for the random generation of β-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium. EMBO J. 4:891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041-1050. [DOI] [PubMed] [Google Scholar]
- 40.Stauber, R. H., and G. N. Pavlakis. 1998. Intracellular trafficking and interactions of the HIV-1 Tat protein. Virology 252:126-136. [DOI] [PubMed] [Google Scholar]
- 41.Tulman, E. R., and D. L. Rock. 2001. Novel virulence and host range genes of African swine fever virus. Curr. Opin. Microbiol. 4:456-461. [DOI] [PubMed] [Google Scholar]
- 42.Viñuela, E. 1987. African swine fever. Nijhoff, Boston, Mass.
- 43.Wen, W., J. L. Meinkoth, R. Y. Tsien, and S. S. Taylor. 1995. Identification of a signal for rapid export of proteins from the nucleus. Cell 82:463-473. [DOI] [PubMed] [Google Scholar]
- 44.Whittaker, G., M. Bui, and A. Helenius. 1996. Nuclear trafficking of influenza virus ribonucleoproteins in heterokaryons. J. Virol. 70:2743-2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wolff, T., G. Unterstab, G. Heins, J. A. Richt, and M. Kann. 2002. Characterization of an unusual importin alpha binding motif in the borna disease virus p10 protein that directs nuclear import. J. Biol. Chem. 277:12151-12157. [DOI] [PubMed] [Google Scholar]
- 46.Yanez, R. J., J. M. Rodriguez, M. L. Nogal, L. Yuste, C. Enriquez, J. F. Rodriguez, and E. Vinuela. 1995. Analysis of the complete nucleotide sequence of African swine fever virus. Virology 208:249-278. [DOI] [PubMed] [Google Scholar]