Abstract
Nipah virus (NiV), a highly pathogenic paramyxovirus, causes a systemic infection in vivo and is able to replicate in cultured cells of many species and organs. Such pantropic paramyxoviruses generally encode fusion (F) proteins with multibasic cleavage sites activated by furin or other ubiquitous intracellular host cell proteases. In contrast, NiV has an F protein with a single arginine (R109) at the cleavage site, as is the case with paramyxoviruses that are activated by trypsin-like proteases only present in specific cells or tissues and therefore only cause localized infections. Unlike these viruses, cleavage of the NiV F protein is ubiquitous and does not require the addition of exogenous proteases in cell culture. To determine the importance of the amino acid sequence at the NiV F protein cleavage site for ubiquitous activation, we generated NiV F proteins with mutations around R109. Surprisingly, neither the exchange of amino acids upstream of R109 nor replacement of the basic residue itself interfered with F cleavage. Thus, R109 is not essential for F cleavage and activation. Our data demonstrate that NiV F-protein activation depends on a novel type of proteolytic cleavage that has not yet been described for any other paramyxovirus F protein. NiV F activation is mediated by a ubiquitous protease that requires neither a monobasic nor a multibasic cleavage site and therefore differs from the furin- or trypsin-like proteases known to activate other ortho- and paramyxovirus fusion proteins.
Nipah virus (NiV) was isolated in 1999 and subsequently identified as the etiological agent responsible for an outbreak of severe respiratory disease and fatal encephalitis in Malaysia and Singapore in pigs and humans (4). Blood vessels appeared to be the key targets of injury in many organs, the central nervous system being the most severely affected in humans. NiV is a negative-stranded RNA virus sharing genome organization and replication strategies with viruses of the family Paramyxoviridae (8). NiV and the closely related Hendra virus (HeV; 70 to 85% sequence homology) have a larger genome and a much longer P protein than other paramyxoviruses, warranting the formation of the new genus “Henipavirus” (19, 40). In contrast to all other paramyxoviruses, NiV and HeV infect a wide range of species, including pigs, cats, and humans (10). Because viral glycoproteins are known to be major determinants for tissue and host tropism, analysis of these proteins is of special interest.
It is known that proteolytic cleavage of the fusion (F) protein of paramyxoviruses at basic amino acid residues (arginine or lysine) is a prerequisite for virus infectivity. In addition to the availability of viral receptors, differences in the distribution of F-protein-activating proteases are crucial factors in determining host range, tissue tropism, and pathogenicity (11, 14, 24, 34, 35). Paramyxovirus F proteins are synthesized as inactive precursors F0 and have to be activated by proteolytic cleavage into the two disulfide-linked subunits F1 and F2, thereby releasing the hydrophobic fusion peptide located at the amino terminus of F1. Two different mechanisms of F-protein activation can be distinguished. F proteins with multiple basic amino acids at the cleavage site are activated by ubiquitous intracellular host cell proteases, such as furin, whereas activation of F proteins with only one basic residue at the cleavage site is mediated by extracellular trypsin-like proteases (TLPs) upon cell surface arrival of the inactive precursors. The mechanism of activation has important implications for virus spread not only in vitro but also in vivo. In most cell cultures, growth of viruses with F proteins containing monobasic cleavage sites is dependent on the addition of trypsin to the medium (20). More important, due to the lack of appropriate TLPs, such as the tryptase Clara or miniplasmin, activation in vivo is usually restricted to the respiratory tract (18, 36). Consequently, paramyxoviruses with monobasic cleavage sites, such as Sendai virus, only cause respiratory infections. In contrast, paramyxoviruses encoding F proteins with multibasic cleavage sites, such as measles virus (MV) or virulent Newcastle disease virus (NDV) strains, can replicate and spread systemically after initial infection of the respiratory tract (for a review see reference 11).
The NiV F protein consists of 546 amino acids with a molecular mass of 60 kDa (8). As for most paramyxoviruses, both surface glycoproteins, the receptor binding protein G and the F protein, are required to mediate pH-independent virus-cell and cell-cell fusion (32). Because the majority of cell cultures tested so far supported NiV fusion, the cellular receptor appears to be widely expressed and activation of the viral F protein seems not to be severely restricted (2). The latter is surprising, because the NiV F protein has only one basic amino acid residue (arginine at position 109 [R109]) amino terminal of a hydrophobic domain which is predicted to be the fusion peptide based on the high level of sequence homology with other paramyxoviruses (Table 1) (8). Therefore, it has to be assumed that the NiV F protein is activated after R109 by trypsin or a TLP, similar to the F proteins with monobasic cleavage sites of avirulent NDV, human parainfluenza virus type I, or Sendai virus (Table 1). However, activation of the NiV F protein in cell culture does not depend on the addition of exogenous trypsin or TLPs (2, 8, 32). In addition, replication and fusion activity is not restricted in vivo. Multinucleated syncytia were found in lung tissue as well as in blood vessels, lymphoid organs, spleen, kidney, and brain (10). This indicates that the cellular receptor is widely expressed and that the NiV F protein is cleaved in most organs. Taken together, these observations indicate that the NiV F protein does not have the structural features typical of other viral fusion proteins activated by ubiquitous proteases. The aim of this study was to analyze the proteolytic activation of the NiV F protein in cell culture and to determine the effects of targeted mutations at the cleavage site on cleavability and fusion activity. Ubiquitous proteolytic activation of the NiV F protein in cell culture was demonstrated by using cell lines either permissive or nonpermissive for NiV-mediated fusion. Furthermore, we proved by amino acid sequencing that cleavage indeed occurred at the predicted cleavage site. The generation of several cleavage site mutants revealed that point mutations around the cleavage site were not able to prevent NiV F activation. Surprisingly, even replacement of R109 itself had no effect on the cleavability. These results indicate that the F protein of NiV is activated by a novel type of ubiquitous proteolytic cleavage that does not require a basic amino acid at the cleavage site.
TABLE 1.
Cleavage and fusion peptides of various paramyxovirusesa
| Virusb | Cleavage peptide (C terminus of F2) | Fusion peptide (N terminus of F1) |
|---|---|---|
| Pneumoviruses | ||
| HRSV | K-K-R-K-R-R | F-L-G-F-L-L-G-V-G-S-A-I |
| BRSV | K-K-R-K-R-R | F-L-G-F-L-L-G-I-G-S-A-V |
| Respiroviruses | ||
| HPIV-1 | D-N-P-Q-T-R | F-F-G-A-V-I-G-T-I-A-L-G |
| HPIV-3 | N-P-R-T-K-R | F-F-G-G-V-I-G-T-I-A-L-G |
| Sendai virus | D-V-P-Q-S-R | F-F-G-A-V-I-G-T-I-A-L-G |
| Rubulaviruses | ||
| Mumps virus | S-R-R-H-K-R | F-A-G-I-A-I-G-I-A-A-L-G |
| Simian virus 5 | T-R-R-R-R-R | F-A-G-V-V-I-G-L-A-A-L-G |
| Avulaviruses | ||
| virulent NDV | G-R-R-Q-K-R | F-I-G-A-I-I-G-G-V-A-L-G |
| avirulent NDV | G-G-R-Q-G-R | L-I-G-A-I-I-G-G-V-A-L-G |
| Morbilliviruses | ||
| MV | S-R-R-H-K-R | F-A-G-V-V-L-A-G-A-A-L-G |
| CDV | G-R-R-Q-R-R | F-A-G-V-V-L-A-G-V-A-L-G |
| Rinderpest virus | S-R-R-H-K-R | F-A-G-V-V-L-A-G-A-A-L-G |
| Henipaviruses | ||
| HeV | L-V-G-D-V-K | L-A-G-V-V-M-A-G-I-A-I-G |
| NiV | L-V-G-D-V-R | L-A-G-V-I-M-A-G-V-A-I-G |
Adapted from Harcourt et al. (8). Boldface letters indicate basic amino acid residues.
HRSV, human respiratory syncytial virus; BRSV, bovine respiratory syncytial virus; HPIV, human parainfluenza virus; CDV, canine distemper virus.
MATERIALS AND METHODS
Cell culture and virus infection.
MDCK (Madin-Darby canine kidney) cells were grown in Eagle's minimal essential medium (MEM; Gibco) containing 10% fetal calf serum (FCS), 100 U of penicillin/ml, and 0.1 mg of streptomycin/ml. Vero (African green monkey kidney), 293 (human embryonic kidney), and HeLa (human cervical cancer) cells were maintained in Dulbecco′s modified MEM supplemented with 10% FCS, penicillin, and streptomycin.
The NiV strain used in this work was isolated from human brain tissue (kindly provided by Pierre Rollin and Tom Ksiazek, Centers for Disease Control and Prevention, Atlanta, Ga.) and propagated in Vero E6 cells. Stock virus was harvested when the cytopathic effect was maximal. For NiV infection of different cell lines, confluent cell monolayers were infected at a multiplicity of infection (MOI) of 0.1. After incubation with virus for 2 h at 37°C and removal of the inocula, the cells were washed twice and were cultured with medium containing 2% FCS at 37°C. All work with live virus was performed under biosafety level 4 conditions.
Plasmid construction and site-specific mutagenesis.
Viral glycoprotein (F and G protein) genes were cloned into a derivative of the replication-deficient murine leukemia virus vector pczCFG (13). DNA fragments spanning the F gene and the G gene of the NiV genome (GenBank accession no. AF212302) were amplified by reverse transcription PCR with primers containing NheI or HindIII sites. After treatment with Klenow enzyme to generate blunt ends, F and G genes were inserted into the SwaI site of pczCFG5-IEGZ to give pczCFG5-NiV F and pczCFG5-NiV G. Due to the lack of NiV F protein-specific antibodies, a tagged version of the protein was established. For this, the nine carboxy-terminal amino acids of the F protein were replaced with amino acids 99 to 107 (YPYDVPDYA) of the human influenza virus hemagglutinin (HA) known as HA-tag. The protein was generated by a recombinant PCR technique (9). Expression level, cleavage, and biological activity of the HA-tagged protein were unchanged compared to those of the parental F protein in transient transfection. All further mutants were based on the HA-tagged NiV F protein. Mutant NiV F protein genes (FR109V, FR109A, FV108A, FD107A, FG106A, FV105A, FL104A, FD103A, FH102A, FL110R, FL110F, and FΔcs) were generated by introduction of site-specific mutations with complementary primers in the double-stranded pczCFG5 plasmids by using the QuikChange site-directed mutagenesis kit (Stratagene). Mutagenic oligonucleotide primers were arranged to bind with 15 to 21 bases on both sites of the mutation site. Sequences of all constructs were confirmed by dideoxy sequencing. Generation of the MV F protein with a monobasic cleavage site (MV Fcm) has been described previously (14).
Surface biotinylation analysis.
Cells grown in 35-mm-diameter dishes were transfected with plasmid DNA encoding parental NiV F, mutant NiV F, or MV Fcm by using the cationic lipid transfection reagent Lipofectamine 2000 (Gibco-BRL), following the instructions of the supplier. Surface labeling with biotin was performed as described previously (16). At 24 h posttransfection, cells were washed three times with cold phosphate-buffered saline (PBS) containing 0.1 mM CaCl2 and 1 mM MgCl2 and were incubated twice for 20 min at 4°C with 2 mg of sulfo-N-hydroxysuccinimidobiotin (Calbiochem)/ml. After biotinylation, cells were washed once with cold PBS containing 0.1 M glycine and three times with cold PBS. Cells were then lysed in 0.5 ml of radioimmunoprecipitation assay (RIPA) buffer (1% Triton X-100, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 0.15 M NaCl, 10 mM EDTA, 10 mM iodoacetamide, 1 mM phenylmethylsulfonyl fluoride, 50 U of aprotinin/ml, 20 mM Tris-HCl, pH 8.5) followed by centrifugation for 20 min at 100,000 × g. NiV F protein was immunoprecipitated from cell lysates with a polyclonal antiserum specific for HA-tagged proteins (Sigma) at a final dilution of 1:250. MV Fcm was precipitated with monoclonal antibody A504 as described previously (16). After incubation with 20 μl of a suspension of protein A-Sepharose CL-4B (Sigma), immune complexes were washed three times with RIPA buffer, suspended in reducing sample buffer for SDS-polyacrylamide gel electrophoresis (PAGE), and separated on a 12% polyacrylamide gel. Proteins were blotted to nitrocellulose by the semidry blot technique. After blocking of nonspecific binding sites, biotinylated cell surface F proteins were detected by incubation with a streptavidin-biotinylated horseradish peroxidase complex (Amersham) diluted 1:2,000 in PBS.
N-terminal amino acid analysis of F1.
MDCK cells cultured in 60-mm-diameter dishes were transfected with the parental F gene. At 24 h posttransfection, cells were lysed in RIPA buffer and F protein was immunoprecipitated as described above. After separation on a 12% SDS gel and blotting to a polyvinylidene chloride (PVDF) membrane, proteins were stained with 0.1% Coomassie brilliant blue R-250 and destained in ethanol/acetic acid (40%/10%) until protein bands were visible. The location of the F1 subunit was determined by parallel immunodetection with monoclonal antibodies directed against the HA-tag (Sigma). The Coomassie-stained F1 band was excised from the PVDF membrane and was submitted for N-terminal amino acid sequencing by automated Edman degradation (WITA, Berlin, Germany).
Fusion assays.
To analyze the biological activity of parental and mutant NiV F proteins, cells were cotransfected with plasmids bearing the genes encoding parental NiV F or NiV F mutants and NiV G. Cell-to-cell fusion was visualized as described earlier (17). Briefly, cells were fixed with ethanol at 24 h posttransfection and were stained with 1:10 diluted Giemsa staining solution. Representative microscopic fields were photographed.
Metabolic labeling.
Twenty-four hours after transfection, MDCK cells transiently expressing parental NiV F or FΔcs protein were incubated for 30 min with medium lacking cysteine and methionine and then metabolically labeled by incubation with medium containing [35S]cysteine and [35S]methionine (Promix; Amersham) at a final concentration of 100 μCi/ml for 10 min. Labeling medium subsequently was replaced by nonradioactive chase medium, and the cells were incubated at 37°C for the times indicated. After radiolabeling, F proteins were immunoprecipitated as described above and were subjected to SDS-PAGE. Dried gels were exposed to Kodak BIOMAX films.
RESULTS
NiV F protein is ubiquitously cleaved in cell culture.
In contrast to other paramyxoviruses encoding F proteins with monobasic cleavage sites, NiV replicates systemically in vivo and induces syncytium formation in many cell lines (2, 10). To demonstrate that the NiV F protein is ubiquitously cleaved in cell culture without the addition of exogenous proteases, we analyzed NiV F protein cleavage in cultured cells either permissive or nonpermissive for NiV-mediated fusion. It had already been shown that Vero cells allow efficient replication of NiV, whereas HeLa cells are nonpermissive (2, 8). To illustrate the ability of permissive and nonpermissive cell lines to support fusion, cells were either infected with NiV or transiently transfected with the NiV glycoproteins. To assess fusion activity after NiV infection, Vero, HeLa, and MDCK cells were infected with NiV under biosafety level 4 conditions and syncytium formation was monitored. As shown in Fig. 1A, Vero and MDCK cells were permissive for NiV infection, whereas HeLa cells did not show any cell-to-cell fusion at 72 h postinfection. To analyze fusion activity in transient expression, different cell lines were cotransfected with the NiV F and G proteins because fusion requires expression of both NiV glycoproteins (2). Here, the result for transfected Vero, HeLa, MDCK, and 293 cells is exemplarily shown. Whereas Vero, MDCK, and 293 cells supported the formation of multinucleated syncytia, HeLa cells were again found to be nonpermissive for F- and G-mediated cell-to-cell fusion (Fig. 1B). Proteolytic cleavage of surface-expressed NiV F protein was detected with a biotinylation assay. For this, transfected cells were labeled with the non-membrane-permeating reagent sulfo-N-hydroxysuccinimidobiotin. After immunoprecipitation, F proteins were subjected to SDS-PAGE and were blotted to nitrocellulose. Surface-expressed biotin-labeled protein was detected with peroxidase-conjugated streptavidin. As shown in Fig. 1C, cleavage of precursor F0 into the subunits F1 and F2 was observed in permissive (Vero, MDCK, 293) and nonpermissive (HeLa) cells. To ensure that nonpermissive cells generated sufficient amounts of correctly cleaved and, thus, biologically active F protein, G- and F-expressing HeLa cells were overlaid with Vero cells. Syncytium formation began to develop only a few hours later (data not shown), supporting the view that nonpermissive cells lack suitable NiV receptors. This result indicates that unlike the viral receptor, the NiV-activating protease is ubiquitously expressed in cell culture.
FIG. 1.
Fusion activity and cleavage of NiV F protein in different cell lines. (A) Vero, HeLa, and MDCK cells were infected at an MOI of 0.1. At 72 h postinfection, syncytium formation was visualized by staining with Giemsa staining solution. Magnification, ×100. (B) Vero, HeLa, MDCK, and 293 cells were cotransfected with the NiV G and F genes. At 24 h posttransfection, cells were fixed and incubated with Giemsa staining solution. Magnification, ×100. (C) At 24 h posttransfection, NiV F-expressing Vero, HeLa, MDCK, and 293 cells were surface labeled with biotin and were lysed. Following immunoprecipitation, samples were subjected to SDS-PAGE under reducing conditions and were blotted to nitrocellulose. Surface-labeled F proteins were visualized with streptavidin-peroxidase and chemiluminescence.
So far, furin or furin-like proteases are the only proteases known to ubiquitously activate F proteins at basic cleavage sites. However, due to the lack of a furin consensus motif at the NiV F protein cleavage site, activation by furin is highly unlikely. To prove this, proteolytic processing of the F protein was analyzed in the presence of the furin inhibitor dec-RVKR-cmk (22). As expected, cleavage was not affected by the inhibitor, clearly demonstrating that furin is not responsible for the ubiquitous NiV F protein activation (data not shown).
NiV F is cleaved at the predicted cleavage site.
Before starting a more detailed analysis in order to elucidate the unusual ubiquitous activation of the monobasic NiV F protein cleavage site, we wanted to ensure that activation indeed occurs at the predicted cleavage site, C terminal of R109. For this, immunoprecipitated NiV F protein was separated on an SDS gel under reducing conditions and was blotted to PVDF. The Coomassie-stained F1 band was excised and subjected to Edman degradation. Eight degradation cycles unambiguously revealed the amino acid sequence LAGVIMAG, confirming that cleavage occurs between R109 and L110.
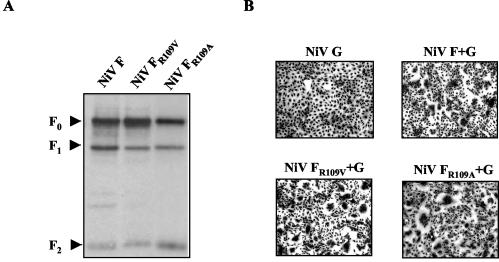
Exchange of R109 affects neither cleavage nor fusion activity of the NiV F protein.
The C-terminal cleavage of R109 suggested that the NiV F protein is activated by a ubiquitous host cell or serum-derived TLP. Consequently, disruption of the monobasic cleavage site by replacing R109 with a nonbasic residue should completely prevent NiV F protein activation. To confirm this hypothesis, we generated a mutant in which R109 was replaced either by alanine or valine (Fig. 2, FR109A and FR109V). However, in contrast to what had been anticipated, both mutants were still cleaved (Fig. 3A, surface biotinylation assay) and were fusion competent (Fig. 3B, fusion assay). Although unexpected, this clearly demonstrates that a basic residue at position 109 is not important for NiV F protein cleavage. Unlike what is known for all other fusion proteins with one basic amino acid residue N terminal of the fusion peptide, cleavage of the NiV F protein does not depend on this basic residue and thus cannot be mediated by a TLP.
FIG. 2.
Schematic diagram of NiV F protein mutants. Boldfaced letters indicate exchanged amino acid residues.
FIG. 3.
Cleavage and biological activity of NiV F proteins with mutations at position 109. (A) Cells were transfected with plasmids carrying either parental F (NiV F), mutant FR109V (NiV FR109V), or mutant FR109A gene (NiV FR109A). Surface biotinylation, immunoprecipitation, and Western blotting was performed as described in the legend to Fig. 1. (B) At 24 h after transfection of the G gene alone (NiV G) or transfection in combination with the parental F (NiV F+G), mutant FR109V (NiV FR109V+G), or mutant FR109A gene (NiV FR109A+G), cell-to-cell fusion was visualized by Giemsa staining. Magnification, ×100.
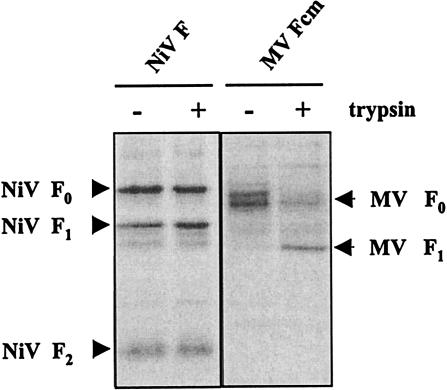
To directly demonstrate the different protease requirements for cleavage of the NiV F protein and cleavage of other paramyxovirus F proteins with monobasic cleavage sites, we compared activation of the NiV F protein with the activation of an MV F protein with a single arginine at the cleavage site (MV Fcm). It was shown previously that MV Fcm requires the addition of exogenous trypsin to become activated. Replication of recombinant viruses carrying the MV Fcm gene has been found to be clearly restricted in vitro and in vivo (14). To analyze cleavage in the absence and presence of trypsin, NiV F and MV Fcm were transiently expressed in MDCK cells. At 6 h posttransfection, the culture supernatant was either replaced by normal medium or by medium containing 1 μg of trypsin per ml. Surface biotinylation was performed at 24 h posttransfection, and F proteins were isolated by immunoprecipitation. Figure 4 shows that precursor F0 in MV Fcm-expressing cells was processed only if trypsin had been added to the culture medium. In contrast, cleavage of the NiV F protein was not affected by the addition of trypsin. Similar results were obtained for all NiV F mutants (data not shown).
FIG. 4.
Influence of trypsin addition on the cleavage of NiV F and MV Fcm. For surface biotinylation, cells were transfected either with parental NiV F (NiV F) or an MV F protein with a monobasic cleavage site (MV Fcm) either in the absence (−) or presence (+) of 1 μg of trypsin/ml. At 24 h posttransfection, surface biotinylation was performed as described in the legend to Fig. 1.
Mutations upstream of R109 do not interfere with activation of the NiV F protein.
To evaluate the importance of the residues upstream of position 109 for NiV F protein cleavage, we replaced each of the seven amino acids up to the next N-glycosylation site (NNT) with alanine (Fig. 2, NiV FV108A, NiV FD107A, NiV FG106A, NiV FV105A, NiV FL104A, NiV FD103A, and NiV FH102A). The surface biotinylation assay revealed that all mutants were efficiently expressed on the cell surface and cleaved (Fig. 5). Consequently, all mutants induced syncytium formation in the fusion assay (data not shown). To demonstrate that the proteolytic activation of mutant F proteins is not mediated by an unusual MDCK-cell-specific protease, cleavage and fusion activity were also analyzed in 293 and Vero cells. As in MDCK cells, all NiV F mutants were activated (data not shown). Obviously, neither single exchange of R109 nor single replacement of any of the seven amino acids in the N terminus of R109 affected cleavage and fusion activity of the NiV F protein. This indicates that the NiV F protein does not require a defined sequence at the C terminus of the F2 subunit for cleavage.
FIG. 5.
Cleavage of NiV F alanine mutants. Cells were transfected with plasmids carrying the F genes indicated. Surface labeling with biotin and immunoprecipitation were performed as described in the legend to Fig. 1.
A polar substitution at position 1 of the fusion peptide does not prevent cleavage but interferes with syncytium formation.
The fusion peptide located at the N terminus of the F1 subunit is highly conserved in structure and sequence among all para- and orthomyxoviruses. Therefore, it must be assumed that the biological activity of NiV F critically depends on the sequence downstream of R109. Viral fusion peptides have been shown to insert into target membranes bridging the two fusing structures. By promoting the negative curvature and lowering the bilayer rupture tension, formation of the fusion pore is accelerated (6). As the amino acid sequence is known to be critical for the function of the fusion peptide, changes in the side chain volume and the hydrophobicity at position 1 are assumed to alter the secondary structure and/or lipid bonding properties, thereby interfering with the fusion process (5, 25). The absolute conservation among all paramyxoviruses (Table 1) clearly indicates that phenylalanine and leucine are the optimal amino acids at position 1 to support fusion. To ensure that the release of the hydrophobic fusion peptide with L110 at the N terminus is also required for NiV-F-mediated fusion, we constructed two mutants in which L110 was replaced either by arginine, a hydrophilic and polar residue, or by phenylalanine, the other hydrophobic residue known to support fusion activity of paramyxovirus F proteins when present at position 1 of the fusion peptide (Fig. 2, mutants NiV FL110R and NiV FL110F). Figure 6 shows that although both mutants were efficiently cleaved, only NiV FL110F was able to induce syncytium formation. This demonstrates that, like other viral fusion proteins, the biological activity of the NiV F protein critically depends on the sequence at the N terminus of the fusion peptide.
FIG. 6.
Proteolytic processing and fusion activity of NiV FL110R and NiV FL110F. (A) Cells transfected with either parental F (NiV F), mutant FL110R (NiV FL110R), or FL110F gene (NiV FL110F) were surface biotinylated. F proteins were immunoprecipitated and detected as described in the legend to Fig. 1. (B) At 24 h after cotransfection of the NiV G gene with parental F (NiV F+G), mutant FL110R (NiV FL110R+G), or the FL110F (NiV FL110F+G) gene, cell-to-cell fusion was visualized by Giemsa staining. Magnification, ×100.
Generation of a cleavage-deficient NiV F mutant.
Because none of the point mutations up- and downstream of the cleavage site had been able to prevent NiV F protein cleavage, we generated a mutant lacking the amino acid residues 104 to 109 (Fig. 2, NiV FΔcs). To analyze protein expression and processing, mutant and parental F proteins were transiently expressed in MDCK cells. Transfected cells were metabolically labeled with [35S]methionine and [35S]cysteine for 10 min (pulse) and were incubated for different times (chase) to allow the proteins to be processed and transported through the secretory pathway. After cell lysis, F proteins were immunoprecipitated, separated on a 12% SDS gel, and subjected to autoradiography. The pulse-chase analysis shown in Fig. 7 revealed that overall expression of the mutant was comparable to that of the parental F protein. After the 10-min pulse, similar amounts of uncleaved precursor F0 were detected. For the parental F protein, first cleavage products were found after a chase period of 45 min, and almost 90% of the protein was cleaved into the subunits F1 and F2 after a chase period of 120 min. In contrast, mutant FΔcs remained completely uncleaved even after long exposure times. Surface biotinylation revealed that FΔcs and the parental F protein were expressed on the cell surface at similar levels and confirmed that the parental F protein, but not FΔcs, was proteolytically processed (data not shown). The fusion assay displayed that FΔcs is not biologically active (Fig. 7B). In contrast to the parental F protein, FΔcs-transfected cells did not show any cell-to-cell fusion. This demonstrates that cleavage of the NiV F protein is a prerequisite for fusion activity and can only be prevented by a larger deletion at the cleavage site.
FIG. 7.
Cleavage and fusion activity of a mutant F protein with a six-amino-acid deletion at the cleavage site (FΔcs). (A) MDCK cells expressing either parental F (NiV F) or mutant FΔcs (NiV FΔcs) were radiolabeled with [35S]Promix for 10 min and then were incubated in chase medium for the times indicated. F proteins were immunoprecipitated from cell lysates, separated on a 12% SDS gel under reducing conditions, and subjected to autoradiography. (B) MDCK cells were transfected with the G gene in combination with the parental F (NiV F+G) or mutant FΔcs gene (NiV FΔcs+G). At 24 h posttransfection, cells were fixed and stained with Giemsa staining solution. Magnification, ×100.
DISCUSSION
All human paramyxoviruses replicating systemically, such as MV or mumps virus, possess an F protein with a multibasic cleavage site. NiV also establishes a systemic infection, but it encodes an F protein with only one basic amino acid residue at the cleavage site, thereby resembling the F proteins of paramyxoviruses that only cause local infections. In vitro, NiV glycoproteins have been shown to induce syncytium formation in many, but not all, cell cultures, and it has been assumed that cell lines not supporting fusion lack a suitable NiV receptor (2). In agreement with this, we found ubiquitous proteolytic cleavage of the NiV F protein in cell lines both permissive and nonpermissive for NiV-mediated fusion. It is well documented that in contrast to multibasic cleavage sites, monobasic cleavage sites cannot be ubiquitously activated by intracellular proteases, such as furin, but rather depend on the activation by extracellular TLPs (11). We clearly confirmed by amino acid sequencing of the NiV F1 N terminus that the F0 precursor protein is cleaved at the predicted cleavage site, the C terminus of the basic residue at position 109, similar to what has been described for the closely related HeV F protein (15). But in contrast to other viral F proteins cleaved after a single arginine, the NiV F protein does not require the addition of proteases for activation, suggesting that the protein has structural features different from those of any other F protein with similar cleavage sites. To define these features on the amino acid level, we generated several cleavage site mutants. Surprisingly, the seven amino acids upstream of the cleavage site as well as R109 itself could be mutated without affecting cleavage or fusion activity. These results clearly indicate that, despite the high level of sequence homology to F proteins of other paramyxoviruses, proteolytic cleavage of the NiV F protein does not depend on the basic residue R109 amino terminal of the hydrophobic fusion peptide sequence, and thus involvement of a ubiquitous TLP in NiV F protein activation can be ruled out.
Mutations altering the number of critical basic residues at the cleavage site always led to changes in protein cleavability. Introduction of a multibasic furin consensus motif normally creates a protein which is ubiquitously activated. For influenza viruses and NDV, multiple studies have shown that such mutations in a viral context result in a higher pathogenicity of the virus (11, 23, 26, 27, 30). In contrast, conversion of a multibasic to a monobasic cleavage site always restricts fusion activation, as has been shown for the F protein of MV. Replication of a recombinant virus with such a mutation is clearly restricted in vitro and in vivo (14). Although there are some reports on ubiquitous activation of viral fusion proteins with monobasic cleavage sites (21, 33), the NiV F protein is the only one known so far which is activated without any single basic residue in this region.
The fact that a polar substitution of L110 (mutant NiV FL110R) but not a conservative replacement by phenylalanine (mutant NiV FL110F) interfered with the ability to induce syncytium formation indicates that position 1 of the NiV fusion peptide must be either a phenylalanine or a leucine to meet the highly conserved requirements for a functional paramyxovirus fusion peptide (Table 1) (1). Thus, for the biological activity the amino acid downstream of the cleavage site is almost invariable. In contrast, NiV F protein proteolytic processing does not rely on stringent sequence requirements at the cleavage site. Whereas ubiquitous activation of other viral fusion proteins depends on a defined consensus motif that allows cleavage by an intracellular Golgi or endoplasmic reticulum protease, such as furin or SKI-1 (7, 12, 22, 31, 37, 38, 39, 41), NiV F protein cleavage is mediated by a ubiquitous protease which does not require a conserved consensus motif around the cleavage site, not even a defined amino acid residue at position 109. Possible candidates mediating such an activation might be metalloproteases, for example, members of the large family of A disintegrin and metalloproteinases (ADAM) proteases. Cleavage of substrates by these proteases does not depend on conserved amino acid sequences directly at the cleavage site but appears to require a stretch (at least 8 to 10 residues long) of presumably unfolded peptide surrounding the target bond (28). Dependence of activation on the overall structure rather than on the exact sequence might explain why only the complete deletion of six amino acids at the cleavage site (mutant FΔcs) interfered with F activation, whereas none of the point mutations up- and downstream of R109 was able to prevent cleavage. The large deletion in the FΔcs mutant likely affects the overall conformation of the cleavage region while single amino acid exchanges have no drastic influence on the structure. Some of the possible candidates for NiV activation, such as ADAM 9, 10, 12, 15, 17, 19, and 33, are widely expressed and have a very broad substrate specificity (for a review see reference 29 and references therein). However, these proteases usually cleave membrane-anchored proteins at cleavage sites in very close vicinity to the cell membrane, a precondition unlikely given the NiV F protein cleavage site. Although the three-dimensional structure of the protein is not yet known, its similarity to other type I fusion proteins with respect to size and the location of hydrophobic domains and heptad repeats indicates that the cleavage peptide is not located in the stalk region near the membrane but in the neck of the protein as has been shown for the cleavage site of the NDV F protein (3).
The observation that addition of trypsin at a concentration which efficiently cleaved F proteins with monobasic cleavage sites could not increase the amount of NiV F protein cleavage products indicates that surface-expressed precursor F0 does not function as a substrate for trypsin. This suggests that the NiV F cleavage region has exceptional structural characteristics that allow activation only by the NiV-activating protease. The hypothesis that the conformational structure of the NiV F protein does not allow cleavage by other F-protein-activating proteases is furthermore supported by a recent finding on a mutant NiV F protein in which a typical furin consensus motif has been introduced upstream of L110 (NiV FcsSRRHKR). Neither endogenous furin nor exogenous trypsin was able to process the multibasic cleavage site in the NiV FcsSRRHKR mutant (S. Diederich, unpublished data).
In summary, our results show that activation of NiV is unique among the members of the Orthomyxoviridae and Paramyxoviridae with respect to sequence requirements and protease usage. Ubiquitous activation of the NiV F protein depends on a novel type of proteolytic cleavage mediated by an enzyme that does not require a basic residue at the cleavage site and thus cannot be a member of the trypsin-like or furin-like protease family.
Acknowledgments
We gratefully acknowledge the technical assistance of Andrea Gumienny and Alan Grolla. We thank Pierre Rollin and Tom Ksiazek for kindly providing the NiV isolate.
This work was supported by a grant from the Deutsche Forschungsgemeinschaft (MA 1886/5-1) to A.M.
REFERENCES
- 1.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell. 3:309-319. [DOI] [PubMed] [Google Scholar]
- 2.Bossart, K. N., L. F. Wang, M. N. Flora, K. B. Chua, S. K. Lam, B. T. Eaton, and C. C. Broder. 2002. Membrane fusion tropism and heterotypic functional activities of the Nipah virus and Hendra virus envelope glycoproteins. J. Virol. 76:11186-11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, L., J. J. Gorman, J. McKimm-Breschkin, L. J. Lawrence, P. A. Tulloch, B. J. Smith, P. M. Colman, and M. C. Lawrence. 2001. The structure of the fusion glycoprotein of Newcastle disease virus suggests a novel paradigm for the molecular mechanism of membrane fusion. Structure (Cambridge) 9:255-266. [DOI] [PubMed] [Google Scholar]
- 4.Chua, K. B., K. J. Goh, K. T. Wong, A. Kamarulzaman, P. S. Tan, T. G. Ksiazek, S. R. Zaki, G. Paul, S. K. Lam, and C. T. Tan. 1999. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 354:1257-1259. [DOI] [PubMed] [Google Scholar]
- 5.Durell, S. R., I. Martin, J. M. Ruysschaert, Y. Shai, and R. Blumenthal. 1997. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion. Mol. Membr. Biol. 14:97-112. [DOI] [PubMed] [Google Scholar]
- 6.Epand, R. M. 2003. Fusion peptides and the mechanism of viral fusion. Biochim. Biophys. Acta 1614:116-121. [DOI] [PubMed] [Google Scholar]
- 7.Hallenberger, S., V. Bosch, H. Angliker, E. Shaw, H. D. Klenk, and W. Garten. 1992. Inhibition of furin-mediated cleavage activation of HIV-1 glycoprotein gp160. Nature 360:358-361. [DOI] [PubMed] [Google Scholar]
- 8.Harcourt, B. H., A. Tamin, T. G. Ksiazek, P. E. Rollin, L. J. Anderson, W. J. Bellini, and P. A. Rota. 2000. Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 271:334-349. [DOI] [PubMed] [Google Scholar]
- 9.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hooper, P., S. Zaki, P. Daniels, and D. Middleton. 2001. Comparative pathology of the diseases caused by Hendra and Nipah viruses. Microbes Infect. 3:315-322. [DOI] [PubMed] [Google Scholar]
- 11.Klenk, H. D., and W. Garten. 1994. Activation cleavage of viral spike proteins by host proteases, p. 241-280. In E. Wimmer (ed.), Cellular receptors for animal viruses. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 12.Lenz, O., J. ter Meulen, H. D. Klenk, N. G. Seidah, and W. Garten. 2001. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc. Natl. Acad. Sci. USA 98:12701-12705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindemann, D., T. Pietschmann, M. Picard-Maureau, A. Berg, M. Heinkelein, J. Thurow, P. Knaus, H. Zentgraf, and A. Rethwilm. 2001. A particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 75:5762-5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maisner, A., B. Mrkic, G. Herrler, M. Moll, M. A. Billeter, R. Cattaneo, and H. D. Klenk. 2000. Recombinant measles virus requiring an exogenous protease for activation of infectivity. J. Gen. Virol. 81:441-449. [DOI] [PubMed] [Google Scholar]
- 15.Michalski, W. P., G. Crameri, L. Wang, B. J. Shiell, and B. Eaton. 2000. The cleavage activation and sites of glycosylation in the fusion protein of Hendra virus. Virus Res. 69:83-93. [DOI] [PubMed] [Google Scholar]
- 16.Moll, M., H. D. Klenk, G. Herrler, and A. Maisner. 2001. A single amino acid change in the cytoplasmic domains of measles virus glycoproteins H and F alters targeting, endocytosis, and cell fusion in polarized Madin-Darby canine kidney cells. J. Biol. Chem. 276:17887-17894. [DOI] [PubMed] [Google Scholar]
- 17.Moll, M., H. D. Klenk, and A. Maisner. 2002. Importance of the cytoplasmic tails of the measles virus glycoproteins for fusogenic activity and the generation of recombinant measles viruses. J. Virol. 76:7174-7186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murakami, M., T. Towatari, M. Ohuchi, M. Shiota, M. Akao, Y. Okumura, M. A. Parry, and H. Kido. 2001. Mini-plasmin found in the epithelial cells of bronchioles triggers infection by broad-spectrum influenza A viruses and Sendai virus. Eur. J. Biochem. 268:2847-2855. [DOI] [PubMed] [Google Scholar]
- 19.Murray, K., P. Selleck, P. Hooper, A. Hyatt, A. Gould, L. Gleeson, H. Westbury, L. Hiley, L. Selvey, B. Rodwell, et al. 1995. A morbillivirus that caused fatal disease in horses and humans. Science 268:94-97. [DOI] [PubMed] [Google Scholar]
- 20.Nagai, Y., and H. D. Klenk. 1977. Activation of precursors to both glycoproteins of Newcastle disease virus by proteolytic cleavage. Virology 77:125-134. [DOI] [PubMed] [Google Scholar]
- 21.Orlich, M., D. Linder, and R. Rott. 1995. Trypsin-resistant protease activation mutants of an influenza virus. J. Gen. Virol. 76(Pt 3):625-633. [DOI] [PubMed] [Google Scholar]
- 22.Ortmann, D., M. Ohuchi, H. Angliker, E. Shaw, W. Garten, and H. D. Klenk. 1994. Proteolytic cleavage of wild type and mutants of the F protein of human parainfluenza virus type 3 by two subtilisin-like endoproteases, furin and Kex2. J. Virol. 68:2772-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Panda, A., Z. Huang, S. Elankumaran, D. D. Rockemann, and S. K. Samal. 2004. Role of fusion protein cleavage site in the virulence of Newcastle disease virus. Microb. Pathog. 36:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peeters, B. P., O. S. de Leeuw, G. Koch, and A. L. Gielkens. 1999. Rescue of Newcastle disease virus from cloned cDNA: evidence that cleavability of the fusion protein is a major determinant for virulence. J. Virol. 73:5001-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiao, H., R. T. Armstrong, G. B. Melikyan, F. S. Cohen, and J. M. White. 1999. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol. Biol. Cell 10:2759-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Romer-Oberdorfer, A., O. Werner, J. Veits, T. Mebatsion, and T. C. Mettenleiter. 2003. Contribution of the length of the HN protein and the sequence of the F protein cleavage site to Newcastle disease virus pathogenicity. J. Gen. Virol. 84:3121-3129. [DOI] [PubMed] [Google Scholar]
- 27.Rott, R. 1992. The pathogenic determinant of influenza virus. Vet. Microbiol. 33:303-310. [DOI] [PubMed] [Google Scholar]
- 28.Schlondorff, J., and C. P. Blobel. 1999. Metalloprotease-disintegrins: modular proteins capable of promoting cell-cell interactions and triggering signals by protein-ectodomain shedding. J. Cell Sci. 112(Pt 21):3603-3617. [DOI] [PubMed] [Google Scholar]
- 29.Seals, D. F., and S. A. Courtneidge. 2003. The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev. 17:7-30. [DOI] [PubMed] [Google Scholar]
- 30.Steinhauer, D. A. 1999. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology 258:1-20. [DOI] [PubMed] [Google Scholar]
- 31.Stieneke-Grober, A., M. Vey, H. Angliker, E. Shaw, G. Thomas, C. Roberts, H. D. Klenk, and W. Garten. 1992. Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J. 11:2407-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tamin, A., B. H. Harcourt, T. G. Ksiazek, P. E. Rollin, W. J. Bellini, and P. A. Rota. 2002. Functional properties of the fusion and attachment glycoproteins of Nipah virus. Virology 296:190-200. [DOI] [PubMed] [Google Scholar]
- 33.Tashiro, M., I. James, S. Karri, K. Wahn, K. Tobita, H. D. Klenk, R. Rott, and J. T. Seto. 1991. Pneumotropic revertants derived from a pantropic mutant, F1-R, of Sendai virus. Virology 184:227-234. [DOI] [PubMed] [Google Scholar]
- 34.Tashiro, M., E. Pritzer, M. A. Khoshnan, M. Yamakawa, K. Kuroda, H. D. Klenk, R. Rott, and J. T. Seto. 1988. Characterization of a pantropic variant of Sendai virus derived from a host range mutant. Virology 165:577-583. [DOI] [PubMed] [Google Scholar]
- 35.Tashiro, M., M. Yamakawa, K. Tobita, H. D. Klenk, R. Rott, and J. T. Seto. 1990. Organ tropism of Sendai virus in mice: proteolytic activation of the fusion glycoprotein in mouse organs and budding site at the bronchial epithelium. J. Virol. 64:3627-3634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tashiro, M., Y. Yokogoshi, K. Tobita, J. T. Seto, R. Rott, and H. Kido. 1992. Tryptase Clara, an activating protease for Sendai virus in rat lungs, is involved in pneumopathogenicity. J. Virol. 66:7211-7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vey, M., W. Schafer, B. Reis, R. Ohuchi, W. Britt, W. Garten, H. D. Klenk, and K. Radsak. 1995. Proteolytic processing of human cytomegalovirus glycoprotein B (gpUL55) is mediated by the human endoprotease furin. Virology 206:746-749. [DOI] [PubMed] [Google Scholar]
- 38.Vincent, M. J., A. J. Sanchez, B. R. Erickson, A. Basak, M. Chretien, N. G. Seidah, and S. T. Nichol. 2003. Crimean-Congo hemorrhagic fever virus glycoprotein proteolytic processing by subtilase SKI-1. J. Virol. 77:8640-8649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volchkov, V. E., H. Feldmann, V. A. Volchkova, and H. D. Klenk. 1998. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc. Natl. Acad. Sci. USA 95:5762-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, L. F., M. Yu, E. Hansson, L. I. Pritchard, B. Shiell, W. P. Michalski, and B. T. Eaton. 2000. The exceptionally large genome of Hendra virus: support for creation of a new genus within the family Paramyxoviridae. J. Virol. 74:9972-9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watanabe, M., A. Hirano, S. Stenglein, J. Nelson, G. Thomas, and T. C. Wong. 1995. Engineered serine protease inhibitor prevents furin-catalyzed activation of the fusion glycoprotein and production of infectious measles virus. J. Virol. 69:3206-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]