Abstract
Intercellular adhesion molecule 1 (ICAM-1) is the cellular receptor for the major group of human rhinovirus serotypes, including human rhinovirus 14 (HRV14) and HRV16. A naturally occurring variant of ICAM-1, ICAM-1Kilifi, has altered binding characteristics with respect to different HRV serotypes. HRV14 binds to ICAM-1 only transiently at physiological temperatures but forms a stable complex with ICAM-1Kilifi. Conversely, HRV16 forms a stable complex with ICAM-1 but does not bind to ICAM-1Kilifi. The three-dimensional structures of HRV14 and HRV16, complexed with ICAM-1, and the structure of HRV14, complexed with ICAM-1Kilifi, have been determined by cryoelectron microscopy (cryoEM) image reconstruction to a resolution of approximately 10 Å. Structures determined by X-ray crystallography of both viruses and of ICAM-1 were fitted into the cryoEM density maps. The interfaces between the viruses and receptors contain extensive ionic networks. However, the interactions between the viruses and ICAM-1Kilifi contain one less salt bridge than between the viruses and ICAM-1. As HRV16 has fewer overall interactions with ICAM-1 than HRV14, the absence of this charge interaction has a greater impact on the binding of ICAM-1Kilifi to HRV16 than to HRV14.
Intercellular adhesion molecule 1 (ICAM-1) is used as a receptor by many pathogens, including the major group of human rhinoviruses (HRVs) (16, 29) and some coxsackieviruses (e.g., CVA13, CVA15, CVA18, CVA20, and CVA21) (34, 57). In addition, when erythrocytes are infected by the malarial parasite, Plasmodium falciparum, they gain the ability to bind cells expressing ICAM-1 (7, 35). ICAM-1 belongs to the immunoglobulin (Ig)-like superfamily (IgSF) (49). Unlike Igs, the five extracellular Ig domains of ICAM-1 (D1 to D5) are unpaired and form an end-to-end tandem chain. The rod-shaped molecule is approximately 180 Å long, with a slight bend between the second and third Ig domains (51, 57). In addition, ICAM-1 also has a transmembrane domain and a short cytoplasmic domain. ICAM-1 is expressed by many cell types, but, unlike expression of ICAM-2 and ICAM-3, the level of ICAM-1 expression is controlled by intercellular factors, such as cytokines that regulate transcription (42, 52). Infection by HRVs up-regulates ICAM-1 expression (38) and causes common cold-like symptoms. A natural variant of ICAM-1, ICAM-1Kilifi, in which Lys29 (AAG) is changed to Met (ATG), is present in certain African populations. This A/T single nucleotide variation also has been reported in other populations (27). Individuals that are homozygous for the Met29 allele have increased susceptibility to cerebral malaria caused by P. falciparum (12, 14).
The structure of the N-terminal two domains of ICAM-1 has been determined previously by X-ray crystallography (5, 10, 29). One β-sheet of an Ig domain contains the A, B, E, and D β-strands, whereas the other β-sheet contains the G, F, and C strands. The first domain of ICAM-1 has been classified as an “intermediate” IgSF domain, which contains an extra strand A′ in the GFC β-sheet (11, 56). The tip of domain D1 is formed by the B-C, D-E, and F-G loops. Lys29 is located at the center of the B-C loop. Domain 2 is classified as a “constant” type IgSF domain and has four potential N-linked glycosylation sites located at Asn103, Asn118, Asn156, and Asn175. The elbow angle between the two domains varies by about 8° between molecules in different crystallographic environments (10, 29).
Picornaviridae are a family of positive, single-stranded RNA viruses, whose genomes are approximately 7 kb long. The viral particles have an external diameter of approximately 310 Å. Their icosahedral capsids are assembled from a total of 60 copies each of the four viral proteins VP1 to VP4. VP1 to VP3 contribute to the external features of the capsid, whereas VP4 is internal and situated at the interface between the protein capsid and the RNA genome. Rhinovirus is one of the nine genera of Picornaviridae (50). The atomic resolution structures of HRV14 and HRV16, as well as other serotypes, have been determined previously (1, 28, 36, 44, 53, 60). The surfaces of these and some other picornaviruses have deep depressions (canyons) surrounding each fivefold vertex. These canyons were predicted to be the site of receptor binding on the assumption that they would be inaccessible to the binding of neutralizing antibodies (44); this assumption was later confirmed for a variety of different picornaviruses (46).
In many picornaviruses, such as polioviruses, rhinoviruses, and coxsackieviruses, there is an unidentified, fatty acid-like molecule (pocket factor) in a hydrophobic pocket within VP1, situated underneath the floor of the canyon (15, 24, 33, 36, 48). In the absence of the pocket factor, the roof of the pocket is collapsed, thus also lowering the floor of the canyon. Small hydrophobic compounds can enter this pocket, displacing the pocket factor if present (2, 19, 47). Filling the hydrophobic pocket with a hydrophobic compound stabilizes the virus and damps the normal dynamic “breathing” of the virion, causing the inhibition of uncoating (30). At the same time, the conformational changes on the floor of the canyon can cause the inhibition of virus attachment to cells (23, 39, 43). Presumably, altering the conformation of the floor of the canyon interferes with the binding of ICAM-1. The overlapping binding sites of the pocket factor and ICAM-1 suggest that the pocket factor and receptor compete for their ability to bind to the virus (36, 43). Thus, when a pocket factor or an antiviral compound is bound in the VP1 pocket, the virus will be stabilized. However, the binding of the receptor presumably displaces the pocket factor and, hence, destabilizes the virus, thus contributing to the initiation of uncoating.
The in vitro binding of soluble ICAM-1 to major-group HRVs at physiological temperatures initiates a sequence of conformational changes in the virion that mimic uncoating of the virus during cell entry and that are characterized by shifts in particle sedimentation from 150S (native virus) to 135S or 80S (26). Such alterations in the virus particle have made structural studies of the virus-receptor complex difficult. However, it has been possible to combine X-ray crystallography and cryoelectron microscopy (cryoEM) image reconstructions by fitting the separately determined virus and receptor crystal structures into cryoEM density. This technique has been used to study the complexes of HRV3, HRV14, and HRV16 with ICAM-1 (29, 58), of poliovirus with CD155 (6, 20), of CVA21 with ICAM-1 (57), of coxsackievirus B3 with the coxsackievirus-adenovirus receptor (21), of echovirus 7 with decay-accelerating factor (22), of HRV2 with very-low-density lipoprotein receptor (25), and of echovirus 1 with the I domain of an integrin (59). ICAM-1, CD155, and the coxsackievirus-adenovirus receptor all have amino-terminal IgSF folds and bind into the canyon with their N-terminal domains. Although the binding site of ICAM-1 in the HRV14 and HRV16 canyons is similar, the residues in the ICAM-1 footprint are different although they maintain some charge complementarity with the residues in the canyons (29).
Here, we report that ICAM-1Kilifi differs from ICAM-1 in its ability to bind to various HRV serotypes. In particular, HRV16, unlike HRV14, does not appear to be able to bind to ICAM-1Kilifi. CryoEM image reconstructions of ICAM-1 complexed with HRV14 and HRV16, as well as of ICAM-1Kilifi complexed with HRV14, have now attained resolutions ranging from 8.6 to 13.1 Å, compared to the previous 28-Å resolution results (5, 29, 37). These cryoEM maps, when fitted with atomic models of the virus and ICAM-1, provide a possible rationalization of the differences between the virus binding properties of ICAM-1 and ICAM-1Kilifi.
MATERIALS AND METHODS
ELISA.
The binding of major-group rhinoviruses HRV14 and HRV16 to ICAM-1 were investigated by capture enzyme-linked immunosorbent assay (ELISA). Soluble forms of either ICAM-1 or ICAM-1Kilifi were expressed as the ICAM-1 ectodomain fused to the Fc region of human IgG as previously described (12). Equivalent concentrations of soluble ICAM-1 or ICAM-1Kilifi in phosphate-buffered saline (PBS) were adsorbed onto the surface of ELISA plates at 4°C overnight, and the plates were blocked with 1% bovine serum albumin (BSA) in PBS for a further hour at 4°C. Plates were washed three times with 0.1% BSA in PBS, and virus (typically, 105 PFU in 50 μl of PBS per well) was bound at 34°C for 1 h. Plates were washed as before, incubated at room temperature with serotype-specific antisera (American Type Culture Collection), and then washed and incubated at room temperature with horseradish peroxidase (HRP)-conjugated secondary antibodies. After washing, bound virus was detected by the activity of HRP on the substrate 1,2-ortho-phenylene-diamine and quantified by determining the absorbance at 490 nm.
Virus neutralization.
The ability of soluble receptor to bind and neutralize virus particles was tested by measuring the reduction in infectious viral titer after exposure to ICAM-1 or ICAM-1Kilifi. Purified virus was incubated with an excess of soluble receptor (estimated to be in the order of 106 molecules per PFU) or serotype-specific antisera (American Type Culture Collection) for 1 h at 34°C, and titers were determined by plaque assays.
Samples for cryoEM.
HRV14 and HRV16 were prepared as previously described (36). ICAM-1 was provided by J. M. Greve (17), and ICAM-1Kilifi was produced as previously described (12).
CryoEM.
HRV16 and HRV14 infectious particles were incubated with ICAM-1 or ICAM-1Kilifi at room temperature for a defined time (Table 1). Room temperature, instead of 4°C, was used to incubate the viruses and ICAM-1 to increase the solubility of the viruses and, hence, to obtain more particles per micrograph. After incubation, the samples were frozen for cryoEM as described previously (57). All the cryoEM micrographs were taken on a Philips FEG300 microscope at a magnification of ×47,000 using 300 kV.
TABLE 1.
CryoEM data collection
| Complex | Virus concn (mg/ml) | No. of receptorsb | Incubation time (min) | No. of micrographs | No. of focal pairs used | No. of boxed particles | No. of particles used | Defocus range (μ) | Final resolution (Å) |
|---|---|---|---|---|---|---|---|---|---|
| HRV16-ICAM-1 | 10 | 3.3 | 85 | 32 | 6 | 7,969 | 5,696 | 0.7-3.2 | 8.6 |
| HRV14-ICAM-1 | 8 | 3.3 | 75 | 14 | 6 | 2,570 | 1,646 | 0.8-3.3 | 13.1 |
| HRV16-ICAM-1Kilifia | 10 | 10.9 | 60 | 7 | 0 | 453 | 96 | 2.0-4.8 | 28.0 |
| HRV14-ICAM-1Kilifi | 8 | 10.9 | 60 | 24 | 3 | 4,052 | 2,831 | 0.9-3.6 | 9.5 |
Because HRV16-ICAM-1Kilifi showed no evidence of bound ICAM-1Kilifi, it was extended to a resolution of only 28 Å.
Per viral binding site in solution.
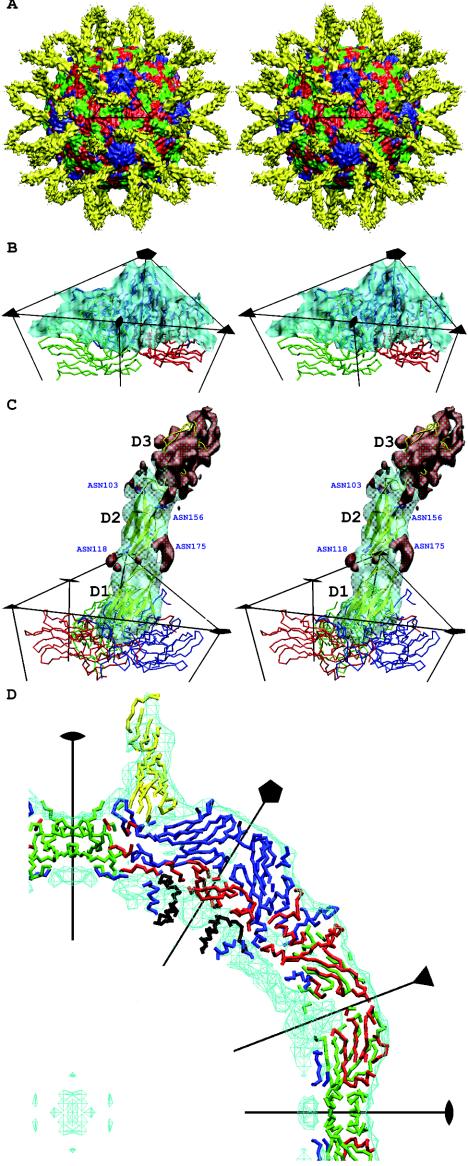
Micrographs were scanned on a Zeiss Phodis SCSI scanner with a step size of 7 μm. Four neighboring pixels were averaged to provide an effective pixel spacing of 14 μm, corresponding to a pixel size of 2.98 Å on the specimen. The selected images were normalized by the program RobEM (R. Ashmore, unpublished program). The out-of-focus distance and the envelope decay factor, the “temperature” factor that determines the decline of the amplitude of the contrast transfer function (CTF) with respect to resolution (4), were estimated with the program CTFest (C. Xiao, unpublished program). This procedure determined the defocus value and decay factor by fitting a Gaussian function to the radially averaged Fourier transform of all the particles on the same micrograph. The corrections were computed by parallelized versions (8, 57) of the image reconstruction programs EMPFT (3) and EM3DR (13). The cryoEM maps were sharpened by using the inverted, previously determined envelope decay factors ranging from 100 to 400 Å2 to emphasize the higher resolution terms. The map resolution (Table 1) was determined by finding where the Fourier shell correlation fell below 0.5 by using Fourier coefficients based only on the density between a radius of 104 and 177 Å, corresponding to the protein shell of the viral capsid plus the first ICAM-1 domain. Atomic coordinates of the crystallographically determined capsid structures of HRV14 (Protein Data Bank[PDB] accession number 4RHV) and HRV16 (PDB accession number 1AYM) were used to compute 10-Å resolution X-ray maps. Fourier terms used to calculate the cryoEM maps were limited to have a resolution of 180 to 10 Å. These cryoEM and X-ray maps were compared within a 110- to 160-Å radius shell (corresponding to most of the protein shell of the capsid) to calibrate the cryoEM map magnification with respect to the well-established scale of the X-ray maps (Fig. 1B and D). This established the pixel size of all the cryoEM maps to be 2.89 Å, rather than 2.98 Å. Difference maps between the scaled cryoEM maps and the corresponding X-ray maps clearly showed the 60 copies of the bound ICAM-1 molecules (Fig. 1).
FIG. 1.
(A) Stereo view of the surface-shaded cryoEM map of HRV16 complexed with ICAM-1 (yellow) and showing VP1 (blue), VP2 (green), and VP3 (red). The black triangle shows the limit of one icosahedral asymmetric unit. (B) Stereo view showing the quality of fit of the HRV16 capsid structure into the cryoEM map surface. VP1 (blue), VP2 (green), and VP3 (red) are represented by a trace of their Cα atoms. The density corresponding to ICAM-1 has been removed. (C) Stereo view of one ICAM-1 molecule bound to HRV16. Oneicosahedral asymmetric unit is outlined in black, showing one copy of the difference density of the ICAM-1 molecule in transparent cyan. ICAM-1 is shown as a yellow ribbon. VP1 (blue), VP2 (green), and VP3 (red) are also shown. The four Asn residues that are N-linked to carbohydrates are drawn and labeled in blue. The densities that have not been fitted with the atomic models for the ICAM-1 domains D1 and D2 are shown in red, which include the densities of the carbohydrates moieties belonging to domain D2, as well as the density of domain D3. (D) The central slab (−10 to 10 Å in the Z axis) of the cryoEM density (cyan) fitted with the appropriate backbone structures of ICAM-1 (yellow), VP1 (blue), VP2 (green), VP3 (red), and VP4 (black).
Fitting of ICAM-1 structures.
Three independent crystal structures of ICAM-1 domains D1 and D2 are available in the PDB, one molecule per asymmetric unit in one crystal form (PDB accession number 1IAM) and two molecules (PDB accession numbers 1IC1-A and 1IC1-B) in another crystal form. These three structures differ primarily in the position of the B-C, D-E, and F-G loops at the tip of domain D1, with a maximum displacement of 11.7 Å in side chain atoms and 5.6 Å in main chain atoms. Three corresponding ICAM-1Kilifi structures were modeled, based on each of the three ICAM-1 structures, by changing Lys29 to Met at the tip of the B-C loop.
The three different sets of coordinates for ICAM-1 and also for ICAM-1Kilifi were fitted independently into the difference maps by using the program EMfit (45). The fitting depends on sumf (the average height of electron density for Cα atoms only or for all atoms), the distance between carbohydrate position and the glycosylated Asn, and the number of atoms in negative density. However, the values of sumf were normalized, not with respect to the somewhat arbitrary highest density (45) but with respect to the root mean square (rms) deviation from the mean density within the protein shell of the virus. The use of rms deviation values instead of the highest density values for normalization is a more rigorous way of comparing different maps, which is required here. In essence, sumf represents the average density taken over all the atoms in the stated molecule. The minimization of the number of atoms fitted into negative density ensured that the ICAM-1 model was placed within the higher-density regions of the difference map and, thus, would be unlikely to clash with the virus structure. Domain D1 was fitted first. Prior to fitting domain D2, the density within a radius of 4.5 Å of each Cα atom in the fitted domain D1 was set to zero. This modified difference map was used to fit domain D2. The position of the four carbohydrate sites in domain D2 (at Asn108, Asn113, Asn156, and Asn175) could be readily identified as protrusions in the ICAM-1 difference density. The Cα atom of each glycosylated residue was restrained to be less than 20 Å from the center of the corresponding carbohydrate site in the map. The N-terminal Cα atom of domain D2 was also restrained to be less than 4 Å from the C-terminal Cα atom of the fitted domain D1. The sumf values for the four viral proteins were calculated by using the known viral coordinates in order to compare the quality of fit of the ICAM-1 domains (Tables 2 and 3).
TABLE 2.
Fitting statistics for ICAM-1a
| Complex coordinate source (PDB) | sumf of domain D1 | sumf of domain D2 | Ratio of D1/D2 sumf values | Distance between Asn Cα and glycosylation sites (Å)
|
|||
|---|---|---|---|---|---|---|---|
| Asn108 | Asn113 | Asn156 | Asn175 | ||||
| HRV16-ICAM-1 | |||||||
| 1IAM | 0.64 | 0.53 | 0.82 | 14.2 | 17.5 | 16.2 | 16.7 |
| 1IC1-A | 0.64 | 0.53 | 0.82 | 13.7 | 17.6 | 16.5 | 17.2 |
| 1IC1-B | 0.64 | 0.52 | 0.82 | 15.1 | 17.3 | 16.6 | 17.3 |
| HRV14-ICAM-1 | |||||||
| 1IAM | 1.06 | 1.01 | 0.95 | 11.6 | 16.4 | 16.1 | 16.9 |
| 1IC1-A | 1.05 | 1.02 | 0.97 | 11.4 | 16.7 | 16.1 | 17.2 |
| 1IC1-B | 1.04 | 1.00 | 0.95 | 11.3 | 16.3 | 16.1 | 17.0 |
| HRV14-ICAM-1Kilifi | |||||||
| 1IAM | 1.04 | 0.87 | 0.83 | 15.9 | 15.5 | 13.3 | 15.8 |
| 1IC1-A | 1.07 | 0.88 | 0.82 | 16.1 | 14.9 | 13.8 | 15.2 |
| 1IC1-B | 1.05 | 0.85 | 0.81 | 16.5 | 15.8 | 12.7 | 15.6 |
The value of sumf is normalized by the rms deviation and represents the average density taken over the Cα atoms of the stated structure (see Materials and Methods).
TABLE 3.
Fitting statistics of the viral proteins VP1 to VP4a
| Protein and region of sequence | HRV16-ICAM-1
|
HRV14-ICAM-1
|
HRV16-ICAM-1Kilifi
|
|||
|---|---|---|---|---|---|---|
| Residue range | sumf | Residue range | sumf | Residue range | sumf | |
| VP1 | ||||||
| Overall | 1-285 | 0.80 | 17-289 | 0.84 | 17-289 | 0.81 |
| N terminus | 17-73 | 0.35 | 17-73 | 0.49 | 17-73 | 0.42 |
| Middle | 74-263 | 0.94 | 74-270 | 0.96 | 74-270 | 0.95 |
| C terminus | 264-285 | 0.75 | 271-289 | 0.62 | 271-289 | 0.62 |
| VP2 | ||||||
| Overall | 10-261 | 0.83 | 8-262 | 0.82 | 8-262 | 0.80 |
| N terminus | 10-31 | 0.47 | 8-31 | 0.49 | 8-31 | 0.50 |
| Middle | 32-253 | 1.01 | 32-253 | 0.86 | 32-253 | 0.84 |
| C terminus | 254-261 | 0.45 | 254-262 | 0.63 | 254-262 | 0.66 |
| VP3 | ||||||
| Overall | 1-238 | 0.82 | 1-236 | 0.78 | 1-236 | 0.83 |
| N terminus | 1-51 | 0.46 | 1-51 | 0.51 | 1-51 | 0.58 |
| Middle | 52-223 | 0.94 | 52-221 | 0.88 | 52-221 | 0.92 |
| C terminus | 224-238 | 0.55 | 222-236 | 0.47 | 222-236 | 0.60 |
| VP4 | ||||||
| Overall | 1-44 | 0.26 | 29-68 | 0.13 | 29-68 | 0.16 |
| N terminus | ||||||
| Middle | ||||||
| C terminus | ||||||
The value of sumf is normalized by the rms deviation and represents the average density taken over the Cα atoms of the stated structure (see Materials and Methods).
A homology-modeled structure of ICAM-1 domain D3 (57) was used for manual fitting into the cryoEM densities. No computational refinement was applied to this fit because the density corresponding to domain D3 was weak.
Elbow angle and twist angle.
The line joining the Cα atoms of Pro28 and Tyr83 was selected to define the long axis of ICAM-1 domain D1. Similarly, the line joining the Cα atoms of Pro115 and Phe185 was used to define the long axis of ICAM-1 domain D2. The elbow angle was defined as being the angle between these two axes (Table 4). In order to determine the twist angle between domains D1 and D2, two additional Cα atoms, Lys40 and Val146, were selected in domains D1 and D2, respectively. The twist angle between the two domains was defined as the angle between the normals of the two planes formed by the Cα atoms of Pro28, Lys40, and Tyr83 in domain D1 and of Pro115, Val146, and Phe185 in domain D2 (Table 4).
TABLE 4.
Elbow angles and twist angles between ICAM-1 domains D1 and D2
| Structure | X-ray determined angles (degrees)
|
EM-fitted angles (degrees)
|
||||||
|---|---|---|---|---|---|---|---|---|
| HRV16-ICAM-1
|
HRV14-ICAM-1
|
HRV14-ICAM-1Kilifi
|
||||||
| Elbow | Twist | Elbow | Twist | Elbow | Twist | Elbow | Twist | |
| 1IAM | 171.2 | 9.9 | 171.2 | 21.1 | 173.1 | 2.92 | 172.2 | 17.2 |
| 1IC1-A | 168.5 | 14.6 | 169.7 | 26.7 | 173.2 | 11.4 | 177.3 | 32.0 |
| 1IC1-B | 176.9 | 9.3 | 171.8 | 34.6 | 173.8 | 11.1 | 173.3 | 17.5 |
Energy minimization.
The rigid-body fitting procedure generated a few ICAM-1 D1 side chain atoms that were in steric collision with virus side chain atoms. Hence, the atoms in the virus-receptor interface were subjected to energy minimization by using the program CHARMM (31), which mostly eliminated steric collisions. No attempt was made to simulate the water environment. The maximum displacement of the side chain atoms incurred by the energy minimization was never greater than 5.8 Å.
Interface analyses.
The interface between the virus and the receptor was defined as being limited by those residues that had atoms less than 4 Å from any opposing atom. Among the three different ICAM-1 structures fitted independently, the structure that had the lowest potential energy was chosen to represent the complex. The buried surface area between the virus and ICAM-1, as well as the number of contact residues, was calculated with the program CNS (9) (Table 5). The potential energies of the interactions between ICAM-1 and virus were calculated with the program CHARMM and included the electrostatic energy, van der Waals energy, and hydrogen bonding energy (Table 5).
TABLE 5.
Overall contact statistics
| Complexa | Buried surface (Å2) | No. of H-bonds | No. of ion pairs | No. of other interactions | Total no. of interactions | Energy (kcal/mol)
|
|||
|---|---|---|---|---|---|---|---|---|---|
| H-bond | Electrostatic interaction | VDW interactionb | Total interaction | ||||||
| HRV14-ICAM-1 | 1,431.2 | 36 | 11 | 37 | 73 | −67.8 | −909.5 | −75.2 | −1,052.6 |
| HRV16-ICAM-1 | 1,274.8 | 27 | 9 | 26 | 53 | −47.6 | −614.5 | −75.2 | −737.3 |
| HRV14-ICAM-1Kilifi | 1,421.8 | 35 | 10 | 39 | 74 | −53.4 | −775.4 | −80.3 | −909.1 |
| HRV16-ICAM-1Kilifi | 1,254.1 | 26 | 7 | 29 | 55 | −48.5 | −508.5 | −92.3 | −643.3 |
ICAM-1Kilifi does not bind to HRV16. Therefore, the results for HRV16-ICAM-1Kilifi were calculated by modeling the Lys29 of ICAM-1 in the HRV16-ICAM-1 complex as a Met.
VDW, van der Waals energy.
RESULTS AND DISCUSSION
Biochemical analysis of receptor binding in vitro.
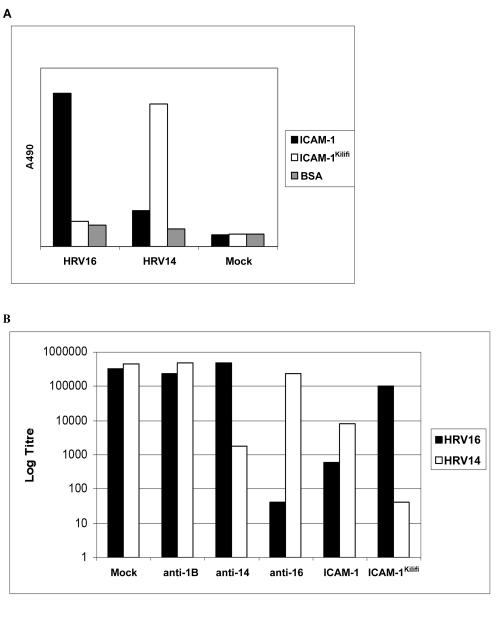
The amount of virus binding to different immobilized receptors was measured by ELISA. These experiments showed that HRV16 bound well to ICAM-1 but did not appear to bind to ICAM-1Kilifi (Fig. 2A). This finding was confirmed by neutralization tests which showed that soluble ICAM-1 neutralized HRV16 but soluble ICAM-1Kilifi did not (Fig. 2B). These results are consistent with an inability to observe ICAM-1Kilifi bound to HRV16 in cryoEM image reconstructions (Table 1).
FIG. 2.
(A) Virus-receptor binding by capture ELISA. Virus was added to wells containing immobilized soluble receptor molecules or BSA (Mock) and detected by using specific primary and HRP-conjugated secondary antibodies. Bound material was visualized by the activity of HRP on the substrate 1,2-ortho-phenylene-diamine and quantified by absorbance at 490 nm. (B) Virus neutralization assays. The effect on the titer of HRV14 or HRV16 is shown after treatment with soluble receptor; antisera against HRV1B (anti-1B), HRV14 (anti-14), or HRV16 (anti-16), or BSA.
Conversely, HRV14 appeared to bind well to immobilized ICAM-1Kilifi but not to ICAM-1 (Fig. 2A). However, in neutralization tests, although HRV14 was neutralized most efficiently by ICAM-1Kilifi, it was also effectively neutralized by ICAM-1 (Fig. 2B).
These findings may be explained by a previous study which showed that HRV14 bound transiently to soluble ICAM-1 and was released from the receptor as the virion underwent the conversion to 135S and 80S particles, as occurs during cell entry (26). Essentially, the greater affinity of ICAM-1 to HRV14 increases the instability of the virus, causing it to convert to 135S and 80S particles which do not bind ICAM-1. Therefore, although the HRV14-ICAM-1 interaction may be a more effective event in terms of uncoating, the converted particle will be released from the immobilized receptor and not be detected by ELISA. In contrast, the binding of HRV14 to ICAM-1Kilifi appeared to form a stable complex without inducing particle conversion and release.
The structure of the virus-receptor complexes.
The cryoEM reconstructions of HRVs complexed with ICAM-1 reported here had a resolution of 8.6 to 13.1 Å (Table 1), whereas earlier results were around 28 Å (5, 29, 37). The improvement is due to using at least 40 times more particles in the present reconstruction and to improved computing techniques, especially for CTF corrections (8). The improved cryoEM maps show surface features (Fig. 1) that increased the accuracy with which the ICAM-1 X-ray structures could be fitted into the cryoEM maps. Although soluble ICAM-1 has five IgSF domains, only the densities of the first two domains and part of the third domain were evident in the cryoEM maps (Fig. 1), which is consistent with earlier results (29). The sumf value of domain D1 fitted into the two HRV14 complexes is about as high as that of the protein shell, suggesting that the ICAM-1 molecule fully occupies all 60 sites around the virus. However, in the complex of HRV16 with ICAM-1, the sumf value of domain D1 is only about 70% of that of the protein shell of the virus, suggesting a lower occupancy of the ICAM-1 molecule (Tables 2 and 3). The sumf values of domain D2 are approximately 0.8, 1.0, and 0.8 of domain D1 in HRV16-ICAM-1, HRV14-ICAM-1, and HRV14-ICAM-1Kilifi, respectively (Table 2). The ratios of the average density of domain D3 compared to domain D1 are approximately 0.7, 0.7, and 0.3 in the three complexes, respectively. The declining density along the length of the ICAM-1 molecule is most probably due to the flexibility of the ICAM-1 molecule. Room temperature, instead of 4°C, had been used to incubate the virus and ICAM-1, which may also have increased the flexibility of the long ICAM-1 molecule (see Materials and Methods). The difference map in which the fitted structure of the D1 and D2 ICAM-1 domains were subtracted from the density shows the carbohydrate moieties, as well as the density of domain D3 (Fig. 1C). The four glycosylation sites in domain D2 have an average distance of 16 Å to their respective Cα atom, similar to values found for carbohydrate sites on Sindbis virus (40). The densities of the glycosylation sites associated with Asn118 and Asn175 on D2 at the hinge region between domains D1 and D2 are in contact with residues from domains D1 and D2 (Fig. 1C). Previous results had shown that glycosylation at Asn175 was critical to proper folding (29). The other two glycosylation sites located at the hinge region between domain D2 and domain D3 have low densities and can barely be detected at the contour level in Fig. 1C, presumably due to greater flexibility at the distal end of the receptor.
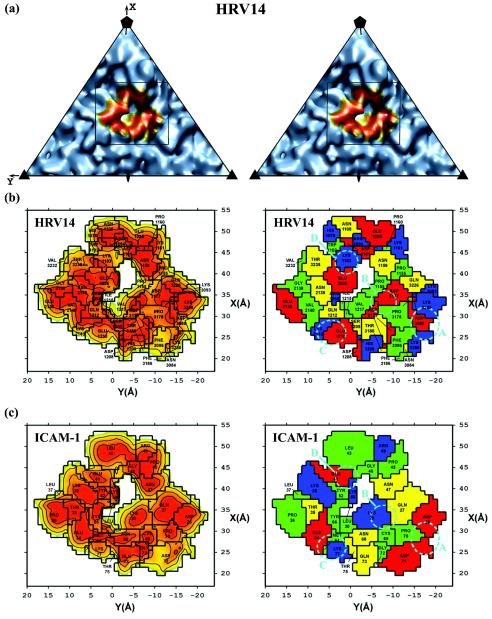
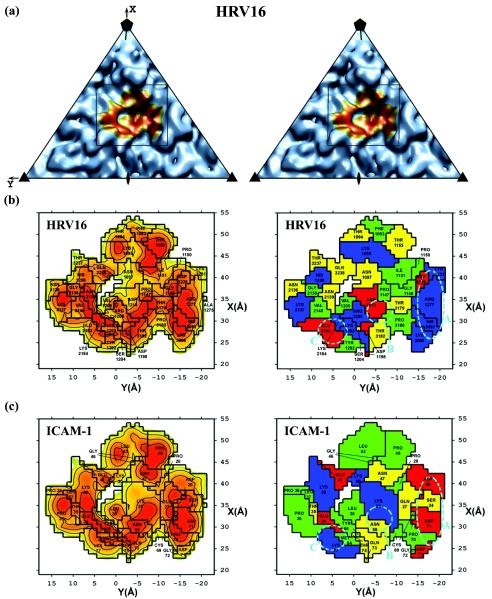
The ICAM-1 domain D1 was found to be situated 3 to 5 Å deeper in the canyon and approximately 1 Å closer to the southern rim of the canyon than reported earlier for both HRV14 and HRV16 (29). There was less than 1° of rotation about the long axis of domain D1 between the present and previous fits. The closer contact between virus and receptor produced a buried surface area of about 1,350 Å2, approximately 1.5 times larger and covering about five more residues (Table 5 and Fig. 3) than was previously found (29). This area is consistent with other virus-receptor interactions and considerably larger than the area of contact between the receptor and its normal cellular ligands (55). A comparison of the three different virus-receptor complexes reported here showed that domain D1 had an rms displacement of 1.4 to 2.0 Å between equivalent Cα atoms. However, for domain D2, the three fitted structures have an rms displacement of 3.9 to 6.8 Å between equivalent Cα atoms. Moreover, domain D2 had a 1° tilt in the HRV16-ICAM-1 complex compared to domain D2 in the HRV14-ICAM-1 and HRV14-ICAM-1Kilifi complexes.
FIG. 3.
Road maps of the ICAM-1 contact area with HRV14 (left page) and HRV16 (right page). Corresponding panels show the same information for the indicated virus. Panel a is a stereo diagram of the viral surface simulated from the crystal structure coordinates within the icosahedral asymmetric unit. The rectangle indicates the area that is enlarged in panel b. The orange region in panel A corresponds to the surface region that makes contact with ICAM-1. On the left side of panels b and c are shown the surface areas of the virus or ICAM-1, respectively, with contours corresponding to separation distances of 2.0 (red), 3.0 (orange), and 4.0 (yellow) Å, representing the closest approach atoms in the receptor and in the virus. On the right side of panels b and c are shown the amino acids involved in the virus-receptor contacts. The residues are coded by their properties; positively charged (blue), negatively charged (red), polar (yellow), and hydrophobic (green) residues are shown. Shown also are the ionic networks A, B, C, and D circled by cyan dashed lines. Whereas panel b shows the surface of the virus, panel c shows the complementary surface of ICAM-1. Note the charge complementarity of the two opposing surfaces.
The elbow angles between domains D1 and D2, calculated from the separately fitted X-ray structures, are within the limits determined by crystallography of the angular range given by the available structures (Table 4). However, the twist angles between domains have a greater variability than in the X-ray structures. This may be due to the long, thin shape of ICAM-1 domains, allowing for considerable variation in rotation about their long axes without showing significant differences in the quality of the fit. The larger error that incurred when fitting domain D2 (see above) into relatively weaker density also may have contributed to the greater variability of the twist angle.
Pocket factor.
In fitting the virus structure into the cryoEM density, it is necessary to determine whether it should be the structure of the virus with or without a pocket factor. Purified and crystallized HRV14 is normally without a pocket factor (1, 19, 47), as presumably the pocket factor does not bind well and has been washed out during the purification process. Thus, the fitting of the HRV14 structure to the cryoEM density of HRV14-ICAM-1 complexes assumed that no pocket factor is present. In contrast, HRV16 usually contains a pocket factor in crystallized preparations (18, 36). However, it has been hypothesized that the binding of ICAM-1 to HRVs displaces the pocket factor (see above). On the other hand, it has also been suggested that the cryoEM results represent an early event where the receptor has not penetrated to the base of the canyon and, thus, has not yet displaced the pocket factor (29). Belnap et al. (6) suggest that the pocket is probably empty in a 21-Å resolution cryoEM map of poliovirus receptor (CD155) bound to poliovirus. This is surprising both because the pocket should be collapsed when empty and because it should be difficult to differentiate a full from an empty pocket at the resolution of their studies. To check whether it is possible to determine the status of the pocket, 8.6-Å resolution maps of HRV14 were calculated by assuming an empty and collapsed conformation, an empty and open conformation, or a full and open conformation of the pocket. These maps were calculated without any attempt to simulate a phasing or amplitude error, yet no significant difference could be detected. Thus, the state of the pocket cannot be differentiated, even at the greatly improved resolution of the present work. Nevertheless, there would be additional side chain steric interference between virus and ICAM-1 structures if the open pocket conformation were assumed. Hence, for the purpose of fitting the virus to the cryoEM density, it was assumed that HRV16 still contains a pocket factor in the expectation that ICAM-1 has not yet reached the floor of the canyon (29).
The absence of the pocket factor in purified preparations of HRV14 (1, 19, 47) presumably contributes to the instability of the virus-ICAM-1 complex, whereas the presence of the pocket factor in purified preparations of HRV16 may contribute to the ability of this serotype to form a stable virus-receptor complex (26).
Ionic and hydrophobic interaction across the virus-receptor interface.
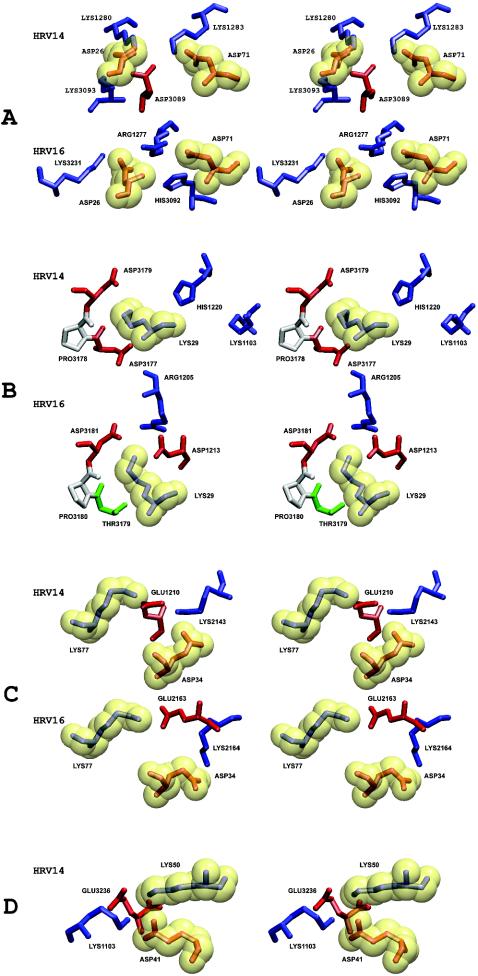
It was shown previously that the interface between ICAM-1 and HRVs has extensive charge complementarity (29). Similar charge complementarity has been observed between other picornaviruses and their receptors (20, 21, 57). However, in the higher-resolution structures presented here, it is apparent that many of these charge interactions are not separate salt bridges but are associated with local regions of ionic networks (Table 6 and Fig. 4). This is also true for the interaction between HRV2 and the very-low-density lipoprotein receptor, for which a 3.5-Å resolution structure shows an ionic network involving a Ca2+ ion (54). Mutations of charged residues of ICAM-1 that are in the HRV14-receptor interface cause a decrease of the binding affinity from 13 to 66% (51). Of the four ion-pair networks that exist at the interface between ICAM-1 and HRV14, there are three equivalent networks in the interface between ICAM-1 and HRV16, suggesting a stronger interaction between ICAM-1 and HRV14 than between ICAM-1 and HRV16.
TABLE 6.
Ion-pair networks formed between ICAM-1 and HRVsa
| Ion-pair group | Residue(s) of ICAM-1 | HRV14
|
HRV16
|
||
|---|---|---|---|---|---|
| Residue | Electrostatic potential energy (kcal/mol) of group | Residue | Electrostatic potential energy (kcal/mol) of group | ||
| A | Asp26, Asp71 | Lys1280 | Arg1277 | ||
| Lys1283 | |||||
| Lys3089 | |||||
| Asp3093 | His3092 | ||||
| Lys3231 | |||||
| −180.4 | −250.0 | ||||
| B | Lys29 | His1220 | Arg1205 | ||
| Asp1213 | |||||
| Asp3177 | |||||
| Asp3179 | Asp3181 | ||||
| −117.5 | −95.5 | ||||
| C | Glu34, Lys77 | Glu1210 | |||
| Lys2143 | |||||
| Glu2163 | |||||
| Lys2164 | |||||
| −160.3 | −106.1 | ||||
| D | Glu41, Lys50 | Lys1103 | |||
| Glu3236 | |||||
| −127.3 | −16.4 | ||||
Equivalent HRV14 and HRV16 residues are aligned. Electrostatic potential energies for each group of interactions are shown.
FIG. 4.
Stereo diagrams showing details of each of the ionic networks (A, B, C, and D) in the virus-receptor interface. Basic (blue), acidic (red), polar (green), and hydrophobic (gray) residues are shown. To differentiate residues in the virus to those in ICAM-1, ICAM-1 residues are enclosed in a transparent yellow van der Waals surface.
Stronger binding of ICAM-1 to HRV14 than to HRV16 was also evident in terms of the surface area of contact and the interaction potential energy (Table 5). ICAM-1 has an approximately 160-Å2 (or 12%) larger contact surface with HRV14 than with HRV16. The additional contact area is mainly due to the additional ionic network (Table 6) and the longer VP1 B-C loop (residue 1088 to residue 1097; residues are numbered starting from 1001, 2001, and 3001 for VP1, VP2, and VP3, respectively) (Fig. 5) in HRV14 compared to HRV16. Similarly, the interaction potential energy was greater for the interface of ICAM-1 with HRV14 than with HRV16 (Table 5). The less than complete occupancy of ICAM-1, when complexed with HRV16 under the same conditions of incubation as were used for the study of the HRV14 complex (Table 1), further supports the weaker binding of receptor to HRV16.
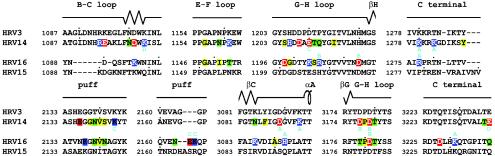
FIG. 5.
Alignment of HRV3, HRV14, HRV15, and HRV16 sequences in the vicinity of the virus-receptor contact areas. The amino acid sequence of HRV14 is closely similar to that of HRV3, as is the sequence of HRV16 to HRV15. The secondary structural elements are shown above the sequences. The “puff” region is a flexible external loop in VP2 (44). Residues in the virus-receptor interface are highlighted with colors based on the residue property. Basic (blue), acidic (red), polar (green), and hydrophobic (yellow) residues are shown. Residues that are involved in forming ionic networks are indicated by the cyan letters of A, B, C, and D, corresponding to the notation used in Fig. 3 and Table 6.
In addition to the electrostatic interactions, there are also extensive hydrophobic interactions between ICAM-1 and HRVs (Table 5). Of special interest is Pro70 of ICAM-1 that is stacked on top of Pro3178 in HRV14 or on top of the equivalent Pro3180 in HRV16. When Pro70 is mutated to almost any other amino acid, the binding between receptor and virus is eliminated (32, 41). Another hydrophobic residue, Phe3086 in HRV14 (Tyr3086 in HRV3), is close to the Pro70-Pro3178 interaction, which may further enhance the local hydrophobic environment. However, a charged residue, Asp3088, is adjacent to the Pro70-Pro3178 interaction in HRV16, which might interfere with the hydrophobic interaction, again suggesting that the binding of ICAM-1 to HRV14 is stronger than that to HRV16.
Mutational studies of Lys29.
Among the seven different charged residues belonging to ICAM-1 that are involved in forming ionic networks with HRVs, Lys29 is of particular interest. This residue is a Met in ICAM-1Kilifi and is the only difference between ICAM-1 and ICAM-1Kilifi. Although the Lys29Met mutation has little effect on ICAM-1 binding to HRV14, binding to HRV16 is eliminated. Similarly, a Lys29Leu mutant of ICAM-1 binds to HRV14, as well as to the closely homologous HRV3 virus (60), but not to HRV15, which has amino acid residues homologous to those in the HRV16-receptor contact regions (41) (Fig. 5). Furthermore, a Lys29Glu mutation has little impact on ICAM-1 binding to HRV3 (32). In the virus-receptor interface, Lys29 is surrounded by Asp3181, Asp1213, and Arg1205 in HRV16 and by Asp3179, Asp3177, His1220, and Lys1103 in HRV14. Thus, replacement of the positively charged Lys29 by a neutral or negatively charged residue might be expected to affect both HRV14 and HRV16. As this is not the case, the absence of a salt bridge to ICAM-1 Lys29 apparently reduces the binding interaction by a far greater proportion of the total binding energy in HRV16 than in HRV14. This result is consistent with the greater number of charge interactions, surface area of contact, and potential energy of interaction that ICAM-1 makes with HRV14 than with HRV16.
Although Lys29 of ICAM-1 makes charge interactions in both the HRV14- and HRV16-receptor complexes, the environment of Lys29 is rather different. Mutation of Lys29 to Met would create a steric problem in the HRV16 complex, where the pocket factor is assumed to be present, but not in the HRV14 complex, where the pocket factor is less well bound. Thus, the apparently stronger attachment of the pocket factor to HRV16 will further contribute to the inability of ICAM-1Kilifi to bind to HRV16.
The biochemical studies reported here (Fig. 2) suggest that ICAM-1 binds more effectively to HRV14 than to HRV16, which is consistent with the cryoEM results presented here. These observations are further supported by mutational studies, surface area calculations, energy calculations, the extent of ionic networks, and studies of the impact of the pocket factor. It would, therefore, seem possible that individuals who are homozygous for ICAM-1Kilifi might be protected from infection by HRV16 and other similar types of HRVs.
Acknowledgments
We thank Richard Kuhn, Carol Post, Tim Baker, Yumin Li, Suchetana (Tuli) Mukhopadhyay, Wei Zhang, and Xing Zhang for helpful discussions and Cheryl Towell and Sharon Wilder for preparation of the manuscript.
This work was support by an NIH grant to M.G.R. (AI 11219), a Keck Foundation Grant to M.G.R. (1419991440) for the purchase of the Philips CM300 FEG microscope, and a reinvestment grant from Purdue University.
REFERENCES
- 1.Arnold, E., and M. G. Rossmann. 1990. Analysis of the structure of a common cold virus, human rhinovirus 14, refined at a resolution of 3.0 Å. J. Mol. Biol. 211:763-801. [DOI] [PubMed] [Google Scholar]
- 2.Badger, J., I. Minor, M. J. Kremer, M. A. Oliveira, T. J. Smith, J. P. Griffith, D. M. A. Guerin, S. Krishnaswamy, M. Luo, M. G. Rossmann, M. A. McKinlay, G. D. Diana, F. J. Dutko, M. Fancher, R. R. Rueckert, and B. A. Heinz. 1988. Structural analysis of a series of antiviral agents complexed with human rhinovirus 14. Proc. Natl. Acad. Sci. USA 85:3304-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker, T. S., and R. H. Cheng. 1996. A model-based approach for determining orientations of biological macromolecules imaged by cryoelectron microscopy. J. Struct. Biol. 116:120-130. [DOI] [PubMed] [Google Scholar]
- 4.Baker, T. S., N. H. Olson, and S. D. Fuller. 1999. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 63:862-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bella, J., P. R. Kolatkar, C. W. Marlor, J. M. Greve, and M. G. Rossmann. 1998. The structure of the two amino-terminal domains of human ICAM-1 suggests how it functions as a rhinovirus receptor and as an LFA-1 integrin ligand. Proc. Natl. Acad. Sci. USA 95:4140-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belnap, D. M., B. M. McDermott, Jr., D. J. Filman, N. Cheng, B. L. Trus, H. J. Zuccola, V. R. Racaniello, J. M. Hogle, and A. C. Steven. 2000. Three-dimensional structure of poliovirus receptor bound to poliovirus. Proc. Natl. Acad. Sci. USA 97:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berendt, A. R., A. McDowall, A. G. Craig, P. A. Bates, M. J. E. Sternberg, K. Marsh, C. I. Newbold, and N. Hogg. 1992. The binding site on ICAM-1 for Plasmodium falciparum-infected erythrocytes overlaps, but is distinct from, the LFA-1 binding site. Cell 68:71-81. [DOI] [PubMed] [Google Scholar]
- 8.Bowman, V. D., E. S. Chase, A. W. Franz, P. R. Chipman, X. Zhang, K. L. Perry, T. S. Baker, and T. J. Smith. 2002. An antibody to the putative aphid recognition site on cucumber mosaic virus recognizes pentons but not hexons. J. Virol. 76:12250-12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brünger, A. T., P. D. Adams, G. M. Clore, W. L. DeLano, P. Gros, R. W. Grosse-Kunstleve, J. S. Jiang, J. Kuszewski, M. Nilges, N. S. Pannu, R. J. Read, L. M. Rice, T. Simonson, and G. L. Warren. 1998. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. Sect. D Biol. Crystallogr. 54:905-921. [DOI] [PubMed] [Google Scholar]
- 10.Casasnovas, J. M., T. Stehle, J. Liu, J. Wang, and T. A. Springer. 1998. A dimeric crystal structure for the N-terminal two domains of intercellular adhesion molecule-1. Proc. Natl. Acad. Sci. USA 95:4134-4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chothia, C., and E. Y. Jones. 1997. The molecular structure of cell adhesion molecules. Annu. Rev. Biochem. 66:823-862. [DOI] [PubMed] [Google Scholar]
- 12.Craig, A., D. Fernandez-Reyes, M. Mesri, A. McDowall, D. C. Altieri, N. Hogg, and C. Newbold. 2000. A functional analysis of a natural variant of intercellular adhesion molecule-1 (ICAM-1Kilifi). Hum. Mol. Genet. 9:525-530. [DOI] [PubMed] [Google Scholar]
- 13.Crowther, R. A. 1971. Procedures for three-dimensional reconstruction of spherical viruses by Fourier synthesis from electron micrographs. Phil. Trans. R. Soc. Lond. B Biol. Sci. 261:221-230. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Reyes, D., A. G. Craig, S. A. Kyes, N. Peshu, R. W. Snow, A. R. Berendt, K. Marsh, and C. I. Newbold. 1997. A high frequency African coding polymorphism in the N-terminal domain of ICAM-1 predisposing to cerebral malaria in Kenya. Hum. Mol. Genet. 6:1357-1360. [DOI] [PubMed] [Google Scholar]
- 15.Filman, D. J., R. Syed, M. Chow, A. J. Macadam, P. D. Minor, and J. M. Hogle. 1989. Structural factors that control conformational transitions and serotype specificity in type 3 poliovirus. EMBO J. 8:1567-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greve, J. M., G. Davis, A. M. Meyer, C. P. Forte, S. C. Yost, C. W. Marlor, M. E. Kamarck, and A. McClelland. 1989. The major human rhinovirus receptor is ICAM-1. Cell 56:839-847. [DOI] [PubMed] [Google Scholar]
- 17.Greve, J. M., C. P. Forte, C. W. Marlor, A. M. Meyer, H. Hoover-Litty, D. Wunderlich, and A. McClelland. 1991. Mechanisms of receptor-mediated rhinovirus neutralization defined by two soluble forms of ICAM-1. J. Virol. 65:6015-6023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hadfield, A. T., W. Lee, R. Zhao, M. A. Oliveira, I. Minor, R. R. Rueckert, and M. G. Rossmann. 1997. The refined structure of human rhinovirus 16 at 2.15 Å resolution: implications for the viral life cycle. Structure 5:427-441. [DOI] [PubMed] [Google Scholar]
- 19.Hadfield, A. T., M. A. Oliveira, K. H. Kim, I. Minor, M. J. Kremer, B. A. Heinz, D. Shepard, D. C. Pevear, R. R. Rueckert, and M. G. Rossmann. 1995. Structural studies on human rhinovirus 14 drug-resistant compensation mutants. J. Mol. Biol. 253:61-73. [DOI] [PubMed] [Google Scholar]
- 20.He, Y., V. D. Bowman, S. Mueller, C. M. Bator, J. Bella, X. Peng, T. S. Baker, E. Wimmer, R. J. Kuhn, and M. G. Rossmann. 2000. Interaction of the poliovirus receptor with poliovirus. Proc. Natl. Acad. Sci. USA 97:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, Y., P. R. Chipman, J. Howitt, C. M. Bator, M. A. Whitt, T. S. Baker, R. J. Kuhn, C. W. Anderson, P. Freimuth, and M. G. Rossmann. 2001. Interaction of coxsackievirus B3 with the full-length coxsackievirus-adenovirus receptor. Nat. Struct. Biol. 8:874-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, Y., F. Lin, P. R. Chipman, C. M. Bator, T. S. Baker, M. Shoham, R. J. Kuhn, M. E. Medof, and M. G. Rossmann. 2002. Structure of decay-accelerating factor bound to echovirus 7: a virus-receptor complex. Proc. Natl. Acad. Sci. USA 99:10325-10329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heinz, B. A., R. R. Rueckert, D. A. Shepard, F. J. Dutko, M. A. McKinlay, M. Fancher, M. G. Rossmann, J. Badger, and T. J. Smith. 1989. Genetic and molecular analyses of spontaneous mutants of human rhinovirus 14 that are resistant to an antiviral compound. J. Virol. 63:2476-2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendry, E., H. Hatanaka, E. Fry, M. Smyth, J. Tate, G. Stanway, J. Santti, M. Maaronen, T. Hyypiä, and D. Stuart. 1999. The crystal structure of coxsackievirus A9: new insights into the uncoating mechanisms of enteroviruses. Structure 7:1527-1538. [DOI] [PubMed] [Google Scholar]
- 25.Hewat, E. A., E. Neumann, J. F. Conway, R. Moser, B. Ronacher, T. C. Marlovits, and D. Blaas. 2000. The cellular receptor to human rhinovirus 2 binds around the 5-fold axis and not in the canyon: a structural view. EMBO J. 19:6317-6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoover-Litty, H., and J. M. Greve. 1993. Formation of rhinovirus-soluble ICAM-1 complexes and conformational changes in the virion. J. Virol. 67:390-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karttunen, A., T. Poyry, O. Vaarala, J. Ilonen, T. Hovi, M. Roivainen, and T. Hyypiä. 2003. Variation in enterovirus receptor genes. J. Med. Virol. 70:99-108. [DOI] [PubMed] [Google Scholar]
- 28.Kim, S., T. J. Smith, M. S. Chapman, M. G. Rossmann, D. C. Pevear, F. J. Dutko, P. J. Felock, G. D. Diana, and M. A. McKinlay. 1989. Crystal structure of human rhinovirus serotype 1A (HRV1A). J. Mol. Biol. 210:91-111. [DOI] [PubMed] [Google Scholar]
- 29.Kolatkar, P. R., J. Bella, N. H. Olson, C. M. Bator, T. S. Baker, and M. G. Rossmann. 1999. Structural studies of two rhinovirus serotypes complexed with fragments of their cellular receptor. EMBO J. 18:6249-6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewis, J. K., B. Bothner, T. J. Smith, and G. Siuzdak. 1998. Antiviral agent blocks breathing of the common cold virus. Proc. Natl. Acad. Sci. USA 95:6774-6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacKerell, A. D. J., B. Brooks, C. L. I. Brooks, L. Nilsson, B. Roux, Y. Won, and M. Karplus. 1998. CHARMM: the energy function and its parameterization with an overview of the program, p. 271-277. In P. v. R. Schleyer (ed.), The encyclopedia of computational chemistry, vol. 1. John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 32.McClelland, A., J. deBear, S. C. Yost, A. M. Meyer, C. W. Marlor, and J. M. Greve. 1991. Identification of monoclonal antibody epitopes and critical residues for rhinovirus binding in domain 1 of ICAM-1. Proc. Natl. Acad. Sci. USA 88:7993-7997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muckelbauer, J. K., M. Kremer, I. Minor, G. Diana, F. J. Dutko, J. Groarke, D. C. Pevear, and M. G. Rossmann. 1995. The structure of coxsackievirus B3 at 3.5 Å resolution. Structure 3:653-667. [DOI] [PubMed] [Google Scholar]
- 34.Newcombe, N. G., P. Andersson, E. S. Johansson, G. G. Au, A. M. Lindberg, R. D. Barry, and D. R. Shafren. 2003. Cellular receptor interactions of C-cluster human group A coxsackieviruses. J. Gen. Virol. 84:3041-3050. [DOI] [PubMed] [Google Scholar]
- 35.Ockenhouse, C. F., R. Betageri, T. A. Springer, and D. E. Staunton. 1992. Plasmodium falciparum-infected erythrocytes bind ICAM-1 at a site distinct from LFA-1, Mac-1, and human rhinovirus. Cell 68:63-69. [DOI] [PubMed] [Google Scholar]
- 36.Oliveira, M. A., R. Zhao, W. Lee, M. J. Kremer, I. Minor, R. R. Rueckert, G. D. Diana, D. C. Pevear, F. J. Dutko, M. A. McKinlay, and M. G. Rossmann. 1993. The structure of human rhinovirus 16. Structure 1:51-68. [DOI] [PubMed] [Google Scholar]
- 37.Olson, N. H., P. R. Kolatkar, M. A. Oliveira, R. H. Cheng, J. M. Greve, A. McClelland, T. S. Baker, and M. G. Rossmann. 1993. Structure of a human rhinovirus complexed with its receptor molecule. Proc. Natl. Acad. Sci. USA 90:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Papi, A., and S. L. Johnston. 1999. Rhinovirus infection induces expression of its own receptor intercellular adhesion molecule 1 (ICAM-1) via increased NF-κB-mediated transcription. J. Biol. Chem. 274:9707-9720. [DOI] [PubMed] [Google Scholar]
- 39.Pevear, D. C., M. J. Fancher, P. J. Felock, M. G. Rossmann, M. S. Miller, G. Diana, A. M. Treasurywala, M. A. McKinlay, and F. J. Dutko. 1989. Conformational change in the floor of the human rhinovirus canyon blocks adsorption to HeLa cell receptors. J. Virol. 63:2002-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pletnev, S. V., W. Zhang, S. Mukhopadhyay, B. R. Fisher, R. Hernandez, D. T. Brown, T. S. Baker, M. G. Rossmann, and R. J. Kuhn. 2001. Locations of carbohydrate sites on Sindbis virus glycoproteins show that E1 forms an icosahedral scaffold. Cell 105:127-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Register, R. B., C. R. Uncapher, A. M. Naylor, D. W. Lineberger, and R. J. Colonno. 1991. Human-murine chimeras of ICAM-1 identify amino acid residues critical for rhinovirus and antibody binding. J. Virol. 65:6589-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roebuck, K. A., and A. Finnegan. 1999. Regulation of intercellular adhesion molecule-1 (CD54) gene expression. J. Leukoc. Biol. 66:876-888. [DOI] [PubMed] [Google Scholar]
- 43.Rossmann, M. G. 1994. Viral cell recognition and entry. Protein Sci. 3:1712-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossmann, M. G., E. Arnold, J. W. Erickson, E. A. Frankenberger, J. P. Griffith, H. J. Hecht, J. E. Johnson, G. Kamer, M. Luo, A. G. Mosser, R. R. Rueckert, B. Sherry, and G. Vriend. 1985. Structure of a human common cold virus and functional relationship to other picornaviruses. Nature (London) 317:145-153. [DOI] [PubMed] [Google Scholar]
- 45.Rossmann, M. G., R. Bernal, and S. V. Pletnev. 2001. Combining electron microscopic with X-ray crystallographic structures. J. Struct. Biol. 136:190-200. [DOI] [PubMed] [Google Scholar]
- 46.Rossmann, M. G., Y. He, and R. J. Kuhn. 2002. Picornavirus-receptor interactions. Trends Microbiol. 10:324-331. [DOI] [PubMed] [Google Scholar]
- 47.Smith, T. J., M. J. Kremer, M. Luo, G. Vriend, E. Arnold, G. Kamer, M. G. Rossmann, M. A. McKinlay, G. D. Diana, and M. J. Otto. 1986. The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science 233:1286-1293. [DOI] [PubMed] [Google Scholar]
- 48.Smyth, M., J. Tate, E. Hoey, C. Lyons, S. Martin, and D. Stuart. 1995. Implications for viral uncoating from the structure of bovine enterovirus. Nat. Struct. Biol. 2:224-231. [DOI] [PubMed] [Google Scholar]
- 49.Springer, T. A. 1990. Adhesion receptors of the immune system. Nature (London) 346:425-434. [DOI] [PubMed] [Google Scholar]
- 50.Stanway, G., T. Hovi, N. J. Knowles, and T. Hyypiä. 2002. Molecular and biological basis of picornavirus taxonomy, p. 17-24. In B. L. Semler and E. Wimmer (ed.), Molecular biology of picornaviruses. ASM Press, Washington, D.C.
- 51.Staunton, D. E., M. L. Dustin, H. P. Erickson, and T. A. Springer. 1990. The arrangement of the immunoglobulin-like domains of ICAM-1 and the binding sites for LFA-1 and rhinovirus. Cell 61:243-254. [DOI] [PubMed] [Google Scholar]
- 52.van de Stolpe, A., and P. T. van der Saag. 1996. Intercellular adhesion molecule-1. J. Mol. Med. 74:13-33. [DOI] [PubMed] [Google Scholar]
- 53.Verdaguer, N., D. Blaas, and I. Fita. 2000. Structure of human rhinovirus serotype 2 (HRV2). J. Mol. Biol. 300:1181-1196. [DOI] [PubMed] [Google Scholar]
- 54.Verdaguer, N., I. Fita, M. Reithmayer, R. Moser, and D. Blaas. 2004. X-ray structure of a minor group human rhinovirus bound to a fragment of its cellular receptor protein. Nat. Struct. Mol. Biol. 11:429-434. [DOI] [PubMed] [Google Scholar]
- 55.Wang, J. 2002. Protein recognition by cell surface receptors: physiological receptors versus virus interactions. Trends Biochem. Sci. 27:122-126. [DOI] [PubMed] [Google Scholar]
- 56.Wang, J., and T. A. Springer. 1998. Structural specializations of immunoglobulin superfamily members for adhesion to integrins and viruses. Immunol. Rev. 163:197-215. [DOI] [PubMed] [Google Scholar]
- 57.Xiao, C., C. M. Bator, V. D. Bowman, E. Rieder, Y. He, B. Hébert, J. Bella, T. S. Baker, E. Wimmer, R. J. Kuhn, and M. G. Rossmann. 2001. Interaction of coxsackievirus A21 with its cellular receptor, ICAM-1. J. Virol. 75:2444-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xing, L., J. M. Casasnovas, and R. H. Cheng. 2003. Structural analysis of human rhinovirus complexed with ICAM-1 reveals the dynamics of receptor-mediated virus uncoating. J. Virol. 77:6101-6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xing, L., M. Huhtala, V. Pietiainen, J. Kapyla, K. Vuorinen, V. Marjomaki, J. Heino, M. S. Johnson, T. Hyypiä, and R. H. Cheng. 2004. Structural and functional analysis of integrin alpha2I domain interaction with echovirus 1. J. Biol. Chem. 279:11632-11638. [DOI] [PubMed] [Google Scholar]
- 60.Zhao, R., D. C. Pevear, M. J. Kremer, V. L. Giranda, J. A. Kofron, R. J. Kuhn, and M. G. Rossmann. 1996. Human rhinovirus 3 at 3.0 Å resolution. Structure 4:1205-1220. [DOI] [PubMed] [Google Scholar]