Abstract
The human polyomavirus JC (JCV) replicates in the nuclei of infected cells. Here we report that JCV virions are efficiently assembled at nuclear domain 10 (ND10), which is also known as promyelocytic leukemia (PML) nuclear bodies. The major capsid protein VP1, the minor capsid proteins VP2 and VP3, and a regulatory protein called agnoprotein were coexpressed from a polycistronic expression vector in COS-7 cells. We found that VP1 accumulated to distinct subnuclear domains in the presence of VP2/VP3 and agnoprotein, while VP1 expressed alone was distributed both in the cytoplasm and in the nucleus. Mutation analysis revealed that discrete intranuclear accumulation of VP1 requires the presence of either VP2 or VP3. However, VP2 or VP3 expressed in the absence of VP1 showed diffuse, not discrete, nuclear localization. The C-terminal sequence of VP2/VP3 contains two basic regions, GPNKKKRRK (cluster 1) and KRRSRSSRS (cluster 2). The deletion of cluster 2 abolished the accumulation of VP1 to distinct subnuclear domains. Deletion of the C-terminal 34 residues of VP2/VP3, including both cluster 1 and cluster 2, caused VP1 to localize both in the cytoplasm and in the nucleus. Using immunoelectron microscopy of cells that coexpressed VP1, VP2/VP3, and agnoprotein, we detected the assembly of virus-like particles in discrete locations along the inner nuclear periphery. Both in oligodendrocytes of the human brain and in transfected cells, discrete nuclear domains for VP1 accumulation were identified as ND10, which contains the PML protein. These results indicate that major and minor capsid proteins cooperatively accumulate in ND10, where they are efficiently assembled into virions.
Progressive multifocal leukoencephalopathy is a fatal demyelinating disorder of the central nervous system caused by infection with the human polyomavirus JC (JCV). Infected oligodendrocytes have enlarged nuclei, in which JCV virions have been identified by electron microscopy as spherical or filamentous structures (40, 46). JCV has shown quite variable distribution patterns in the nucleus, as follows. The spherical virions, which are quite uniform in size, may form clusters in which they are perfectly aligned in a crystalloid array. Filamentous viruses may be aligned in strands or spirals, as if they are stretched along matrix-like structures. There are also many cells in which spherical and/or filamentous forms are scattered randomly in the nucleus. Based on extensive observation of a patient's brain, it has been postulated that the first virus progeny usually appear in the vicinity of the nuclear membrane (29). However, it is largely unknown how JCV replicates in the nucleus.
JCV has a double-stranded genomic DNA in a capsid that is likely composed of the major capsid protein VP1 and the minor capsid proteins VP2 and VP3 in an appropriate ratio. Its fine structure seems similar to those of genetically analogous simian virus 40 (SV40) and mouse polyomavirus (PyV), which have been analyzed by crystallography (9, 25, 41, 42). During the late stage of the virus replication cycle, the capsid proteins of these polyomaviruses are synthesized in the cytoplasm and then transported to the nucleus. JCV VP1 has a functionally weak nuclear localization signal (NLS), KRK-RK, and thus requires other viral proteins for nuclear transport (38); in contrast, in SV40 and PyV, each capsid protein contains an effective NLS (7, 8, 10, 22, 30). Despite the presence of independent NLSs, however, the major and minor capsid proteins of SV40 and PyV are believed to associate in the cytoplasm and then be cotransported to the nucleus (3, 5, 14, 15, 19, 23). Similarly, JCV VP1 may be cotransported with the minor capsid proteins VP2 and/or VP3. However, the expression of JCV VP2/VP3 has not yet been detected with specific antibodies, and the functions of JCV VP2/VP3 are unknown.
How and where in the nucleus are the JCV virions assembled? Recently, it has been reported that L1 and L2 capsid proteins of human papillomavirus type 33 (HPV33) and bovine papillomavirus accumulate in nuclear domain 10 (ND10), which is also known as the promyelocytic leukemia (PML) nuclear bodies (12, 17, 18), suggesting that virion assembly occurs at this domain. If so, is there any subnuclear domain that, during JCV infection, supports the efficient assembly of virions? If it exists, how does JCV VP1, which has a weak NLS, accumulate in this subnuclear domain? VP1 may be transported from the cytoplasm to the nucleus in association with VP2 and/or VP3 and may soon cotranslocate to the subnuclear domain. If so, does VP2 or VP3 have an effective targeting signal? Is the signal distinct from an NLS? For the polyomaviruses, many reports describe the transport of the capsid proteins from the cytoplasm to the nucleus (3, 5, 7, 8, 10, 14, 15, 19, 22, 23, 30). However, few studies have focused on what subsequently occurs after entry into the nucleus.
For this study, therefore, we investigated the functions of the capsid proteins, especially for intranuclear accumulation and subsequent assembly into virions. JCV, by its nature, replicates slowly and inefficiently in culture systems, and this has been a major barrier to studying the capsid proteins. Unlike the case for SV40, initial studies reported that JCV late RNAs encoding the capsid proteins were not detected until 10 days after infection of primary human fetal glial cells (11), and the VP1 protein was not detected until 2 weeks after infection of a human neuroblastoma cell line, IMR-32 (1). To overcome this difficulty, we established a eukaryotic expression system to produce virus-like particles under the control of the powerful SRα promoter (44). A viral genome fragment encoding VP1, VP2, VP3, and a regulatory protein called agnoprotein was placed downstream of the SRα promoter, and a polycistronic expression vector, AVP231-SRα, was constructed (37, 38). The viral proteins were expressed in COS-7 cells, in which the promoter functions most efficiently in the presence of SV40 T antigen. This system enabled us to analyze the capsid proteins at 3 days posttransfection, and the mechanisms of nuclear transport and intranuclear accumulation were investigated. We also identified nuclear domains relating to virion assembly in oligodendrocytes of a human brain with progressive multifocal leukoencephalopathy.
MATERIALS AND METHODS
Plasmids.
The JCV Tokyo-1 strain has been described elsewhere (28, 32, 38) (GenBank accession number AF030085). The expression vectors AVP231-SRα and VP1-SRα contain genomic fragments of JCV Tokyo-1 (37, 38). The sequences of the oligonucleotide primers used for this study will be provided upon request.
(i) MUT-1 to MUT-7.
Mutant vectors MUT-1 through MUT-7 were derived from AVP231-SRα by mutating the ATG start codons for agnoprotein, VP2, and VP3 in all possible combinations. First, a viral genomic fragment (nucleotides [nt] 375 to 1720; 1,446 bp) including ATG start codons for all three proteins was cloned into PstI and EcoRI sites of pUC19, resulting in PstEco1446-pUC. Each of the ATG start codons in the insert was mutated to TTG by use of a QuikChange site-directed mutagenesis kit (Stratagene). After mutagenesis, the sequences of the entire 1,446-bp fragments were confirmed, and they were used to replace the corresponding PstI-EcoRI fragment in AVP231-SRα.
(ii) MUT-8 to MUT-11.
MUT-8 through MUT-11 bear a TAA stop mutation in either of two codons in the C-terminal region of VP2/VP3. Within PstEco1446-pUC, a sense codon was replaced with a TAA stop codon (STOP-1 or STOP-2; the locations are shown in Fig. 8A) by use of a QuikChange site-directed mutagenesis kit (Stratagene). For the construction of MUT-9 and MUT-11, the ATG start codon for agnoprotein was also mutated to TTG. After mutagenesis, the sequences of the entire 1,446-bp fragments were confirmed, and then the fragments replaced the corresponding fragment in AVP231-SRα.
FIG. 8.
Immunocytochemistry of cells transfected with truncation mutant vectors (MUT-8 to MUT-11). (A and B) Double staining of VP1 and VP2/VP3 (A) and of VP1 and agnoprotein (B) in cells transfected with MUT-8. (C) Double staining of VP1 and VP2/VP3 in cells transfected with MUT-9. (D and E) Double staining of VP1 and VP2/VP3 (D) and of VP1 and agnoprotein (E) in cells transfected with MUT-10. (F) Double staining of VP1 and VP2/VP3 in cells transfected with MUT-11. VP1 was visualized with Alexa fluor 488 (green), and VP2/VP3 or agnoprotein was visualized with Alexa fluor 568 (red). In panels D and F, truncated VP2/VP3 was not detected because of the deletion of the antigenic sequence for the anti-VP2/VP3C antibody.
(iii) VP2/VP3, VP2/M120L, VP3, Del-1, Del-2, and Del-3.
For the construction of expression vectors for VP2/VP3 and VP3, the coding sequences of VP2/VP3 (nt 524 to 1558) and VP3 (nt 881 to 1558) were amplified by PCR. The sequences of the PCR products were confirmed, and they were cloned into PstI and KpnI sites of the pcDL-SRα296 expression vector (44). For the construction of VP2/M120L, the ATG start codon for VP3 in the plasmid VP2/VP3 was mutated to TTG. After sequencing, the fragment was cloned into the pcDL-SRα296 expression vector. For the construction of Del-1, the 27 nt encoding cluster 1 (see Fig. 7) were deleted from VP2/VP3 by use of a QuikChange site-directed mutagenesis kit (Stratagene). For Del-2, the C-terminal 27 nt encoding cluster 2 (Fig. 7) were deleted by PCR. Both of these deletions were introduced for the construction of Del-3. The mutated fragments or the PCR products were sequenced and then cloned into the pcDL-SRα296 expression vector.
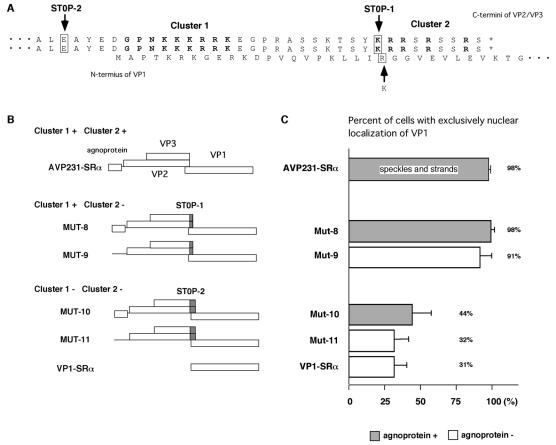
FIG. 7.
Roles of VP2/VP3 C-terminal sequence in nuclear localization of VP1. (A) C-terminal sequences of VP2 and VP3 and overlapping N-terminal sequence of VP1. STOP-1 and STOP-2 show the locations where TAA stop codons replaced the sense codons of VP2/VP3 in the MUT-8 to MUT-11 vectors derived from AVP231-SRα. STOP-1, a mutation of the first codon of cluster 2, also changes the 22nd amino acid of VP1 from arginine to lysine. (B) Structures of mutant vectors containing STOP-1 (MUT-8 and MUT-9) and STOP-2 (MUT-10 and MUT-11). MUT-8 and MUT-9 encode cluster 1, but not cluster 2. MUT-10 and MUT-11 lack the C-terminal 34 residues of VP2/VP3, including both cluster 1 and cluster 2. MUT-8 and MUT-10 encode agnoprotein, but MUT-9 and MUT-11 do not. (C) Percentages of cells showing exclusively nuclear localization in cells transfected with MUT-8 to MUT-11. Speckled or stranded intranuclear localization patterns were not seen in cells transfected with these mutant vectors.
Cells and transfection.
COS-7 cells were incubated at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. COS-7 cells were transfected by the use of Lipofectamine Plus (GIBCO BRL) according to the manufacturer's procedure. Approximately 150 ng of DNA was transfected into 104 cells, and the cells were harvested 3 days after transfection.
Antibodies.
A rabbit polyclonal antibody against the potential BC loop structure of VP1 (anti-VP1BC antibody) has been described previously (38). An antibody against the potential HI loop structure of VP1 (anti-VP1HI antibody) (38, 39) was provided by K. Nagashima (Hokkaido University). A rabbit polyclonal antibody for VP2/VP3 (anti-VP2/VP3C antibody) was prepared against the C-terminal sequence RKEGPRASSKTSYKR, and an antibody for agnoprotein (anti-agnoC antibody) was prepared against the C-terminal sequence CKRQKHSGLTQQTYSA. An anti-PML antibody was obtained from MBL (Nagoya, Japan).
Immunohistochemistry.
JCV-infected human brain tissue was obtained from a patient with progressive multifocal leukoencephalopathy after autopsy at Aizawa Hospital (Matsumoto, Japan). Paraffin-embedded thin sections were deparaffinized with xylol and then rehydrated in 90, 70, and 50% ethanol. To expose the antigenic epitopes, we treated the sections in autoclaves for 20 min in antigen-unmasking solution (Vector Laboratories, Burlington, Calif.), washed them for 15 min with phosphate-buffered saline (PBS), and then used them for immunostaining in the manner described below.
Immunocytochemistry.
COS-7 cells were grown on tissue culture glass slides (Falcon) treated with poly-l-lysine. Transfected cells were washed with PBS, fixed in 4% paraformaldehyde in PBS for 15 min at room temperature, and then permeabilized with 0.5% Triton X-100 in PBS for 20 min at room temperature. After fixation and permeabilization, the cells were rinsed with PBS containing 0.05% Tween 20 and then blocked with PBS containing 5% normal goat serum at room temperature for 30 min. The cells were incubated with primary antibodies for 1 h, washed in PBS containing 0.05% Tween 20, and further incubated with the appropriate secondary antibody, either Alexa fluor 488-conjugated anti-rabbit immunoglobulin G (IgG) or Alexa fluor 568-conjugated anti-rabbit IgG. After three washes in PBS, the samples were mounted in VectaShield (Vector Laboratories). Fluorescent signals were observed and captured with a TCS-SP confocal laser scanning microscope (Leica, Heidelberg, Germany). The confocal images were sequentially acquired and superposed with Adobe Photoshop software.
Immunoelectron microscopy.
Cells transfected with AVP231-SRα were fixed at 72 h posttransfection in 4% paraformaldehyde in 0.1 M phosphate buffer, pH 7.4, for 15 min. The cells were dehydrated and embedded in LR White resin. Ultrathin sections were prepared and mounted on nickel grids. After incubation with 10% normal goat serum for 10 min, the sections were incubated overnight at 4°C with the anti-VP1HI antibody. After being washed with PBS, the sections were incubated with a mixture of a goat anti-rabbit IgG conjugated to 15-nm-diameter gold particles (1:30) for 30 min at room temperature. The sections were then washed with water and stained with uranyl acetate. The sections were examined with a Hitachi H-7100 electron microscope.
RESULTS
VP1 accumulates in distinct subnuclear domains in the presence of VP2/VP3 and agnoprotein.
To study the mechanism by which JCV replicates in the nucleus, we established a eukaryotic expression system to produce virus-like particles under the control of the powerful SRα promoter (37, 38). The expression vector AVP231-SRα contains a polycistronic fragment encoding agnoprotein, VP1, VP2, and VP3 (Fig. 1A). The major capsid protein VP1 and the minor capsid proteins VP2 and VP3 are encoded in an overlapping manner downstream of agnoprotein. Since the coding sequence of VP3 is identical to two-thirds of the C-terminal sequence of VP2, VP2 and VP3 will be described as VP2/VP3 when they are not distinguished.
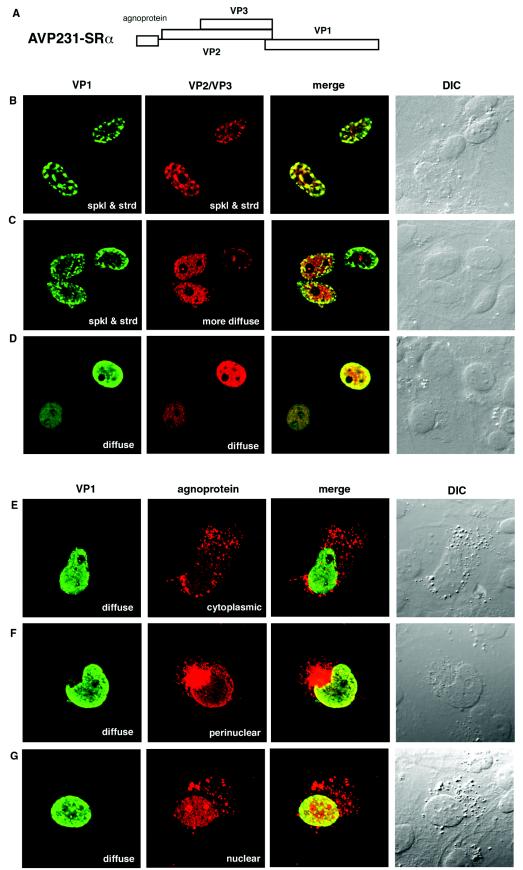
FIG.1.
Localization of VP1, VP2/VP3, and agnoprotein in COS-7 cells transfected with AVP231-SRα. (A) Schematic illustration of AVP231-SRα expression vector. AVP231-SRα encodes a polycistronic fragment for agnoprotein, VP1, VP2, and VP3. (B to G) Immunocytochemistry of transfected cells. COS-7 cells transfected with AVP231-SRα were fixed at 3 days posttransfection. VP1 was visualized with Alexa fluor 488 (green), and VP2/VP3 or agnoprotein was visualized with Alexa fluor 568 (red). (B to D) Double staining of VP1 and VP2/VP3. The intranuclear distribution of VP1 and VP2/VP3 was variable. Both VP1 and VP2/VP3 show speckled staining patterns in panel B, there is a speckled staining of VP1 and a more diffuse staining of VP2/VP3 in panel C, and both show diffuse staining patterns in panel D. (E to G) Double staining of VP1 and agnoprotein. Agnoprotein shows a cytoplasmic localization pattern in panel E, a perinuclear inclusion in panel F, and a predominantly nuclear localization in panel G. spkl & strd, speckled and stranded staining patterns.
The distribution of VP1 was examined by confocal microscopy of cells transfected with AVP231-SRα (Fig. 1B to G). In 98% of VP1-positive cells, VP1 was localized exclusively to the nucleus. There were two distinct intranuclear distribution patterns of VP1. Some cells showed a discrete accumulation of VP1 in speckles or strands, suggesting that VP1 localized to distinct subnuclear structures such as nuclear bodies or the nuclear matrix (Fig. 1B and C), while other cells showed a rather diffuse distribution of VP1 over large parts of the nucleus, excluding the nucleolus (Fig. 1D to G). Generally, the VP1 density was higher at the periphery of the nucleus and lower in the center (Fig. 1B to G). By the use of double staining, VP2/VP3 was also detected in the nucleus (Fig. 1B to D). The intranuclear distribution of VP2/VP3 was similar to that of VP1, but not identical. In some cells, VP2/VP3 was colocalized with VP1 in speckles or strands (Fig. 1B), while in other cells, VP2/VP3 was distributed more diffusely than VP1 (Fig. 1C). There were also cells in which both VP1 and VP2/VP3 were distributed diffusely (Fig. 1D). These observations may indicate that the JCV capsid proteins dynamically alter their locations in the nuclei of living cells.
The interactive localization of VP1 and agnoprotein was also examined (Fig. 1E to G). Since the expression level of agnoprotein is lower than that of VP1, only about 10% of VP1-positive cells were also positive for agnoprotein. In 70% of double-positive cells, agnoprotein was localized predominantly in the cytoplasm. Some cells showed diffuse and granular cytoplasmic distribution patterns of agnoprotein (Fig. 1E), while other cells showed large perinuclear inclusions (Fig. 1F). Agnoprotein was sometimes densely colocalized with VP1 at the margins of the nucleus, possibly the nuclear membrane or nuclear lamina (Fig. 1F), suggesting a potential role for agnoprotein in nuclear entry of VP1. In 30% of double-positive cells, agnoprotein was distributed diffusely in the nucleus, with dense localization in the nucleoli (Fig. 1G). When agnoprotein was seen in the nucleus, VP1 was usually distributed diffusely, not discretely. Thus, agnoprotein may change its localization in living cells, possibly associated with the intranuclear localization of VP1.
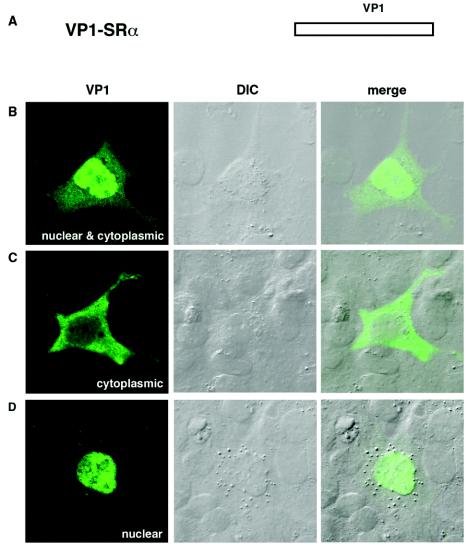
To test whether the localization of VP1 depends on other late proteins, we transfected cells with VP1-SRα, an expression vector encoding only VP1 (Fig. 2A). In these cells, VP1 distribution was markedly more cytoplasmic (Fig. 2B to D). In about 69% of the cells, VP1 was distributed both in the nucleus and in the cytoplasm (Fig. 2B). Some cells showed more VP1 in the cytoplasm than in the nucleus (Fig. 2C). Only about 31% of VP1-positive cells showed exclusively nuclear localization; moreover, none of the cells showed distinct VP1 staining as speckles or strands (Fig. 2D). The different VP1 distribution in cells transfected with AVP231-SRα versus those transfected with VP1-SRα indicates that the localization of VP1 is highly dependent on the presence of VP2, VP3, or agnoprotein.
FIG. 2.
Localization of VP1 in COS-7 cells transfected with VP1-SRα. (A) Schematic illustration of VP1-SRα expression vector. VP1-SRα carries only the coding sequence for VP1. (B to D) Localization of VP1. COS-7 cells transfected with VP1-SRα were analyzed by immunocytochemistry. The localization of VP1 was visualized with Alexa fluor 488 (green). The VP1 distribution was variable among the cells: VP1 localized both in the cytoplasm and in the nucleus in panel B, predominantly in the cytoplasm in panel C, and in the nucleus in panel D.
The presence of either VP2 or VP3 is essential for accumulation of VP1 in distinct subnuclear structures.
To determine which protein is responsible for the exclusively nuclear localization of VP1 and for VP1 accumulation in distinct subnuclear structures, we prepared vectors to express VP2, VP3, and agnoprotein in all possible combinations by mutating the ATG translation start codons to TTG within AVP231-SRα. The resulting mutant vectors (MUT-1 through MUT-7) are shown schematically in Fig. 3A. The vectors were divided into four groups based on the presence or absence of the minor capsid proteins VP2 and VP3. Group 1 (AVP231-SRα and MUT-1) encodes both VP2 and VP3. Group 2 (MUT-2 and MUT-3) encodes VP2 but not VP3 while group 3 (MUT-4 and MUT-5) encodes VP3 but not VP2. Group 4 (MUT-6, MUT-7, and VP1-SRα) encodes neither VP2 nor VP3. Within the C-terminal sequence of VP2/VP3, there are two in-frame ATG methionine codons (nt 1208 to 1210 and nt 1406 to 1409). To avoid expression of the C-terminal partial sequence of VP2 and VP3, we also mutated these ATGs to TTG in MUT-6 and MUT-7. Each group includes vectors encoding agnoprotein (AVP231-SRα, MUT-2, MUT-4, and MUT-6) and those lacking agnoprotein (MUT-1, MUT-3, MUT-5, MUT-7, and VP1-SRα).
FIG. 3.
Effects of VP2, VP3, and agnoprotein on nuclear localization of VP1. (A) Schematic illustration of MUT-1 to MUT-7 mutant vectors. The mutant vectors were derived from AVP231-SRα by mutating the ATG initiation codons for agnoprotein, VP2, and/or VP3 to TTG. Oblique lines on the schematic illustration for MUT-6 and MUT-7 indicate a mutation of ATG codons in frame within the VP2/VP3 sequence. Based on the presence or absence of VP2 and VP3, the mutant vectors were divided into four groups (group 1 to group 4). (B) Percentages of cells showing exclusively nuclear localization of VP1. Cells were transfected with each vector, and VP1 localization was examined by immunofluorescence. The numbers of cells showing exclusively nuclear localization of VP1 were calculated as percentages of the total numbers of VP1-positive cells (range, 300 to 1,200), and the averages and standard deviations from three or more independent experiments are shown.
Each of the mutant vectors was transfected into cells, and VP1 localization was examined. In cells transfected with mutant vectors from groups 1, 2, and 3, the VP1 protein was exclusively localized to the nucleus in 95 to 98, 95, and 93 to 98% of VP1-positive cells, respectively. In contrast, in cells transfected with mutant vectors from group 4, only 31 to 34% of cells showed exclusively nuclear staining patterns. The percentage of cells showing exclusively nuclear staining was slightly higher in mutants encoding agnoprotein than in those lacking agnoprotein (Fig. 3B).
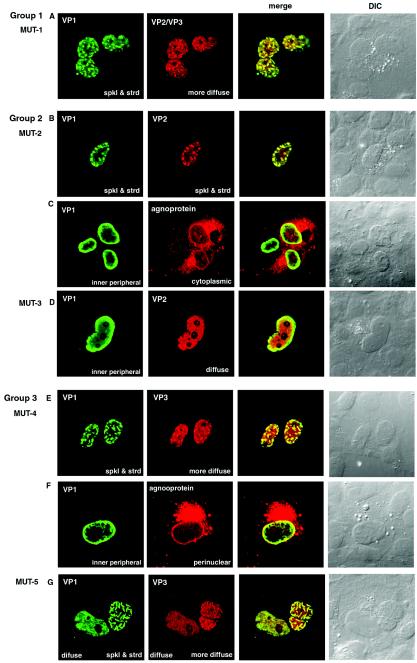
Representative cells transfected with each mutant vector are shown in Fig. 4 and 5. In cells transfected with vectors from group 1 (Fig. 4A), group 2 (Fig. 4B to D), and group 3 (Fig. 4E to G), the distribution patterns of VP1, VP2/VP3, and agnoprotein were basically similar to those transfected with AVP231-SRα. The VP1 protein was predominantly localized to the nucleus in the presence of either VP2 or VP3, and VP1 accumulated in distinct subnuclear structures. The discrete intranuclear accumulation of VP1 was seen more clearly in cells expressing VP3 (cells transfected with vectors from group 3) than in those expressing VP2 (cells transfected with vectors from group 2). In both types of cells, strong VP1 staining was occasionally seen at the nuclear membrane and/or inner nuclear periphery (Fig. 4C, D, and F). Agnoprotein was usually seen predominantly in the cytoplasm (Fig. 4C and F). The intranuclear accumulation of VP1 was clearer in cells transfected with expression vectors encoding agnoprotein than in those lacking agnoprotein.
FIG.4.
Immunocytochemistry of cells transfected with mutant vectors (MUT-1 to MUT-5). (A) Localization of VP1 and VP2/VP3 in cells transfected with MUT-1. (B and C) Double staining of VP1 and VP2 (B) and of VP1 and agnoprotein (C) in cells transfected with MUT-2. (D) Double staining of VP1 and VP2 in cells transfected with MUT-3. (E and F) Double staining of VP1 and VP3 (E) and of VP1 and agnoprotein (F) in cells transfected with MUT-4. (G) Double staining of VP1 and VP3 in cells transfected with MUT-5. VP1 was visualized with Alexa fluor 488 (green), and VP2, VP3, and agnoprotein were visualized with Alexa fluor 568 (red). spkl & strd, speckled and stranded staining patterns.
FIG. 5.
Immunocytochemistry of cells transfected with mutant vectors (MUT-6 and MUT-7). (A and B) Double staining of VP1 and VP2/VP3 (A) and of VP1 and agnoprotein (B) in cells transfected with MUT-6. (C) Double staining of VP1 and VP2/VP3 in cells transfected with MUT-7.
In contrast, cells transfected with group 4 vectors yielded VP1 distribution patterns similar to the predominantly cytoplasmic pattern for VP1-SRα. Even in cells with an exclusively nuclear localization of VP1, no distinct intranuclear staining patterns of speckles or strands were seen (Fig. 5). In cells transfected with MUT-6, agnoprotein was seen predominantly in the cytoplasm, occasionally as a perinuclear inclusion. Cells that were positive for both VP1 and agnoprotein tended to have enlarged nuclei (Fig. 5B).
Therefore, the presence of either VP2 or VP3 is essential for the exclusively nuclear localization of VP1 and its accumulation in distinct subnuclear domains. Agnoprotein is not essential for this localization. However, agnoprotein can facilitate the nuclear localization of VP1, since the percentage of cells presenting exclusively nuclear localization of VP1 was consistently higher in the presence of agnoprotein than in its absence.
VP2 and VP3 diffusely localize to the nucleus in the absence of VP1.
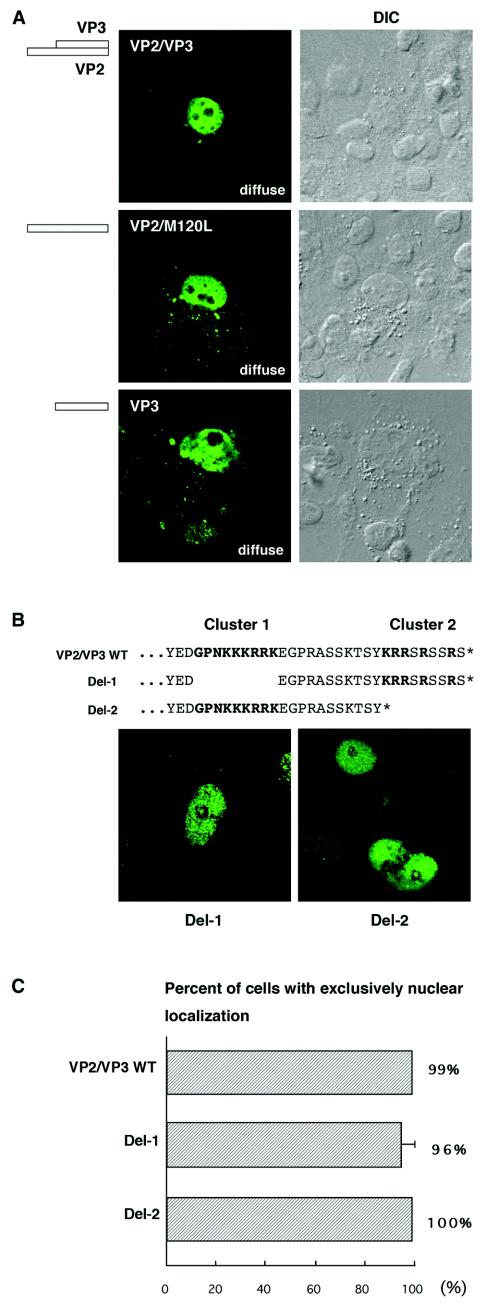
How do VP2 and VP3 promote the nuclear transport of VP1 and its accumulation in distinct subnuclear structures? During HPV33 infection, L2 first accumulates in ND10 and later recruits L1 to the same domains (17). In the case of JCV, we assumed that VP1 and VP2/VP3 translocate as a complex to distinct subnuclear structures, using a targeting signal that is possibly carried by VP2 and VP3. If so, VP2 or VP3 itself should show staining patterns of speckles or strands when expressed independently. To test this hypothesis, we expressed VP2 and VP3 in COS-7 cells in the absence of VP1. However, although VP2/VP3 was exclusively localized to the nucleus in 99% of positive cells, no speckled or stranded staining patterns were seen (Fig. 6A). To further examine the distribution of VP2 alone, we mutated the ATG start codon for VP3, which corresponds to the 120th residue of VP2, to TTG within VP2/VP3, resulting in VP2/M120L. To see the distribution of VP3 alone, we also expressed VP3 in the absence of VP2. However, VP2/M120L and VP3 consistently showed a diffuse distribution in the nucleus (Fig. 6A). Therefore, an accumulation in distinct subnuclear structures is a cooperative action between VP1 and VP2/VP3. Neither VP2 nor VP3, when expressed alone, accumulates discretely in the nucleus.
FIG.6.
Localization of VP2/VP3, VP2, VP3, and their deletion mutants. (A) Expression vectors encoding VP2 and VP3 (VP2/VP3), VP2 alone (VP2/M120L), or VP3 alone (VP3) were transfected into COS-7 cells. The localization of VP2/VP3, VP2/M120L, and VP3 was examined by immunocytochemistry. (B) Two clusters of basic amino acids, GPNKKKRRK (cluster 1) and KRRSRSSRS (cluster 2), in the C-terminal sequence of VP2/VP3. The asterisk indicates the C terminus. Deletion mutants Del-1 and Del-2 were constructed for these clusters, and the localization of the mutant proteins was examined by immunocytochemistry. Both Del-1 and Del-2 were diffusely localized to the nucleus. (C) Percentages of cells showing exclusively nuclear localization of VP2/VP3, Del-1, and Del-2.
We next tested the NLS of VP2/VP3. In the C-terminal sequence of VP2/VP3, there are two clusters that are rich in basic amino acids, GPNKKKRRK (residues 316 to 324 and 197 to 205 in JCV VP2 and VP3, respectively) and KRRSRSSRS (residues 336 to 344 and 217 to 225 in JCV VP2 and VP3, respectively). They will be described as cluster 1 and cluster 2, respectively (Fig. 6B). The cluster 1 sequence is nearly identical to the NLS of SV40 VP2/VP3, GPNKKKRKL (residues 198 to 206 and 316 to 324 in SV40 VP2 and VP3, respectively) (10). The cluster 2 sequence is nearly identical to the C-terminal nine residues, which include the SV40 DNA-binding domain, KRR-R-R (13) (see Fig. 11). To test whether cluster 1 or cluster 2 is responsible for the nuclear transport of JCV VP2/VP3, we prepared deletion mutants Del-1 and Del-2, which lack cluster 1 and cluster 2, respectively. When these deletion mutants were expressed in cells, about 96% of Del-1-expressing cells and 100% of Del-2-expressing cells still showed exclusively nuclear staining patterns (Fig. 6B and C). However, both Del-1 and Del-2 had dramatically reduced staining with the anti-VP2/VP3C antibody. The number of stained cells was reduced to approximately 1/10 that with wild-type VP2/VP3. Although we also constructed Del-3, lacking both cluster 1 and cluster 2, the Del-3 mutant was not detected with the anti-VP2/VP3C antibody. Similar results were obtained for deletion mutants derived from VP2/M120L and VP3. Therefore, even in the absence of either cluster 1 or cluster 2, both VP2 and VP3 are readily transported to the nucleus. However, we cannot conclude how VP2 and VP3 are distributed in the absence of both clusters. Cluster 1 and cluster 2 may be essential to stabilize VP2 and VP3 or to maintain antigenic structures of the C-terminal sequence.
FIG. 11.
Comparison of C-terminal sequences of VP2/VP3 among polyomaviruses. JCV has a unique sequence in the C-terminal region of VP2/VP3. Compared with the BK virus (BKV) and SV40 sequences, eight amino acids are absent between two clusters that are rich in basic amino acids (cluster 1 and cluster 2) in the JCV sequence. Compared with the PyV sequence, an additional 20 amino acids are present in the C terminus in the JCV sequence. Functional domains (a to c) that were shown to be important for virion production are underlined. In PyV and SV40, a region important for the interaction with VP1 lies upstream of the region shown (9, 21), and it has also been reported for the C terminus of the SV40 sequence (20).
Cluster 2 of VP2/VP3 is essential for accumulation of VP1 in distinct subnuclear structures but not for its transport to the nucleus.
To test whether the C-terminal sequence of VP2/VP3 is required for the nuclear transport of VP1 and for its accumulation in distinct subnuclear structures, we constructed mutant vectors from AVP231-SRα to place a stop codon before cluster 1 or cluster 2. MUT-8 and MUT-9 have a TAA stop codon (STOP-1) in place of the first amino acid of cluster 2 (Fig. 7A and B). Since STOP-1 overlaps the VP1 coding sequence in a different reading frame, the 22nd residue of VP1 was also mutated, from arginine to lysine. Agnoprotein is encoded by MUT-8, but not by MUT-9. When MUT-8 and MUT-9 were transfected into COS-7 cells, 98 and 91%, respectively, of VP1-positive cells showed exclusively nuclear staining patterns (Fig. 7C and 8A to C). However, unlike AVP231-SRα, MUT-8 and MUT-9 yielded no distinct intranuclear localization patterns of speckles or strands (Fig. 8A to C). A double-staining analysis revealed that VP2/VP3 lacking cluster 2 was also distributed diffusely in the nucleus (Fig. 8A and C). The distribution of agnoprotein was similar in cells transfected with MUT-8 and in those transfected with AVP231-SRα (Fig. 8B). Thus, cluster 2 is essential for capsid proteins to accumulate in distinct subnuclear domains but not for their transport from the cytoplasm to the nucleus.
MUT-10 and MUT-11 encode a TAA stop signal (STOP-2) in place of the 34th residue from the C terminus of VP2/VP3 (Fig. 7A and B). Thus, cells transfected with MUT-10 or MUT-11 express VP1 and VP2/VP3 lacking both cluster 1 and cluster 2. Agnoprotein is encoded by MUT-10, but not by MUT-11. When these vectors were transfected into cells, VP1 localization was more cytoplasmic, as in cells transfected with VP1-SRα. Exclusively nuclear localization of VP1 was seen in only 44 and 32% of VP1-positive cells transfected with MUT-10 and MUT-11, respectively (Fig. 7C and 8D to F). Distinct speckled or stranded intranuclear staining patterns were not seen. Truncated VP2/VP3 was not detected because the antigenic sequence for the VP2/VP3C antibody lay within the deleted C-terminal 34 residues (Fig. 8D and F). Agnoprotein was, as usual, detected predominantly in the cytoplasm in cells transfected with MUT-10 (Fig. 8E). These results indicate that the C-terminal 34 residues of VP2/VP3 are essential for the exclusively nuclear localization of VP1 and for its accumulation in distinct subnuclear structures. Consistent with other results, agnoprotein is not essential, but it likely facilitates the efficient nuclear localization of VP1.
Capsid proteins are assembled into virions at ND10.
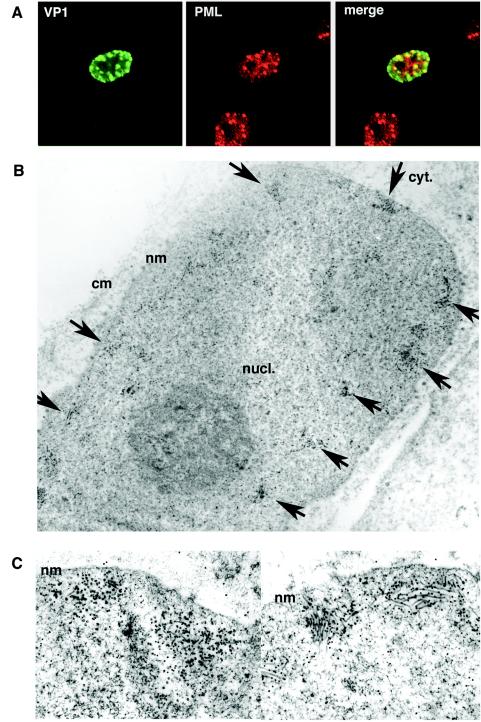
To determine where in the nucleus the capsid proteins accumulated, we double stained cells transfected with AVP231-SRα with antibodies for VP1 and PML, a major component of ND10. In the nucleus, VP1 and PML were colocalized along the inner nuclear periphery, presenting speckled staining patterns (Fig. 9A). Thus, VP1 accumulates in ND10. The cells were also analyzed by immunoelectron microscopy with an antibody against VP1. Of 25 VP1-positive cells examined, 10 cells showed distinct clusters of colloidal gold at the inner periphery of the nucleus (Fig. 9B), while 15 cells showed a diffuse nuclear distribution of colloidal gold (data not shown). All 10 cells with discrete clusters of colloidal gold showed the assembly of numerous virus-like particles with spherical and/or filamentous forms (Fig. 9B and C). Since both colloidal gold and virus-like particles were seen only in discrete locations and not in other parts of the nucleus, it is highly likely that the capsid proteins first accumulate in ND10 and then are assembled into virions. In contrast, in cells transfected with VP1-SRα, colloidal gold was distributed more diffusely both in the cytoplasm and in the nucleus. Although >25 VP1-positive cells were examined, the assembly of virus-like particles was not detected (data not shown).
FIG. 9.
Capsid proteins accumulate at ND10 for assembly into virus-like particles. (A) Localization of VP1 at ND10. Cells transfected with AVP231-SRα were double stained for VP1 and PML, a component of ND10. VP1 was visualized with Alexa fluor 488 (green), and PML was visualized with Alexa fluor 568 (red). The two proteins were colocalized, showing speckled staining patterns at the inner periphery of the nucleus. (B) Immunoelectron microscopy of cells transfected with AVP231-SRα by use of an antibody against VP1. Colloidal gold spheres (15-nm diameter), indicating VP1 molecules, were clustered at multiple locations along the inner periphery of the nucleus (arrows). (C) Assembling virus-like particles with both spherical and filamentous structures were seen in association with clusters of colloidal gold. nucl., nucleus; cyt., cytoplasm; nm, nuclear membrane; cm, cellular membrane.
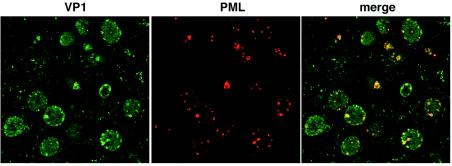
We finally investigated whether the capsid proteins also accumulate in ND10 in oligodendrocytes from the human brain. Human brain tissue was obtained from an autopsied patient with progressive multifocal leukoencephalopathy and was double stained with antibodies for VP1 and PML. In enlarged nuclei of infected oligodendrocytes, colocalization of VP1 and PML was detected at the inner nuclear periphery, consistent with the results for transfected COS-7 cells (Fig. 10). Therefore, these results consistently indicate that major and minor capsid proteins cooperatively accumulate in ND10 and that ND10 is the location where the capsid proteins are efficiently assembled into virions.
FIG. 10.
VP1 localization at ND10 in oligodendrocytes from the human brain. Human brain tissue obtained from an autopsied patient with progressive multifocal leukoencephalopathy was double stained for VP1 and PML. VP1 was visualized with Alexa fluor 488 (green), and PML was visualized with Alexa fluor 568 (red). The two proteins were colocalized in speckles along the inner nuclear periphery, consistent with the results for transfected COS-7 cells.
DISCUSSION
We have shown in this paper that JCV virions are assembled at ND10. The cooperation of VP1 with VP2 or VP3 is essential for localization at ND10, since neither VP1 nor VP2/VP3 accumulated in ND10 when it was expressed alone. The C-terminal sequence of VP2/VP3 is important for efficient nuclear transport as well as for the accumulation in ND10.
JCV VP2/VP3 may have two independent domains that are functional as NLSs. Since the NLS of JCV VP1, KRK-RK, is not efficient at nuclear transport (38), the VP2/VP3 NLSs may have markedly important roles if VP1 and VP2/VP3 are cotransported to the nucleus. The cluster 1 sequence, GPNKKKRRK, is nearly identical to the NLS of SV40 VP2/VP3, GPNKKKRKL (10) (Fig. 11). However, in SV40 VP3, a single amino acid substitution in a basic residue of the NLS abolished its nuclear localization (10, 33), while in JCV, a deletion of the entire cluster 1 did not abolish the exclusively nuclear localization of VP2/VP3 (Fig. 6). We assume that, in JCV, cluster 1 and cluster 2 provide redundant NLS functions, but this possibility is not yet proven, since a mutant VP2/VP3 lacking both clusters (Del-3) could not be detected with the anti-VP2/VP3C antibody.
The VP1 interactive domain of JCV VP2/VP3 has not been analyzed. However, it may be analogous to that of PyV VP2/VP3, which has been mapped to the polyomavirus family consensus sequence based on X-ray crystallography (9) and biochemical analysis (3). SV40 VP2/VP3 is reported to interact with VP1 at the analogous sequence (21). Although for SV40 the C-terminal 13 residues, KARHKRRNRSSRS, have also been reported to interact with VP1 (20), the JCV cluster 2 sequence, KRRSRSSRS, which is nearly identical to the C-terminal 9 residues of the SV40 sequence, was not required for VP1 to localize exclusively to the nucleus (Fig. 7 and 8). Therefore, cluster 2 is not essential for the JCV VP1-VP2/VP3 interaction. In contrast, a deletion of the C-terminal 34 residues of VP2/VP3 caused VP1 to become less efficient at nuclear transport. This was likely due to defective nuclear targeting of the mutant VP2/VP3 rather than a defective VP1-VP2/VP3 interaction, as the sequence analogous to the VP1 interactive domain of PyV was retained while cluster 1 and cluster 2 were deleted.
The cluster 2 sequence, KRRSRSSRS, contains residues identical to those in the DNA-binding domain mapped in SV40 VP2/VP3, KRR-R-R (13) (Fig. 11). Interestingly, we have found that cluster 2 is necessary, but not sufficient, for discrete accumulation at ND10 (Fig. 7 and 8). For SV40, it has been reported that T-antigen-directed DNA replication is restricted to ND10 (45). For HPV, it has also been reported that translocation of the capsid proteins can be partly coupled with DNA amplification at ND10 to promote virion assembly (43). Similarly, in JCV, if viral genomic DNA replicates at ND10, then recruitment of the VP1-VP2/VP3 complex would be quite reasonable, and cluster 2 may play roles not only in targeting to ND10 but also in further encapsidation of the viral genomic DNA.
The JCV agnoprotein was not essential for the nuclear transport of VP1, but it may facilitate nuclear entry of the capsid proteins. In the presence of agnoprotein, a higher percentage of cells consistently showed exclusively nuclear staining of VP1 than in its absence (Fig. 3 and 7). Agnoprotein was also colocalized with VP1 at the margins of the nucleus, possibly at the nuclear membrane or nuclear lamina (Fig. 1, 4, and 5). For SV40, it has been reported that VP1 was delayed during nuclear transport and localized to the periphery of the nucleus in the absence of agnoprotein (6, 36). The SV40 agnoprotein was also suggested to interact with VP1 pentamers and to prevent their aberrant aggregation in the cytoplasm, which also results in facilitated nuclear transport (2, 26). Although the functions of the JCV agnoprotein remain largely unclear, its multiple functions may include the efficient entry of VP1-VP2/VP3 complexes into the nucleus.
The nucleus has distinct substructures in the interchromatin space, which are collectively called the nuclear matrix and nuclear bodies (27, 34). Especially ND10, or the PML nuclear body, is known as a site for the transcription and replication of many DNA viruses (4). Viral gene products, including herpes simplex virus ICP0, cytomegalovirus IE1, Epstein-Barr virus BZLF1, and E1 and E2 of HPV, have been reported to associate with ND10 and to reconstruct its structures (16, 35). Contrasting with these early regulatory proteins, L1 and L2 of HPV are rare examples of capsid (structural) proteins that associate with ND10. Unlike JCV and other polyomaviruses, HPV L1 and L2 separately translocate to the nucleus; the minor capsid protein L2 first accumulates in ND10 and later recruits L1 to the same domain (12, 17). Although JCV is distinct from HPV in its manner of intranuclear accumulation of the capsid proteins, we have shown the first evidence that the JCV capsid proteins accumulate in ND10 for efficient assembly into virions. The data are strikingly important for understanding the JCV replication cycle and the pathogenesis of the virus-induced demyelinating disorder progressive multifocal leukoencephalopathy.
In conclusion, the major and minor JCV capsid proteins cooperatively accumulate in ND10 for assembly into virions. Recently, the dynamic movement of ND10 and of DNA replication foci has been demonstrated in living cells (24, 31). We hope to learn the dynamics of JCV VP1, VP2/VP3, and agnoprotein in association with moving nuclear components in future studies.
Acknowledgments
We thank Miyoko Ojima and Masako Kawano (Instrument Analysis Center, Tokyo Medical and Dental University) for assistance with electron microscopy.
This study was supported in part by the Prion Disease and Slow Virus Infection Research Committee of the Ministry of Health, Labor, and Welfare of Japan, the Japanese Science and Technology Corporation, and the Japanese Society for the Promotion of Science.
Footnotes
This paper is in memory of Gerald L. Stoner for his great support and encouragement of this project.
REFERENCES
- 1.Akatani, K., M. Imai, M. Kimura, K. Nagashima, and N. Ikegami. 1994. Propagation of JC virus in human neuroblastoma cell line IMR-32. J. Med. Virol. 43:13-19. [DOI] [PubMed] [Google Scholar]
- 2.Barkan, A., R. C. Welch, and J. E. Mertz. 1987. Missense mutations in the VP1 gene of simian virus 40 that compensate for defects caused by deletions in the viral agnogene. J. Virol. 61:3190-3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barouch, D. H., and S. C. Harrison. 1994. Interactions among the major and minor coat proteins of polyomavirus. J. Virol. 68:3982-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borden, K. L. 2002. Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22:5259-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai, X., D. Chang, S. Rottinghaus, and R. A. Consigli. 1994. Expression and purification of recombinant polyomavirus VP2 protein and its interactions with polyomavirus proteins. J. Virol. 68:7609-7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carswell, S., and J. C. Alwine. 1986. Simian virus 40 agnoprotein facilitates perinuclear-nuclear localization of VP1, the major capsid protein. J. Virol. 60:1055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, D., J. I. D. Haynes, J. N. Brady, and R. A. Consigli. 1992. Identification of a nuclear localization sequence in the polyomavirus capsid protein VP2. Virology 191:978-983. [DOI] [PubMed] [Google Scholar]
- 8.Chang, D., J. I. D. Haynes, J. N. Brady, and R. A. Consigli. 1992. The use of additive and subtractive approaches to examine the nuclear localization sequence of the polyomavirus major capsid protein VP1. Virology 189:821-827. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X. S., T. Stehle, and S. C. Harrison. 1998. Interaction of polyomavirus internal protein VP2 with the major capsid protein VP1 and implications for participation of VP2 in viral entry. EMBO J. 17:3233-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clever, J., and H. Kasamatsu. 1991. Simian virus 40 Vp2/3 small structural proteins harbor their own nuclear transport signal. Virology 181:78-90. [DOI] [PubMed] [Google Scholar]
- 11.Daniel, A. M., and R. J. Frisque. 1993. Transcription initiation sites of prototype and variant JC virus early and late messenger RNAs. Virology 194:97-109. [DOI] [PubMed] [Google Scholar]
- 12.Day, P. M., R. B. Roden, D. R. Lowy, and J. T. Schiller. 1998. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J. Virol. 72:142-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dean, D. A., P. P. Li, L. M. Lee, and H. Kasamatsu. 1995. Essential role of the Vp2 and Vp3 DNA-binding domain in simian virus 40 morphogenesis. J. Virol. 69:1115-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delos, S. E., T. P. Cripe, A. D. Leavitt, H. Greisman, and R. L. Garcea. 1995. Expression of the polyomavirus minor capsid proteins VP2 and VP3 in Escherichia coli: in vitro interactions with recombinant VP1 capsomeres. J. Virol. 69:7734-7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delos, S. E., L. Montross, R. B. Moreland, and R. L. Garcea. 1993. Expression of the polyomavirus VP2 and VP3 proteins in insect cells: coexpression with the major capsid protein VP1 alters VP2/VP3 subcellular localization. Virology 194:393-398. [DOI] [PubMed] [Google Scholar]
- 16.Everett, R. D. 2001. DNA viruses and viral proteins that interact with PML nuclear bodies. Oncogene 20:7266-7273. [DOI] [PubMed] [Google Scholar]
- 17.Florin, L., C. Sapp, R. E. Streeck, and M. Sapp. 2002. Assembly and translocation of papillomavirus capsid proteins. J. Virol. 76:10009-10014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Florin, L., F. Schafer, K. Sotlar, R. E. Streeck, and M. Sapp. 2002. Reorganization of nuclear domain 10 induced by papillomavirus capsid protein l2. Virology 295:97-107. [DOI] [PubMed] [Google Scholar]
- 19.Forstova, J., N. Krauzewicz, S. Wallace, A. J. Street, S. M. Dilworth, S. Beard, and B. E. Griffin. 1993. Cooperation of structural proteins during late events in the life cycle of polyomavirus. J. Virol. 67:1405-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gharakhanian, E., and H. Kasamatsu. 1990. Two independent signals, a nuclear localization signal and a Vp1-interactive signal, reside within the carboxy-35 amino acids of SV40 Vp3. Virology 178:62-71. [DOI] [PubMed] [Google Scholar]
- 21.Gordon-Shaag, A., O. Ben-Nun-Shaul, V. Roitman, Y. Yosef, and A. Oppenheim. 2002. Cellular transcription factor Sp1 recruits simian virus 40 capsid proteins to the viral packaging signal, ses. J. Virol. 76:5915-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishii, N., N. Minami, E. Y. Chen, A. L. Medina, M. M. Chico, and H. Kasamatsu. 1996. Analysis of a nuclear localization signal of simian virus 40 major capsid protein Vp1. J. Virol. 70:1317-1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishii, N., A. Nakanishi, M. Yamada, M. H. Macalalad, and H. Kasamatsu. 1994. Functional complementation of nuclear targeting-defective mutants of simian virus 40 structural proteins. J. Virol. 68:8209-8216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonhardt, H., H. P. Rahn, P. Weinzierl, A. Sporbert, T. Cremer, D. Zink, and M. C. Cardoso. 2000. Dynamics of DNA replication factories in living cells. J. Cell Biol. 149:271-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liddington, R. C., Y. Yan, J. Moulai, R. Sahli, T. L. Benjamin, and S. C. Harrison. 1991. Structure of simian virus 40 at 3.8-A resolution. Nature 354:278-284. [DOI] [PubMed] [Google Scholar]
- 26.Margolskee, R. F., and D. Nathans. 1983. Suppression of a VP1 mutant of simian virus 40 by missense mutations in serine codons of the viral agnogene. J. Virol. 48:405-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matera, A. G. 1999. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 9:302-309. [DOI] [PubMed] [Google Scholar]
- 28.Matsuda, M., M. Jona, K. Yasui, and K. Nagashima. 1987. Genetic characterization of JC virus Tokyo-1 strain, a variant oncogenic in rodents. Virus Res. 7:159-168. [DOI] [PubMed] [Google Scholar]
- 29.Mazlo, M., and I. Tariska. 1980. Morphological demonstration of the first phase of polyomavirus replication in oligodendroglia cells of human brain in progressive multifocal leukoencephalopathy (PML). Acta Neuropathol. 49:133-143. [DOI] [PubMed] [Google Scholar]
- 30.Moreland, R. B., and R. L. Garcea. 1991. Characterization of a nuclear localization sequence in the polyomavirus capsid protein VP1. Virology 185:513-518. [DOI] [PubMed] [Google Scholar]
- 31.Muratani, M., D. Gerlich, S. M. Janicki, M. Gebhard, R. Eils, and D. L. Spector. 2002. Metabolic-energy-dependent movement of PML bodies within the mammalian cell nucleus. Nat. Cell Biol. 4:106-110. [DOI] [PubMed] [Google Scholar]
- 32.Nagashima, K., K. Yamaguchi, K. Yasui, and H. Ogiwara. 1981. Progressive multifocal leukoencephalopathy. Neuropathology and virus isolation. Acta Pathol. Jpn. 31:953-961. [PubMed] [Google Scholar]
- 33.Nakanishi, A., D. Shum, H. Morioka, E. Otsuka, and H. Kasamatsu. 2002. Interaction of the Vp3 nuclear localization signal with the importin alpha 2/beta heterodimer directs nuclear entry of infecting simian virus 40. J. Virol. 76:9368-9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nickerson, J. 2001. Experimental observations of a nuclear matrix. J. Cell Sci. 114:463-474. [DOI] [PubMed] [Google Scholar]
- 35.Regad, T., and M. K. Chelbi-Alix. 2001. Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20:7274-7286. [DOI] [PubMed] [Google Scholar]
- 36.Resnick, J., and T. Shenk. 1986. Simian virus 40 agnoprotein facilitates normal nuclear location of the major capsid polypeptide and cell-to-cell spread of virus. J. Virol. 60:1098-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shishido, Y., S. Nukuzuma, J. Mukaigawa, S. Morikawa, K. Yasui, and K. Nagashima. 1997. Assembly of JC virus-like particles in COS7 cells. J. Med. Virol. 51:265-272. [PubMed] [Google Scholar]
- 38.Shishido-Hara, Y., Y. Hara, T. Larson, K. Yasui, K. Nagashima, and G. L. Stoner. 2000. Analysis of capsid formation of human polyomavirus JC (Tokyo-1 strain) by a eukaryotic expression system: splicing of late RNAs, translation and nuclear transport of major capsid protein VP1, and capsid assembly. J. Virol. 74:1840-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shishido-Hara, Y., and K. Nagashima. 2001. Synthesis and assembly of polyomavirus virions, p. 149-177. In K. Khalili and G. L. Stoner (ed.), Human polyomaviruses: molecular and clinical perspectives. John Wiley & Sons, Inc., New York, N.Y.
- 40.Silverman, L., and L. J. Rubinstein. 1965. Electron microscopic observations on a case of progressive multifocal leukoencephalopathy. Acta Neuropathol. (Berlin) 5:215-224. [DOI] [PubMed] [Google Scholar]
- 41.Stehle, T., S. J. Gamblin, Y. Yan, and S. C. Harrison. 1996. The structure of simian virus 40 refined at 3.1 A resolution. Structure 4:165-182. [DOI] [PubMed] [Google Scholar]
- 42.Stehle, T., and S. C. Harrison. 1997. High-resolution structure of a polyomavirus VP1-oligosaccharide complex: implications for assembly and receptor binding. EMBO J. 16:5139-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Swindle, C. S., N. Zou, B. A. Van Tine, G. M. Shaw, J. A. Engler, and L. T. Chow. 1999. Human papillomavirus DNA replication compartments in a transient DNA replication system. J. Virol. 73:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takebe, Y., M. Seiki, J. Fujisawa, P. Hoy, K. Yokota, K. Arai, M. Yoshida, and N. Arai. 1988. SR alpha promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol. Cell. Biol. 8:466-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang, Q., P. Bell, P. Tegtmeyer, and G. G. Maul. 2000. Replication but not transcription of simian virus 40 DNA is dependent on nuclear domain 10. J. Virol. 74:9694-9700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zu Rhein, G. M., and S.-M. Chou. 1965. Particles resembling papova viruses in human cerebral demyelinating disease. Science 11:1477-1479. [DOI] [PubMed] [Google Scholar]