Abstract
The coronavirus spike protein (S) forms the distinctive virion surface structures that are characteristic of this viral family, appearing in negatively stained electron microscopy as stems capped with spherical bulbs. These structures are essential for the initiation of infection through attachment of the virus to cellular receptors followed by fusion to host cell membranes. The S protein can also mediate the formation of syncytia in infected cells. The S protein is a type I transmembrane protein that is very large compared to other viral fusion proteins, and all except a short carboxy-terminal segment of the S molecule constitutes the ectodomain. For the prototype coronavirus mouse hepatitis virus (MHV), it has previously been established that S protein assembly into virions is specified by the carboxy-terminal segment, which comprises the transmembrane domain and the endodomain. We have genetically dissected these domains in the MHV S protein to localize the determinants of S incorporation into virions. Our results establish that assembly competence maps to the endodomain of S, which was shown to be sufficient to target a heterologous integral membrane protein for incorporation into MHV virions. In particular, mutational analysis indicated a major role for the charge-rich carboxy-terminal region of the endodomain. Additionally, we found that the adjacent cysteine-rich region of the endodomain is critical for fusion of infected cells, confirming results previously obtained with S protein expression systems.
Coronaviruses are a family of enveloped RNA viruses responsible for a variety of respiratory, enteric, neurologic, and other diseases in mammalian and avian hosts. In humans, two coronaviruses are known to cause upper respiratory tract infections, while a third human coronavirus is the recently discovered causative agent of severe acute respiratory syndrome.
Coronaviruses contain a relatively small number of structural proteins. The most prominent among these is the spike glycoprotein (S), which protrudes from the virion surface and forms peplomers or spike structures that interact with host receptors and mediate virus-cell and cell-cell fusion. The narrow host range of most coronaviruses, which generally infect only one or just a few species, resides almost entirely in the specificity of the interactions between S proteins and their corresponding host cell receptors (21, 24, 41, 43). For the prototype coronavirus mouse hepatitis virus (MHV), the S protein is a 180-kDa, N-glycosylated, type I integral membrane protein. In MHV-A59 (the strain used for this study), the amino-terminal ectodomain of S is made up of 1,263 of the 1,324 amino acid residues of the molecule (29). The remaining residues of S form the transmembrane domain and the endodomain, which are integrated within the viral envelope, the principal constituent of which is the 25-kDa triple-spanning membrane protein (M). A third membrane-bound component, the small envelope protein (E), is minor in both size (10 kDa) and stoichiometry relative to other virion structural proteins. Expression studies of the formation of virus-like particles (VLPs) (1, 3, 7, 42) and the engineering of viral mutants (8, 17, 26, 34) have revealed a critical role for the E protein, in conjunction with the M protein, during virion morphogenesis. Remarkably, the E protein is not absolutely essential for MHV viability (26); in contrast, for the porcine coronavirus transmissible gastroenteritis virus, disruption of the E gene is lethal (8, 34).
Although it is indispensable for virion infectivity, the S protein is not an obligatory participant in virion assembly. This was first indicated by key early work showing that MHV-infected cells treated with tunicamycin assembled virions lacking spikes (22, 36). Analyses of coimmunoprecipitated complexes from infected cells or from cells expressing subsets of viral proteins revealed that oligomerization of the S protein precedes its availability for assembly, but that after a lag, S is trapped by association with newly synthesized M protein (31, 33). M is thus the central organizer for virion assembly, as it also associates with itself (13) and with the nucleocapsid (N) protein (15, 25, 30, 39). The generation of coronavirus VLPs formed by the coexpression of just the M and E proteins (1, 3, 7, 42) provided an additional avenue for exploring the rules underlying S protein assembly into virions. The exchange of S protein domains between the distantly related MHV and feline infectious peritonitis virus (FIPV) led to the demonstration that the incorporation of S into VLPs was determined solely by the transmembrane domain and the endodomain of the S molecule (19). An extension of this principle to entire virions enabled the construction of the interspecies chimeric viruses fMHV (24), and very recently, mFIPV (21). Each of these viruses displays the ectodomain of a heterologous S protein in place of its own and as a consequence has switched its host cell species specificity.
We have performed a genetic dissection of the transmembrane domain and endodomain of the MHV S protein to localize the determinants for its incorporation into virions. Our results establish that assembly competence maps to the endodomain of S, and in particular, that the charge-rich carboxy terminus of the endodomain makes the major contribution to assembly. In addition, we have found that the amino-terminal cysteine-rich region of the endodomain is critical for fusion of infected cells, confirming results previously obtained with S protein expression systems (2, 5).
MATERIALS AND METHODS
Cells and viruses.
Wild-type MHV strain A59 and recombinants were propagated in mouse 17 clone 1 (17Cl1) or L2 cells; plaque assays and plaque purification of mutant recombinants were carried out in mouse L2 cells. The interspecies chimeric viruses fMHV (24) and fMHV.v2 (20) were grown in feline AK-D fetal lung cells.
Plasmid constructs.
Donor RNA transcription vectors used for this study were derived from pMH54 (Fig. 1), which encodes an RNA containing a 468-nucleotide (nt) 5′ segment of the MHV genome fused, via a short polylinker, to codon 28 of the hemagglutinin-esterase (HE) gene and all of the 3′ end of the MHV genome that follows thereafter (24). As noted previously, MHV segments within pMH54 correspond exactly to the sequence of our laboratory wild-type strain of MHV-A59, with the exception of: (i) coding-silent changes introduced to create RsrII and AvrII sites in the HE and S genes, respectively (24); (ii) coding-silent changes introduced to eliminate a HindIII site in the S gene (16); and (iii) an SbfI (Sse8387I) site created 12 nt downstream of the S gene stop codon (24).
FIG. 1.
Transcription vectors for synthesis of donor RNAs used in the construction of transmembrane domain and endodomain mutants. (A) Plasmids pHKP1 and pMH54P4 were derived from pMH54 (24), as detailed in Materials and Methods. In each plasmid schematic the arrow denotes the T7 promoter, and the restriction sites shown are those relevant to plasmid construction; all restriction sites, except for those in parentheses, are unique in the plasmid in which they appear. The solid rectangle in pMH54P4 indicates the 27-nt deletion. The expanded section of pHKP1 shows the elements encoded by the Hook gene: the signal peptide (sig), the influenza virus HA epitope tag, the anti-hapten single-chain antibody (sFv) fragment, two copies of the Myc epitope tag, and the transmembrane domain (Tm) and endodomain (Ed) from PDGFR. Also indicated are the TRSs preceding the Hook and S genes. HE, 99-nt remnant of the HE gene. Mutated derivatives of each parent plasmid were constructed by the insertion of PCR-generated fragments into the SalI-AscI and PshAI-SbfI intervals of pHKP1 and pMH54P4, respectively. (B) DNA sequences of the four junctions that are labeled in panel A. 1 and 2, the upstream and downstream boundaries of the Hook gene insertion in pHKP1, including the introduced TRS for the Hook open reading frame and relevant restriction sites; 4, the PshAI site and 27-nt deletion created in pMH54P4 between the S gene ectodomain and transmembrane domain; 3, the wild-type counterpart of junction 4 in pMH54. Underlined nucleotides are non-MHV sequences, including base changes created to introduce restriction sites. Shown beneath the DNA sequences are translations of portions of the 1b, Hook, and S open reading frames (with asterisks denoting stop codons); for pMH54 and pMH54P4, amino acid residues in bold are in the transmembrane domain of the S protein.
To examine the requirements for incorporation of an exogenous surface protein into MHV virions, we expanded pMH54 to include a segment of gene 1b and a chimeric reporter gene, designated Hook (6). The resulting vector, pHKP1 (Fig. 1A), was assembled through a series of intermediate constructs from cloned cDNAs and PCR products that were generated to provide necessary restriction sites and transcription elements. In pHKP1, the RsrII site of pMH54 is retained as part of the polylinker and is followed by an in-frame fusion to the 3′-most 1,248-nt segment of gene 1b, an MHV transcription-regulating sequence (TRS) (Fig. 1B); the 1,071-nt EcoRV-XhoI fragment of pHook-1 (Invitrogen) (6); and a 99-nt remnant of the 3′ end of the HE gene. Elsewhere, pHKP1 is identical to pMH54. For the construction of mutants of pHKP1, the region encoding the platelet-derived growth factor receptor (PDGFR) transmembrane domain and endodomain of the Hook protein was excised with SalI and AscI (Fig. 1A). This segment was then replaced with various wild-type or mutated components from the MHV S gene, which were generated by splicing overlap extension-PCR (23) followed by restriction with SalI and AscI.
To incorporate mutations directly into the gene region encoding the transmembrane domain and endodomain of the MHV S protein (in a vector devoid of the Hook gene), we constructed a second derivative of pMH54. In this plasmid, pMH54P4 (Fig. 1A), splicing overlap extension-PCR between the MluI and SbfI sites of the S gene was used to create a coding-silent PshAI site together with a 27-nt deletion encompassing nine codons at the boundary of the S protein ectodomain and transmembrane domain (Fig. 1B). Mutations were then transferred from a given pHKP1-derived plasmid by PCR amplification of the relevant region of the Hook gene, using primers that rebuilt the missing nine codons of pMH54P4. The resulting PCR fragment (blunt at the 5′ end) was restricted with PstI (for which the restriction site is contained within that for SbfI) at its 3′ end and was inserted in place of the PshAI-SbfI fragment of pMH54P4.
All manipulations of DNA were performed by standard methods (37). The compositions of all constructs were checked by restriction analysis; all cloned cDNA precursors, PCR-generated segments, and newly created junctions of each plasmid were verified by automated DNA sequencing.
Mutant construction by targeted RNA recombination.
The Hook gene was incorporated into the MHV genome and mutations were created in the transmembrane domain and endodomain of either Hook or MHV S by targeted RNA recombination, with fMHV or fMHV.v2 as the recipient virus, as described previously (20, 24-26). Briefly, monolayers of feline AK-D cells were infected with fMHV for 2 to 3 h at 37°C and then gently suspended by treatment with a low concentration of trypsin. Capped synthetic donor RNA transcripts were generated with a T7 polymerase transcription kit (mMessage mMachine; Ambion) by using PacI-truncated plasmid templates. Donor RNAs (5 to 10 μg) were transfected into 1.2 × 107 infected cells by electroporation with a Gene Pulser II electroporation apparatus (Bio-Rad), with two consecutive pulses at settings of 975 μF and 0.3 kV. The infected and transfected AK-D cells were then overlaid onto murine L2 cell monolayers, and released progeny virus was harvested when the formation of syncytia or a cytopathic effect was apparent in the murine cells at 24 to 72 h postinfection at 37°C.
Recombinant candidates were purified by two rounds of plaque titration on L2 cell monolayers at 37°C. Each purified recombinant candidate was subsequently used to infect a 10-cm2 monolayer of 17Cl1 cells, from which total RNA was extracted at 8 h postinfection by use of Ultraspec reagent (Biotecx). Reverse transcription of RNAs was carried out with a random hexanucleotide primer and avian myeloblastosis virus reverse transcriptase (Life Sciences) under standard conditions (37). To ascertain the presence of incorporated genes and mutations, we performed PCR amplifications of the cDNAs by using primer pairs flanking the relevant regions of the genome. PCRs were performed with AmpliTaq polymerase (Roche) under standard conditions of 30 s at 94°C, 45 s at 52°C, and 1 min at 72°C, repeated for 30 cycles. The products were analyzed directly by agarose gel electrophoresis and purified with Quantum prep columns (Bio-Rad) prior to automated sequencing. The virus referred to as the wild type throughout this work is Alb240, a well-characterized wild-type recombinant that was previously reconstructed by targeted recombination between fMHV and pMH54 donor RNA (20, 26).
Virus purification.
Hook recombinant viruses were purified by polyethylene glycol precipitation followed by two cycles of equilibrium centrifugation on preformed gradients of 0 to 50% potassium tartrate, which was osmotically counterbalanced with 30 to 0% glycerol, in a buffer of 50 mM Tris-maleate (pH 6.5) and 1 mM EDTA. Gradients were centrifuged at 111,000 × g in a Beckman SW41 rotor at 4°C for 16 to 24 h. After each equilibrium centrifugation step, viral bands were collected from the gradients, diluted with 50 mM Tris-maleate (pH 6.5)-100 mM NaCl-1 mM EDTA, and pelleted onto glycerol cushions by centrifugation for 2 h at 151,000 × g in a Beckman SW41 rotor at 4°C. In some initial experiments, the final glycerol cushion was omitted, and a further purification step (with all operations performed at 4°C) was added as follows. Gradient-purified virions were resuspended in 100 μl of magnesium- and calcium-free phosphate-buffered saline (PBS), pH 7.4, and were separated by chromatography on a 30- by 0.9-cm Superdex 200 column (Amersham Pharmacia). The column was eluted with PBS, and the eluant was collected in 20 1-ml fractions, with monitoring of the absorbance at 260 nm. Virions appeared as the first peak in flowthrough fractions 7 through 10. These fractions were pooled and concentrated by ultracentrifugation prior to further analysis.
Western blotting.
Confluent monolayers (10 cm2) of 17Cl1 cells were infected with wild-type MHV or the constructed mutants or were mock infected, and then the cells were incubated at 37°C. At 8 h postinfection, the monolayers were washed twice with PBS and lysed by the addition of 600 μl of a buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1.0% Nonidet P-40, 0.7 μg of pepstatin/ml, 1.0 μg of leupeptin/ml, 1.0 μg of aprotinin/ml, and 0.5 mg of Pefabloc SC (Roche)/ml. The lysates were held for 5 min on ice and were then clarified by centrifugation. Samples of either purified virions or infected cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis through 10% polyacrylamide (for Hook or N protein analysis) or 8% polyacrylamide (for S protein analysis) gels and were transferred to a polyvinylidene difluoride membrane. Blots were probed with one of the following antibodies: an anti-hemagglutinin (HA) epitope tag antibody (monoclonal antibody 12CA5; Roche), rabbit antisera raised against a bacterially expressed maltose binding protein-MHV N protein fusion, or goat anti-S protein antiserum AO4 (generously provided by Kathryn Holmes). Bound primary antibodies were visualized by use of a chemiluminescent detection system (ECL; Amersham).
Immunofluorescence.
L2 cells grown in 4.2-cm2 wells on glass slides were infected with wild-type MHV (Alb240), mutant SΔ20, or mutant SΔC at a multiplicity of infection of 0.1 to 0.5 PFU/cell or else were mock infected, and the cells were then incubated at 37°C. At 16 h postinfection, the cells were fixed with 2% paraformaldehyde in PBS, blocked in PBS containing 100 mM glycine, and permeabilized with 50% acetone-50% methanol at −20°C. The cells were blocked with PBS containing 0.2% Tween 20, 3% immunoglobulin G (IgG)-free bovine serum albumin, and 3% normal goat serum prior to incubation with anti-MHV N primary antibodies. The cells were then washed with PBS containing 0.2% Tween 20, incubated with a fluorescein isothiocyanate-conjugated goat anti-rabbit IgG F(ab′)2 secondary antibody (Roche), and counterstained with 0.01% Evans blue. Specimens were viewed under a Zeiss Axioskop2 fluorescence microscope with an Orca-ER charge-coupled device camera.
RESULTS
Generation of two different classes of recombinant viruses.
To systematically explore which segments and residues of the MHV S protein transmembrane domain and endodomain govern the incorporation of the S protein into virions, we constructed two different sets of mutants by targeted RNA recombination. For the first class of recombinants, a heterologous gene encoding an engineered transmembrane protein (6), designated Hook, was inserted into the MHV genome to serve as a surrogate for the S protein. For the second class of recombinants, mutations originally made in the Hook gene were transferred directly into the S gene in an otherwise wild-type background (lacking the Hook gene). We took this dual approach for two principal reasons. First, we expected that Hook would serve as a reporter for S protein incorporation into virions. Since Hook was not required for virus viability, it would tolerate mutations that might be lethal in the intact S protein. Second, Hook could be used to separate requirements for incorporation into virions from other possible essential functions of the S protein transmembrane domain and endodomain, such as a role in fusion.
To generate Hook recombinants, we constructed a donor RNA transcription vector, pHKP1, as described in Materials and Methods. In this vector, the Hook gene was inserted downstream of a segment of the viral polymerase 1b gene, replacing genes 2a and HE (Fig. 1A). This site was chosen because it was previously shown to allow a strong level of expression of a repositioned MHV M gene in the mutant MHV-MSEN (14). Moreover, MHV gene 2a and the HE gene (a pseudogene in strain A59) are each nonessential (28, 38), and the deletion of both has no detectable effect on the tissue culture phenotype of the virus (10). The Hook gene TRS and its upstream context (Fig. 1B) were patterned directly after the 1b-M junction in MHV-MSEN (14). Downstream of the Hook gene, pHKP1 retained the wild-type context of the S gene TRS as well as a wild-type S gene. The Hook gene (Fig. 1A, top) codes for a chimeric protein consisting of a murine Ig κ-chain signal peptide, the HA epitope tag, an anti-hapten single-chain antibody fragment, two tandem copies of the Myc epitope tag, and the transmembrane domain and endodomain of PDGFR (6). Hook gene transmembrane domain and endodomain variants were created by shuttling PCR-generated SalI-AscI fragments into pHKP1, replacing the PDGFR region and one of the Myc epitopes.
To generate S gene recombinants, we constructed pMH54P4 (Fig. 1A), a derivative of pMH54 in which a small deletion in the S gene transmembrane domain-encoding segment was created in order to accommodate a unique PshAI site (Fig. 1B). This arrangement facilitated the transfer of mutations from pHKP1-derived vectors and the simultaneous restoration of the deleted portion of the transmembrane domain by shuttling PCR-generated blunt SbfI fragments, as described in Material and Methods.
Hook and S gene mutants were constructed by targeted RNA recombination, a reverse genetics system for coronaviruses that has been used extensively to manipulate the structural genes of MHV (9, 10-14, 16, 17, 20, 21, 24-26, 32, 35) and that allows the efficient recovery of mutants with extremely defective phenotypes (20, 25, 26). This method makes use of the high rate of homologous recombination that occurs during coronavirus infection, even with transfected donor RNAs, and it couples this property to the selective power resulting from the stringent species specificity that many coronaviruses exhibit in tissue culture. To generate recombinants, we transfected pHKP1-derived or pMH54P4-derived donor RNAs into feline cells that were infected with fMHV (24), a chimeric mutant containing the ectodomain of the S protein of FIPV in place of the corresponding region of the MHV S protein (Fig. 2). Owing to this substitution, fMHV is capable of growth only in feline cells. We then isolated recombinants that had regained the MHV S ectodomain, as a result of a homologous crossover event either in gene 1b (for pHKP1 donor RNAs) or in the HE gene (for pMH54P4 donor RNAs), by selecting for progeny virus that had regained the ability to grow in mouse cells (Fig. 2). All mutant viruses were plaque purified, and the presence of incorporated genes and mutations was confirmed by RT-PCR and sequencing as described in Materials and Methods.
FIG. 2.
Construction of Hook protein and S protein mutants by targeted RNA recombination with the interspecies chimera fMHV (24), which contains the ectodomain-encoding region of the FIPV S gene (hatched rectangle) and is consequently able to grow in feline cells, but not in murine cells. Viruses expressing the Hook protein or mutants thereof, in addition to the wild-type S protein, were obtained by selection for recombination between fMHV and donor RNAs transcribed from plasmids derived from pHKP1. Single crossover events in the 3′-most 1.2 kb of gene 1b generated recombinants that had obtained the exogenous Hook gene and had simultaneously reacquired the MHV S ectodomain, which conferred the ability to grow in murine cells. Corresponding constructs expressing mutants of the S protein, in the absence of the Hook protein, were obtained by selection for recombination between fMHV and donor RNAs transcribed from plasmids derived from pMH54P4. In this case, single crossover events in the 3′-most 1.2 kb of the HE gene generated recombinants that had reacquired the MHV S ectodomain as well as the transmembrane domain or endodomain mutation(s), which (if not lethal) conferred the ability to grow in murine cells. In each case, the star represents a mutation or set of mutations.
Endodomain of MHV S protein mediates incorporation of Hook protein into virions.
Initially, we constructed viral recombinants expressing four variants of the Hook protein. These variants all had the same Hook protein ectodomain as that encoded by pHKP1 (Fig. 1), but each had a unique carboxy terminus (Fig. 3A). Mutants HK-M and HK-P contained the entire transmembrane domain and the endodomain from the MHV S protein and PDGFR, respectively. HK-M(Ed−) contained the MHV S protein transmembrane domain with just a three-amino-acid remnant of the 38-residue endodomain. HK-P(Ed+), on the other hand, contained the PDGFR transmembrane domain joined to the full endodomain of the MHV S protein. These four recombinants expressed their individual chimeric Hook proteins to comparable extents, as detected by a Western blot analysis of infected cell lysates (Fig. 3B). We noted that each of the Hook protein derivatives had a slower migration rate by sodium dodecyl sulfate-polyacrylamide gel electrophoresis than was expected for their predicted molecular masses of 36.0, 32.2, 35.3, and 38.3 kDa, for HK-M, HK-M(Ed−), HK-P, and HK-P(Ed+), respectively, but this was consistent with the anomalously slow migration shown previously for the parent construct (6).
FIG. 3.
MHV S protein endodomain mediates the incorporation of Hook protein into MHV virions. (A) Amino acid composition of the carboxy termini of four Hook protein constructs. The four types of mutants were HK-M, containing the transmembrane domain and endodomain from the MHV S protein; HK-M(Ed−), identical to HK-M with a truncated endodomain; HK-P, containing the transmembrane domain and endodomain from PDGFR (as in the original Hook construct [Fig. 1]); and HK-P(Ed+), identical to HK-P but with the PDGFR endodomain replaced with the endodomain of the MHV S protein. Transmembrane domain residues are shown in bold; underlined residues are part of the adjacent Myc epitope tag in the ectodomain. (B and C) Western blots of lysates from cells infected with Hook protein mutants (B) or of purified virions of Hook protein mutants (C). The Hook protein was detected with an antibody for the HA epitope tag; the amounts of lysate or virion samples analyzed were normalized to contain equivalent amounts of N protein, as detected by an anti-N antibody. (D) Plaques formed by viruses encoding the four variant Hook proteins. Wild-type MHV and the mutants HK-M, HK-M(Ed−), HK-P, and HK-P(Ed+) were titrated on mouse L2 cell monolayers, stained with neutral red at 44 h postinfection, and photographed 8 h later.
We next determined which of the Hook protein derivatives were incorporated into MHV virions that were extensively purified by two rounds of centrifugation in glycerol-tartrate gradients followed by size exclusion chromatography. As expected, the HK-M protein was incorporated into MHV virions (Fig. 3C), in accord with a previous demonstration that attachment of the FIPV S ectodomain to the MHV S transmembrane domain and endodomain allowed its assembly into MHV virions (24). In contrast, the entirely heterologous HK-P protein, with its transmembrane domain and endodomain derived from PDGFR, was not incorporated into virions. Interestingly, the nearly complete truncation of the MHV S endodomain in the HK-M(Ed−) protein abolished the ability to assemble into virions, whereas the transfer of the MHV S endodomain to replace its PDGFR counterpart in the HK-P(Ed+) protein conferred the ability to assemble into virions (Fig. 3C).
These results showed that the 38 amino acid residues of the carboxy-terminal endodomain of the MHV S protein are sufficient for the assembly of a heterologous membrane protein into virions. Consistent with this finding, recombinants that incorporated the Hook protein into virions formed somewhat smaller plaques (Fig. 3D) and grew to lower titers (data not shown) than did the wild type or recombinants that excluded the Hook protein. This suggests that assembly-competent variants of the Hook protein compete with the S protein for a saturable number of incorporation sites in virions.
Surprisingly, when we attempted to transfer the same set of four transmembrane domain and endodomain variants to the MHV S protein (in a background devoid of the Hook gene), only the counterpart of HK-M, i.e., a virus bearing the reconstructed wild-type S protein, yielded viable viral recombinants in multiple independent trials. Thus, despite the fact that a chimeric Hook protein [HK-P(Ed+)] containing the PDGFR transmembrane domain and the MHV S endodomain was incorporated into virions, this composition was not sufficient for S protein function. Therefore, although the S protein transmembrane domain is not required for assembly, it must contain sequence-specific determinants that participate in some other essential role in viral replication, possibly fusion, as has been suggested in one expression study (5).
The charged region of the S protein endodomain plays the major role in determining assembly into virions.
To examine in more detail the features of the S protein endodomain that govern assembly, we constructed a series of deletion mutations. The 38-residue endodomain comprises two subregions, an 18-residue cysteine-rich segment and a 27-residue charge-rich segment, which partially overlap each other (Fig. 4A). Initial experiments showed that a short carboxy-terminal truncation of 1, 3, 6, or 8 amino acids entering the charge-rich segment had no effect on Hook incorporation or S protein function (data not shown). Similarly, removal of the 12 carboxy-terminal residues of the HK-M protein (mutant HKΔ12) did not diminish its ability to assemble into virions (Fig. 4B). However, larger truncations of 15, 17, 20, and 22 amino acids effectively abolished the incorporation of HK-M protein into virions. Moreover, the addition of eight random residues to the HKΔ20 mutant, creating a protein (HKΔ20+) of the same size as HKΔ12, did not restore its ability to assemble into virions. This indicated that the residues contained within the span of amino acids 13 through 22 from the carboxy terminus of the molecule make a key contribution to the assembly of the S protein into virions. In contrast, a deletion of almost the entire cysteine-rich segment (mutant HKΔC) only partially decreased the incorporation of HK-M protein into virions (Fig. 4B), suggesting that this region plays a minor role in assembly.
FIG. 4.
Effect of partial deletions of the S protein endodomain on the incorporation of Hook protein into virions. (A) Amino acid composition of the carboxy termini of Hook protein or S protein constructs. M and M(Ed−) were as described in the legend for Fig. 3. All other mutants were derivatives of M with deletions of various numbers of endodomain residues, and in one case (Δ20+), a substitution of eight heterologous residues. Scores at the right indicate whether the corresponding Hook mutant (HK) incorporated the Hook protein into virions or whether the corresponding S protein mutant (S) was viable. nd, not determined. (B) Western blots of purified virions of Hook protein mutants or of lysates from cells infected with Hook protein mutants. The Hook protein was detected with an antibody for the HA epitope tag; the amounts of virions analyzed were normalized to contain equivalent amounts of N protein, as determined by staining with Coomassie blue.
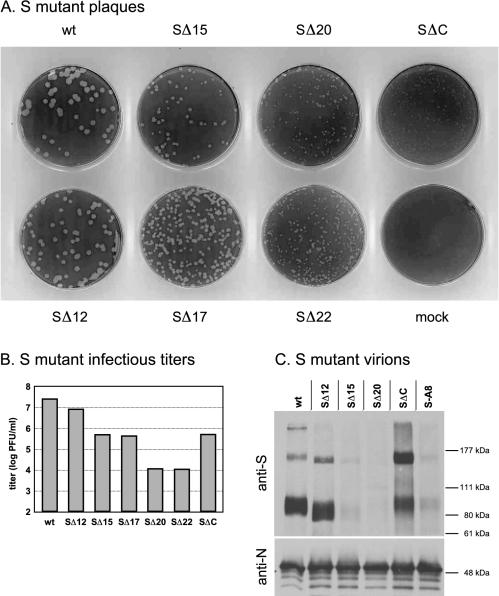
When these deletion mutations were transferred from Hook gene constructs into the MHV S gene in the absence of Hook, a broader range of functional effects was observed (Fig. 5). Most of the same mutations that reduced the virion assembly of the Hook protein to levels beneath detection, and that thus might have been expected to be lethal in the S protein, actually gave rise to viruses that were viable, albeit significantly impaired. This apparent greater impact of a given mutation in the Hook protein compared to its effect in the S protein likely stemmed from two sources. First, to be incorporated into virions, a mutant Hook protein had to compete with wild-type S protein. Second, the detection of the viability of an S protein mutant was more sensitive because viral titers many orders of magnitude below that of the wild type could be measured. The progressive truncation of residues from the carboxy terminus of the S protein resulted in a decrease in both plaque sizes (Fig. 5A) and viral infectious titers (Fig. 5B), and this loss of function occurred in discrete steps. The removal of the first 12 carboxy-terminal amino acids of the S endodomain (mutant SΔ12) had only a minimal phenotypic effect, consistent with the results obtained with the corresponding HKΔ12 mutant. However, the truncation of an additional three or five amino acids (to form mutants SΔ15 and SΔ17) resulted in a marked decrease in plaque size and a >50-fold drop in infectious titer compared to the wild type. A second transition occurred upon truncation of still another three or five amino acids (to form mutants SΔ20 and SΔ22). This resulted in a further decrease in plaque size and a further drop in infectious titers, to >40-fold lower levels than those obtained with the SΔ15 and SΔ17 mutants or 2,000-fold lower levels than that of the wild type. The sharp transitions that occurred between the SΔ12 and SΔ15 mutations and between the SΔ17 and SΔ20 mutations (Fig. 5B) suggest critical roles for the amino acids HQD and DEY, respectively. This finding more clearly delineated the importance of the interval of amino acids 13 through 22 from the carboxy terminus, which was previously identified as being pivotal for endodomain function in the Hook protein deletion analysis (Fig. 4B).
FIG. 5.
Effect of partial deletions of the S protein endodomain on viability of S protein mutants and incorporation of S protein into virions. (A) Plaques formed by viruses containing the S protein endodomain mutations listed in Fig. 4A. Wild-type MHV and mutants were titrated on mouse L2 cell monolayers, stained with neutral red at 44 h postinfection, and photographed 8 h later. (B) Comparison of maximal infectious titers of passage 3 stocks of S protein mutants. Monolayers of 17Cl1 cells were inoculated at a multiplicity of infection of 0.01 PFU per cell and were harvested when the cultures exhibited a complete cytopathic effect (at 40 h postinfection for the wild type, SΔ12, SΔ15, and SΔ17 viruses and at 72 h postinfection for the SΔ20, SΔ22, and SΔC viruses). (C) Western blots of purified virions of S protein mutants. The S protein was detected with an anti-S antibody; the amounts of virion samples analyzed were normalized to contain equivalent amounts of N protein, as detected by an anti-N antibody.
As noted above (Fig. 3), the truncation of 13 more residues beyond those truncated in SΔ22 resulted in a complete loss of function and was therefore lethal [mutant S-M(Ed−), equivalent to SΔ35]. The separate internal deletion of the same 13 residues, which constitute most of the cysteine-rich region, had an unusual effect with respect to the results obtained with the S protein set of carboxy-terminal endodomain truncations. Plaques of this mutant, SΔC, were as tiny as those of the most severely affected terminal truncation mutants, SΔ20 and SΔ22 (Fig. 5A). However, SΔC plaques had a cloudy appearance that was reminiscent of that of plaques of the nonfusogenic strain MHV-2 (44). In addition, the SΔC mutant grew much better than did SΔ20 and SΔ22, producing infectious titers at levels equivalent to those of SΔ15 and SΔ17 (Fig. 5B). These characteristics suggest that the defect in SΔC is qualitatively different from the effects caused by the carboxy-terminal truncations.
A direct examination of the virion assembly competence of S proteins containing endodomain deletions gave results consistent with the trends seen in plaque sizes and infectious titers for the carboxy-terminal truncation mutants (Fig. 5C). The S protein in the SΔ12 mutant was incorporated into virions at levels only slightly lower than those of the wild type, while S protein incorporation dropped significantly in the SΔ15 mutant and was barely detectable in the SΔ20 mutant. In contrast, S protein incorporation into virions of the SΔC mutant was reduced only minimally, to a degree similar to that of the SΔ12 mutant. This indicated that the altered plaque morphology and infectious titer of the SΔC virus were only partially due to decreased assembly of the S protein into virions, and it suggested that the SΔC mutation created some other defect in S protein function. In addition, we observed that the ratio of cleaved (90 kDa) to uncleaved (180 kDa) S protein in the SΔC mutant was markedly lower than that in the wild type or the terminally truncated S mutants (Fig. 5C). To check that this change did not result from some extraneous mutation in the S protein cleavage site (2, 29, 40), we sequenced a 200-amino-acid region of the S gene ectodomain encompassing the cleavage site in the SΔC mutant and confirmed that it was identical to the wild-type sequence.
To more closely evaluate the contributions of particular amino acids in the charge-rich segment of the endodomain, we constructed several point mutants in which individual residues or multiples of residues were changed to alanines (Fig. 6A). Our choice of residues was guided by the previous truncation results and by consideration of the most conserved elements of the aligned S protein endodomains of group 2 coronaviruses. No single point mutation of a charged amino acid to alanine had any detectable effect on the incorporation of the Hook protein into virions (Fig. 6B, mutants HK-A1 through HK-A5), nor were recombinants harboring the corresponding S gene mutations detectably impaired (data not shown). Curiously, the mutation of a highly conserved valine to alanine (mutant HK-A9) reduced the assembly competence of the Hook protein, although the deletion of the same amino acid (in mutant HKΔ12) had shown no effect. Combining charged residue-to-alanine mutations significantly reduced the incorporation of the Hook protein into virions (mutations HK-A6 through HK-A8). However, a phenotypic effect was observed for the S protein counterpart of only one of these, mutant S-A8, which had a slightly smaller plaque size than the wild type (Fig. 6C) and a reduced incorporation of S protein into virions, to a level comparable to that observed with mutant SΔ15 (Fig. 5C). It is noteworthy that the mutations in S-A8 (and HK-A8) abolished the charged residues among the two amino acid triplets, HQD and DEY, that had been identified as being critical in the truncation experiments. Taken together, these results indicate that no individual charged amino acid is essential for endodomain function. Additive effects were observed for the removal of multiple charged side chains, but none of these had as dramatic an effect on Hook protein incorporation or S mutant viability as the larger endodomain truncations.
FIG. 6.
Effect of point mutations in S protein endodomain on incorporation of Hook protein into virions or viability of S protein mutants. (A) Amino acid compositions of carboxy termini of Hook protein or S protein constructs. M is as described in the legend for Fig. 3. For all other mutants, only the mutated residue(s) is indicated. Scores at the right indicate whether the corresponding Hook mutant (HK) incorporated the Hook protein into virions or whether the corresponding S protein mutant (S) was viable. (B) Western blots of purified virions of Hook protein mutants or of lysates from cells infected with Hook protein mutants. The Hook protein was detected with an antibody for the HA epitope tag; the amounts of virions analyzed were normalized to contain equivalent amounts of N protein, as determined by staining with Coomassie blue. The HK-M and HK-M(Ed−) virion controls shown here are the same as those shown in Fig. 4A. (C) Plaques formed by mutant S-A8 and the wild type. Viruses were titrated on mouse L2 cell monolayers, stained with neutral red at 48 h postinfection, and photographed 8 h later.
Involvement of the cysteine-rich region of the S protein endodomain in cell-cell fusion.
The unusual plaque morphology of the SΔC mutant, as well as the retention of assembly competence by its Hook protein counterpart, HKΔC, prompted us to examine the ability of SΔC to induce cell-cell fusion during viral infection. Tissue culture growth of the SΔC mutant was rarely observed to give rise to syncytium formation in monolayers of either L2 or 17Cl1 cells, even at late times postinfection. No such defect was seen with any of the deletion or point mutants of the endodomain charge-rich region. With several other recombinant viruses harboring point mutations in the endodomain cysteine-rich region, we noted dramatically delayed syncytium formation (R. Ye and P. S. Masters, unpublished data), but with SΔC the block appeared to be nearly absolute. This was most clearly demonstrated by immunofluorescence analysis (Fig. 7). A low-multiplicity infection of L2 cells with SΔC resulted in widespread infection in the monolayer by 16 h postinfection, as judged by expression of the viral N protein. However, the fusion of adjacent cells into small syncytia of four to eight cells was only occasionally observed. In contrast, the more severely defective SΔ20 mutant, although it had a much lower viral yield than either SΔC or the wild type, induced syncytium formation comparable to that of the wild type.
FIG.7.
Immunofluorescence analysis of cells infected with wild-type MHV, the SΔ20 mutant, or the SΔC mutant. Infected or mock-infected L2 cell monolayers were stained at 16 h postinfection with an anti-N antibody, as described in Materials and Methods, and were viewed at a magnification of ×200. Left panels, fluorescence microscopy; right panels, differential interference contrast images of the same fields. The amino acid sequences of the carboxy termini of the S proteins of the three viruses are shown at the bottom.
DISCUSSION
In the work reported here we have shown that the incorporation of the MHV S protein into virions is dependent upon the presence of the endodomain of the S molecule. More specifically, the carboxy-terminal, charge-rich region of the endodomain makes the major contribution to the selective inclusion of the S protein into virions. The adjacent cysteine-rich region of the endodomain makes a minor contribution to assembly, and it also plays a critical role in cell-cell fusion. The transmembrane domain of the S protein is not required for assembly, but it performs some other function that is essential for infectivity.
Part of our investigation took advantage of a surrogate for the S protein, a chimeric transmembrane protein designated Hook, that we could manipulate without harming virion infectivity. The Hook gene was inserted at a genomic site between gene 1b and the S gene, replacing two nonessential genes (Fig. 1), in a position and context that was previously shown to support the expression of an essential viral structural protein in a rearranged MHV genome (14). The resulting abundant expression of the Hook protein demonstrated that this site can be used successfully for the expression of heterologous genes in MHV, thereby increasing the repertoire of this virus as a vector. It has previously been shown that the green fluorescent protein gene can be inserted into the MHV genome in place of gene 4 (9, 16). Very recently, de Haan and coworkers surveyed a large number of additional loci for the insertion of foreign genes, and they achieved stable expression of Renilla or firefly luciferase, or both, from many of these sites (12).
We observed that the HK-M protein, a chimeric construct linking the Hook ectodomain to the transmembrane domain and endodomain of the MHV S protein, was specifically incorporated into virions (Fig. 3). This finding was expected on the basis of previous results from VLP systems (19) as well as from the construction of the two interspecies chimeric viruses fMHV (24), in which the FIPV S ectodomain is joined to the transmembrane domain and endodomain of MHV S, and mFIPV (21), in which the MHV S ectodomain is joined to the transmembrane domain and endodomain of FIPV S. However, although the MHV and FIPV S ectodomains are highly divergent, there is some limited homology between them, especially within their carboxy-terminal portions. In addition, there may be functionally homologous regions between the two ectodomains that do not possess extensive primary sequence conservation. Thus, the possible participation of some part of the coronavirus S protein ectodomain in assembly could not previously be strictly ruled out. Since the Hook ectodomain is totally unrelated to the coronavirus S ectodomain, the assembly competence of the HK-M protein formally proved that only the MHV transmembrane domain and endodomain contain a determinant sufficient for incorporation into MHV virions. We then more precisely mapped this determinant through the construction of additional chimeras and found that it could be localized solely to the endodomain.
Our further dissection of the 38-amino-acid MHV S endodomain was based on its demarcation into two partially overlapping subdomains, an amino-terminal cysteine-rich region and a carboxy-terminal charge-rich region (Fig. 4A). This layout, which was previously noted by others (2, 5), is conserved among all coronaviruses, despite there being little primary endodomain sequence conservation across coronavirus groups, and in some cases, within groups. Throughout this report, we have defined the boundaries of the transmembrane domain and the endodomain on the basis of hydrophobicity, largely in accord with the scheme described by Chang et al. (5); however, note that another report has considered most of the cysteine-rich region to be part of the transmembrane domain (2). It remains to be experimentally determined which residues are buried in the membrane bilayer (and this topology may vary between S protein in virions and S protein at the plasma membrane). Nevertheless, this difference in the assignment of boundaries does not affect comparisons of our conclusions with those of other investigators (2, 5).
Based on our results, integrated with those from many prior studies, our working model of the S protein endodomain is that the main role of the charge-rich region occurs in virion assembly and that this is accomplished through S protein-M protein interactions. Conversely, the main role of the cysteine-rich region is in cell-cell membrane fusion, and this is accomplished through S protein-S protein interactions. The most distinctive feature of the charge-rich region is its hydrophilicity. It contains 7 charged and 12 polar residues in a 27-amino-acid span, with a net charge of −4 (with the inclusion of the negative charge of the carboxy terminus). Deletions extending 20 or 22 residues into this region abolished HK-M protein incorporation into virions (Fig. 4) and impaired S protein incorporation sufficiently to reduce the viral yield by >3 orders of magnitude (Fig. 5). However, we could define no single essential charged residue through the construction of point mutants, and even clusters of point mutations were not as detrimental as smaller deletions had been (Fig. 6). This suggests that multiple contacts exist between this endodomain region and its interacting partner, presumably the M protein. Moreover, although charged residues are the most salient characteristic of this subdomain, it will be necessary to investigate more completely the contributions of other types of residues. The association of S protein with M protein has been examined by coimmunoprecipitation of complexes from infected cells or from cells expressing both proteins (31, 33). An analysis of such complexes in the VLP system has begun to localize the regions of M required for this interaction (11). We are now seeking to establish genetic selections to reveal contacts between the S endodomain charge-rich region and the M protein in virions. However, such an approach is complicated by the absolutely central role of the M protein in viral assembly and the multiplicity of protein-protein interactions in which M participates.
At later times during infection, the fraction of newly synthesized S protein that has migrated to the plasma membrane without assembling into virions mediates syncytium formation between infected and adjacent uninfected cells. Although fusion is primarily the responsibility of the ectodomain (4, 18, 27), the cooperation of part of the endodomain is also required. The cysteine-rich region of the S protein endodomain comprises 9 cysteines in a span of 18 amino acids, 7 of which overlap the amino-terminal end of the charge-rich region (Fig. 4). Because they face the reducing environment of the cytoplasm, it seems unlikely that the cysteines participate in intramolecular or intermolecular disulfide bonds, and it is also improbable that they are involved in RNA binding. It is known that at least one of the cysteines is modified by palmitoylation (2, 40). The involvement of this subdomain in cell-cell fusion was discovered by Bos and coworkers through expression studies with chimeric S proteins containing the endodomain of the vesicular stomatitis virus G protein in place of various intervals of the S protein endodomain (2). An analysis of point mutants further suggested a dominant role in fusion for one of the residues in the triplet of cysteines at the amino-terminal end of the cysteine-rich region. A subsequent expression study by Chang and coworkers (5), which also examined chimeric constructs and point mutations, more quantitatively addressed the role of various cysteine residues. This study similarly suggested a strong contribution to fusion by the first residue in the cysteine triplet. In addition, both studies concluded that there was no detectable role of the charge-rich region in cell-cell fusion. Our results are consistent with most of these observations.
The striking fusion-negative phenotype of the SΔC mutant (Fig. 5A and 7), the first coronavirus bearing such an endodomain mutation, establishes the fact that the significance of the cysteine-rich region, previously inferred from expression studies, carries through to the intact virus. It is intriguing that the deletion of most of the cysteine-rich region only minimally affected the assembly into virions (Fig. 4B, mutant HKΔC, and Fig. 5C, mutant SΔC). This may indicate that the cysteine-rich region loops out of the endodomain structure in such a way that its absence does not grossly alter the distance between the charge-rich region and its interacting partner. It is also noteworthy that the critical cysteine triplet is retained in the SΔC mutant, suggesting that its presence, although possibly necessary for fusion, is not sufficient by itself.
Although the SΔC mutant is severely defective in the induction of cell-cell fusion, it is able to form infectious virions, indicating that its capability for virus-cell fusion is at least partially intact. This may suggest that there is a basic mechanistic difference between the two types of fusion events. Alternatively, we cannot rule out the possibility that the almost complete failure of the SΔC protein to cause syncytium formation results from a block in its transport to the plasma membrane. This seems unlikely, because all of the previously described cysteine-rich region mutants, including the chimeras, were shown to be expressed at the cell surface (2, 5). Studies that are currently under way may further elucidate the role of the cysteine-rich region in cell-cell membrane fusion.
Acknowledgments
We thank Lili Kuo for providing extensive critical advice throughout the course of this work, and we are grateful to Kelley Hurst for valuable assistance in immunofluorescence experiments. We thank Kathryn Holmes, University of Colorado Health Sciences Center, for generously providing an anti-S antibody. We are indebted to Kathleen McDonough for her considerable help with microscopy, and we acknowledge the use of the DAI Microscopy Core Facility. We also thank the Molecular Genetics Core Facility of the Wadsworth Center for oligonucleotide synthesis and DNA sequencing. We are grateful to Stuart Lehrman of the Wadsworth Center Computerized Photography and Illustration Unit for his expert assistance in the preparation of figures.
This work was supported by Public Health Service grants AI 39544 and AI 45695 from the National Institutes of Health.
REFERENCES
- 1.Baudoux, P., C. Carrat, L. Besnardeau, B. Charley, and H. Laude. 1998. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 72:8636-8643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bos, E. C. W., L. Heijnen, W. Luytjes, and W. J. M. Spaan. 1995. Mutational analysis of the murine coronavirus spike protein: effect on cell-to-cell fusion. Virology 214:453-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bos, E. C. W., W. Luytjes, H. van der Meulen, H. K. Koerten, and W. J. M. Spaan. 1996. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology 218:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bosch, B. J., R. van der Zee, C. A. M. de Haan, and P. J. M. Rottier. 2003. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 77:8801-8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, K. W., Y. W. Sheng, and J. L. Gombold. 2000. Coronavirus-induced membrane fusion requires the cysteine-rich domain in the spike protein. Virology 269:212-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chesnut, J. D., A. R. Baytan, M. Russell, M.-P. Chang, A. Bernard, I. H. Maxwell, and J. P. Hoeffler. 1996. Selective isolation of transiently transfected cells from a mammalian cell population with vectors expressing a membrane anchored single-chain antibody. J. Immunol. Methods 193:17-27. [DOI] [PubMed] [Google Scholar]
- 7.Corse, E., and C. E. Machamer. 2003. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology 312:25-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Curtis, K. M., B. Yount, and R. S. Baric. 2002. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 76:1422-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das Sarma, J., E. Scheen, S. H. Seo, M. Koval, and S. R. Weiss. 2002. Enhanced green fluorescent protein expression may be used to monitor murine coronavirus spread in vitro and in the mouse central nervous system. J. Neurovirol. 8:381-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Haan, C. A. M., P. S. Masters, X. Shen, S. Weiss, and P. J. M. Rottier. 2002. The group-specific murine coronavirus genes are not essential, but their deletion, by reverse genetics, is attenuating in the natural host. Virology 296:177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Haan, C. A. M., M. Smeets, F. Vernooij, H. Vennema, and P. J. M. Rottier. 1999. Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein. J. Virol. 73:7441-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Haan, C. A. M., L. van Genne, J. N. Stoop, H. Volders, and P. J. M. Rottier. 2003. Coronaviruses as vectors: position dependence of foreign gene expression. J. Virol. 77:11312-11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Haan, C. A. M., H. Vennema, and P. J. M. Rottier. 2000. Assembly of the coronavirus envelope: homotypic interactions between the M proteins. J. Virol. 74:4967-4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Haan, C. A. M., H. Volders, C. A. Koetzner, P. S. Masters, and P. J. M. Rottier. 2002. Coronaviruses maintain viability despite dramatic rearrangements of the strictly conserved genome organization. J. Virol. 76:12491-12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Escors, D., J. Ortego, H. Laude, and L. Enjuanes. 2001. The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability. J. Virol. 75:1312-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer, F., C. F. Stegen, C. A. Koetzner, and P. S. Masters. 1997. Analysis of a recombinant mouse hepatitis virus expressing a foreign gene reveals a novel aspect of coronavirus transcription. J. Virol. 71:5148-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer, F., C. F. Stegen, P. S. Masters, and W. A. Samsonoff. 1998. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 72:7885-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallagher, T. M., C. Escarmis, and M. J. Buchmeier. 1991. Alteration of the pH dependence of coronavirus-induced cell fusion: effect of mutations in the spike glycoprotein. J. Virol. 65:1916-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godeke, G. J., C. A. de Haan, J. W. Rossen, H. Vennema, and P. J. M. Rottier. 2000. Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. J. Virol. 74:1566-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goebel, S. J., B. Hsue, T. F. Dombrowski, and P. S. Masters. 2004. Characterization of the RNA components of a putative molecular switch in the 3′ untranslated region of the murine coronavirus genome. J. Virol. 78:669-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haijema, B. J., H. Volders, and P. J. M. Rottier. 2003. Switching species tropism: an effective way to manipulate the feline coronavirus genome. J. Virol. 77:4528-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holmes, K. V., E. W. Dollar, and L. S. Sturman. 1981. Tunicamycin resistant glycosylation of a coronavirus glycoprotein: demonstration of a novel type of viral glycoprotein. Virology 115:334-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horton, R. M., and L. R. Pease. 1991. Recombination and mutagenesis of DNA sequences using PCR, p. 217-247. In M. J. McPherson (ed.), Directed mutagenesis, a practical approach. IRL Press, New York, N.Y.
- 24.Kuo, L., G.-J. Godeke, M. J. B. Raamsman, P. S. Masters, and P. J. M. Rottier. 2000. Retargeting of coronavirus by substitution of the spike glycoprotein ectodomain: crossing the host cell species barrier. J. Virol. 74:1393-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuo, L., and P. S. Masters. 2002. Genetic evidence for a structural interaction between the carboxy termini of the membrane and nucleocapsid proteins of mouse hepatitis virus. J. Virol. 76:4987-4999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuo, L., and P. S. Masters. 2003. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 77:4597-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo, Z., and S. R. Weiss. 1998. Roles in cell-to-cell fusion of two conserved hydrophobic regions in the murine coronavirus spike protein. Virology 244:483-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luytjes, W., P. J. Bredenbeek, A. F. H. Noten, M. C. Horzinek, and W. J. M. Spaan. 1988. Sequence of mouse hepatitis virus A59 mRNA2: indications for RNA recombination between coronaviruses and influenza C virus. Virology 166:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luytjes, W., L. S. Sturman, P. J. Bredenbeek, J. Charite, B. A. M. van der Zeijst, M. C. Horzinek, and W. J. M. Spaan. 1987. Primary structure of the glycoprotein E2 of coronavirus MHV-A59 and identification of the trypsin cleavage site. Virology 161:479-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Narayanan, K., and S. Makino. 2001. Cooperation of an RNA packaging signal and a viral envelope protein in coronavirus RNA packaging. J. Virol. 75:9059-9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen, V.-P., and B. Hogue. 1997. Protein interactions during coronavirus assembly. J. Virol. 71:9278-9284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ontiveros, E., T. S. Kim, T. M. Gallagher, and S. Perlman. 2003. Enhanced virulence mediated by the murine coronavirus, mouse hepatitis virus strain JHM, is associated with a glycine at residue 310 of the spike glycoprotein. J. Virol. 77:10260-10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Opstelten, D.-J. E., M. J. B. Raamsman, K. Wolfs, M. C. Horzinek, and P. J. M. Rottier. 1995. Envelope glycoprotein interactions in coronavirus assembly. J. Cell Biol. 131:339-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ortego, J., D. Escors, H. Laude, and L. Enjuanes. 2002. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 76:11518-11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Phillips, J. J., M. M. Chua, E. Lavi, and S. R. Weiss. 1999. Pathogenesis of chimeric MHV4/MHV-A59 recombinant viruses: the murine coronavirus spike protein is a major determinant of neurovirulence. J. Virol. 73:7752-7760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rottier, P. J. M., M. C. Horzinek, and B. A. M. van der Zeijst. 1981. Viral protein synthesis in mouse hepatitis virus strain A59-infected cells: effects of tunicamycin. J. Virol. 40:350-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Schwarz, B., E. Routledge, and S. G. Siddell. 1990. Murine nonstructural protein ns2 is not essential for virus replication in transformed cells. J. Virol. 64:4784-4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sturman, L. S., K. V. Holmes, and J. Behnke. 1980. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 33:449-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sturman, L. S., C. S. Ricard, and K. V. Holmes. 1985. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: activation of cell-fusing activity of virions by trypsin and separation of two different 90K cleavage fragments. J. Virol. 56:904-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan, K., B. D. Zelus, R. Meijers, J. H. Liu, J. M. Bergelson, N. Duke, R. Zhang, A. Joachimiak, K. V. Holmes, and J. H. Wang. 2002. Crystal structure of murine sCEACAM1a[1,4]: a coronavirus receptor in the CEA family. EMBO J. 21:2076-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vennema, H., G.-J. Godeke, J. W. A. Rossen, W. F. Voorhout, M. C. Horzinek, D.-J. E. Opstelten, and P. J. M. Rottier. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15:2020-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wentworth, D. E., and K. V. Holmes. 2001. Molecular determinants of species specificity in the coronavirus receptor aminopeptidase N (CD13): influence of N-linked glycosylation. J. Virol. 75:9741-9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamada, Y. K., K. Takimoto, M. Yabe, and F. Taguchi. 1997. Acquired fusion activity of a murine coronavirus MHV-2 variant with mutations in the proteolytic cleavage site and the signal sequence of the S protein. Virology 227:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]