Abstract
Adenovirus serotype 5 (Ad5) vectors containing Ad B-group fibers have become increasingly popular as gene transfer vectors because they efficiently transduce human cell types that are relatively refractory to Ad5 infection. So far, most B-group fiber-containing vectors have been first-generation vectors, deleted of E1 and/or E3 genes. Transduction with these vectors, however, results in viral gene expression and is associated with cytotoxicity and immune responses against transduced cells. To circumvent these problems, we developed fiber-chimeric Ad vectors devoid of all viral genes that were produced either by the homologous recombination of first-generation vectors or by using the Cre/lox-based helper virus system. In this study we compared early steps of infection between first-generation (35-kb genome) and Ad vectors devoid of all viral genes with genome sizes of 28 kb and 12.6 kb. All vectors possessed an Ad35-derived fiber knob domain, which uses CD46 as a primary attachment receptor. Using immortalized human hematopoietic cell lines and primary human CD34-positive hematopoietic cells, we found that the Ad genome size did not affect the efficiency of virus attachment to and internalization into cells. Furthermore, independently of the genome length and structure, all vectors migrated to the nucleus through late endosomal and lysosomal cellular compartments. However, the vector containing the short 12.6-kb genome was unable to efficiently escape from endosomes and deliver its DNA into the nucleus. Moreover, compared to other vectors, these Ad particles were less stable and had an abnormal capsid protein composition, including a lack of capsid-stabilizing protein IX. Our data indicate that the size and structure of the packaged viral genomes can affect the integrity of Ad particles, which in turn results in lower infectivity of Ad vectors.
Adenoviruses (Ad) have been used as a tool to efficiently deliver transgenes into a wide variety of cell types in vitro and in vivo. Most Ad vectors generated so far are based on human group C Ad serotype 5 (Ad5). Ad5 binds to cells via interaction between the viral fiber protein and its cellular receptor, the coxsackie and Ad receptor (CAR) (1, 32). However, many important therapeutic targets, including hematopoietic stem cells, dendritic cells, and primary tumor cells, express only low levels of CAR and are, therefore, inefficiently infected by Ad5-based vectors (20, 31, 45, 50). Chimeric Ad5 vectors possessing fiber proteins derived from subgroup B Ad serotypes such as Ad11, Ad16, Ad35, or Ad50 have become increasingly popular as gene transfer vectors because they can efficiently deliver genes to cell types that are refractory to Ad5 infection (11, 28, 33, 36, 37, 40, 47). Recently, it has been demonstrated that most group B Ads utilize CD46 as a primary attachment receptor (10, 35, 41, 52). The binding of chimeric Ad5-based vectors possessing group B Ad35 or Ad7 fibers to cellular attachment receptor(s) was found to predetermine incoming virus particles to an intracellular trafficking route that is different from that of unmodified Ad5 vectors (26, 27, 38). While Ad5 efficiently escaped from the endosomal environment early after infection, Ad particles with Ad35 fibers remained in late endosomal and lysosomal compartments and used these compartments to achieve localization to the nucleus. However, a significant number of these particles appeared to be retrogradely transported and deposited at the cell surface.
First-generation Ad vectors deleted of E1 and E3 cause significant cytotoxicity due to the expression of viral genes in infected cells (16). In vivo, the expression of viral genes from first-generation Ad vectors results in the elimination of transduced cells by cytotoxic T lymphocytes and, therefore, the extinction of transgene expression. To reduce the cytotoxic effects of Ads and achieve long-term transgene expression in vivo, vectors devoid of all viral genes, so-called “gutless” or helper-dependent (HD) vectors, were developed. In HD vectors, the entire coding region of the Ad genome is replaced with heterologous DNA containing the transgene of interest and inert stuffer DNA. HD vectors conferred efficient gene transfer in vitro with dramatically reduced cytotoxicity and prolonged therapeutic transgene expression after in vivo application (16).
Recently, investigators developed Ad5 vectors with Ad35 fibers containing small genomes (up to 16 kb) devoid of all viral genes generated though homologous recombination between two first-generation Ad genomes (ΔAd) (5, 6, 36, 42). The yields of these ΔAd vectors produced in 293 cells were 5 to 10 times lower than those of full-length vectors. Overall, ΔAd5 vectors transduced human hematopoietic cell lines less efficiently than first-generation vectors. Furthermore, after serial amplification and determining the titers of HD vectors with different genome sizes, Parks and Graham (29) found that yields of vectors with genome sizes less than about 75% of the wild-type genome were roughly 50% lower than yields of vectors with full-length genomes. Although this study demonstrated a disadvantage in the amplification of Ads with shortened genomes, it did not definitively demonstrate which step of the Ad life cycle was deficient for these viruses. To achieve efficient infection, Ad must undergo attachment, internalization, endosome escape, intracellular trafficking, nuclear pore docking, genome translocation into the nucleus, viral replication, and packaging. A deficiency in any of these steps may result in reduced vector yields during virus amplification.
To investigate the effect of genome size on the infectivity of Ad vectors, we compared in this study the early steps of infection between first-generation (35-kb) vectors and Ad vectors devoid of all viral genes with genome sizes of 28 kb (HD vectors) and 12.6 kb (ΔAd vectors). All these vectors possess an Ad35-derived fiber knob domain, which recognizes CD46 as a primary attachment receptor. Using immortalized human hematopoietic cell lines and primary human CD34-positive hematopoietic cells, we found that regardless of the genome size, cellular uptake of all three vectors was equally efficient. Furthermore, all three vectors migrated to the nucleus within late endosomal or lysosomal cellular compartments. However, in contrast to first-generation and HD vectors, ΔAd particles containing the 12.6-kb genome were unable to efficiently escape from endosomes and deliver their DNA into the nucleus. We also found that ΔAd particles were less stable and possessed abnormal capsid protein composition, including a lack of the capsid-stabilizing protein IX.
MATERIALS AND METHODS
Cells and viruses.
Human leukemic cell lines MO7e (18), HEL (19), and K562 (ATCC 45506) were maintained in RPMI 1640 medium containing 10% fetal calf serum (FCS), 2 mM l-glutamine, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. For culturing MO7e cells, 0.1 ng of granulocyte-macrophage colony stimulating factor (Immunex, Seattle, Wash.) per ml was added to the medium. Primary human CD34+-enriched cells were purified from cord blood by MiniMACS LS+ separation columns and a CD34-progenitor cell isolation kit (Miltenyi Biotec, Auburn, Calif.), according to the manufacturer's protocol. Aliquots of cells were stored in liquid nitrogen. Sixteen hours before the experiment, cells were recovered from frozen stocks and incubated overnight in Iscove modified Dulbecco's medium, supplemented with 20% FCS, 10−4 M β-mercaptoethanol, 100 μg of DNaseI per ml, 2 mM glutamine, 10 U of IL-3 per ml, 50 ng of stem cell factor per ml, and 10 ng of thrombopoietin per ml. The purity of CD34+ cell preparations was verified by flow cytometry and was consistently greater than 90%.
For propagation of Ad vectors, 293 cells (Microbix, Toronto, Canada) were maintained in Dulbecco's modified Eagle's medium, 10% FCS, and 2 mM glutamine. All Ad vectors used in this study possess identical green fluorescent protein (GFP) gene expression cassettes driven by the mouse stem cell virus (MSCV) promoter. Ad5/35 (first-generation 35-kb genomes) and Δ5/35 (deleted for all viral genes; 12.6-kb genome) were described in an earlier study where they were named AdAAV-b and ΔAdAAVF35, respectively (36). To generate the HD Ad5/35 vector (28-kb genome), a protocol described elsewhere was modified (15). The helper vector was based on pHPBG containing an Ad5 E1/E3-deleted genome with lox sites flanking the packaging signal. In pHPBG, the sequence corresponding to the Ad5 fiber knob domain was replaced with the sequence encoding the Ad35 fiber knob domain by homologous recombination in Escherichia coli (7). The resulting plasmid pHPBG-F35 was transfected into 293 cells and the helper virus AdHPBGF35 was rescued, amplified, and purified by using standard Ad propagation and purification techniques (6). To obtain the HD GFP-expressing Ad vector, the red fluorescent protein expression cassette in pHCARedFPneo was replaced with an MSCV-GFP expression cassette by homologous recombination in E. coli. The resulting plasmid pHCA-MSCV-GFP contained the Ad 5′ inverted terminal repeat and packaging signal, the 3′ inverted terminal repeat, a MSCV-GFP expression cassette identical to those in Ad5/35 and Δ5/35, and stuffer DNA from the human X chromosome to obtain a final genome length of 28 kb. To produce the HD, GFP-expressing vector HD5/35, pHPBGF35 and pHCA-MSCV-GFP were cotransfected into C7-CRE cells (15) by using the calcium phosphate coprecipitation technique. HD5/35 was amplified and purified as described elsewhere (15). The contamination of Δ5/35 and HD5/35 preparations with first-generation or helper vector was measured by quantitative Southern blotting (6). For all experiments described, only Δ5/35 and HD5/35 vector stocks containing less than 2% of contaminating first-generation or helper virus was used. Ad genome titers were determined by quantitative Southern blotting. For each Ad vector preparation, virion DNA was extracted from purified viral particles and was run on agarose gels in serial (twofold) dilutions with standard DNA of known concentrations (preparatively purified Ad5/35 DNA). After transfer onto Hybond N+ nylon membranes (Amersham, Piscataway, N.J.), filters were hybridized with a 32P-labeled DNA probe (0.7-kb fragment, corresponding to the GFP gene of pEGFP-1; Clontech, Palo Alto, Calif.), and DNA concentrations were measured by PhosphorImager. These values were used to calculate the genome titer for each virus stock used. For each Ad vector used in this study, at least two independent virus stocks were produced and characterized by determining PFU titers on 293 cells and genome titers by Southern blotting.
Labeling of Ads with Cy3- or Cy2-fluorochromes.
To label Ad capsids with Cy3 (red) and Cy2 (green) fluorochromes (Cy3 and Cy2 Bifunctional Reactive Dyes; Amersham Pharmacia Biotech, Little Chalfont, United Kingdom), we used the manufacturer's protocol without modifications. The ratio between the volumes of Ad and labeling reagent was 1:9. Labeled viruses were dialyzed against 10 mM Tris-HCl (pH 7.5), 10 mM MgCl2, and 10% glycerol solution at 4°C overnight to remove unincorporated chemicals. The concentrations of dye-labeled viruses were determined by quantitative Southern blotting as described above.
Attachment and internalization assays.
The attachment and internalization studies were based on a protocol published elsewhere (39). For these assays, 3.5 × 105 MO7e cells were incubated for 1 h on ice with equal amounts of Ad particles, corresponding to a multiplicity of infection (MOI) of 8,000 genomes per cell in 100 μl of ice-cold adhesion buffer (Dulbecco's modified Eagle's medium supplemented with 2 mM MgCl2, 1% bovine serum albumin [BSA], and 20 mM HEPES). Cells were then pelleted by centrifugation at 1,000 × g for 4 min and washed two times with 0.5 ml of ice-cold phosphate-buffered saline (PBS). After the last washing step, the cells were pelleted at 1,500 × g, the supernatant was removed, cells were lysed, and total cellular DNA was extracted as described previously. To determine the fraction of internalized Ad particles, cells were incubated on ice for 1 h with the corresponding virus, washed with PBS as described above, resuspended in 100 μl of adhesion buffer, and then incubated at 37°C for 30 min. Following this incubation, cells were diluted threefold with cold 0.05% trypsin-0.5 mM EDTA solution and incubated at 37°C for an additional 5 to 10 min. Finally, the cells were pelleted at 1,500 × g for 5 min, the supernatant was removed, and total cellular DNA was extracted. Subsequently, a Southern blot analysis was performed to evaluate the attachment and internalization efficiencies for different Ad vectors on MO7e cells.
Ad infection of cells and analysis of Ad trafficking.
A total of 3 × 105 MO7e, HEL, K562, or primary human CD34+-enriched cells were infected at the indicated MOIs (expressed as viral genomes per cell) in 400 μl of growth medium. Cells were incubated for 6 h at 37°C. Then, virus-containing media was removed; cells were washed once with PBS and incubated in normal medium for 24 h before analysis by flow cytometry. To analyze intracellular trafficking of Ad vectors, MO7e cells (3 ×105 cells per well) were pulse-infected with Cy-3- or Cy-2-labeled Ads in a total volume of 100 μl of medium containing 1010 virus particles per ml. After incubation at 37°C for 15 min, virus-containing media was removed, and cells were incubated in growth medium for the indicated period of time before fixation in a methanol-acetone mixture (1:1, vol/vol). For analysis of Ad attachment, cells were incubated with viruses for 15 min at 37°C, washed with cold PBS, and immediately fixed for further analysis. For analysis of Ad particle localization to late endosomal or lysosomal compartments, fixed cells were incubated with primary polyclonal rabbit anti-cathepsin B antibody (Ab-3) (1/40 dilution; Oncogene, Boston, Mass.) for 1 h at 37°C. The binding of primary antibody was developed with a 1/200 dilution of Alexa Fluor 488-conjugated goat anti-rabbit secondary antibody (Molecular Probes, Eugene, Ore.) at room temperature for 30 min and visualized on a Leica fluorescence microscope or on a confocal microscope. To visualize cell nuclei for confocal microscopy analyses, samples were stained with ToPro dye (Molecular Probes, Eugene, Ore.). To visualize cellular centrosomes, samples were stained with anti-γ-tubulin primary mouse monoclonal antibody (Sigma, St. Louis, Mo.). The binding of primary antibodies was developed with Alexa Fluor 488-conjugated rat anti-mouse antibody and analyzed by confocal microscopy. Multiple images for each experimental condition were obtained and representative pictures are shown.
Electron microscopy (EM) studies.
For analysis of virus distribution, MO7e cells were infected with Ad vectors at an MOI of 2,000 genomes per cell. At the indicated time points, cells were fixed with 2% glutaraldehyde in PBS with subsequent fixation in 1% OsO4-phosphate buffer. Cells were then embedded in Medcast (Ted Pella, Redding, Calif.) and ultrathin sections were stained with uranyl acetate and lead citrate. Processed grids were evaluated and photomicrographed with a Phillips 410 electron microscope, operated at 80 kV (magnification, ×21,000). For each particular Ad vector, the intracellular distribution was analyzed by counting at least 100 virus-containing cells.
Southern blot analysis.
Extraction of genomic DNA, labeling of DNA fragments, and hybridization were performed as described earlier (6, 30).
Cellular membrane permeabilization assay.
Ad vector-mediated cell membrane permeabilization assays were performed as described elsewhere (48) without any modifications. Briefly, 5 × 106 to 7 × 106 MO7e cells in 15 ml of medium were incubated with [3H]choline chloride (2 μCi/ml; NEN Life Science Products, Inc., Boston, Mass.) at 37°C for 90 min. Cells were then chilled on ice and washed three times with ice-cold virus binding buffer (10 mM HEPES-buffered saline [pH 7.2], 0.2% BSA, 1 mM CaCl2, 1 mM MgCl2, 50 mM NaN3). Then, 4 × 1010 virus particles were added to 5 × 105 cells, and the samples were incubated on ice for 1 h to allow for virus attachment. Unattached virus particles were then washed with virus binding buffer, and cells were resuspended in 400 μl of cell permeabilization buffers (50 mM MES [morpholineethanesulfonic acid], 0.2% BSA, 1 mM CaCl2, 1 mM MgCl2, 50 mM NaN3) with pH values varying from 7.0 to 5.0. After the incubation of cells at 37°C for 1 h, cells were pelleted by centrifugation, and the amount of radioactive choline released into the supernatant was measured by using a scintillation counter. Cells incubated with the permeabilization buffers in the absence of viruses were used to measure nonspecific choline release. The percentage of choline release, mediated by Ad particles, was calculated by using the following formula: [(amount of choline released with Ad − amount of choline released without Ad)/amount of choline released without Ad] × 100. The data presented are the results obtained from two independent experiments performed in quadruplicate.
RESULTS
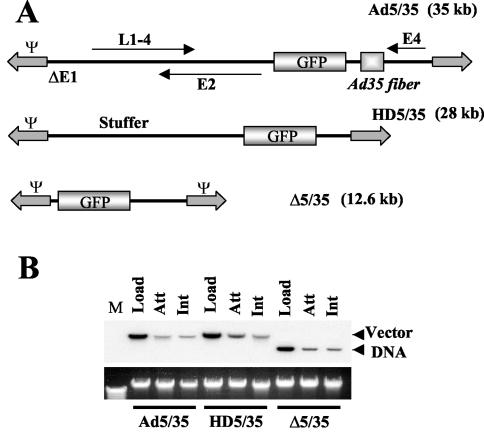
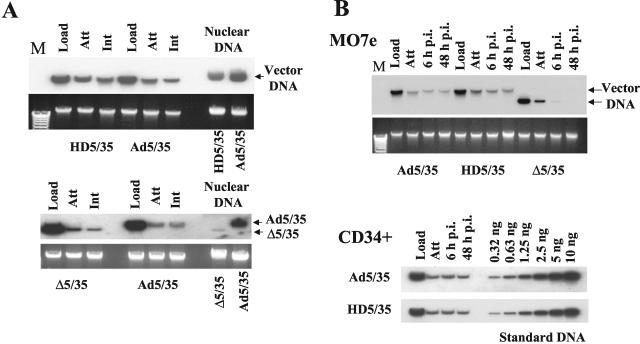
Ad cell attachment and internalization efficiency are comparable for vectors that differ in genome length and structure. To analyze the influence of genome size and structure on the efficiency of Ad attachment and internalization, a set of Ad vectors with different genome sizes was used (Fig. 1A). All of these vectors possess a GFP reporter gene driven by the MSCV promoter. Ad5/35 is a first-generation Ad vector with a 35-kb genome (36). HD5/35 (28-kb genome) is an HD vector with stuffer DNA instead of viral genes. HD5/35 was produced according to standard protocols for HD Ads (15, 34) by using a helper virus possessing an Ad35 fiber knob domain instead of the Ad5 fiber knob. Δ5/35 has a 12.6-kb genome and was produced by homologous recombination between two first-generation Ad vectors as described previously (36). The attachment and internalization of these vectors were analyzed on MO7e cells, a growth factor-dependent, CD34-positive, human erythroleukemia cell line that is often used as a model to study gene transfer into human hematopoietic cells (24). Southern blot analysis demonstrated that when MO7e cells were exposed to equal doses of Ad particles of all three vectors (Fig. 1B, Load), comparable amounts of Ad particles attached to and were internalized into cells (Fig. 1B, Att and Int, respectively). These data demonstrate that primary Ad attachment to cell surface receptors and subsequent virus particle internalization are comparable for vectors with different genome length and structure.
FIG. 1.
(A) Schematic representation of genome organizations of fiber-chimeric Ad vectors. (B) Comparative analysis of Ad vector attachment and internalization on MO7e cells. Equal amounts of indicated Ad vectors were mixed with MO7e cells and one portion of cells was lysed immediately (upper panel, Load lanes), while the others were processed to determine the levels of Ad attachment or internalization by Southern blotting as described in Materials and Methods. Att, attachment; Int, internalization. The lower panel shows the agarose gel before Southern blotting to demonstrate equivalent DNA loads. The membrane was hybridized with a GFP gene-specific 32P-labeled probe, and vector DNA was visualized by autoradiography. M, molecular weight marker. Corresponding vector DNA bands are indicated by arrows.
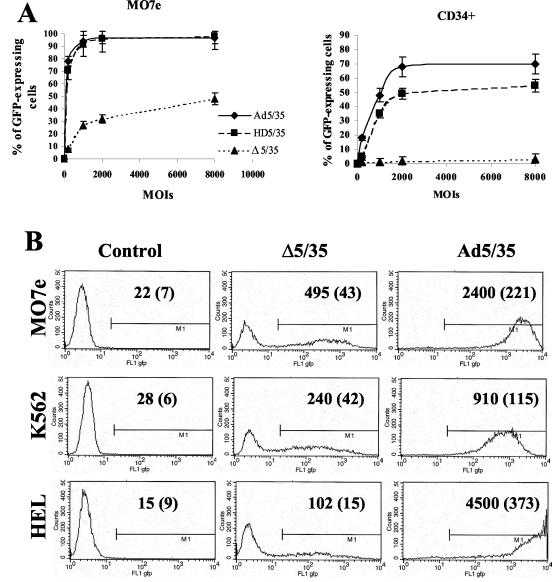
Percentages of GFP-expressing cells and GFP levels vary between vectors with different genome length and structure and between leukemia cell lines and primary human CD34+ cells.
Our previous studies have shown that the efficiency of Ad5/35- and Δ5/35-mediated gene transfer varied over 200-fold in different human cell lines (4, 5, 36). To extend this study and analyze whether the infectivity of Ad vectors depends on the length of viral genomes or cell types used, we infected several human leukemia cell lines (MO7e, K562, and HEL) and primary human CD34+ cells with Ad5/35, HD5/35, and Δ5/35 vectors (Fig. 2). In an initial study that compared the efficiency of Ad vector-mediated gene transfer into these cell lines, we found that both first-generation and HD vectors infected MO7e cells at comparable levels (Fig. 2A). Infection of CD34+ cells with HD5/35, however, was less efficient and did not reach the Ad5/35 transduction level, even at high MOIs (up to 8,000 virus particles per cell). As shown previously, MO7e cell infection with Δ5/35 was less efficient than with Ad5/35 (36) (Fig. 2A). However, it was unexpected that Δ5/35 would be completely unable to confer transgene expression in primary human CD34-positive cells. To extend this observation, we infected two additional leukemia cell lines (K562 and HEL cells) with the Δ5/35 vector (Fig. 2B). Overall, as with MO7e cells, Δ5/35-mediated GFP expression was clearly detectable in these cell lines, albeit at a lower level than Ad5/35. Surprisingly, there was no direct correlation between the gene transfer efficiencies of the vectors on these cell lines. For example, while Ad5/35 infected HEL cells twice as efficiently as MO7e cells (Fig. 2B), the Δ5/35 infection of HEL cells was fivefold less efficient than infection of MO7e cells. Considering that the attachment and internalization of all vectors were comparable, our data suggest that the intracellular environment affects the efficiency of the postinternalization steps of Ad infection. The dramatic variation in infectivity between Ad5/35 and Δ5/35, and especially between HD5/35 and Δ5/35 vectors on MO7e and primary hematopoietic CD34+ cells (Fig. 2A), suggests that vectors with genomes ∼30% the size of full-length genomes have a reduced ability to complete postinternalization steps during cell infection. This block appears to be more pronounced in primary human CD34-positive cells than in the immortalized cell lines.
FIG. 2.
Transduction efficiency of different cell lines varies significantly for Ad vectors with different genome sizes. (A) Immortalized MO7e or primary human CD34-positive cells were infected at the indicated MOIs (virus particles per cell), and the percentage of GFP-expressing cells was analyzed 24 h later by flow cytometry (n= 6). (B) Immortalized human MO7e, K562, or HEL cells were infected at an MOI of 200 virus particles per cell with Δ5/35 or Ad5/35 vectors. At 24 h postinfection, the transduction efficiency was analyzed by flow cytometry. Mean GFP fluorescence intensities and standard deviations (in parentheses) are indicated on the corresponding histograms (n = 4). In the control settings (Control) cells were incubated with virus dilution buffer only.
Inefficient gene transfer with Δ5/35 vector is due to inefficient delivery of viral DNA to the nucleus.
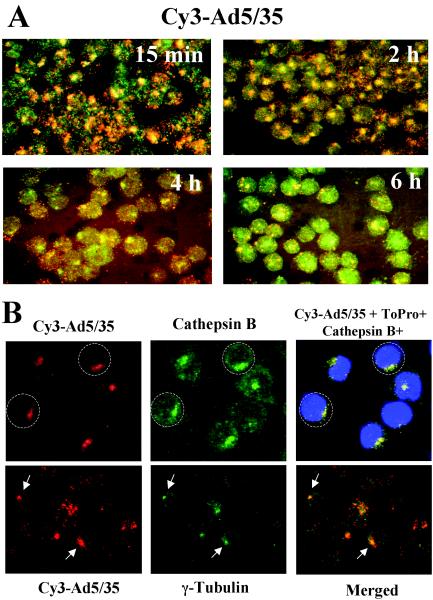
Our previous studies on human epithelial HeLa cells showed that after binding of Ad35-fiber-containing vectors to the primary attachment receptor and subsequent internalization, these capsid-chimeric Ad5/35 vectors are rapidly transported into cathepsin B-containing (late endosomal and lysosomal) compartments, thus selecting an intracellular trafficking route that is different from that of unmodified Ad5 vectors (38). To test whether our findings on HeLa cells are also applicable for hematopoietic cell lines, we infected MO7e cells with a Cy3-labeled Ad5/35 vector (Fig. 3). At 15 min and 2, 4, and 6 h after infection, cells were fixed, and the colocalization of virus particles with cathepsin B-containing cellular compartments was analyzed (Fig. 3A). This analysis revealed that, similar to HeLa cells, Ad35 fiber knob-possessing particles are efficiently transported into late endosomal and lysosomal cellular compartments of MO7e cells shortly after internalization (Fig. 3A, 15-min time point). Maximum colocalization of Ad5/35 particles with cathepsin B-containing cellular compartments was observed 2 h postinfection. By 4 h postinfection, colocalization of Ad particles with cathepsin B declined, suggesting that the Ad particles are released from these compartments and either transported back to the cell surface (retrograde transport [38]), degraded within lysosomes, or have escaped into the cytoplasm. Apparently, by 6 h postinfection, the intracellular organization of cathepsin B-containing compartments has changed, and significant colocalization of virus particles, as found at 2 h postinfection, is no longer observed. Notably, at 6 h postinfection, GFP expression becomes detectable in Ad5/35-infected MO7e cells by fluorescence microscopy and flow cytometry (data not shown), suggesting that at least some of the Ad genomes were translocated into the nucleus.
FIG. 3.
Intracellular trafficking of capsid-modified vectors in hematopoietic MO7e cells. (A) Kinetics of Cy3-labeled Ad5/35 vector (red) colocalization with and escape from the cathepsin B-containing late endosomal and lysosomal cellular compartments (green). Cells were pulse-infected with Cy3-Ad5/35 virus for 15 min at 37°C and allowed to incubate further for the indicated periods. Then, cells were fixed, permeabilized, and stained with anti-cathepsin B-specific primary antibody. The staining of primary antibody was detected with Alexa Fluor 488-conjugated secondary antibody (green), and images of cells were taken on a Leica fluorescence microscope. To determine the colocalization of virus with cathepsin B-positive intracellular compartments, images of the same areas were taken in red and green consecutively. Note thepronounced clustering of Ad in cathepsin B-positive compartments at 2 h postinfection. (B) Trafficking of Ad to the perinuclear space within cathepsin B-positive endosomal compartments (upper panel) and colocalization of virus clusters with MTOC in MO7e cells (lower panel). Cells were infected with Cy-3 labeled Ad5/35 virus (red) and 2 h later fixed, permeabilized, and stained with anti-cathepsin B or γ-tubulin antibodies. Binding of primary antibodies was detected with Alexa Fluor 488 secondary antibodies (green). To visualize nuclei, cells were additionally stained with ToPro dye (blue). Confocal images in three channels were superimposed on a Leica confocal microscope. Representative fields are shown.
Migration of Ad particles towards the nucleus within endosomes or as free particles occurs along the cellular microtubule network (43). This minus-end-directed transport results in the accumulation of virus particles around microtubule organization centers (MTOCs) located in proximity to the nucleus, thereby facilitating subsequent docking of Ad capsids to the nuclear pore complexes and translocation of virus genomes into the nucleus (12). To test whether the areas of accumulated Cy-3-labeled virus particles seen within infected cells at 2 h postinfection represents virus accumulation in the perinuclear space, we stained virus-infected cells with anti-cathepsin B antibody and ToPro dye (blue) to visualize nuclei. Subsequent confocal microscopic analysis demonstrated (Fig. 3B, upper panel) that, by 2 h postinfection, virus accumulated within cathepsin B-positive late endosomal and lysosomal vesicles localized at the perinuclear space. To test whether virus-containing vesicles accumulate at MTOC, we stained infected cells with anti-γ-tubulin antibody to visualize centrosomes (8). Confocal microscopic analysis (Fig. 3B, lower panel) showed that accumulations of Ad particles in the perinuclear space overlapped with γ-tubulin-positive staining of centrosomes. In summary, this demonstrates that in human hematopoietic MO7e cells, the Ad5/35 vector travels toward MTOC and the nucleus within late-endosomal or lysosomal cathepsin B-containing vesicles.
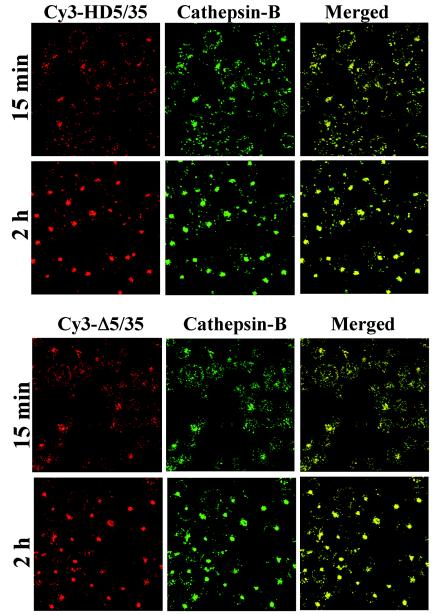
After we had established the route of intracellular trafficking of Ad5/35 in hematopoietic cells, we analyzed trafficking of Cy3-labeled HD5/35 and Δ5/35 vectors, which differ in viral genome length and structure from the first-generation virus. Using confocal microscopic analysis, we found that both Cy3-labeled HD5/35 and Δ5/35 vectors colocalized efficiently with cathepsin B-positive endosomal compartments as early as 15 min postinfection (Fig. 4). Similar to Ad5/35, HD5/35 and Δ5/35 particles by 2 h postinfection form large aggregates within cells, which colocalize with cathepsin B. These data demonstrate that the selection of intracellular trafficking route is not affected by the length or structure of the virus genome and that both HD5/35 and Δ5/35 vectors efficiently enter cathepsin-B-positive intracellular compartments and use them for translocation to the nucleus.
FIG. 4.
Intracellular trafficking of Cy3-labeled HD5/35 and Δ5/35 vectors in MO7e cells. Cells were infected with indicated Ad vectors (red) for 15 min and 2 h. Then, cells were fixed and stained to detect colocalization of virus with cathepsin B-positive endosomal compartments (green) as described in the legend of Fig. 3A. Confocal images of cells were taken in both green and red channels. Colocalization of virus with cathepsin B-positive endosomes appears as yellow. Representative fields are shown.
Although useful, the data obtained from confocal microscopic analysis of intracellular virus trafficking did not explain the differences in infectivity of Ad5/35 and Δ5/35 on MO7e cells (Fig. 2A). Moreover, these analyses do not allow for a quantitative assessment of vector DNA delivery into the nucleus. To evaluate the efficiency of Ad genome delivery into the nucleus for different vectors, we utilized Southern blot analysis. MO7e cells were infected with equal doses of Ad5/35, HD5/35, and Δ5/35 vectors, and 8 h later, nuclei were purified and total nuclear DNA was extracted as described previously (39). Southern blot analysis of vector DNA associated with the nuclei of infected cells demonstrated that while the amounts of Ad5/35 and HD5/35 vector genomes were comparable, significantly fewer Δ5/35 genomes were present in the nuclear fraction of cells infected with this virus (Fig. 5A). Using Southern blotting, we also analyzed the stability of viral DNA in infected cells over a period of 48 to 96 h in MO7e and CD34-positive primary cells (Fig. 5B). This analysis demonstrated that both Ad5/35 and HD5/35 genomes were stable within infected cells and present in comparable amounts over the time of analysis. However, the amount of Δ5/35 genomes gradually declined within infected cells and became barely detectable by 96 h postinfection. Taken together, these data demonstrate that both in MO7e and CD34-positive cells, HD5/35 was as efficient as Ad5/35 in delivering viral genomes into the nuclei of infected cells. However, Δ5/35 virus was unable to efficiently deliver its DNA into the nucleus and, instead, was rapidly degraded.
FIG. 5.
Delivery of viral DNA into the nucleus and its stability in infected cells over time. (A) MO7e cells were infected with the indicated Ad vectors for 8 h. Then, nuclei were extracted and nuclear DNA was purified for Southern blot analysis. To ensure that equivalent virus doses were used for initial cell infection and that all vectors were able to attach to and internalize into cells with similar efficiencies, at the corresponding time points duplicates of infected cells were lysed, and total cellular DNA was extracted and used for Southern blot analysis. The lower panel shows the agarose gel before Southern blotting to demonstrate equivalent DNA loads. The membrane was hybridized with a GFP gene-specific 32P-labeled probe, and vector DNA was visualized by autoradiography. Corresponding vector DNA bands are indicated by arrows. Note that although the Δ5/35 and Ad5/35 vectors were able to attach to and internalize into cells with similar efficiencies, the amount of vector DNA associated with nuclei was dramatically lower for Δ5/35 compared to Ad5/35. (B) MO7e or human primary CD34-positive cells were infected with the indicated Ad vectors as described in Materials and Methods. At the indicated time points, cells were lysed and total cellular DNA was purified for Southern blot analysis. Note that HD5/35 vector is maintained in infected cells as efficiently as Ad5/35 vector. However, Δ5/35 vector DNA is rapidly removed from the infected cells and is not detectable at 48 h post infection by Southern blotting. Serial dilutions of Ad5/35 plasmid DNA of known concentration was applied on the agarose gel as standard DNA. Att, attachment; Int, internalization; Load, the amount of virus vector used for cell infection; M, molecular weight marker.
Endosomal escape is the limiting step for cell infection with Ad vector possessing a shortened genome.
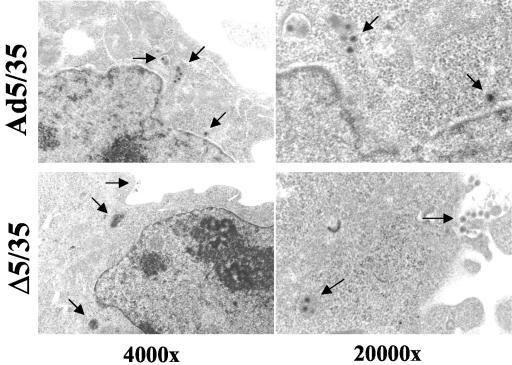
After internalization into the cell, Ad particles escape from endosomes, are transported in the cytoplasm, dock to nuclear pores, get disassembled, and translocate viral genomes into the nucleus. The inability to undergo one or several of these steps can cause the observed low ΔAd5/35 infectivity. To further analyze the steps of virus infection leading to inefficient nuclear transport of Δ5/35 genomes, we infected MO7e cells with Δ5/35 and Ad5/35 vectors and 2 h postinfection assessed intracellular virus distribution by EM (38). Semiquantitative analysis of intracellular virus particle distribution revealed that although a significant amount of Ad5/35 virions was seen in endosomes (47%), about 11% of virus particles were found free in the cytoplasm, and another 30% of virions were localized within a distance of three virion diameters to the nucleus (also called perinuclear space [13, 25]) (Table 1 and Fig. 6). In contrast, only about 2% of Δ5/35 particles (4 virus particles out of 179 particles counted) were found in the cytoplasm, and only 1 particle was seen in the perinuclear space. About 63% of Δ5/35 virions were found within electron-dense endosomes or at the cell periphery, suggesting that Δ5/35 virus is unable to efficiently escape from endosomes.
TABLE 1.
Intracellular localization of Ad5/35 and Δ5/35 virions in MO7e cells 2 h postinfectiona
| Compartment (at 2 h p.i.) | Ad5/35 virions (%) | Δ5/35 virions (%) |
|---|---|---|
| Endosome | 47 | 63 |
| Cytoplasm | 11 | 2 |
| Perinuclear space | 30 | 1 |
| Cell periphery | 12 | 34 |
The total number of counted Ad5/35 virions was 154; the total number of counted Δ5/35 virions was 179. p. i., postinfection.
FIG. 6.
EM analysis of intracellular distribution for Δ5/35 and Ad5/35 vectors in MO7e cells 2 h after virus infection. Virus particles are marked by arrows.
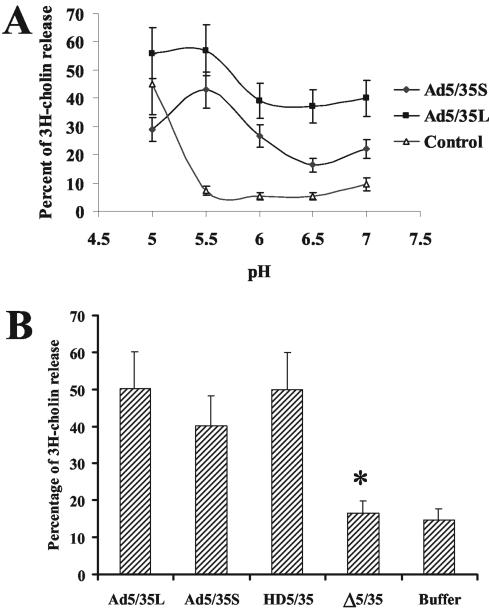
Efficient virus escape from the endosomes requires the membrane-permeabilizing activity of the Ad penton protein (25, 44, 48, 49). This activity, however, is exerted only upon Ad particle exposure to the acidic endosomal environment and after the interaction of the penton protein with cellular integrins (13, 14). To analyze the ability of the penton base proteins of Ad5/35, HD5/35, or Δ5/35 vectors to permeabilize cellular membranes, we incubated [3H]choline pulse-labeled MO7e cells with equal virus doses in the presence of acidic pH buffers and quantitated the amounts of [3H]choline released from cells as described previously (48). This analysis demonstrated that the peak of cell membrane permeabilization for both long-shafted (39) and short-shafted Ad5/35 vectors occurs at pH 5.5 (Fig. 7A). Our data are in agreement with previously published studies by Miyazawa and other investigators (19, 26, 27), demonstrating that, unlike CAR-interacting Ad5 vectors, which permeabilize cellular membranes at pH 6.0, the non-CAR-interacting Ad7 virus and an Ad5/7 fiber-chimeric vector permeabilized cellular membranes under more acidic conditions (pH 5.5). Importantly, our analysis also revealed that while the permeabilizing activity of the penton base protein in HD5/35 vector is comparable to that of Ad5/35 vectors, this activity is dramatically reduced for Δ5/35 (Fig. 7B). This finding corroborates the data analyzing virus distribution within infected cells by EM, suggesting that endosome escape is the limiting step in Δ5/35 cell infection.
FIG. 7.
Efficiency of [3H]choline release from pulse-chase labeled cells (membrane lysis) at different pH values by Ad5/35 vectors with different fiber shaft lengths (A) and following exposure to HD5/35 and Δ5/35 vectors (B). In panel A, cells were labeled with [3H]choline and exposed to equal doses of viruses at various pH levels as described in Materials and Methods. The amount of [3H]choline released from cells was quantified by a scintillation counter. Note that the Ad5/35 vector with a short fiber shaft (Ad5/35S) permeabilized cells less efficiently than its long-shafted counterpart (Ad5/35L). In panel B, [3H]choline-labeled cells were exposed to equal doses of Ad vectors, and the virus cell permeabilizing activity was analyzed at pH 5.5. Control, percentage of the total cell radioactivity released after incubation with different pH buffers only. n = 4.
Long Ad genomes confer integrity to Ad capsid.
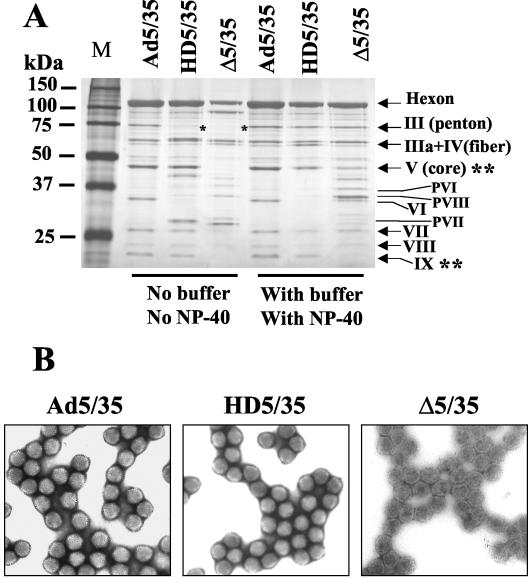
Earlier studies by Laver et al. (17) have shown that the exposure of Ad particles to low-salt acidic conditions results in the spontaneous loss of penton base capsomers. It has also been shown that this spontaneous virus disassembly can be prevented by the addition of small amounts (up to 0.05%) of the nonionic detergents Triton X-100 or NP-40 to the virus (9). Therefore, the exposure of Ad particles to an acidic pH in low-salt buffers and analysis of the virion protein composition may provide important information on the general integrity and stability of virus capsids possessing genomes of different sizes. To examine the effect of acidic pH buffers and NP-40 addition on the protein composition of our vectors, we produced Ad5/35, HD5/35, and Δ5/35 vectors with or without the addition of NP-40 during the last cell lysis step before virus purification and with or without buffering CsCl solutions during virus ultracentrifugation. The protein composition of virus preparations was analyzed by polyacrylamide gel electrophoresis and silver nitrate staining of gels. This analysis showed that both HD5/35 and Δ5/35 vectors are more vulnerable than Ad5/35 to acidic conditions and readily lose capsid penton proteins (Fig. 8A). It is also apparent that Δ5/35 particles lack the core protein V and the capsid-stabilizing protein IX, while HD5/35 particles contain the same amount of these proteins as Ad5/35. The addition of NP-40 during virus purification and the buffering of CsCl solutions during ultracentrifugation prevented the loss of penton proteins and stabilized both the HD5/35 and Δ5/35 vector capsids. However, even under optimal purification conditions, Δ5/35 virus particles possessed less core protein V as well as mature and precursor forms of pVI, pVII and pVIII, while they lack the capsid-stabilizing protein IX. Such significant differences in virus capsid protein composition between Δ5/35 and other vectors should result in detectable morphological abnormalities of the virus capsid. To evaluate the morphology of virus particles, we performed EM analysis of fixed, negatively stained vector particles (Fig. 8B). While the icosahedral structure of Ad particles was preserved for Ad5/35 and HD5/35, Δ5/35 particles displayed a lighter electron density, possessed slightly irregular, uneven shapes, and lacked visible hexon capsomers on the virion surface. Based on these data, we concluded that because of their altered virion structure, Δ5/35 particles are unable to properly respond to the intraendosomal environment during the virus disassembly process. This may result in an inability to escape into the cytoplasm and to deliver viral genomes into the nucleus.
FIG. 8.
Ad capsid protein composition after exposure to low-salt acidic conditions with and without NP-40 treatment and EM analysis of Ad particles. (A) The indicated Ad vectors were purified without buffering CsCl and dialysis solutions (pH 5.0) and exposure to 0.05% NP-40 (No buffer/No NP-40) or with HEPES-buffered CsCl and dialysis solutions (pH 7.4) and exposure to 0.05% NP-40 (With buffer/With NP-40). Equal amounts of vectors were then loaded onto sodium dodecyl sulfate-polyacrylamide protein gel. Following electrophoresis, virus capsid protein composition was visualized by staining with silver nitrate. M, molecular weight marker. Note that Ad5/35 has the same capsid protein composition and that Δ5/35 has less pV core proteins and completely lacks the capsid-stabilizing protein pIX (indicated by double asterisks). HD5/35 and Δ5/35 vectors lose pIII penton proteins when exposed to acidic environment (indicated by single asterisks). (B) Visualization of Ad5/35, HD5/35, and Δ5/35 virus particles by EM following their negative-contrast staining with uranyl acetate. Note the abnormal morphology of Δ5/35 virus particles compared to other vectors.
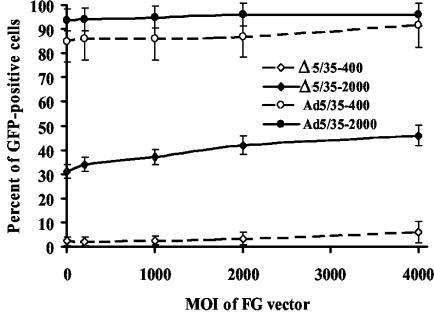
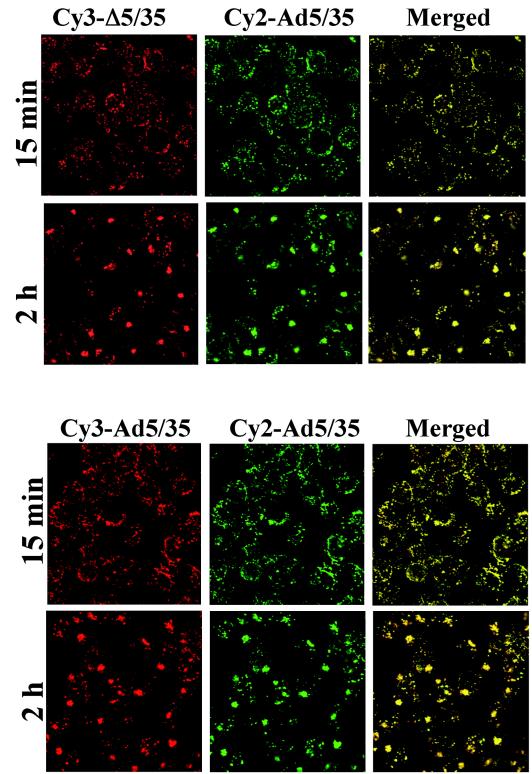
To test whether coinfection with first-generation vector particles can restore in trans the ability of Δ5/35 particles to efficiently escape endosomes, we coinfected MO7e cells with Δ5/35 (containing the GFP gene) and increasing amounts of Ad5/35-β-gal (containing the β-galactosidase gene [37]) (Fig. 9). This analysis demonstrated that even when up to 70% of the total virus dose constituted the Ad5/35-β-gal vector, there was only a modest increase in the percentage of GFP-expressing cells, and there was no increase in mean GFP fluorescent intensity (data not shown) of cells transduced with Δ5/35. To confirm that Ad5/35-β-gal and Δ5/35 virus particles enter the same intracellular compartments, we infected MO7e cells with Cy3-labeled Δ5/35 (red) and Cy2-labeled Ad5/35-β-gal (green) vectors simultaneously. Confocal microscopy analysis revealed the efficient colocalization of Δ5/35 and Ad5/35-β-gal virus particles, confirming that both vectors enter the same endosomal compartments in infected cells (Fig. 10). The inability of Ad5/35-β-gal virus to restore Δ5/35 infectivity suggests that efficient endosome lysis provided in trans by fully infectious Ad5/35 vector is insufficient to allow for improvement of Δ5/35 transduction.
FIG. 9.
First-generation capsid-modified Ad vector is unable to improve the infectivity of Δ5/35 vector on MO7e cells. MO7e cells were infected with Δ5/35 or Ad5/35 vectors expressing GFP at the indicated MOIs (virus particles per cell) together with increasing amounts of first-generation Ad5/35-β-galactosidase virus (FG). The percentage of GFP-expressing cells was analyzed at 24 h postinfection by flow cytometry (n = 4).
FIG. 10.
Intracellular trafficking of Cy3-labeled Δ5/35 (red) and Cy2-labeled Ad5/35-β-gal vectors in MO7e cells. Cells were coinfected with indicated Ad vectors for 15 min and 2 h. Then, cells were fixed and stained to detect the colocalization of viruses within cellular endocytic compartments. Confocal images of cells were taken in both green and red channels. To confirm that both viruses labeled with different dyes are capable of entering the same endocytic compartments, MO7e cells were coinfected with Δ5/35 and Ad5/35-β-gal vectors (lower panels). Colocalization of two viruses appears as yellow. Representative fields are shown.
Taken together, our data indicate that the Δ5/35 vector possessing small 12-kb genomes undergoes the initial attachment and internalization steps of infection with an efficiency comparable to that of the first-generation or HD vector possessing large genomes. However, unlike these vectors, Δ5/35 that lacks mature core proteins and possesses altered capsids is unable to complete its trafficking and disassembly program by properly responding to intraendosomal signals. This results in its inability to efficiently escape from endocytic vesicles and deliver its DNA into the nucleus.
DISCUSSION
In this paper, we analyzed the effect of viral genome size on the infectivity of capsid-modified Ad vectors. We compared early steps of infection between first- generation (35-kb genome) and Ad vectors devoid of all viral genes with genome sizes of 28 kb and 12.6 kb. All vectors possess Ad35-derived fiber knob domains, which recognize CD46 as a primary attachment receptor (10, 35, 41, 52). Using immortalized human hematopoietic cell lines and primary human CD34-positive hematopoietic cells, we found that the Ad genome size did not affect the efficiency of virus attachment to and internalization into the cell. Our data also demonstrated that, independently of the length of the genome, all vectors migrated to the nucleus through late endosomal or lysosomal cathepsin B-containing cellular compartments. However, the vector containing the 12.6-kb genome was unable to efficiently escape from endosomes and deliver its DNA into the nucleus. Moreover, ΔAd particles with 12.6-kb genomes were less stable and possessed an abnormal capsid protein composition, including a lack of capsid-stabilizing protein IX, compared to other vectors.
Packaged Ad5 DNA exists inside the capsid in a supercoiled structure associated with core proteins VII, V, and IVa2, forming the virus chromatin. In contrast to polyomavirus and papillomavirus, Ad genomes are not packaged with cellular histones. According to a model proposed by Wong and Hsu (51), Ad core proteins (VII and V) bind to seven defined, ∼200-bp regions of Ad5 DNA. The DNA regions between the core binding sites form characteristic supercoiled loop domains. About eight loops can be accommodated in an Ad5 virion as demonstrated by EM. In the 12.6-kb genome, the potential core binding sites are deleted, and the total amount of core proteins is reduced. It is unclear whether a compact structure similar to that of the full-length genome can be formed. It is important to note that while our HD5/35 vector also lacked these putative core protein-binding regions, its infectivity on hematopoietic cell lines was not dramatically reduced compared to first-generation Ad5/35 vector (Fig. 2A). These data are in agreement with previous findings that HD vectors with genome sizes equal or larger than 27 kb can efficiently transduce a variety of cell types in vitro and in vivo (16, 29). These data also suggest that, although present in a wild-type Ad genome, these 200-bp core protein-binding cis elements are not absolutely required to confer Ad infectivity.
Ad assembly is a multistep process. Hexon proteins polymerize to form capsomers, which join with other structural proteins, including the penton base and fiber protein, to form empty capsids (18, 23). These empty capsids (with a density of 1.29 g/cm3) also contain precursors of proteins VI and VIII and can be easily distinguished from mature virions (density, 1.34 g/cm3) in CsCl equilibrium gradients. The viral genome is thought to be inserted into the empty capsids along with DNA-associated core proteins. The final maturation step of Ad capsid assembly is mediated by the viral protease (13). The densities of both the HD5/35 and Δ5/35 vectors were distinct from the density of the fist-generation vector. Following equilibrium centrifugation of vector stocks in a CsCl gradient, the HD5/35 vector is recovered in a fraction with a density of 1.327 g/cm3, while the Δ5/35 vector banded at a density of 1.30 to 1.31 g/cm3. Although these differences in buoyant densities between virus particles can be partially explained by the different lengths of vector genomes, our analyses demonstrated differences in both their capsid protein composition and overall morphology (Fig. 8). It is apparent that the Ad particle core has to be filled with DNA of sufficient size so that, together with core proteins, the mature icosahedral virion structure can be formed and stabilized. To this end, the lack of capsid-stabilizing protein IX within virus particles may be one of the factors responsible for the abnormal morphology of Δ5/35 virions. According to the present model, protein IX is buried within large cavities of the icosahedral facet of Ad (2, 3, 46). These cavities are created by hexon molecules symmetrically arranged on each facet. Although pIX stabilizes a group of nine hexons within the icosahedral facet, it does not participate in the stabilization of peripentonal hexons, which are readily lost after virion exposure to low-salt acidic conditions. Clearly, this, together with the reduced amounts and immature state of the core proteins and the absence of pIX in the Δ5/35 virions, further diminishes their ability to properly respond to intracellular stimuli during early steps of infection.
Several lines of evidence indicate that an Ad vector possessing shortened genomes is deficient in the ability to escape from cellular endosomes. Southern blot analysis of vector DNA delivery into the nucleus demonstrated that the Δ5/35 vector delivers its genomes with reduced efficiency compared to vectors with large genomes (Fig. 5A). We also found that Δ5/35 virus particles cannot efficiently lyse cellular membranes when exposed to acidic pHs. Finally, direct visualization of virus particles within infected cells by EM and semiquantitative analysis of the distribution of virus particles among different intracellular compartments clearly demonstrated that Δ5/35 vector is retained within cellular endosomes. The Ad endosome escape is an essential step in the virus disassembly program, ensuring that viral genomic DNA is efficiently delivered into the cell nucleus. The inability of Ad to escape from endosomes results in retrograde virus transport and deposition at the cell surface or its degradation within cellular lysosomes, a phenomenon that is more pronounced for Ad vectors containing Ad35 fibers. The virus-encoded protease L3/p23 was found to be critical for the virus endosome escape step (13). Located in the internal cavities at ∼10 copies per virion and bound to DNA, L3/p23 protease degrades capsid protein pVI and enables the capsid to escape from endosomes and virus uncoating at the nucleus membrane. Importantly, incorrectly assembled particles of mutant Ad (ts1) that lack the functional L3/p23 protease but possess genomes of wild-type length are unable to release fibers and escape from endosomes (14). One can speculate that Δ5/35 vector particles, with a smaller DNA content than first-generation or HD vectors, may incorporate less of the virus protease into the virion or fail to provide the spatial arrangement in proximity to pVI, which is required for its proteolytic processing.
Although it was previously thought that Ad penton protein interaction with cellular integrins and exposure to acidic pHs are sufficient for efficient virus escape from endosomes, recent data by Wang et al. (48) demonstrated that the signaling downstream of virus-interacting integrins can dramatically affect the efficiency of endosome release. They found that on CS-1 melanoma cells, expressing both the αvβ3 and the αvβ5 integrins, mutations introduced into the cytoplasmic tail domain of β5 but not into β3 integrin can block virus-mediated endosome lysis. While it has been previously demonstrated that Ad can utilize both αvβ5 and αvβ3 integrins for productive cell infection, the preferential use of β5 integrins on CS-1 cells by Ad is not fully understood. It is clear, however, that each cell type infected by Ad in vitro or in vivo provides a unique environment for virus infection since different types of integrins are expressed on cell surfaces. Furthermore, differential use of primary cellular attachment receptors by Ad belonging to different subgroups adds to the complexity of virus-host cell interactions and eventually affects the apparent infectivity of Ad vectors. Miyazawa et al. (26) have found that replacing Ad5 fiber protein within Ad5-based vector with Ad7-derived fibers retains chimeric Ad5/7 vector for longer times within endosomal cellular compartments compared to parental virus. Notably, the kinetics and route of intracellular trafficking were comparable between the chimeric Ad5/7 vector and the unmodified Ad7 virus, indicating that the interaction between the fiber knob domain and the cellular attachment receptor determines the intracellular trafficking route of Ad. We subsequently showed that the replacement of the Ad5 fiber knob with that of Ad35 was sufficient to modify the intracellular trafficking of capsid-chimeric Ad5/35 vector. Although efficient at transducing human hematopoietic cells, dendritic cells, and other primary human cell types (which are poorly infected with parental unmodified Ad5 vector) on human HeLa cells, Ad5/35 vector was less efficient than Ad5 (38). It is of interest to note that the vector with a small genome packaged into wild-type unmodified Ad5 capsid was more infectious compared to its first-generation counterpart on human KB cells (21, 22) than Δ5/35 compared to Ad5/35 on hematopoietic MO7e cells. Moreover, Δ5 vector infectivity could be restored by the addition in trans of sufficient amounts of first-generation virus. Our data on hematopoietic cells demonstrated that fully infectious Ad5/35-β-gal particles were unable to rescue the Δ5/35 deficiency in cell transduction in trans (Fig. 9). Clearly, the differences between Δ5 and Δ5/35 in the usage of cellular integrins (KB versus MO7e cells) or primary attachment receptors (CAR versus CD46), as well as cell type-specific factors may be accounted for by the observed differences in infectivity of these viruses on different cell lines. Because Δ5/35 infectivity varies between different cell types, being least infectious on primary human CD34+ hematopoietic cells and most infectious on immortalized human MO7e and K562 cells, it is clear that the intraendosomal block during virus infection is not absolute. However, the cellular factors and mechanisms involved in modulating Ad trafficking in a cell type-specific manner are yet to be characterized.
Ad infection is a multistep process of highly coordinated virus interactions with the cell, ultimately resulting in efficient delivery of the virus genome into the nucleus. Development of Ad-based vectors has been aimed towards improving certain features of naturally existing Ads in order to adjust their properties for a desired application. Our study helps us to better understand how genome size affects the infectivity of Ad. These data provide new insights into Ad-host cell interaction and may help in developing improved gene transfer vectors based on Ad.
Acknowledgments
We thank Daniel Stone for helpful discussion.
This work was supported by grants from the NIH (R01 CA80192, P30 DK47754, and HL-00-008) and the Cystic Fibrosis Foundation.
REFERENCES
- 1.Bergelson, J. M., J. A. Cunningham, G. Droguett, E. A. Kurt-Jones, A. Krithivas, J. S. Hong, M. S. Horwitz, R. L. Crowell, and R. W. Finberg. 1997. Isolation of a common receptor for Coxsackie B viruses and adenoviruses 2 and 5. Science 275:1320-1323. [DOI] [PubMed] [Google Scholar]
- 2.Burnett, R. M. 1985. The structure of the adenovirus capsid. II. The packing symmetry of hexon and its implications for viral architecture. J. Mol. Biol. 185:125-143. [DOI] [PubMed] [Google Scholar]
- 3.Burnett, R. M., M. G. Grutter, and J. L. White. 1985. The structure of the adenovirus capsid. I. An envelope model of hexon at 6 A resolution. J. Mol. Biol. 185:105-123. [DOI] [PubMed] [Google Scholar]
- 4.Carlson, C. A., D. M. Shayakhmetov, and A. Lieber. 2002. An adenoviral expression system for AAV rep78 using homologous recombination. Mol. Ther. 6:91-98. [DOI] [PubMed] [Google Scholar]
- 5.Carlson, C. A., D. M. Shayakhmetov, and A. Lieber. 2002. Restoration of a functional open reading frame by homologous recombination between two adenoviral vectors. Mol. Ther. 6:99-105. [DOI] [PubMed] [Google Scholar]
- 6.Carlson, C. A., D. S. Steinwaerder, H. Stecher, D. M. Shayakhmetov, and A. Lieber. 2002. Rearrangements in adenoviral genomes mediated by inverted repeats. Methods Enzymol. 346:277-292. [DOI] [PubMed] [Google Scholar]
- 7.Chartier, C., E. Degryse, M. Gantzer, A. Dieterle, A. Pavirani, and M. Mehtali. 1996. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J. Virol. 70:4805-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dammermann, A., A. Desai, and K. Oegema. 2003. The minus end in sight. Curr. Biol. 13:R614-R624. [DOI] [PubMed] [Google Scholar]
- 9.Everitt, E., A. de Luca, and Y. Blixt. 1992. Antibody-mediated uncoating of adenovirus in vitro. FEMS Microbiol. Lett. 77:21-27. [DOI] [PubMed] [Google Scholar]
- 10.Gaggar, A., D. M. Shayakhmetov, and A. Lieber. 2003. CD46 is a cellular receptor for group B adenoviruses. Nat. Med. 9:1408-1412. [DOI] [PubMed] [Google Scholar]
- 11.Gao, W., P. D. Robbins, and A. Gambotto. 2003. Human adenovirus type 35: nucleotide sequence and vector development. Gene Ther. 10:1941-1949. [DOI] [PubMed] [Google Scholar]
- 12.Greber, U. F., M. Suomalainen, R. P. Stidwill, K. Boucke, M. W. Ebersold, and A. Helenius. 1997. The role of the nuclear pore complex in adenovirus DNA entry. EMBO J. 16:5998-6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greber, U. F., P. Webster, J. Weber, and A. Helenius. 1996. The role of the adenovirus protease on virus entry into cells. EMBO J. 15:1766-1777. [PMC free article] [PubMed] [Google Scholar]
- 14.Greber, U. F., M. Willetts, P. Webster, and A. Helenius. 1993. Stepwise dismantling of adenovirus 2 during entry into cells. Cell 75:477-486. [DOI] [PubMed] [Google Scholar]
- 15.Hartigan-O'Connor, D., C. Barjot, R. Crawford, and J. S. Chamberlain. 2002. Efficient rescue of gutted adenovirus genomes allows rapid production of concentrated stocks without negative selection. Hum. Gene Ther. 13:519-531. [DOI] [PubMed] [Google Scholar]
- 16.Kochanek, S., G. Schiedner, and C. Volpers. 2001. High-capacity “gutless” adenoviral vectors. Curr. Opin. Mol. Ther. 3:454-463. [PubMed] [Google Scholar]
- 17.Laver, W. G., N. G. Wrigley, and H. G. Pereira. 1969. Removal of pentons from particles of adenovirus type 2. Virology 39:599-604. [DOI] [PubMed] [Google Scholar]
- 18.Leibowitz, J., and M. S. Horwitz. 1975. Synthesis and assembly of adenovirus polypeptides. III. Reversible inhibition of hexon assembly in adenovirus type 5 temperature-sensitive mutants. Virology 66:10-24. [DOI] [PubMed] [Google Scholar]
- 19.Leopold, P. L., B. Ferris, I. Grinberg, S. Worgall, N. R. Hackett, and R. G. Crystal. 1998. Fluorescent virions: dynamic tracking of the pathway of adenoviral gene transfer vectors in living cells. Hum. Gene Ther. 9:367-378. [DOI] [PubMed] [Google Scholar]
- 20.Li, Y., R. C. Pong, J. M. Bergelson, M. C. Hall, A. I. Sagalowsky, C. P. Tseng, Z. Wang, and J. T. Hsieh. 1999. Loss of adenoviral receptor expression in human bladder cancer cells: a potential impact on the efficacy of gene therapy. Cancer Res. 59:325-330. [PubMed] [Google Scholar]
- 21.Lieber, A., C. Y. He, I. Kirillova, and M. A. Kay. 1996. Recombinant adenoviruses with large deletions generated by Cre-mediated excision exhibit different biological properties compared with first-generation vectors in vitro and in vivo. J. Virol. 70:8944-8960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lieber, A., D. S. Steinwaerder, C. A. Carlson, and M. A. Kay. 1999. Integrating adenovirus-adeno-associated virus hybrid vectors devoid of all viral genes. J. Virol. 73:9314-9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maizel, J. V., Jr., D. O. White, and M. D. Scharff. 1968. The polypeptides of adenovirus. II. Soluble proteins, cores, top components and the structure of the virion. Virology 36:126-136. [DOI] [PubMed] [Google Scholar]
- 24.Mantel, C., P. Hendrie, and H. E. Broxmeyer. 2001. Steel factor regulates cell cycle asymmetry. Stem Cells 19:483-491. [DOI] [PubMed] [Google Scholar]
- 25.Meier, O., and U. F. Greber. 2003. Adenovirus endocytosis. J. Gene Med. 5:451-462. [DOI] [PubMed] [Google Scholar]
- 26.Miyazawa, N., R. G. Crystal, and P. L. Leopold. 2001. Adenovirus serotype 7 retention in a late endosomal compartment prior to cytosol escape is modulated by fiber protein. J. Virol. 75:1387-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyazawa, N., P. L. Leopold, N. R. Hackett, B. Ferris, S. Worgall, E. Falck-Pedersen, and R. G. Crystal. 1999. Fiber swap between adenovirus subgroups B and C alters intracellular trafficking of adenovirus gene transfer vectors. J. Virol. 73:6056-6065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizuguchi, H., and T. Hayakawa. 2002. Adenovirus vectors containing chimeric type 5 and type 35 fiber proteins exhibit altered and expanded tropism and increase the size limit of foreign genes. Gene 285:69-77. [DOI] [PubMed] [Google Scholar]
- 29.Parks, R. J., and F. L. Graham. 1997. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packaging. J. Virol. 71:3293-3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peeters, M. J., G. A. Patijn, A. Lieber, L. Meuse, and M. A. Kay. 1996. Adenovirus-mediated hepatic gene transfer in mice: comparison of intravascular and biliary administration. Hum. Gene Ther. 7:1693-1699. [DOI] [PubMed] [Google Scholar]
- 31.Rebel, V. I., S. Hartnett, J. Denham, M. Chan, R. Finberg, and C. A. Sieff. 2000. Maturation and lineage-specific expression of the coxsackie and adenovirus receptor in hematopoietic cells. Stem Cells 18:176-182. [DOI] [PubMed] [Google Scholar]
- 32.Roelvink, P. W., A. Lizonova, J. G. Lee, Y. Li, J. M. Bergelson, R. W. Finberg, D. E. Brough, I. Kovesdi, and T. J. Wickham. 1998. The coxsackievirus-adenovirus receptor protein can function as a cellular attachment protein for adenovirus serotypes from subgroups A, C, D, E, and F. J. Virol. 72:7909-7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakurai, F., H. Mizuguchi, and T. Hayakawa. 2003. Efficient gene transfer into human CD34+ cells by an adenovirus type 35 vector. Gene Ther. 10:1041-1048. [DOI] [PubMed] [Google Scholar]
- 34.Sandig, V., R. Youil, A. J. Bett, L. L. Franlin, M. Oshima, D. Maione, F. Wang, M. L. Metzker, R. Savino, and C. T. Caskey. 2000. Optimization of the helper-dependent adenovirus system for production and potency in vivo. Proc. Natl. Acad. Sci. USA 97:1002-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Segerman, A., J. P. Atkinson, M. Marttila, V. Dennerquist, G. Wadell, and N. Arnberg. 2003. Adenovirus type 11 uses CD46 as a cellular receptor. J. Virol. 77:9183-9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shayakhmetov, D. M., C. A. Carlson, H. Stecher, Q. Li, G. Stamatoyannopoulos, and A. Lieber. 2002. A high-capacity, capsid-modified hybrid adenovirus/adeno-associated virus vector for stable transduction of human hematopoietic cells. J. Virol. 76:1135-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shayakhmetov, D. M., Z. Y. Li, S. Ni, and A. Lieber. 2002. Targeting of adenovirus vectors to tumor cells does not enable efficient transduction of breast cancer metastases. Cancer Res. 62:1063-1068. [PubMed] [Google Scholar]
- 38.Shayakhmetov, D. M., Z. Y. Li, V. Ternovoi, A. Gaggar, H. Gharwan, and A. Lieber. 2003. The interaction between the fiber knob domain and the cellular attachment receptor determines the intracellular trafficking route of adenoviruses. J. Virol. 77:3712-3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shayakhmetov, D. M., and A. Lieber. 2000. Dependence of adenovirus infectivity on length of the fiber shaft domain. J. Virol. 74:10274-10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shayakhmetov, D. M., T. Papayannopoulou, G. Stamatoyannopoulos, and A. Lieber. 2000. Efficient gene transfer into human CD34+ cells by a retargeted adenovirus vector. J. Virol. 74:2567-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sirena, D., B. Lilienfeld, M. Eisenhut, S. Kalin, K. Boucke, R. R. Beerli, L. Vogt, C. Ruedl, M. F. Bachmann, U. F. Greber, and S. Hemmi. 2004. The human membrane cofactor CD46 is a receptor for species B adenovirus serotype 3. J. Virol. 78:4454-4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steinwaerder, D. S., C. A. Carlson, and A. Lieber. 1999. Generation of adenovirus vectors devoid of all viral genes by recombination between inverted repeats. J. Virol. 73:9303-9313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suomalainen, M., M. Y. Nakano, S. Keller, K. Boucke, R. P. Stidwill, and U. F. Greber. 1999. Microtubule-dependent plus- and minus end-directed motilities are competing processes for nuclear targeting of adenovirus. J. Cell Biol. 144:657-672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svensson, U. 1985. Role of vesicles during adenovirus 2 internalization into HeLa cells. J. Virol. 55:442-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tillman, B. W., T. D. de Gruijl, S. A. Luykx-de Bakker, R. J. Scheper, H. M. Pinedo, T. J. Curiel, W. R. Gerritsen, and D. T. Curiel. 1999. Maturation of dendritic cells accompanies high-efficiency gene transfer by a CD40-targeted adenoviral vector. J. Immunol. 162:6378-6383. [PubMed] [Google Scholar]
- 46.van Oostrum, J., and R. M. Burnett. 1985. Molecular composition of the adenovirus type 2 virion. J. Virol. 56:439-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogels, R., D. Zuijdgeest, R. van Rijnsoever, E. Hartkoorn, I. Damen, M. P. de Bethune, S. Kostense, G. Penders, N. Helmus, W. Koudstaal, M. Cecchini, A. Wetterwald, M. Sprangers, A. Lemckert, O. Ophorst, B. Koel, M. van Meerendonk, P. Quax, L. Panitti, J. Grimbergen, A. Bout, J. Goudsmit, and M. Havenga. 2003. Replication-deficient human adenovirus type 35 vectors for gene transfer and vaccination: efficient human cell infection and bypass of preexisting adenovirus immunity. J. Virol. 77:8263-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, K., T. Guan, D. A. Cheresh, and G. R. Nemerow. 2000. Regulation of adenovirus membrane penetration by the cytoplasmic tail of integrin β5. J. Virol. 74:2731-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wickham, T. J., P. Mathias, D. A. Cheresh, and G. R. Nemerow. 1993. Integrins alpha v beta 3 and alpha v beta 5 promote adenovirus internalization but not virus attachment. Cell 73:309-319. [DOI] [PubMed] [Google Scholar]
- 50.Wickham, T. J., E. Tzeng, L. L. Shears II, P. W. Roelvink, Y. Li, G. M. Lee, D. E. Brough, A. Lizonova, and I. Kovesdi. 1997. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J. Virol. 71:8221-8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong, M. L., and M. T. Hsu. 1989. Linear adenovirus DNA is organized into supercoiled domains in virus particles. Nucleic Acids Res. 17:3535-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, E., S. A. Trauger, L. Pache, T. M. Mullen, D. J. von Seggern, G. Siuzdak, and G. R. Nemerow. 2004. Membrane cofactor protein is a receptor for adenoviruses associated with epidemic keratoconjunctivitis. J. Virol. 78:3897-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]