Abstract
Rotavirus is the most important cause of viral gastroenteritis and dehydrating diarrhea in young children. Rotavirus nonstructural protein 4 (NSP4) is an enterotoxin that was identified as an important agent in symptomatic rotavirus infection. To identify cellular proteins that interact with NSP4, a two-hybrid technique with Saccharomyces cerevisiae was used. NSP4 cDNA, derived from the human rotavirus strain Wa, was cloned into the yeast shuttle vector pGBKT7. An intestinal cDNA library derived from Caco-2 cells cloned into the yeast shuttle vector pGAD10 was screened for proteins that interact with NSP4. Protein interactions were confirmed in vivo by coimmunoprecipitation and immunohistochemical colocalization. After two-hybrid library screening, we repeatedly isolated cDNAs encoding the extracellular matrix (ECM) protein laminin-β3 (amino acids [aa] 274 to 878) and a cDNA encoding the ECM protein fibronectin (aa 1755 to 1884). Using deletion mutants of NSP4, we mapped the region of interaction with the ECM proteins between aa 87 and 145. Deletion analysis of laminin-β3 indicated that the region comprising aa 726 to 875 of laminin-β3 interacts with NSP4. Interaction of NSP4 with either laminin-β3 or fibronectin was confirmed by coimmunoprecipitation. NSP4 was present in infected enterocytes and in the basement membrane (BM) of infected neonatal mice and colocalized with laminin-β3, indicating a physiological interaction. In conclusion, two-hybrid screening with NSP4 yielded two potential target proteins, laminin-β3 and fibronectin, interacting with the enterotoxin NSP4. The release of NSP4 from the basal side of infected epithelial cells and the subsequent binding to ECM proteins localized at the BM may signify a new mechanism by which rotavirus disease is established.
Rotavirus is the most important cause of a severe, life-threatening viral gastroenteritis and dehydrating diarrhea in young children (22, 36). Mortality rates are low in developed countries, but about 440,000 children <5 years of age die each year in developing countries because of an infection with rotavirus (63). The high incidence of rotavirus disease, especially in developing countries, calls for the development of effective vaccines. However, efforts to develop safe and useful rotavirus vaccines are continuing, since use of the first vaccine routinely prescribed in the United States was suspended in 1999 because of a possible association with intussusception (55).
The nonstructural protein NSP4 encoded by rotavirus gene 10 has been shown to play a key role in viral morphogenesis (1, 2, 21, 47). In the membrane of the endoplasmic reticulum (ER), NSP4 functions as a receptor for newly formed virus particles that are translocated across the ER membrane (1). Increasing lines of evidence show that NSP4 is involved in the pathogenicity of rotavirus infection and that it acts as a viral enterotoxin (3, 30, 50, 72). When administered to young mice, NSP4 from different rotavirus groups or a synthetic peptide corresponding to amino acids (aa) 114 to 135 was able to induce age-dependent diarrhea (3, 30, 50, 72). NSP4 is thought to mediate phospholipase C-dependent cell signaling after being secreted from infected cells by increasing intracellular calcium levels leading to chloride secretion in adjacent uninfected cells (3, 20, 52, 79). Studies with cystic fibrosis knockout mice imply that the age-dependent diarrhea-inducing properties of NSP4 are due to age-dependent changes in calcium-mediated anion permeability (52). Recent studies indicate that a specific cleavage product of NSP4 (aa 112 to 175) is actively secreted from infected cells and that NSP4 is released from the apical side of infected epithelial cells via an atypical pathway that bypasses the Golgi apparatus and involves lipid microdomains called rafts (71, 87). Consistent with the proposed apical secretion of NSP4, only apical but not basolateral administration of NSP4 was found to cause a reduction in transepithelial electrical resistance and redistribution of zonula occludens-1 in MDCK-1 cells (76). On the other hand, NSP4 is able to cause chloride secretion when added to either the apical (luminal) or basal (submucosal) surface of mouse mucosal sheets (3), indicating that NSP4 is possibly also active at the basal side of the intestinal epithelium.
Laminin and fibronectin are multifunctional proteins with diverse biological activities. They can influence cell adhesion, growth, morphology, differentiation, migration, and agglutination as well as the assembly of the extracellular matrix (ECM) (19, 66). Members of the laminin family of glycoproteins are major constituents of basement membranes (BMs), ECMs found in close contact with individual cells and cell layers. Each laminin is a heterotrimer assembled from α, β, and γ chain subunits, secreted and incorporated into cell-associated ECMs. Laminins can be found in association with other BM compoents (collagens, proteoglycans, and nidogens) and with a variety of other macromolecules including fibronectin and growth factors (80, 86). Through these interactions, laminins contribute to cell differentiation, cell shape and movement, maintenance of tissue phenotypes, and promotion of tissue survival. Laminin-5, a heterotrimer of α2, β3, and γ2 chain subunits, can bind to α3β1, α6β4, and α6β1 integrins (15, 46, 57) and is distributed in BMs in several epithelial tissues including the intestine (17, 42). Fibronectin is a large multidomain protein that can exist in multiple forms that arise from alternative splicing of a single pre-mRNA (24, 38). Fibronectin mediates a wide variety of cellular interactions with the ECM and plays important roles in cell adhesion, migration, growth, and differentiation (62). Each monomer consists of three types of repeating units (termed FN repeats): type I, type II, and type III. These repeats account for approximately 90% of the fibronectin sequence and are also found in other molecules (62, 64). Fibronectin can be a ligand for many members of the integrin receptor family, including α5β1, α4β1, α4β7, and α9β1 (66), and it has several heparin binding sites (4, 13, 34).
In this study, we used a two-hybrid system with Saccharomyces cerevisiae to identify proteins that interact with NSP4. After screening, we repeatedly isolated cDNAs encoding the ECM proteins laminin-β3 and fibronectin. The region of NSP4 responsible for interaction with both proteins was mapped between aa 87 to 145. Binding of NSP4 to laminin-β3 and fibronectin was further confirmed by coimmunoprecipitation in rotavirus-infected Caco-2 cells and colocalization in the BMs of neonatal mice infected with rotavirus. These findings might reveal a novel pathway via which NSP4 is involved in rotavirus-induced diarrhea.
MATERIALS AND METHODS
Two-hybrid screening.
The Matchmaker System 3 two-hybrid assay using S. cerevisiae (Clontech) (18, 25) was used to detect interactions between NSP4 and cellular proteins. An intestinal cDNA library derived from Caco-2 cells cloned into the yeast shuttle vector pGAD10 was obtained from C. Mitchelmore and O. Norén, Copenhagen, Denmark. The construct used as bait corresponded to the NSP4 protein of human rotavirus (Wa strain). The NSP4 bait lacked the hydrophobic N terminus (aa 1 to 86) and was designated NSP4ΔN (nucleotides [nt] 300 to 750 and corresponding to aa 87 to 175). To obtain the pGBKT7-NSP4ΔN bait, reverse transcription-PCR was performed with RNA from rotavirus strain Wa-infected MA104 monkey kidney cells. NSP4ΔN was cloned with an NSP4 forward primer (5′-GGTACCATGGAGCAGGTTACTACAAAAGACG-3′) based on the NSP4 Wa coding sequence (nt 300 to 321; National Center for Biotechnology Information (NCBI) data accession number AF093200) (16) and a reverse primer (5′-TCCCCCGGGTCACATTAAGACCGTTCCT-3′) based on the SA11 coding sequence (nt 730 to 750) (11). With these primers, an NcoI restriction site was introduced at the 5′ (underlined) end, and an SmaI restriction site was introduced at the 3′ end to allow in-frame insertion into the plasmid pGBKT7 (Clontech) containing the GAL4 DNA binding domain.
S. cerevisiae strain AH109 (Clontech) containing the four reporter genes ADE2, HIS3, MEL1, and lacZ was cotransfected with the pGBKT7-NSP4ΔN bait plasmid and the pGAD10 Caco-2 cDNA library by the lithium acetate method (27, 73). AH109 cells were grown on plates containing medium lacking tryptophan and leucine (Trp− Leu−) to select for the presence of both the bait and the library plasmids. A total of 2 × 106 of these double transformants were then grown on dropout medium deprived of tryptophan, leucine, histidine and adenine (Trp− Leu− His− Ade−) to select for protein-protein interactions. Transformants that grew were assayed for α-galactosidase activity on 5-bromo-4-chloro-3-indolylphosphate (X-α-Gal) plates according to the manufacturer's instructions (Clontech). Plasmids were extracted from blue yeast clones and were then transfected into Escherichia coli DH5α. Bacteria were grown on medium with ampicillin to select for bacteria containing library plasmids. The plasmid DNA was then isolated and sequenced with an ABI 310 sequencer with a primer identical to nt 788 to 820 of the pGAD10 cloning vector and a primer reverse complementary to nt 862 to 893 of the pGAD10 vector as a forward and reverse primer, respectively. Homology searches were performed using the National Center for Biotechnology Information database with Mega BLAST. All positive constructs were rescreened for their ability to grow on complete dropout medium when transfected together with the NSP4 bait or with the empty pGBKT7 plasmid to verify potential interactions and to eliminate false positives.
Serum and monoclonal antibodies.
For immunostaining and immunoprecipitation experiments, we used the following antibodies: a NSP4 polyclonal rabbit hyperimmune serum directed against aa 114 to 135 of the NSP4 SA11 strain and designated anti-NSP4(114-135) (10), a rabbit polyclonal serum directed against aa 120 to 147 of SA11 NSP4 and designated anti-NSP4(120-147) (87), a rabbit hyperimmune serum raised against SA11 rotavirus particles (10), the rabbit polyclonal anti-laminin-β3 (catalogue no. H300; Santa Cruz), the rabbit polyclonal anti-fibronectin (Sigma), and the goat polyclonal anti-laminin-β3 (catalogue no. C-19; Santa Cruz). Peroxidase-conjugated secondary antibodies were purchased from DAKO.
Immunohistochemistry.
Tissue was obtained from 7-day-old BALB/c mice that were inoculated intragastrically with 2 × 104 focus-forming units (FFU) of the EDIM rotavirus strain as previously described (10). At different time points after infection, mice were sacrificed and tissue from the small intestine was fixed for immunohistochemistry as previously described (10). Deparaffinized and rehydrated tissue sections from neonatal control mice and mice infected with the murine EDIM strain (10) were incubated overnight at 4°C with the following rabbit polyclonal antibodies: anti-NSP4(114-135) (directed against aa 114 to 135; 1:1,000 dilution), anti-NSP4(120-147) (directed against aa 120 to 147; 1:1,000 dilution), anti-SA11 (1:1,500 dilution), or anti-laminin-β3 (1:500 dilution). Immunohistochemistry was carried out as described previously (10, 69); however, antigen unmasking for laminin-β3 was carried out by heating the sections for 10 min in 0.01 M sodium citrate (pH 6.0) at 100°C and by additional treatment with protease XXIV (Sigma) for 10 min at 37°C. The slides were analyzed by microscopy with a Nikon Eclipse 800 microscope and Leica IM-500 software.
Infection of Caco-2 cells, coimmunoprecipitation, and Western blotting.
Human colonic adenocarcinoma Caco-2 cells (Numico Research, Wageningen, The Netherlands) were grown and maintained in 75-cm2 tissue culture flasks in Dulbecco's modified Eagle's medium (Gibco BRL) with glutamax-1, pyridoxine, 0.11g of sodium pyruvate/liter, 4.5 g of glucose/liter, 10% fetal calf serum (Integro), 100 U of penicillin (Sigma)/ml, 100 μg of streptomycin (Sigma)/ml, and nonessential amino acids (BioWhittaker Europe, Verviers, Belgium) at 37°C and 5% CO2 in a humidified atmosphere. One day prior to infection, Caco-2 cells were seeded in 175-cm2 tissue culture flasks at 2.6 × 105 cells per cm2.
The human rotavirus strain Wa was obtained from M. Koopmans (National Institute for Public Health and the Environment, Bilthoven, The Netherlands) and was activated using 10 mg of trypsin (Sigma)/ml at 37°C for 1 h. Rotavirus was diluted to 2 × 102 FFU/ml (corresponding to a multiplicity of infection of 0.5) in serum-free Dulbecco's modified Eagle's medium. Cells were infected with rotavirus at 37°C and 5% CO2 in a humidified atmosphere. After 15 h of incubation, cells were washed in phosphate-buffered saline (PBS) and harvested into Tris lysis buffer (20 mM Tris [pH 7.5], 100 mM NaCl, 0.5% NP-40, 0.5 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml, and 10 μg of soybean trypsin inhibitor/ml). Cells were swelled on ice for 10 min and then lysed by 10 passages through a 23-gauge needle. The cellular homogenate was centrifuged at 10,000 × g for 10 min. Lysates (corresponding to 2.35 × 106 cells) were immunoprecipitated with anti-NSP4 antibody for 2 h at 4°C, and then 25 μl of protein A-Sepharose CL-4B beads (Pharmacia) was added to the mixture and incubated for another 2 h at 4°C. Beads coupled to immune complexes were washed three times with Tris lysis buffer. The immune complexes were then suspended in 3× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and analyzed by SDS-PAGE with 7.5 or 10% polyacrylamide and Western blotting. Proteins were transferred to nitrocellulose membranes (Nitran; Schleicher & Schuell, Dassell, Germany) and were consecutively incubated with anti-NSP4 rabbit polyclonal antibodies (directed against aa 114 to 135; 1:1,000 dilution) and anti-fibronectin (1:500 dilution) or anti-laminin-β3 goat polyclonal antibody (1:500 dilution) and peroxidase-conjugated anti-rabbit or anti-goat secondary antibodies (1:2,000 dilution). Bands were revealed by enhanced chemiluminescence, according to the instructions of the manufacturer (Amersham).
Deletion analysis of NSP4 and laminin-β3.
Five NSP4 deletion fragments and five laminin-β3 deletion clones were obtained by PCR. For all forward primers used to create deletion mutants of NSP4, an NcoI site (underlined in the sequences below) was added at the beginning of the forward primers to direct in-frame subcloning in the pGBKT7 plasmid.
The NSP4 clones NSP4(87-114), NSP4(87-145), NSP4(113-145), NSP4(145-175), and NSP4(113-175 were obtained using the following primers: 5′-GGTACCATGGAGCAGGTTACTACAAAAGACG-3′ and 5′-ATCAATCATCTCCAGCTGACGTCTC-3′ (nt 301 to 321 and 359 to 383), GGTACCATGGA GCAGGTTACTACAAAAGACG-3′ and 5′-CCTTCGACATATCTATAACG-3′ (nt 301 to 321 and 461 to 480), 5′-ACGCCATGGTGATTGATAAACTAACTACTCG-3′ and 5′-CCTTCGACATATCTATAACG-3′(nt 376 to 397 and 461 to 480), 5′-ATAGCCATGGCGAAGGAATTCAATCAGAA-3′ and 5′-TCAACC TCTCACATGGATGC-3′(nt 475 to 493 and 558 to 577), and 5′-ACGCCATGGTGATTGATAAACTAACTAC TCG-3′ and 5′-TCAACCTCTCACATGGATGC-3′ (nt 376 to 397 and 558 to 577), based on NCBI database accession number AF093200 (16). Primers for human laminin-β3 were based on the sequence with NCBI database accession number NM_000228 (26). The laminin-β3 fragments corresponding to aa regions 274 to 377 (nt 903 to 1212), 368 to 497 (nt 1185 to 1572), 488 to 618 (nt 1545 to 1935), 605 to 739 (nt 1896 to 2298), and 726 to 875 (nt 2259 to 2706) were obtained by PCR with the following oligonucleotides: 5′-GAATTCGTCCACGATGTCTGTGTCTGC-3′ and 5′-GGTCTCCTGAATGGAAGCTCC-3′, 5′-GAATTCGGAGCTTCCATTCAGGAGACC-3′ and 5′-CTGGTTGCACTGTGGGCTGAG-3′, 5′-GAATTCCCTCAGCCCACAGTGCAACCA-3′ and 5′-CTCCAGCCCAGGCCCTGACCA-3′, 5′-GAATTCACCGCCAGCCTGTGGTCAGGG-3′ and 5′-GCGCGAGCTGTCGGAGACCTG-3′, and 5′-GAATTCCAGGCTGCTCAGCAGGTCTCC-3′ and 5′CCTGGGTCTCCAAGCGCTGGG-3′, respectively. An EcoRI site (underlined) was added at the beginning of the forward primers to direct in-frame insertion in the pGADT7 plasmid containing the GAL4 activation domain. All constructs were controlled by DNA sequencing and expression of the corresponding fusion proteins in yeast cells.
We investigated the interaction of the fragmented NSP4 proteins with a pGAD10 laminin-β3 library clone (D19), the fragmented laminin-β3 clones in the PGADT7 vector, and the pGAD10 fibronectin library clone with yeast cells. Qualitative and quantitative results were obtained by assessing the ability of yeast to grow in the absence of tryptophan, leucine, histidine, and adenine (Trp− Leu− His− Ade−), the appearance of blue colonies in the presence of X-α-Gal, and the α-galactosidase activity of yeast grown in liquid medium with the chromogenic substrate PNP-α-Gal (p-nitrophenyl-α-d-galactopyranoside; Sigma-Aldrich) according to the manufacturer's instructions (Clontech).
Statistical analysis.
Statistical analysis of all data was performed using Student's t test for unpaired data (two tailed). Data were expressed as the mean ± the standard error of the mean, and P values of ≤0.05 were considered statistically significant.
RESULTS
Identification of proteins binding to NSP4.
To identify host cell proteins that interact with the NSP4 enterotoxin of rotavirus, we used the yeast two-hybrid approach. NSP4 cDNA derived from the human rotavirus strain Wa was cloned into yeast shuttle vector pGBKT7. We used the region spanning aa 87 to 175 of NSP4 (NSP4ΔN) to screen an intestinal Caco-2 cell cDNA library, since the first 86 aa of NSP4 contain three hydrophobic domains. These domains could possibly interfere with the two-hybrid system, since the fusion proteins have to travel into the nucleus of the yeast cell where transactivation of the reporter genes occurs. Additionally, many functionally properties of NSP4 are located between aa 112 and 175 (2, 3, 29, 87). The intestinal cDNA library derived from Caco-2 cells was screened for proteins that might interact with NSP4ΔN. The Caco-2 cell line has enterocyte-like characteristics and can be infected by the human rotavirus Wa strain. In total, 238 transformants were able to activate transcription of the reporter genes (MEL1, HIS3, and ADE2). These transformants grew in the absence of histidine and adenine and showed blue colonies in the presence of X-α-Gal substrate. After rescreening, restriction analysis, and sequencing, 23 clones fused to the activating domain of GAL4 were identified. Twenty-two of these clones encoded a 1,172-aa protein identified as laminin-β3 (accession number NM_000228) (26). These clones were derived from three independent clones. The common overlapping sequence of these clones corresponded to aa 274 to 878 (nt 903 to 2716) of laminin-β3. One clone encoded aa 1755 to 1884 of a protein identified as fibronectin (nt 5532 to 5919; NCBI database accession number NM_002026).
NSP4 interacts with laminin-β3 and fibronectin during rotavirus infection in Caco-2 cells.
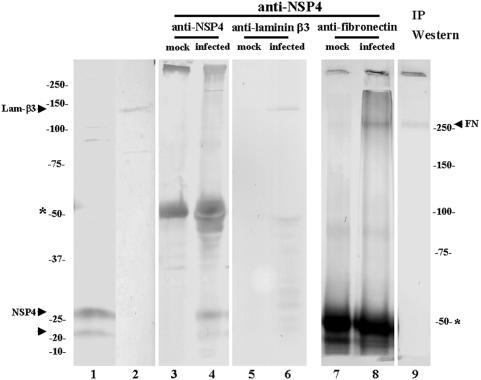
To establish that NSP4 associates in vivo with laminin-β3 and fibronectin, we performed coimmunoprecipitation studies. Cell lysates from mock-infected or rotavirus Wa-infected Caco-2 cells were prepared in a mild Tris lysis buffer. NSP4 (fully glycosylated and nonglycosylated forms of 28 and 20 kDa, respectively), laminin-β3 (140 kDa), and fibronectin (250 kDa) can be detected by Western blotting in lysates from infected (NSP4) and mock-infected (laminin-β3 and fibronectin) Caco-2 cells with specific antibodies (Fig. 1, lanes 1, 2, and 9). NSP4 was immunoprecipitated with hyperimmune anti-NSP4 serum in lysates from infected Caco-2 cells but not from mock-infected Caco-2 cells (Fig. 1, lanes 3 and 4). After immunoprecipitation with anti-NSP4 serum, both fibronectin and laminin-β3 could be detected in coimmunoprecipitated material from infected cells by Western blotting with specific goat and rabbit polyclonal antisera (Fig. 1, lanes 6 and 8). No coimmunoprecipitation of NSP4 and fibronectin or NSP4 and laminin-β3 was observed in lysates from mock-infected Caco-2 cells (Fig. 1B, lanes 5 and 7). Therefore, we demonstrated that NSP4 interacts with both fibronectin and laminin-β3 during rotavirus infection in Caco-2 cells.
FIG. 1.
Coimmunoprecipitation of NSP4 and laminin-β3 or NSP4 and fibronectin protein in infected Caco-2 cells. NSP4 (fully glycosylated form, 28 kDa; nonglycosylated form, 20 kDa (lane 1), laminin-β3 (Lamβ-3, 140 kDa) (lane 2) and fibronectin (FN, 250 kDa) (lane 9) were detected by Western blotting of lysates from, respectively, infected NSP4 and mock-infected Caco-2 cells (laminin-β3 and fibronectin) with specific antibodies. Lysates from infected or mock-infected Caco-2 cells were subjected to immunoprecipitation with protein A-Sepharose beads loaded with anti-NSP4 antibody (lanes 3 to 8). Immunoprecipitates were analyzed by SDS-PAGE and anti-NSP4 (lanes 3 and 4), anti-laminin-β3 (lanes 5 and 6), or anti-fibronectin (lanes 7 and 8) Western blotting. The bands around 50 kDa (asterisk; lanes 3 to 8) correspond to the immunoglobulin heavy chain. The immune complexes were analyzed with a 7.5% (lanes 7 to 9) or 10% (lanes 1 to 6) polyacrylamide gel.
NSP4 colocalizes with the enterocyte basement membrane during rotavirus infection in suckling mice.
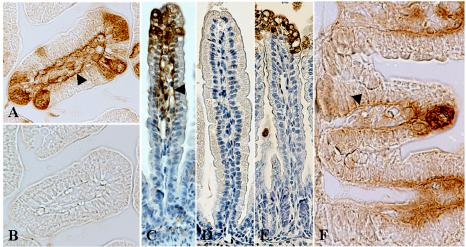
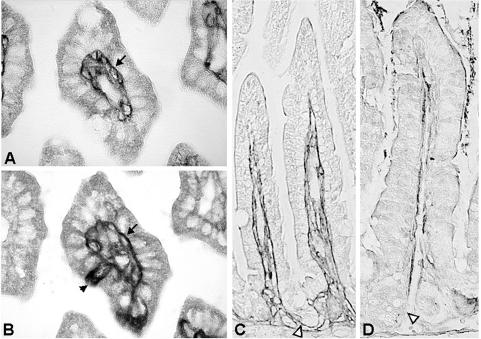
As NSP4 interacts with laminin-β3 and fibronectin, proteins that are localized in the ECM, we wanted to investigate whether NSP4 could be detected in the ECM in vivo. We used embedded tissue from 11-day-old BALB/c mice that had been inoculated with 2 × 104 FFU of the EDIM rotavirus strain or tissue from control mice that were mock infected with PBS. In this mouse model that we previously described, virus replication occurs in two peaks at 1 and 4 days postinfection (dpi), and mice develop diarrhea over a period of 5 days (10). The onset of diarrhea is 1 dpi, with the highest percentage of diarrhea and maximal severity of diarrheal illnes at 3 dpi. In infected mice at 1 (data not shown) and 4 dpi, anti-NSP4(114-135) rabbit polyclonal serum stained infected epithelial cells of the small intestine (jejunum) (Fig. 2A and C). The antiserum, however, also stained the BM of infected cells on the villi, as well as the BM of noninfected cells further down the villus (Fig. 2C). When infected cells low on the villus were observed, we could detect NSP4 in the BM of lower villus and crypt cells (Fig. 2F). Preimmune serum (obtained before antigen boosting with NSP4) (Fig. 2B) did not stain an adjacent section of infected tissue, indicating that NSP4 staining was specific. With control mice, no staining was observed with the NSP4 antiserum 4 days after mock infection with PBS (Fig. 2D). Similar to the anti-NSP4(114-135) rabbit serum, a rabbit polyclonal serum against SA11 NSP4 (directed against aa 120 to 147) (87), also stained the BMs of uninfected as well as infected epithelial cells in infected mice (data not shown). A rabbit polyclonal antiserum against SA11 rotavirus stained only infected cells and not the BM (Fig. 2E), indicating that rotavirus structural proteins are not present in the BM and that BM staining was specific for the NSP4 antibodies. Using adjacent immunohistochemical sections, NSP4 could be colocalized with laminin-β3 in the BM of small intestinal epithelial cells (Fig. 3). During rotavirus infection in neonatal mice, laminin-β3 was detected in the BMs of small intestinal epithelium at 4 dpi (Fig. 3A). In the adjacent section (Fig. 3B), rotavirus-infected cells were detected with the anti-NSP4 antibody, whereas the BMs of infected and noninfected cells were also stained. During development, laminin-5 (or its single subunits) is initially present in the BM associated with both the villus and crypt epithelia. Expression in the adult human small intestine is eventually restricted mainly to the villus region, with only weak expression in the BM of the upper crypt region (37, 40, 42). In agreement with these data, laminin-β3 was detected in the BM associated with villus as well as crypt epithelium in 11-day-old mice (Fig. 3C). In 28-day-old mice that are not susceptible to rotavirus-induced diarrhea (68), laminin-β3 expression appeared decreased and was only weak in the BMs of the villus and upper crypt region (Fig. 3D). These observations suggest that NSP4 is released on the basal side from infected epithelial cells in neonatal mice and binds to laminin-β3 in the ECM. We also tested several anti-fibronectin antibodies to stain the BM. When paraffin-embedded tissue was used, all the antibodies failed to stain the BM. In contrast, when frozen tissue sections were used, numerous studies previously described expression of fibronectin in the BM of small intestinal tissue in different species (5-7, 37, 39, 67, 75). These studies point out that fibronectin is found predominantly in the crypt region while expression gradually decreases towards the tips of the villi. Therefore, it is apparent that NSP4, similar to laminin-β3, also colocalizes with fibronectin in the ECM, although we could not detect fibronectin by our methods.
FIG. 2.
NSP4 associates with the epithelial BM in mouse jejunum during rotavirus infection in neonatal mice. (A and C) With the anti-NSP4(114-135) rabbit polyclonal serum, NSP4 protein was detected in epithelial cells at the tips of intestinal villi at 4 dpi and at the epithelial BM of infected and noninfected cells (arrowheads). (B) Localization of NSP4 at the BM was not observed with preimmune serum. (D) NSP4 protein expression was not detected in control mice. (E) An antibody that recognizes rotavirus structural proteins stained infected epithelial cells but failed to stain the epithelial BMs. (F) NSP4 could occasionally be detected in the BMs of lower villus and crypt cells (arrowhead). Panels A and B are cross-sectional slides; magnification, ×400. Panels C to F are longitudinal slides; magnification, ×200 (C to E) and ×600 (F).
FIG. 3.
Colocalization of NSP4 with laminin-β3 in the BMs of small intestinal epithelial cells during rotavirus infection in neonatal mice. (A) Laminin-β3 was detected in the BMs of small intestinal epithelia at 4 dpi (arrow). (B) In the adjacent section, rotavirus-infected cells were detected with anti-NSP4 antibody (arrowhead), whereas the BMs of infected and noninfected cells were also stained (arrow). (C) Laminin-β3 was detected in the BM associated with the villus and crypt epithelium (open arrowhead) in 11-day-old mice. (D) In 28-day-old mice, laminin-β3 expression was decreased in the BM of the villus region, weakly present in the upper crypt region, and not observed in the lower crypt region (open arrowhead). Panels A and B are cross-sectional slides; magnification, ×600. Panels C and D are longitudinal slides; magnification, ×200.
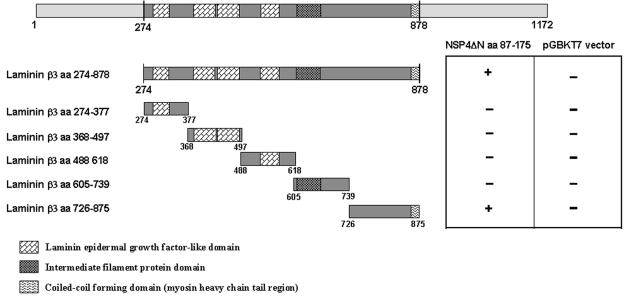
Characterization of the interacting domains of NSP4, laminin-β3, and fibronectin.
The laminin-β3 insert region (aa 274 to 878) contains four laminin-type epidermal growth factor-like domains (aa 277 to 304, 378 to 428, 431 to 478, and 533 to 570), an intermediate filament protein domain (aa 627 to 686), and a small region of the coiled-coil-forming domain (myosin tail; aa 868 to 878) per NCBI Conserved Domain Database accession number NM_000228 (26) (Fig. 4). To evaluate which regions or domains of laminin-β3 interact with NSP4, we constructed five different deletion fragments of the laminin-β3 clones and tested them against NSP4ΔN in the yeast two-hybrid screening (Fig. 4). The readout of the interaction between the various laminin-β3 deletion mutants with NSP4ΔN was the growth of yeasts harboring the two plasmids in His− Ade− medium and the development of a blue color in the presence of X-α-Gal substrate (Fig. 4). As a negative control, the laminin-β3 clones were also screened against the pGBKT7 plasmid without NSP4 as bait. Only laminin-β3 (aa 274 to 878) or the laminin-β3 clone (aa 726 to 875) interacted with NSP4. None of the other constructs encoding a fragment of laminin-β3 was able to interact with NSP4 (Fig. 4). This indicates that NSP4 interacts with the region between aa 726 and 875 of laminin-β3.
FIG. 4.
Binding domain analysis of laminin-β3 with NSP4ΔN. Laminin-β3 insert of clone D19 (aa 274 to 875) was fragmented into five subclones by PCR. Bars represent truncated laminin-β3 inserts fused to the GAL4 activation domain. Amino acid positions are indicated. Yeast strain AH109 was cotransformed with the laminin-β3 clones and pGBKT7-NSP4ΔN. The interaction was assessed by checking for blue colonies in the presence of X-α-Gal. Functional and structural domains of laminin-β3 are presented in boxes.
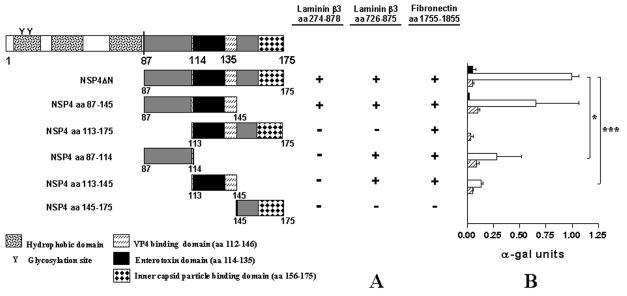
The NSP4ΔN bait (aa 87 to 175) includes several previously described functional domains including an amphipatic α-helical region (aa 95 to 137) involved in the formation of a coiled-coil domain, oligomerization, and membrane-destabilizing activities of NSP4 (56, 77, 78); an enterotoxic peptide sequence (aa 114 to 135) domain that was shown to induce diarrhea in young mice and potentate chloride secretion (3) and seems to be involved in the direct inhibitory effect of NSP4 on the Na-d-glucose symporter (SGLT1) (29); a VP4 binding site (aa 112 to 146) (2); and a region that functions as a receptor for inner capsid particles (aa 156 to 175) (59). Using PCR, we constructed five mutants of NSP4. To map the sites of NSP4 involved in binding, we subcloned these mutants in the pGBKT7 plasmid to obtain deletion mutants of NSP4 fused to the DNA binding domain of the yeast transcription factor GAL4 and screened them against the GAL4-activating domain fusion proteins laminin-β3 (aa 274 to 878), laminin-β3 (aa 726 to 875), and fibronectin in a yeast-two-hybrid screening assay (Fig. 5A). The interaction of the five NSP4 deletion clones with laminin-β3 (aa 274 to 878), laminin-β3 (aa 726 to 875), or fibronectin was assayed on His− Ade− selection medium in the presence of X-α-Gal substrate through selection by growth and the development of blue colonies (Fig. 5A). Interactions were confirmed by a quantitative α-galactosidase assay based on the induction of the Mel1 gene (Fig. 5B). NSP4ΔN was used as a positive control. Deletion of aa 87 to 114 of NSP4 in the NSP4(113-175) subclone abolished or suppressed the interaction of NSP4 with laminin-β3 (aa 274 to 878) and laminin-β3 (aa 726 to 875), respectively (Fig. 5). The laminin-β3 (aa 726 to 875) fragment was able to interact with the NSP4-truncated clones NSP4(87-114) and NSP4(113-145). These interactions, however, were significantly suppressed compared to the interactions with NSP4ΔN and NSP4(87-145). Interaction of laminin-β3 (aa 726 to 875) with NSP4 was much stronger than with the full-length laminin-β3 clone. This could be due to the fact that the laminin-β3 subclones were constructed in the pGADT7 high-copy-number vector instead of the pGAD10 low-copy-number vector used for the construction of the Caco-2 cDNA library. On the other hand, however, shorter fragments frequently give stronger interactions than longer fragments when the two-hybrid system is used, which can be caused by differences in folding of the fusion proteins (33, 65).
FIG. 5.
Binding domain analysis of NSP4 with laminin-β3 (aa 274 to 878), laminin-β3 (aa 726 to 875), and fibronectin (aa 1755 to 1885). (A) NSP4ΔN was fragmented into five subclones by PCR. Bars represent truncated NSP4 clones fused to the GAL4 binding domain. Amino acid positions are indicated. Yeast strain AH109 was cotransformed with the laminin-β3 clones and pGBKT7-NSP4ΔN. The interactions were assessed by checking for blue colonies in the presence of X-α-Gal. Functional and structural domains of NSP4 are presented in boxes. (B) Quantitative analysis of the interactions were obtained from independent yeast cotransformants assayed with p-nitrophenyl-α-d-galactosidase as a substrate. α-Galactosidase activity was expressed in units and calculated by the following formula: (OD410 × Vf × 1,000)/[OD600 × T × (ɛ × b) × Vi], where OD410 is the absorbance of the reaction mixture measured by optical density at 410 nm, OD600 is the optical cell density of the culture at 600 nm, T is the reaction time in minutes, Vf is the total volume (in milliliters) of the assay, Vi is the volume of culture medium supernatant added, ɛ is the molar absorbtivity for p-nitrophenol, and b is the light path in centimeters. Solid, open, and striped bars correspond to interactions of laminin-β3 (D19 clone), laminin-β3 (aa 726 to 875), and fibronectin (clone D22) with the NSP4 constructs, respectively, indicated. Standard errors of the mean are shown for the results of three independent experiments. *, P < 0.05; ***, P < 0.001 by Student's t test.
Interaction of NSP4 with fibronectin was abolished only when the C-terminal clone NSP4(145-175) was used in two-hybrid screening (Fig. 5). These data indicate that for the interaction of NSP4 with laminin-β3 and fibronectin, aa 87 to 145 are sufficient and that for binding to laminin-β3, aa 87 to 114 are most important.
DISCUSSION
To date, several cellular partners of NSP4 have been identified. NSP4 has been shown to interact with the ER-associated lectin-like chaperone calnexin during infection in MA104 cells (48). For this interaction, glycosylation of NSP4 was important and glucose trimming was necessary, indicating that the N-terminal domain of NSP4 is involved and that binding to calnexin is of a carbohydrate nature. NSP4 has a direct inhibiting effect on the Na-d-glucose symporter (SGLT1), indicating that NSP4 inhibits water resorption and thus is crucial for rotavirus pathogenesis (29). However, whether or not the inhibition involves direct binding of NSP4 to SGLT1 has yet to be confirmed. Finally, NSP4 has been identified along microtubule-like structures, where it disrupts microtubule-mediated membrane trafficking from the ER to the Golgi complex (84). Although several NSP4-interacting proteins have thus far been identified, none revealed more insight into the mechanism of NSP4-induced chloride secretion.
In the present work, we searched for cellular proteins that interact with the rotavirus protein NSP4 to gain more insight into rotavirus pathogenesis. For this purpose, we used the two-hybrid technique with yeast cells and screened a human Caco-2 cell cDNA library fused to the GAL4 activation domain with aa 87 to 175 of NSP4 fused to the DNA binding domain of GAL4. As NSP4-interacting cDNA clones, laminin-β3 and fibronectin were isolated. The interaction of NSP4 with the ECM proteins laminin-β3 and fibronectin was confirmed by coimmunoprecipitation of infected Caco-2 cell lysates. The interaction with laminin-β3 was further established by colocalization of NSP4 with laminin-β3 in the BMs of infected neonatal mice. Deletion analysis showed that aa 87 to 145 of NSP4 were sufficient for interaction with laminin-β3 and fibronectin. For binding to laminin-β3, aa 87 to 114 of NSP4 were the most important. Interaction of NSP4 with laminin-β3 and fibronectin was abolished only when the C-terminal clone NSP4(145-175) was used in the two-hybrid screening, indicating that this C-terminal region is not involved in NSP4 binding. The laminin-β3 clones encoded several conserved domains. NSP4 was found to interact only with a truncated laminin-β3 fragment (aa 726 to 875) containing the N-terminal part of a domain involved in the formation of the triple-stranded α-helical coil-coiled structure that represents the rod of the long arm of laminins (32, 82).
The fibronectin insert (aa 1755 to 1884) contains the last 57 aa of type III FN12 and the first 72 aa of FN13 (74). This region of fibronectin, the HepII region, is known to be involved in cell binding and cell spreading (8, 83) and binds to heparan sulfate proteoglycans like syndecan-4 (4, 13, 34, 83) and tenascin-C (31) and integrins α4β1 and αIIbβ3 (4, 13, 34, 49, 54). The fibronectin insert found with the two-hybrid system contains all six basic residues previously shown to be crucial for heparin binding (13) and thus represents the principal heparin binding site of fibronectin. By binding to the HepII region in fibronectin, syndecan-4 has been shown to function as a coreceptor in integrin signaling (83). It might be possible that NSP4, through binding to fibronectin, could interfere with this interaction and influence integrin signaling. Such a mechanism was observed for tenascin-C, as binding of tenascin-C interfered with fibronectin-syndecan-4 interactions and replaced syndecan-4, thereby influencing cell signaling and proliferation (31). The fibronectin type III domain is a very common constituent of animal proteins, and its main functions are to mediate protein-protein interactions and to act as a linker (14). Fibronectin type III domains have been found in intracellular, extracellular, and membrane-spanning proteins and are additionally found in many bacterial extracellular glycohydrolases (14, 41).
Rotavirus-induced disease is thought to be caused by a combination of factors (70), including a reduction in epithelial surface area, replacement of mature enterocytes by immature (crypt-like) cells (10, 60), an osmotic effect resulting from incomplete absorption of carbohydrates from the intestinal lumen in combination with bacterial fermentation of these nonabsorbed compounds, secretion of intestinal fluid and electrolytes through activation of the enteric nervous system (43), and the secretory effect of NSP4, which is thought to act as a viral enterotoxin (3). During rotavirus infection, there is an enhanced turnover of epithelial cells that is thought to result in the presence of immature cells on the villi (10, 60). Since laminin-5 and fibronectin are proteins involved in cell differentiation and cellular migration (5, 6, 62, 67, 75), it is possible that NSP4 could modulate migration and/or differentiation of epithelial cells via binding to fibronectin and laminin. The mechanism contributing to diarrheal disease and involving NSP4 is thought to occur through release of NSP4 or its cleavage product NSP4(112-175 from infected cells and through binding to an as-yet-unidentified apical membrane receptor. Binding would directly or indirectly trigger a signal transduction pathway to enhance chloride secretion, eventually resulting in diarrhea (3, 23, 51, 87). This secretory mechanism, however, is thought to act in the intestinal crypts (44). It is still not clear if and how NSP4 reaches the intestinal crypts. However, it seems unlikely that this occurs via the luminal route, since continuous abundant secretion from the crypts results in a hydrodynamic flow against which NSP4 has to diffuse (44). We observed a BM localization of NSP4 and colocalization with laminin-β3 in the ECM during rotavirus infection of neonatal mice. BM localization was found underneath infected as well as noninfected cells. Our data, therefore, suggest that NSP4 is also released from the basal sides of infected enterocytes. We presently can only speculate whether NSP4 or a cleavage product is released from the basal side of infected cells. However, since we detected NSP4 in vivo at the BM with antibodies against aa 114 to 135 and 120 to 147, it can be concluded that the released peptide sequence at least contains the enterotoxic domain. After basal release, NSP4 would bind to the ECM proteins laminin-β3 and fibronectin. Since we could detect NSP4 in the BM of lower villus cells and occasionally underneath epithelial cells in the crypts, it is temping to speculate that NSP4 travels towards the crypt region where it can induce fluid secretion via a route involving the ECM. In concurrence with the hypothesis stated above and with our results, Ball and coworkers found that NSP4 is able to cause chloride secretion when added to the submucosal (basal) surface of mouse mucosal sheets (3).
Recent studies indicate that rotavirus infection is not confined to the intestine but instead leaves the intestine and enters the circulatory system (9, 53). Rotavirus has been detected in cerebral spinal fluid (35, 45, 58, 61, 85), liver and kidney (28), extraintestinal lymphoid tissue (12), and the sera of infected children and animals (9, 81). By in situ reverse transcription-PCR, rotavirus was also found in the heart, where it was mainly localized to endothelial cells (53). It was suggested that rotavirus, analogous to what has been found for reovirus, enters the circulatory system through M cells that are present in the epithelium overlying Peyer's patches (9, 53). Our data also imply that NSP4 could get into the circulatory system, as it is apparently at least partly secreted into the tissue underlying the enterocytes. If NSP4 is also able to enter the circulatory system, it is likely that it does so through release from the basal side of infected epithelial cells.
In summary, our results show that NSP4 binds to the ECM molecules laminin-β3 and fibronectin in vitro and in vivo. Binding to the ECM and localization at the BM imply that NSP4 is released from the basal side of infected epithelial cells. From our data, the significance of the deposition of NSP4 in the BM is still unclear, but it could possibly point to a new mechanism by which rotavirus disease is established. The involved mechanisms, however, remain to be defined.
Acknowledgments
We are very grateful to C. Mithelmore and O. Norén for donating the Caco-2 cDNA library. We thank Marion Koopmans for kindly providing the rotavirus Wa strain, and Mary Estes for donating the NSP4 (120-147) antibody. We also thank Jan Dekker for critical reading of the manuscript.
This work was supported by grants from the Sophia Foundation for Medical Research (SSWO), The Netherlands Digestive Diseases Foundation (MLDS), and The Netherlands Society for Scientific Research (NWO).
REFERENCES
- 1.Au, K. S., W. K. Chan, J. W. Burns, and M. K. Estes. 1989. Receptor activity of rotavirus nonstructural glycoprotein NS28. J. Virol. 63:4553-4562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Au, K. S., N. M. Mattion, and M. K. Estes. 1993. A subviral particle binding domain on the rotavirus nonstructural glycoprotein NS28. Virology 194:665-673. [DOI] [PubMed] [Google Scholar]
- 3.Ball, J. M., P. Tian, C. Q. Zeng, A. P. Morris, and M. K. Estes. 1996. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science 272:101-104. [DOI] [PubMed] [Google Scholar]
- 4.Barkalow, F. J., and J. E. Schwarzbauer. 1991. Localization of the major heparin-binding site in fibronectin. J. Biol. Chem. 266:7812-7818. [PubMed] [Google Scholar]
- 5.Beaulieu, J. F. 1992. Differential expression of the VLA family of integrins along the crypt-villus axis in the human small intestine. J. Cell Sci. 102:427-436. [DOI] [PubMed] [Google Scholar]
- 6.Beaulieu, J. F., P. H. Vachon, and S. Chartrand. 1991. Immunolocalization of extracellular matrix components during organogenesis in the human small intestine. Anat. Embryol. (Berlin) 183:363-369. [DOI] [PubMed] [Google Scholar]
- 7.Berghem, L. E., and K. J. Johanson. 1983. Effect of 137Cs gamma radiation on the fibronectin content in basement membrane of mouse small intestine. Acta Radiol. Oncol. 22:389-393. [DOI] [PubMed] [Google Scholar]
- 8.Bloom, L., K. C. Ingham, and R. O. Hynes. 1999. Fibronectin regulates assembly of actin filaments and focal contacts in cultured cells via the heparin-binding site in repeat III13. Mol. Biol. Cell 10:1521-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blutt, S. E., C. D. Kirkwood, V. Parreno, K. L. Warfield, M. Ciarlet, M. K. Estes, K. Bok, R. F. Bishop, and M. E. Conner. 2003. Rotavirus antigenaemia and viraemia: a common event? Lancet 362:1445-1449. [DOI] [PubMed] [Google Scholar]
- 10.Boshuizen, J. A., J. H. Reimerink, A. M. Korteland-Van Male, V. J. Van Ham, M. P. Koopmans, H. A. Buller, J. Dekker, and A. W. Einerhand. 2003. Changes in small intestinal homeostasis, morphology, and gene expression during rotavirus infection of infant mice. J. Virol. 77:13005-13016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Both, G. W., L. J. Siegman, A. R. Bellamy, and P. H. Atkinson. 1983. Coding assignment and nucleotide sequence of simian rotavirus SA11 gene segment 10: location of glycosylation sites suggests that the signal peptide is not cleaved. J. Virol. 48:335-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown, K. A., and P. A. Offit. 1998. Rotavirus-specific proteins are detected in murine macrophages in both intestinal and extraintestinal lymphoid tissues. Microb. Pathog. 24:327-331. [DOI] [PubMed] [Google Scholar]
- 13.Busby, T. F., W. S. Argraves, S. A. Brew, I. Pechik, G. L. Gilliland, and K. C. Ingham. 1995. Heparin binding by fibronectin module III-13 involves six discontinuous basic residues brought together to form a cationic cradle. J. Biol. Chem. 270:18558-18562. [DOI] [PubMed] [Google Scholar]
- 14.Campbell, I. D., and C. Spitzfaden. 1994. Building proteins with fibronectin type III modules. Structure 2:333-337. [DOI] [PubMed] [Google Scholar]
- 15.Carter, W. G., M. C. Ryan, and P. J. Gahr. 1991. Epiligrin, a new cell adhesion ligand for integrin alpha 3 beta 1 in epithelial basement membranes. Cell 65:599-610. [DOI] [PubMed] [Google Scholar]
- 16.Chang, K. O., Y. J. Kim, and L. J. Saif. 1999. Comparisons of nucleotide and deduced amino acid sequences of NSP4 genes of virulent and attenuated pairs of group A and C rotaviruses. Virus Genes 18:229-233. [DOI] [PubMed] [Google Scholar]
- 17.Cheng, Y. S., M. F. Champliaud, R. E. Burgeson, M. P. Marinkovich, and P. D. Yurchenco. 1997. Self-assembly of laminin isoforms. J. Biol. Chem. 272:31525-31532. [DOI] [PubMed] [Google Scholar]
- 18.Chien, C. T., P. L. Bartel, R. Sternglanz, and S. Fields. 1991. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA 88:9578-9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colognato, H., and P. D. Yurchenco. 2000. Form and function: the laminin family of heterotrimers. Dev. Dyn. 218:213-234. [DOI] [PubMed] [Google Scholar]
- 20.Dong, Y., C. Q. Zeng, J. M. Ball, M. K. Estes, and A. P. Morris. 1997. The rotavirus enterotoxin NSP4 mobilizes intracellular calcium in human intestinal cells by stimulating phospholipase C-mediated inositol 1,4,5-trisphosphate production. Proc. Natl. Acad. Sci. USA 94:3960-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Estes, M. K. 2001. Rotaviruses and their replication, p. 1747-1785. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 22.Estes, M. K., and J. Cohen. 1989. Rotavirus structure and function. Microbiol. Rev. 53:410-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Estes, M. K., and A. P. Morris. 1999. A viral enterotoxin. A new mechanism of virus-induced pathogenesis. Adv. Exp. Med. Biol. 473:73-82. [PubMed] [Google Scholar]
- 24.ffrench-Constant, C. 1995. Alternative splicing of fibronectin—many different proteins but few different functions. Exp. Cell Res. 221:261-271. [DOI] [PubMed] [Google Scholar]
- 25.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 26.Gache, Y., M. Allegra, C. Bodemer, A. Pisani-Spadafora, Y. de Prost, J. P. Ortonne, and G. Meneguzzi. 2001. Genetic bases of severe junctional epidermolysis bullosa presenting spontaneous amelioration with aging. Hum. Mol. Genet. 10:2453-2461. [DOI] [PubMed] [Google Scholar]
- 27.Gietz, D., A. St. Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilger, M. A., D. O. Matson, M. E. Conner, H. M. Rosenblatt, M. J. Finegold, and M. K. Estes. 1992. Extraintestinal rotavirus infections in children with immunodeficiency. J. Pediatr. 120:912-917. [DOI] [PubMed] [Google Scholar]
- 29.Halaihel, N., V. Liévin, J. M. Ball, M. K. Estes, F. Alvarado, and M. Vasseur. 2000. Direct inhibitory effect of rotavirus NSP4(114-135) peptide on the Na+-d-glucose symporter of rabbit intestinal brush border membrane. J. Virol. 74:9464-9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Horie, Y., O. Nakagomi, Y. Koshimura, T. Nakagomi, Y. Suzuki, T. Oka, S. Sasaki, Y. Matsuda, and S. Watanabe. 1999. Diarrhea induction by rotavirus NSP4 in the homologous mouse model system. Virology 262:398-407. [DOI] [PubMed] [Google Scholar]
- 31.Huang, W., R. Chiquet-Ehrismann, J. V. Moyano, A. Garcia-Pardo, and G. Orend. 2001. Interference of tenascin-C with syndecan-4 binding to fibronectin blocks cell adhesion and stimulates tumor cell proliferation. Cancer Res. 61:8586-8594. [PubMed] [Google Scholar]
- 32.Hunter, I., T. Schulthess, M. Bruch, K. Beck, and J. Engel. 1990. Evidence for a specific mechanism of laminin assembly. Eur. J. Biochem. 188:205-211. [DOI] [PubMed] [Google Scholar]
- 33.Ikeshima-Kataoka, H., J. B. Skeath, Y. Nabeshima, C. Q. Doe, and F. Matsuzaki. 1997. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature 390:625-629. [DOI] [PubMed] [Google Scholar]
- 34.Ingham, K. C., S. A. Brew, and D. H. Atha. 1990. Interaction of heparin with fibronectin and isolated fibronectin domains. Biochem. J. 272:605-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iturriza-Gomara, M., I. A. Auchterlonie, W. Zaw, P. Molyneaux, U. Desselberger, and J. Gray. 2002. Rotavirus gastroenteritis and central nervous system (CNS) infection: characterization of the VP7 and VP4 genes of rotavirus strains isolated from paired fecal and cerebrospinal fluid samples from a child with CNS disease. J. Clin. Microbiol. 40:4797-4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kapikian, A. Z., and R. M. Chanock. 1990. Rotaviruses, p. 1353-1404. In B. N. Fields, D. M. Knipe, R. M. Chanock, M. S. Hirsch, J. L. Melnick, and B. Roizman (ed.), Virology, 2nd ed. Raven Press, New York, N.Y.
- 37.Korhonen, M., M. Ormio, R. E. Burgeson, I. Virtanen, and E. Savilahti. 2000. Unaltered distribution of laminins, fibronectin, and tenascin in celiac intestinal mucosa. J. Histochem. Cytochem. 48:1011-1020. [DOI] [PubMed] [Google Scholar]
- 38.Kosmehl, H., A. Berndt, and D. Katenkamp. 1996. Molecular variants of fibronectin and laminin: structure, physiological occurrence and histopathological aspects. Virchows Arch. 429:311-322. [DOI] [PubMed] [Google Scholar]
- 39.Laurie, G. W., C. P. Leblond, and G. R. Martin. 1982. Localization of type IV collagen, laminin, heparan sulfate proteoglycan, and fibronectin to the basal lamina of basement membranes. J. Cell Biol. 95:340-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leivo, I., T. Tani, L. Laitinen, R. Bruns, E. Kivilaakso, V. P. Lehto, R. E. Burgeson, and I. Virtanen. 1996. Anchoring complex components laminin-5 and type VII collagen in intestine: association with migrating and differentiating enterocytes. J. Histochem. Cytochem. 44:1267-1277. [DOI] [PubMed] [Google Scholar]
- 41.Little, E., P. Bork, and R. F. Doolittle. 1994. Tracing the spread of fibronectin type III domains in bacterial glycohydrolases. J. Mol. Evol. 39:631-643. [DOI] [PubMed] [Google Scholar]
- 42.Lu, W., K. Miyazaki, H. Mizushima, and N. Nemoto. 2001. Immunohistochemical distribution of laminin-5 gamma2 chain and its developmental change in human embryonic and foetal tissues. Histochem. J. 33:629-637. [DOI] [PubMed] [Google Scholar]
- 43.Lundgren, O., A. T. Peregrin, K. Persson, S. Kordasti, I. Uhnoo, and L. Svensson. 2000. Role of the enteric nervous system in the fluid and electrolyte secretion of rotavirus diarrhea. Science 287:491-495. [DOI] [PubMed] [Google Scholar]
- 44.Lundgren, O., and L. Svensson. 2001. Pathogenesis of rotavirus diarrhea. Microbes Infect. 3:1145-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lynch, M., B. Lee, P. Azimi, J. Gentsch, C. Glaser, S. Gilliam, H. G. Chang, R. Ward, and R. I. Glass. 2001. Rotavirus and central nervous system symptoms: cause or contaminant? Case reports and review. Clin. Infect. Dis. 33:932-938. [DOI] [PubMed] [Google Scholar]
- 46.Marchisio, P. C., O. Cremona, P. Savoia, G. Pellegrini, J. P. Ortonne, P. Verrando, R. E. Burgeson, R. Cancedda, and M. De Luca. 1993. The basement membrane protein BM-600/nicein codistributes with kalinin and the integrin α6β4 in human cultured keratinocytes. Exp. Cell Res. 205:205-212. [DOI] [PubMed] [Google Scholar]
- 47.Meyer, J. C., C. C. Bergmann, and A. R. Bellamy. 1989. Interaction of rotavirus cores with the nonstructural glycoprotein NS28. Virology 171:98-107. [DOI] [PubMed] [Google Scholar]
- 48.Mirazimi, A., M. Nilsson, and L. Svensson. 1998. The molecular chaperone calnexin interacts with the NSP4 enterotoxin of rotavirus in vivo and in vitro. J. Virol. 72:8705-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohri, H., J. Tanabe, K. Katoh, and T. Okubo. 1996. Identification of a novel binding site to the integrin αIIbβ3 located in the C-terminal heparin-binding domain of human plasma fibronectin. J. Biol. Chem. 271:15724-15728. [DOI] [PubMed] [Google Scholar]
- 50.Mori, Y., M. A. Borgan, N. Ito, M. Sugiyama, and N. Minamoto. 2002. Diarrhea-inducing activity of avian rotavirus NSP4 glycoproteins, which differ greatly from mammalian rotavirus NSP4 glycoproteins in deduced amino acid sequence in suckling mice. J. Virol. 76:5829-5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morris, A. P., and M. K. Estes. 2001. Microbes and microbial toxins: paradigms for microbial-mucosal interactions. VIII. Pathological consequences of rotavirus infection and its enterotoxin. Am. J. Physiol. Gastrointest. Liver Physiol. 281:G303-G310. [DOI] [PubMed] [Google Scholar]
- 52.Morris, A. P., J. K. Scott, J. M. Ball, C. Q. Zeng, W. K. O'Neal, and M. K. Estes. 1999. NSP4 elicits age-dependent diarrhea and Ca2+-mediated I− influx into intestinal crypts of CF mice. Am. J. Physiol. 277:G431-G444. [DOI] [PubMed] [Google Scholar]
- 53.Mossel, E. C., and R. F. Ramig. 2003. A lymphatic mechanism of rotavirus extraintestinal spread in the neonatal mouse. J. Virol. 77:12352-12356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mould, A. P., A. Komoriya, K. M. Yamada, and M. J. Humphries. 1991. The CS5 peptide is a second site in the IIICS region of fibronectin recognized by the integrin α4β1. Inhibition of α4β1 function by RGD peptide homologues. J. Biol. Chem. 266:3579-3585. [PubMed] [Google Scholar]
- 55.Murphy, T. V., P. M. Gargiullo, M. S. Massoudi, D. B. Nelson, A. O. Jumaan, C. A. Okoro, L. R. Zanardi, S. Setia, E. Fair, C. W. LeBaron, M. Wharton, J. R. Livengood, and J. R. Livingood. 2001. Intussusception among infants given an oral rotavirus vaccine. N. Engl. J. Med. 344:564-572. [DOI] [PubMed] [Google Scholar]
- 56.Newton, K., J. C. Meyer, A. R. Bellamy, and J. A. Taylor. 1997. Rotavirus nonstructural glycoprotein NSP4 alters plasma membrane permeability in mammalian cells. J. Virol. 71:9458-9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Niessen, C. M., F. Hogervorst, L. H. Jaspars, A. A. de Melker, G. O. Delwel, E. H. Hulsman, I. Kuikman, and A. Sonnenberg. 1994. The α6β4 integrin is a receptor for both laminin and kalinin. Exp. Cell Res. 211:360-367. [DOI] [PubMed] [Google Scholar]
- 58.Nishimura, S., H. Ushijima, H. Shiraishi, C. Kanazawa, T. Abe, K. Kaneko, and Y. Fukuyama. 1993. Detection of rotavirus in cerebrospinal fluid and blood of patients with convulsions and gastroenteritis by means of the reverse transcription polymerase chain reaction. Brain Dev. 15:457-459. [DOI] [PubMed] [Google Scholar]
- 59.O'Brien, J. A., J. A. Taylor, and A. R. Bellamy. 2000. Probing the structure of rotavirus NSP4: a short sequence at the extreme C terminus mediates binding to the inner capsid particle. J. Virol. 74:5388-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Osborne, M. P., S. J. Haddon, A. J. Spencer, J. Collins, W. G. Starkey, T. S. Wallis, G. J. Clarke, K. J. Worton, D. C. Candy, and J. Stephen. 1988. An electron microscopic investigation of time-related changes in the intestine of neonatal mice infected with murine rotavirus. J. Pediatr. Gastroenterol. Nutr. 7:236-248. [DOI] [PubMed] [Google Scholar]
- 61.Pang, X. L., J. Joensuu, and T. Vesikari. 1996. Detection of rotavirus RNA in cerebrospinal fluid in a case of rotavirus gastroenteritis with febrile seizures. Pediatr. Infect. Dis. J. 15:543-545. [DOI] [PubMed] [Google Scholar]
- 62.Pankov, R., and K. M. Yamada. 2002. Fibronectin at a glance. J. Cell Sci. 115:3861-3863. [DOI] [PubMed] [Google Scholar]
- 63.Parashar, U. D., E. G. Hummelman, J. S. Bresee, M. A. Miller, and R. I. Glass. 2003. Global illness and deaths caused by rotavirus disease in children. Emerg. Infect. Dis. 9:565-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Patel, R. S., E. Odermatt, J. E. Schwarzbauer, and R. O. Hynes. 1987. Organization of the fibronectin gene provides evidence for exon shuffling during evolution. EMBO J. 6:2565-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Piron, M., P. Vende, J. Cohen, and D. Poncet. 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 17:5811-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Plow, E. F., T. A. Haas, L. Zhang, J. Loftus, and J. W. Smith. 2000. Ligand binding to integrins. J. Biol. Chem. 275:21785-21788. [DOI] [PubMed] [Google Scholar]
- 67.Quaroni, A., K. J. Isselbacher, and E. Ruoslahti. 1978. Fibronectin synthesis by epithelial crypt cells of rat small intestine. Proc. Natl. Acad. Sci. USA 75:5548-5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ramig, R. F. 1988. The effects of host age, virus dose, and virus strain on heterologous rotavirus infection of suckling mice. Microb. Pathog. 4:189-202. [DOI] [PubMed] [Google Scholar]
- 69.Renes, I. B., J. A. Boshuizen, D. J. Van Nispen, N. P. Bulsing, H. A. Buller, J. Dekker, and A. W. Einerhand. 2002. Alterations in Muc2 biosynthesis and secretion during dextran sulfate sodium-induced colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 282:G382-G389. [DOI] [PubMed] [Google Scholar]
- 70.Salim, A. F., A. D. Phillips, and M. J. Farthing. 1990. Pathogenesis of gut virus infection. Baillieres Clin. Gastroenterol. 4:593-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sapin, C., O. Colard, O. Delmas, C. Tessier, M. Breton, V. Enouf, S. Chwetzoff, J. Ouanich, J. Cohen, C. Wolf, and G. Trugnan. 2002. Rafts promote assembly and atypical targeting of a nonenveloped virus, rotavirus, in Caco-2 cells. J. Virol. 76:4591-4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sasaki, S., Y. Horie, T. Nakagomi, M. Oseto, and O. Nakagomi. 2001. Group C rotavirus NSP4 induces diarrhea in neonatal mice. Arch. Virol. 146:801-806. [DOI] [PubMed] [Google Scholar]
- 73.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 74.Sharma, A., J. A. Askari, M. J. Humphries, E. Y. Jones, and D. I. Stuart. 1999. Crystal structure of a heparin- and integrin-binding segment of human fibronectin. EMBO J. 18:1468-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simon-Assmann, P., O. Lefebvre, A. Bellissent-Waydelich, J. Olsen, V. Orian-Rousseau, and A. De Arcangelis. 1998. The laminins: role in intestinal morphogenesis and differentiation. Ann. N. Y. Acad. Sci. 859:46-64. [DOI] [PubMed] [Google Scholar]
- 76.Tafazoli, F., C. Q. Zeng, M. K. Estes, K. E. Magnusson, and L. Svensson. 2001. NSP4 enterotoxin of rotavirus induces paracellular leakage in polarized epithelial cells. J. Virol. 75:1540-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Taylor, J. A., J. A. O'Brien, and M. Yeager. 1996. The cytoplasmic tail of NSP4, the endoplasmic reticulum-localized non-structural glycoprotein of rotavirus, contains distinct virus binding and coiled coil domains. EMBO J. 15:4469-4476. [PMC free article] [PubMed] [Google Scholar]
- 78.Tian, P., J. M. Ball, C. Q. Zeng, and M. K. Estes. 1996. The rotavirus nonstructural glycoprotein NSP4 possesses membrane destabilization activity. J. Virol. 70:6973-6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tian, P., M. K. Estes, Y. Hu, J. M. Ball, C. Q.-Y. Zeng, and W. P. Schilling. 1995. The rotavirus nonstructural glycoprotein NSP4 mobilizes Ca2+ from the endoplasmic reticulum. J. Virol. 69:5763-5772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Timpl, R., and J. C. Brown. 1996. Supramolecular assembly of basement membranes. Bioessays 18:123-132. [DOI] [PubMed] [Google Scholar]
- 81.Ushijima, H., K. Q. Xin, S. Nishimura, S. Morikawa, and T. Abe. 1994. Detection and sequencing of rotavirus VP7 gene from human materials (stools, sera, cerebrospinal fluids, and throat swabs) by reverse transcription and PCR. J. Clin. Microbiol. 32:2893-2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Utani, A., M. Nomizu, R. Timpl, P. P. Roller, and Y. Yamada. 1994. Laminin chain assembly. Specific sequences at the C terminus of the long arm are required for the formation of specific double- and triple-stranded coiled-coil structures. J. Biol. Chem. 269:19167-19175. [PubMed] [Google Scholar]
- 83.Woods, A., R. L. Longley, S. Tumova, and J. R. Couchman. 2000. Syndecan-4 binding to the high affinity heparin-binding domain of fibronectin drives focal adhesion formation in fibroblasts. Arch. Biochem. Biophys. 374:66-72. [DOI] [PubMed] [Google Scholar]
- 84.Xu, A., A. R. Bellamy, and J. A. Taylor. 2000. Immobilization of the early secretory pathway by a virus glycoprotein that binds to microtubules. EMBO J. 19:6465-6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoshida, A., T. Kawamitu, R. Tanaka, M. Okumura, S. Yamakura, Y. Takasaki, H. Hiramatsu, T. Momoi, M. Iizuka, and O. Nakagomi. 1995. Rotavirus encephalitis: detection of the virus genomic RNA in the cerebrospinal fluid of a child. Pediatr. Infect. Dis. J. 14:914-916. [PubMed] [Google Scholar]
- 86.Yurchenco, P. D., and J. J. O'Rear. 1994. Basement membrane assembly. Methods Enzymol. 245:489-518. [DOI] [PubMed] [Google Scholar]
- 87.Zhang, M., C. Q. Zeng, A. P. Morris, and M. K. Estes. 2000. A functional NSP4 enterotoxin peptide secreted from rotavirus-infected cells. J. Virol. 74:11663-11670. [DOI] [PMC free article] [PubMed] [Google Scholar]