Abstract
Homologous recombination between different species of alphaherpesviruses has been described between herpes simplex viruses 1 and 2 but has not yet been observed between other alphaherpesviruses. In the present study we chose to assess to what extent in vitro recombination can occur between members of a well-defined group of closely related viruses such as ruminant alphaherpesviruses. At 24 h after infection of epithelial bovine kidney cells with a double-deleted mutant of bovine herpesvirus 1 (BoHV-1) (containing green fluorescent protein and red fluorescent protein genes) and different ruminant alphaherpesviruses, four types of progeny viruses were detected and distinguished according to their phenotype. Frequent recombination events between identical or different strains of BoHV-1 were observed (up to 30%), whereas only two BoHV-1/BoHV-5 recombinants were identified, and no recombinants between BoHV-1 and less closely related caprine and cervine herpesviruses were detected. Restriction analysis of the genomes of the two BoHV-1/BoHV-5 recombinants showed different genetic backgrounds. One possessed a restriction pattern close to BoHV-1, whereas the other one was close to BoHV-5. This exhaustive analysis of each combination of coinfection in a unique situation of five closely related alphaherpesviruses revealed the importance of a high degree of genetic relatedness and similar parental virus growth kinetics for successful interspecific recombination.
Bovine herpesvirus 1 (BoHV-1), a member of the Alphaherpesvirinae subfamily, is a major viral pathogen of cattle. Infection usually goes together with various clinical manifestations such as infectious bovine rhinotracheitis, infectious pustular vulvovaginitis, infectious pustular balanoposthitis, abortion, and generalized systemic infection (44, 60). BoHV-1 isolates were classified into subtype 1 (BoHV-1.1) and BoHV-1.2 according to distinct restriction enzyme profiles of the genomes (17). Due to the significant losses in the cattle industry, Europe has initiated a control program based on the use of marker vaccines deleted in the glycoprotein E (gE) gene. These marker vaccines, either inactivated or live attenuated, allow differentiation between vaccinated and infected cattle (67). In this context, two potential risks need to be accounted for: infection of cattle with heterologous ruminant alphaherpesviruses closely related to BoHV-1 and interspecific recombination between BoHV-1 and related viruses, which could hamper infectious bovine rhinotracheitis eradication programs with BoHV-1 live marker vaccines. Infection of cattle with heterologous ruminant alphaherpesviruses has been demonstrated (41, 54, 59, 62, 63), whereas there is no evidence of interspecific recombination between ruminant alphaherpesviruses.
BoHV-5, caprine herpesvirus 1 (CpHV-1), and cervine herpesvirus 1 (CvHV-1) and CvHV-2 are related to BoHV-1 and are able to cross the species barrier to infect cattle. BoHV-5 is responsible for fatal meningoencephalitis in calves (19, 40). CpHV-1 causes enteritis and generalized infection in neonates. Although most infections in adults are subclinical, CpHV-1 can induce vulvovaginitis, balanoposthitis, or abortion (3, 27, 57). CvHV-1, which is widespread in free-living and farmed red deer, was first isolated in 1982 from an outbreak of ocular disease in a red deer farm in Scotland (25). CvHV-2 was isolated from reindeer in Finland, and serological evidence of infection with a virus related to BoHV-1 has been reported in reindeer in the United States and Canada (14-16). Although all of these viruses considerably differ in their virulence and pathogenicity, they are closely related both genetically (46, 47, 66) and antigenically (35, 43). Moreover, all of these viruses establish, in their specific hosts, a latent infection in a similar manner to that of BoHV-1 (6, 15, 45, 68).
Some experiments have shown that the related herpesviruses described above are able to cross the species barrier and establish infection in heterologous animal species. For example, CpHV-1 can infect cattle, but reactivation of latent CpHV-1 has not been reported yet in cattle, although viral CpHV-1 DNA has been detected in cattle trigeminal ganglia (54). Experimental infection of goats with BoHV-1 clearly showed that this virus is able to infect the heterologous host and establish a latent infection (54). BoHV-1 has also been isolated from a naturally infected goat (62). Cattle was refractory to CvHV-1 but was successfully infected with CvHV-2 by intranasal challenge (41, 59). Red deer could be infected after a BoHV-1 challenge, whereas experimental infection of reindeer with BoHV-1 failed (41). Considering the resistance of cattle to CvHV-1 infection, this virus has not been included in coinfection experiments.
Genetic recombination is a molecular process enabling the creation of new combinations of genetic materials through pairing and shuffling of related DNA sequences. This process functions to maintain chromosomal integrity through recombinational repair and also generates genetic diversity. Four different types of recombination have been described: (i) homologous recombination, which makes use of DNA sequence homology to recognize recombining partners; (ii) site-specific recombination which occurs between DNA molecules sharing little to no sequence homology; (iii) transposition, which occurs for defined DNA sequences (transposable elements) that are recognized by transposon-encoded proteins; and (iv) illegitimate recombination, in which neither sequence homology nor specific sequences can be identified (65). Homologous and illegitimate recombinations are used by herpesviruses (65). Recombination between herpesviruses was first demonstrated in 1955 when wild-type herpes simplex virus 1 (HSV-1) was recovered from mixed inoculations between pairs of temperature-sensitive mutants (70). Since then, herpesvirus recombination has been studied both in vitro and in vivo between distinguishable strains of HSV-1 or HSV-2 (4, 5, 26, 42, 64), pseudorabies virus (PrV) (10, 21, 24), feline herpesvirus 1 (20), BoHV-1 (39, 51, 52), and varicella-zoster virus (12). Timbury and Subak-Sharpe (61) first showed, with HSV-1 and HSV-2, that interspecific recombination could occur between alphaherpesviruses. Moreover, interspecific recombinants such as, for example, BoHV-5 expressing BoHV-1 gC and BoHV-1 expressing BoHV-5 gC, were constructed and showed modified neurotropism (8, 34). However, these artificial recombinants did not emerge from coinfections.
The ability of related ruminant alphaherpesviruses to circulate in the ruminant population is a major threat for the BoHV-1 eradication scheme. Indeed, when animals are coinfected with BoHV-1 and a related ruminant alphaherpesvirus, recombination could generate new viruses increasing the complexity of the eradication scheme. Therefore, in the present study, we chose to assess in vitro the risk of recombination between BoHV-1.2 and a group of four related ruminant alphaherpesviruses (BoHV-1.1, BoHV-5, CpHV-1, and CvHV-2) that could naturally encounter BoHV-1.2. The present study aims to determine to what extent recombination could occur according to the phylogenic distance in a group of closely related viruses such as ruminant alphaherpesviruses related to BoHV-1.
The use of green fluorescent protein (GFP) and red fluorescent protein (RFP) as recombination markers, restriction enzyme profiles, and monoclonal antibodies (MAbs) allowed us to detect in vitro interspecific recombinants between BoHV-1 and BoHV-5 for the first time from coinfections and, especially, to extensively assess recombination between related ruminant alphaherpesviruses.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
The herpesviruses used in the present study are listed in Table 1. They were propagated in Madin-Darby bovine kidney (MDBK; ATCC CCL-22) cells grown in Earle minimal essential medium (MEM; Invitrogen S.A., Merelbeke, Belgium) supplemented with PS (penicillin [5,000 U/ml] and streptomycin [5,000 μg/ml]; Invitrogen S.A., Merelbeke, Belgium) and 5% heat-inactivated fetal bovine serum (BioWhittaker, Verviers, Belgium). For transfections, bovine pharyngeal cell line 244 (KOP-R [provided by the Veterinary Medecine, Insel Riems, Germany, collection of cell lines]) were used. KOP-R cells were grown in Dulbecco modified Eagle medium supplemented with PS and 10% heat-inactivated fetal bovine serum. The construction of recombinant viruses BoHV-1.2/ΔgC-GFP, BoHV-1.2/ΔgI-RFP, and BoHV-1.2/ΔgC-GFP-ΔgI-RFP is described below.
TABLE 1.
Ruminant alphaherpesvirus strains and mutants used in this study
Viral stocks were produced by infection of confluent MDBK cells at a multiplicity of infection (MOI) of 0.1 in MEM supplemented with PS and 3% heat-inactivated horse serum (BioWhittaker). When the cytopathic effect (CPE) reached 90%, the culture medium was removed and clarified by centrifugation (1,500 × g) for 20 min. The supernatants were divided into aliquots, frozen at −80°C, and titrated by plaque assay on MDBK cells as previously described (31).
The BH35 MAb is directed against BoHV-1 gE (2). MAb 1507 recognizes BoHV-1 gC (36). MAb 2F12 recognizes the BoHV-1 gE/gI complex (32). Two BoHV-5 specific MAbs (1624 and 2915) were used.
Construction of plasmids.
All cloning procedures were performed according to the method of Sambrook et al. (48). Plasmid piegC contains a HindIII-NcoI fragment isolated from the EcoRI D fragment of genomic BoHV-1 DNA (nucleotides [nt] 15007 to 18212 of the BoHV-1 genome sequence, accession number AJ004801). The HindIII-NcoI fragment contains the BoHV-1 gC ORF (complement of nt 18209 to 16683) from strain Schönböken (obtained from O. C. Straub, Tübingen, Germany) cloned into pie (30). The region from nt 16626 to 17465 was removed from plasmid piegC by cleavage with BsrGI. The residual plasmid was blunt ended and used for the integration of the blunt-ended 1,640-bp AseI-AflII fragment from pEGFP-N1 (Clontech) that contains the entire GFP expression unit. In the resulting plasmid pΔgC-GFP, the transcription of the GFP open reading frame (ORF) occurs in the same direction as the gC transcription in the BoHV-1 genome (see also Fig. 1). Plasmid pPromi (30) contains sequences that enable homologous recombination downstream and upstream gI. Plasmid pPromiRFP was obtained by integration of the RFP ORF, isolated after cleavage of pDsRed-1 (Clontech) with BglII and NotI into the BglII and NotI-cleaved BoHV-1 recombination vector pPromi (30). In the plasmid pPromiRFP, the transcription of the RFP ORF is under control of the murine cytomegalovirus immediate-early 1 promoter in the same direction as the gI transcription in the BoHV-1 genome (see also Fig. 1).
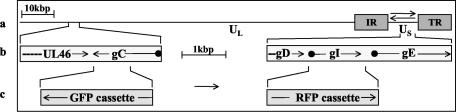
FIG. 1.
Construction of GFP and RFP expressing BoHV-1. (a) Schematic representation of the BoHV-1 genome. Unique long (UL) and unique short (US) segments and the internal (IR) and terminal repeat (TR) sequences are marked. Arrows indicate isomerization of the US segment. (b) Location of the genes used for integration of GRP and RFP expression cassettes. The regions between nt 15007 and 18211 encoding gC and UL46 (partial) and between nt 119500 and 123501 encompassing genes for gD (partial), gI, and gE are enlarged. Arrowheads indicate the direction and termination of transcription, and black dots indicate transcription start sites. (c) Interruption of the gC ORF by the GFP expression cassette and replacement of the gI ORF by the RFP expression cassette. The exchanged sequences are indicated. Arrows mark the direction of GFP and RFP transcription. Cassettes are not drawn to scale. The numbering of nucleotides and location of the BoHV-1 genes correspond to the complete genome sequence (accession number AJ004801).
Construction of recombinant virus.
KOP-R cells were cotransfected with 5 μg of recombination plasmid and 1 μg of purified BoHV-1 DNA as previously described (18). Progeny viruses from the culture supernatants were titrated on KOP-R cells. Cultures were incubated under a 0.6% agarose overlay until plaques appeared. Infected autofluorescent cells were isolated by aspiration, resuspended in culture medium, frozen at −70°C, and then thawed, and recombinant viruses were further plaque purified to homogeneity. The correct insertion of the respective expression cassettes was verified by Southern blot hybridization of HindIII-digested DNA isolated from infected cells. BoHV-1.2/ΔgC-GFP was isolated after cotransfection of BoHV-1/Aus12 DNA (18) with pΔgC-GFP, and BoHV-1.2/ΔgI-RFP was generated by cotransfection of BoHV-1/80-221 DNA (18) with pPromiRFP. Cotransfection of BoHV-1/ΔgC-GFP DNA with pPromiRFP led to the isolation of BoHV-1.2/ΔgC-GFP-ΔgI-RFP.
Interspecific coinfections.
Five experiments of coinfection were performed (BoHV-1.2/ΔgC-GFP-ΔgI-RFP with BoHV-1.1, BoHV-5, CvHV-2, CpHV-1, and BoHV-1.2 wt as a control). Monolayers of MDBK cells prepared in 24-well plates were coinfected with the different couples of viruses at an MOI of 10. Virus attachment was allowed for 2 h at 4°C. Cells were then further incubated at 37°C. At 2 h after the temperature shift, the cells were washed twice with MEM and further incubated at 37°C in 1 ml of MEM supplemented with PS and 2% heat-inactivated horse serum. At 24 h after the temperature shift (when the monolayers showed extensive CPE), the culture medium was removed, clarified twice by centrifugation (1,000 × g), divided into aliquots, and stored at −80°C. Temporally separated coinfections with BoHV-1, CvHV-2, and CpHV-1 were carried out as previously described with BoHV-1 (39).
Isolation and screening of progeny viruses resulting from coinfections.
An aliquot of the supernatant from each situation of coinfection was diluted serially in MEM containing PS to identify the appropriate dilution for individual plaque isolation. After mild sonication, each dilution was used to infect MDBK monolayers cultured in six-well plates (Multiwell; Becton Dickinson). After a 1-h incubation at 37°C, the supernatant was removed and cell monolayers were overlaid with MEM containing PS, 1% (wt/vol) agarose (Agar Bacteriological; Oxoid) and 10% bovine serum containing BoHV-1 neutralizing antibodies. After 72 h of incubation, individual plaques were picked and propagated by inoculation of MDBK cells grown in 24-well plates with MEM containing PS and 2% horse serum. For each coinfection, 50 progeny viruses were propagated. This experiment was repeated three times. When the CPE reached 50%, cells infected by each virus were observed with a TCS SP confocal microscope (Leica) in order to classify progeny viruses. After recombination, progeny viruses were characterized as parental (GFP+/RFP+ and GFP−/RFP−) or recombinant (GFP+/RFP− and GFP−/RFP+). When the CPE reached 90%, supernatant aliquots were stored at −80°C.
Sequence analysis.
The following previously published sequences were used in the present study: BoHV-1 complete genome (NCBI accession number NC001847), BoHV-5 complete genome (GenBank accession number AY261359) (11), HSV-1 complete genome (NCBI accession number NC001806), and HSV-2 complete genome (EMBL accession number Z86099) (13). The sequences were compared by using the Stretcher program (which finds the best global alignment between two sequences) (http://www.be.embnet.org/EMBOSS/).
Virus growth analysis.
MDBK cells were infected with the respective viruses at MOIs of 0.1 and 5. After 2 h at 4°C, prewarmed medium was added, and cells were further incubated for 2 h at 37°C to allow virus penetration. The inoculum was then removed, and the cells were washed twice with MEM and overlaid with fresh MEM supplemented with PS and 2% heat-inactivated horse serum. Immediately thereafter and after 6, 12, 24, 36, and 48 h (MOI = 0.1) and 6, 12, 15, 18, and 24 h (MOI = 5) of incubation at 37°C, the culture medium was removed and clarified twice by centrifugation (1,000 × g). Viral titers were determined by plaque assays on MDBK cells.
Penetration kinetics.
The penetration kinetics of parental viruses were determined as described previously (38) by using low-pH inactivation of extracellular virions at different times after a shift of infected cells from 4 to 37°C.
Immunofluorescence assay.
In order to develop a specific immunofluorescence assay for BoHV-1.2 and BoHV-5, MAbs were first selected by flow cytometric analysis as previously described (28).
An immunofluorescence assay was performed as described by Schynts et al. (51) with minor modifications. Briefly, MDBK cells grown on glass coverslips (Assistent) were infected with the different viruses and incubated for 48 h in MEM containing 4% HS and 0.6% carboxymethyl cellulose. The coverslips with individual plaques were fixed in phosphate-buffered saline containing 2% (wt/vol) paraformaldehyde and incubated with undiluted hybridoma supernatant or 1,000-fold-diluted ascitic fluid in phosphate-buffered saline containing 10% fetal bovine serum. Bound antibodies were revealed by fluorescein isothiocyanate-conjugated rabbit immunoglobulins anti-mouse immunoglobulin G (2 μg/ml; Dako) for RFP recombinant viruses, whereas GFP recombinant viruses were revealed by Alexa Fluor 568-conjugated rabbit immunoglobulins anti-mouse immunoglobulin G (2 mg/ml; Molecular Probes). Coverslips were mounted by using the Prolong Antifade kit (Molecular Probes Europe BV, Leiden, The Netherlands).
Preparation of extracellular virion DNA.
MDBK cells were cultured in six-well plates, infected with viruses at an MOI of 5 in MEM supplemented with PS and 2% heat-inactivated horse serum, and incubated at 37°C for 30 h. For the preparation of virion DNA, infected cell culture medium was clarified twice by centrifugation (1,000 × g), and virions were pelleted by ultracentrifugation (100,000 × g) of the supernatant for 2 h at 4°C. Pellets were resuspended in TE (10 mM Tris [pH 8.0], 1 mM EDTA) and gently mixed with an equal volume of molten (50°C) 2% low-melting-point preparative-grade agarose (Bio-Rad) in TE and cast into plug molds (Bio-Rad) so that each agarose plug contained virions derived from the supernatant of one well of a six-well plate.
Restriction enzyme analysis.
Restriction endonucleases (HindIII, EcoRI, and BamHI) were purchased from New England Biolabs, England, United Kingdom. Digestions and analysis of digestion products by pulsed-field gel electrophoresis were carried out as previously described (53). SmartLadder (1 kb; Eurogentec), Pulse Marker (0.1 to 200 kb; Sigma), and Pulse Marker (50 to 1,000 kb; Sigma) were used as molecular mass markers.
RESULTS
Isolation of recombinant viruses between BoHV-1 and related ruminant alphaherpesviruses.
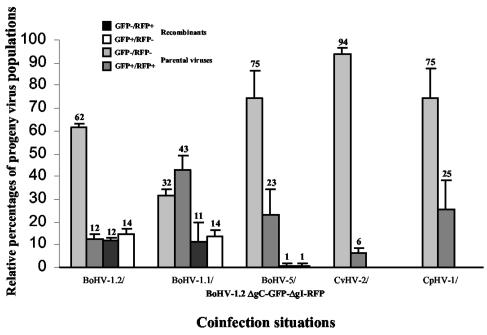
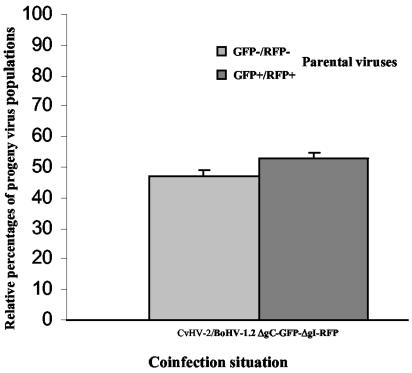
To assess recombination between BoHV-1.2 and four related alphaherpesviruses (BoHV-1.1, BoHV-5, CvHV-2, and CpHV-1), MDBK cells were coinfected with both BoHV-1.2/ΔgC-GFP-ΔgI-RFP (GFP+/RFP+), and wild-type viruses (GFP−/RFP−). Coinfection with BoHV-1.2 wild type was performed as a control. After an incubation of 24 h, 50 individual progeny viruses were isolated from the supernatant and characterized as parental (GFP−/RFP− and GFP+/RFP+) or recombinant (GFP+/RFP− and GFP−/RFP+) viruses. This methodology was used to determine the relative proportions, expressed as a percentage of the total number of isolates, of the four possible progeny populations (parental and recombinant) (Fig. 2). Between BoHV-1.2 and BoHV-1.2, as well as between BoHV-1.2 and BoHV-1.1, ca. 25% of progeny viruses were characterized as recombinants (GFP+/RFP− and GFP−/RFP+), confirming previous results that demonstrated frequent recombination events between strains of BoHV-1 (39, 52) (Fig. 2). In contrast, recombinant viruses between BoHV-1 and another alphaherpesvirus species, such as BoHV-5, were not numerous. Indeed, only two recombinant isolates (of 150 viruses) were isolated (Fig. 2), one of which was GFP+/RFP−, whereas the other was GFP−/RFP+. This observation is the first evidence of interspecific recombination between ruminant alphaherpesviruses. In contrast, we did not isolate interspecific recombinants between BoHV-1.2 and CvHV-2 and between BoHV-1.2 and CpHV-1 (Fig. 2).
FIG. 2.
Relative percentages of progeny virus populations obtained after coinfections of BoHV-1.2/ΔgC-GFP-ΔgI-RFP and different alphaherpesviruses (CpHV-1, CvHV-2, BoHV-5, BoHV-1.1, and BoHV-1.2). The standard deviations of three independent experiments are indicated by vertical lines.
Except with BoHV-1.2/BoHV-1.1 coinfection where GFP+/RFP+ progeny viruses were numerous (43%), a majority of the progeny viruses were GFP−/RFP− as wild-type viruses. This observation is obvious with BoHV-1.2/CvHV-2 coinfection, where GFP−/RFP− viruses represented 94% of progeny viruses (Fig. 2).
Temporal separation of infections did not allow recombination between BoHV-1 and CvHV-2.
In view of previous work dealing with parameters influencing recombination, it is worth mentioning that sequence homologies and simultaneity of infections are of primary importance (21, 39). As reported above, we did not detect any recombinant virus between BoHV-1 and CvHV-2, as well as CpHV-1 (Fig. 2). Since the absence of detection of interspecific recombinants could be due to phase displacement between their viral cycles, we decided to assess the penetration and growth kinetics of parental viruses, two parameters that can drastically influence the encounter of the different viral genomes in cells.
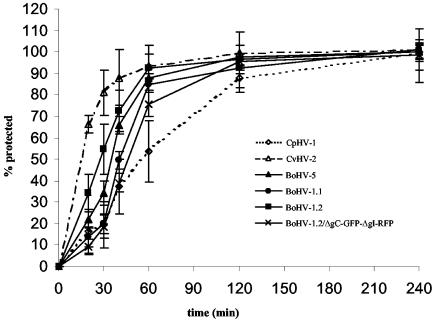
Although BoHV-5 and BoHV-1.2/ΔgC-GFP-ΔgI-RFP entered cells at a rate similar to wt BoHV-1.1 and 1.2, CvHV-2 and CpHV-1 exhibited different rates of viral penetration (Fig. 3). After 20 min, when ca. 20 to 30% of the bovine herpesviruses had entered the cells, almost 70% of CvHV-2 had become protected against low-pH inactivation (Fig. 3). After 60 min, when ca. 85% of the bovine and cervine herpesviruses had entered, only 55% of CpHV-1 had become protected (Fig. 3). Although its penetration behavior was similar, BoHV-1.2 globally entered cells faster than BoHV-1.1. The penetration rates of mutant BoHV-1.2/ΔgC-GFP-ΔgI-RFP are slightly lower than the one of wild-type BoHV-1.
FIG. 3.
Penetration kinetics of six ruminant alphaherpesviruses. To determine the rate of entry of these viruses, penetration kinetics were established by the low-pH inactivation method. The standard deviations of three independent experiments are indicated by vertical lines.
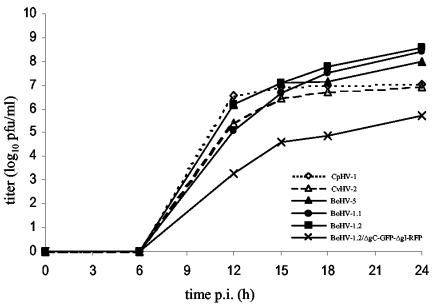
Virus growth experiments were conducted to analyze the kinetics of parental viruses. At the indicated time points after infection at an MOI of 5 (Fig. 4) and at an MOI of 0.1 (data not shown), progeny viruses were titrated on MDBK cells to provide equal growth conditions for all viruses. BoHV-1.2/ΔgC-GFP-ΔgI-RFP exhibited a growth deficiency (Fig. 4). Although the duration of eclipse phase and increase in virus titer were similar for all viruses obtained, titers were lower for the mutant virus. The highest titers were detected for BoHV-1.2, and intermediate titers were detected for cervine and caprine herpesviruses (Fig. 4).
FIG. 4.
Growth analysis of six ruminant alphaherpesviruses (MOI = 5). To analyze the in vitro replication of these alphaherpesviruses, MDBK cells were infected at an MOI of 5. Extracellular virus titers were determined by plaque assay on MDBK cells.
According to these results, time-delayed infections were carried out in order to increase the probability to generate recombinant viruses between BoHV-1 and CvHV-2 and CpHV-1. A time interval of 2 h was selected between infection with BoHV-1.2/ΔgC-GFP-ΔgI-RFP and CvHV-2, which entered cells rapidly. This delay between infections drastically decreased the percentages of GFP−/RFP− progeny viruses. However, even if percentages of GFP+/RFP+ and GFP−/RFP− viruses were nearly equal, we did not detect any recombinant virus (Fig. 5). A time interval between BoHV-1 and CpHV-1 infections also did not allow isolation of interspecific recombinant viruses (data not shown). In conclusion, temporal separation of less closely related ruminant alphaherpesviruses is not sufficient to allow the detection of interspecific recombinants.
FIG. 5.
Relative percentages of progeny virus populations obtained after infection of MDBK cells with BoHV-1.2/ΔgC-GFP-ΔgI-RFP and superinfection with CvHV-2 2 h later. The standard deviations of three independent experiments are indicated by vertical lines.
Characterization of recombinant viruses arising from BoHV-1 and BoHV-5 coinfections with MAbs.
Different specific MAbs were used in order to show the double origin of the two recombinant viruses issued from BoHV-1/BoHV-5 coinfections, MAbs BH35, 1507, and 2F12 are directed against BoHV-1 gE, gC, and gE/gI complex, respectively, whereas MAbs 2915 and 1625 recognize gC and a not-yet-identified antigen of BoHV-5. Selected MAbs were tested for their specificity by immunofluorescence labeling revealed by fluorescence-activated cell sorting (Table 2) and subsequently used for the analysis of the two recombinant viruses. The first recombinant (R1), which contained the GFP expression unit, did not react with MAbs specific for BoHV-1, but did react with the BoHV-5-specific MAb 1625. In contrast, cells infected with the second recombinant (R2), possessing the RFP expression unit, were clearly stained by BH35, specific for BoHV-1 gE, and also by 2915, specific for BoHV-5 gC. As expected, R1 did not react with MAb 2915 since GFP was inserted in place of gC in parental virus BoHV-1.2/ΔgC-GFP−ΔgI-RFP. Taken together, these results prove the mixed character (BoHV-1/BoHV-5) of R1 and R2, respectively.
TABLE 2.
Reactivity of selected MAbs against related herpesviruses as determined by fluorescence-activated cell sorting analysis and immunofluorescence
| Immunization virus | MAb designation | Reactivitya against virus:
|
|||
|---|---|---|---|---|---|
| BoHV-1.2 Aus12 | BoHV-5 N569 | R1 (GFP) | R2 (RFP) | ||
| BoHV-1 | BH35 | + | − | − | + |
| 1507 | + | − | − | − | |
| 2F12 | + | − | − | − | |
| BoHV-5 | 2915 | − | + | − | + |
| 1625 | − | + | + | − | |
R1 (GFP) and R2 (RFP) were from BoHV-1.2/ΔgC-GFP-ΔgI-RFP/BoHV-5 coinfection.
Characterization of interspecific recombinant viruses by restriction analysis.
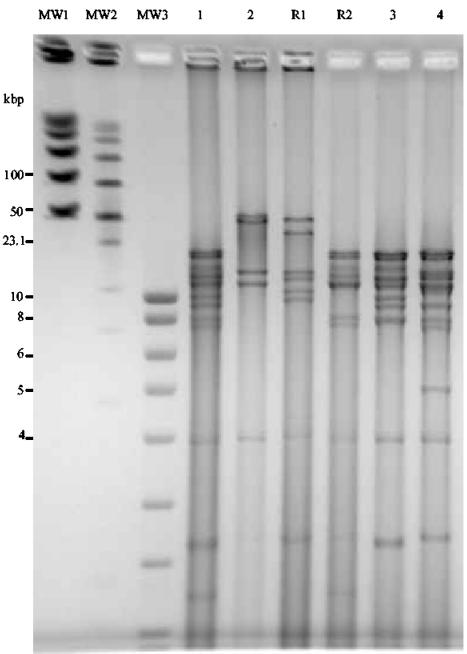
In order to further characterize R1 and R2, we chose to compare restriction enzyme cleavage patterns of R1 and R2 to those of parental viruses. Three restriction enzymes were selected: HindIII (Fig. 6), EcoRI, and BamHI (data not shown). The restriction fragment pattern of recombinant R1 (track R1) was close to that of BoHV-5 (track 2), whereas the fragment pattern of R2 (track R2) resembled the HindIII fragments generated from the DNA of BoHV-1.2/ΔgC-GFP-ΔgI-RFP, BoHV-1.2/ΔgC-GFP, or BoHV-1.2/ΔgI-RFP (track 1, 3, or 4, respectively). Comparable results were obtained by using EcoRI or BamHI (data not shown). Consequently, virus R1, containing the GFP gene, had a restriction pattern similar to BoHV-5, whereas virus R2 (GFP−/RFP+) had a pattern close to BoHV-1, suggesting that R1 had a BoHV-5 genetic background with integration of the GFP gene coming from BoHV-1.2/ΔgC-GFP-ΔgI-RFP and that R2 was generated by integration of BoHV-5 sequences into BoHV-1 (Fig. 7).
FIG. 6.
Cleavage patterns of the DNAs of BoHV-1.2 (lane 1), BoHV-5 (lane 2), recombinant 1 (GFP) (R1 from BoHV-1.2/ΔgC-GFP-ΔgI-RFP/BoHV-5 coinfection), recombinant 2 (RFP) (R2 from BoHV-1.2/ΔgC-GFP-ΔgI-RFP/BoHV-5 coinfection), BoHV-1.2/ΔgC-GFP (lane 3), and BoHV-1.2/ΔgI-RFP (lane 4). The DNAs were digested with HindIII and fragments were separated by PFGE. Fragments were visualized by ethidium bromide. Positions of molecular weight markers (MW) are indicated.
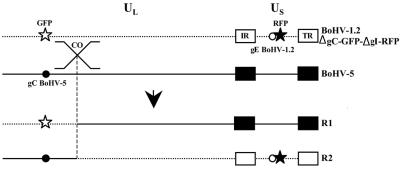
FIG. 7.
Location of potential single crossover in interspecific recombinant viruses (R1 and R2). Schematic representation of the potential single crossover (co) between BoHV-1.2/ΔgC-GFP-ΔgI-RFP and BoHV-5. Unique long (UL) and unique short (US) segments and the internal (IR) and terminal repeat (TR) are marked. Black and white stars indicate location of RFP and GFP expression cassettes in the genomes, respectively. Black and white dots indicate location of gC and gE of BoHV-5 and BoHV-1.2, respectively. The observed single crossover is most likely located close to the GFP marker for both R1 and R2.
DISCUSSION
We present here the first evidence of in vitro interspecific recombination between BoHV-1 and BoHV-5 after coinfections. Two recombinant viruses were isolated among progeny viruses resulting from coinfections between these viral species. In contrast, we did not detect any recombinant virus between BoHV-1 and other closely related viruses such as CvHV-2 and CpHV-1. This result probably reflects the lower nucleic acid sequence homology between BoHV-1 and CvHV-2 or CpHV-1 compared to that of BoHV-1 and BoHV-5 (82.3%) (11, 46, 47). Since genomes of CpHV-1 and CvHV-2 have not been completely sequenced, phylogenetic trees of ruminant alphaherpesviruses were based on gB and gD nucleotide sequences analysis by either the neighbor-joining or the parsimony methods. It revealed that BoHV-5 is the most closely related virus to the BoHV-1.1 and BoHV-1.2 cluster (identity of 98.1% for gB) and that CpHV-1 is the most distantly related (89% for gB) (46). CvHV-2 appeared to be more closely related to BoHV-1 (92.3% for gB) and BoHV-5 than to CpHV-1 (88.2% for gB) (46). However, under the conditions described here, although unlikely, recombination between these viruses and BoHV-1 cannot be completely ruled out. In addition, 26% of recombinant viruses were isolated from coinfections with the same or different subtypes of BoHV-1, a finding which is in agreement with previously published results (39, 52).
Because two crossovers between GFP and RFP markers, for example, can produce recombinant viruses with parental phenotypes (GFP+/RFP+ and GFP−/RFP−), the question of “invisible recombinants” needs to be addressed, especially in coinfections involving CvHV-2 and CpHV-1. Even if double crossover cannot be ruled out, there is little likelihood of their existence between distantly located markers, respectively, situated in the unique long (at the beginning) and the unique short genome segments. Indeed, as previously described in HSV-1 and HSV-2 (5), these events are less frequent than simple crossovers, which are undetectable between selected markers under the conditions described here. Moreover, in a previous study (39), the decrease in the frequency of the recombinant phenotype was strongly linked to the disappearance of mixed concatemeric DNA (mixed concatemers result from recombination events between two parental BoHV-1 genomes). Nevertheless, since virus screening was applied only on viable progeny viruses in the present study, without complete sequencing of their genomes, further investigations are needed to completely address the issue of “invisible recombinants.” It is also of particular interest to investigate interspecific recombination at the level of replication by the study of concatemers, especially since this methodology has been successfully used previously to assess recombination between alphaherpesviruses (39, 55). Furthermore, studying concatemers avoids bias introduced in the detection of recombinants by nonviable recombinants and dominance between progeny viruses.
The lack of detection of interspecific recombinants could be due to a strong stimulation of alpha/beta interferon (IFN-α/β), especially IFN-β, in the context of a high MOI. However, it probably did not influence the emergence of recombinants. First, with a high MOI, all of the cells were infected at the same time, allowing a single viral cycle. Consequently, IFN-β could not influence the generation of new viruses. Second, our experimental protocol included further dilutions of the progeny viruses (by 10−6), and IFN was also diluted by the same way. Its concentration was therefore assumed to be very low. Moreover, BoHV-1 and related viruses are relatively resistant to IFN-β (1, 9, 49).
Specific MAbs analysis coupled to restriction analysis clearly demonstrated the double nature of the two BoHV-1/BoHV-5 recombinant viruses. Indeed, the first one possesses a BoHV-5 pattern containing a BoHV-5 epitope and a GFP gene from BoHV-1 in place of gC, whereas the second one has a BoHV-1 pattern containing a BoHV-5 gC epitope and an RFP gene in place of gI. This shows that the generated recombinants possess characters deriving from both parental viruses with, apparently, only short stretches of “foreign” DNA transferred between their genomes. The survival of mutants with relatively short distances between two potential crossovers might be favored. It is noteworthy that all of the recombinants (BoHV-1.2/1.2wt and BoHV-1.2/1.1 included [data not shown]) differ in the particular combination of their phenotypes as previously described in studies dealing with HSV-1/HSV-2 interspecific recombinants (22, 23, 72). However, according to restriction patterns and antibody analysis, single crossover could be located near the GFP marker (Fig. 7). To definitely assess the possibility of double or multiple crossovers along the genome in the generation of recombinant viruses, partial sequencing of parental (slightly different from published genomes) and complete sequencing of recombinant viruses are then required.
The method described here was used for a first assessment of recombination between related alphaherpesviruses. It allowed us to analyze more than 1,000 progeny viruses in a short period of time. Five types of coinfection involving BoHV-1.2 were carried out under identical experimental conditions. This is the first time that recombination has been studied among a group of genetically related alphaherpesviruses that can naturally coinfect the same animal. This group of cross-related viruses is unique among alphaherpesviruses. Previously, recombination has been studied in vitro and in vivo between HSV-1 and HSV-2, two closely related human alphaherpesviruses (22, 23, 61, 72), and between HSV-1 and BoHV-2 or HSV-1 and PrV (23). Consequently, the present study completes the previous studies. Recombination was assessed between viruses showing lower and higher sequence homologies than those observed between HSV-1 and HSV-2 and between HSV-1 and BoHV-2. Interspecific recombinant viruses were isolated between HSV-1 and HSV-2, whereas no recombination events were reported between HSV-1 and BoHV-2 or PrV (22, 23, 61, 72). Despite the higher degree of nucleic acid homology (BoHV-1 and BoHV-5, 82.3%; HSV-1 and HSV-2, 75%), BoHV-5 did not recombine more with BoHV-1 than HSV-1 did with HSV-2. More interestingly, our results show that with a couple of ruminant viruses that are less closely related (BoHV-1/CvHV-2 and BoHV-1/CpHV-1), recombinants events cannot be detected. As observed previously with RNA viruses (29, 71), sequence homology between potentially recombining genomes is a very important physical constraint concerning homologous recombination. However, even if it is probably the main factor influencing recombination, other factors that could be implied should also be considered, particularly those affecting the distribution of different viruses to common target cells and thereby limiting or increasing the likelihood of cellular coinfections. Coinfections could be blocked by host factors (i.e., immune response that keeps virus populations small enough to prevent multiple infection of any individual cell), host cell genetic factors that block the entry of more than one virus particle into a cell, and viral factors. In vivo, some of these factors include (i) the dose of the inoculated viruses, (ii) the distance between inoculation sites, (iii) the time interval between inoculation of the first and the second virus, and (iv) the genes in which the mutations are located (21, 39). It has recently been reported that the time interval drastically influences the rise of BoHV-1 recombinant viruses in vitro (39). Indeed, a short time interval of 2 or 4 h already influenced the percentages of recombinant and parental progeny viruses (39). Since the penetration and growth kinetics of some parental viruses are very different, especially for CvHV-2, which enters cells very rapidly, a certain influence of these parameters on the recombination events can be assumed. Nevertheless, in the case of coinfection involving BoHV-1 and CvHV-2, an increase in the time interval did not overcome the barrier to interspecific recombination.
Penetration and growth kinetics in bovine epithelial cells showed significant differences among parental viruses. The alphaherpesvirus envelope contains five glycoproteins with defined roles in viral penetration (33, 50). BoHV-1 cell binding is principally mediated by gC but also by gB and gD (7, 33, 50, 56). At least four different viral glycoproteins (gB, gD, gH, and gL) and a gD receptor (HveC, a cell adhesion molecule more commonly called nectin 1) are required for the BoHV-1-induced membrane fusion that enables viral entry and cell fusion (7, 33, 50, 56). Because very little is known about gD receptors presented on bovine epithelial cells or about glycoproteins of related ruminant alphaherpesviruses, we can only postulate the implication of these elements in penetration and growth kinetic differences. Another possible explanation could reside in viral cycles, which show differences between related ruminant alphaherpesviruses as described, for example, with CpHV-1, the gD of which is expressed as a late protein unlike BoHV-1 (V. Keuser, B. Detry, F. Schynts, P.-P. Pastoret, A. Vanderplasschen, and E. Thiry, unpublished data).
Since the isolation of BoHV-1/BoHV-5 interspecific recombinants in the laboratory and the in vitro and in vivo generation of HSV-1/HSV-2 interspecific recombinants was relatively easy, there is no reason per se why such recombinants could not arise under natural conditions. The generation of interspecific recombinant viruses during an infection could have important epidemiological consequences when a mixed infection with different viruses is possible, as in the case with BoHV-1. Indeed, all of the related ruminant herpesviruses described above are able to cross the species barrier and establish infection in heterologous animal species so that they can encounter each other naturally (41, 54, 59, 62, 63). Thus far, there is no convincing evidence that any interspecific recombinant has been isolated. Moreover, under the experimental conditions used here, interspecific recombinants have been isolated from mixed infections at high MOIs (10), and such a situation is unlikely to occur under natural conditions. However, it is important to stress that a single BoHV-1 interspecific recombinant, keeping virulence and acquiring characteristics of a related alphaherpesvirus, i.e., gE− genotype of BoHV-1 vaccinal strains, would be enough to severely impair control programs based on vaccination. In this context, future research on BoHV-1/BoHV-5 recombinants in vivo will be of particular interest to evaluate the risk generated by these new viruses in the field.
All of the viruses selected in this study establish, in their specific hosts, a latent infection in a similar manner as BoHV-1 (6, 15, 45, 68). Latency could influence recombination in vivo. Indeed, interspecific recombination could occur during primary coinfection but also after reactivation and reexcretion of one or both viruses. Latency increases the likelihood of cellular coinfection, which enables recombination between viruses. Nevertheless, since marked differences in invasiveness and virulence have been described between viruses, different patterns of distribution of viral DNA in tissue can occur and probably influence the likelihood of coinfection. Moreover, it was demonstrated that a single host can support the latent infection of two distinguishable BoHV-1 strains (69) and that a single neuron can be dually infected with HSV-1 and varicella-zoster virus (58). Additional investigations are needed to assess the impact of latency on interspecific recombination. For example, it will be of particular interest to study the distribution and consequent probabilities of coinfection of related alphaherpesviruses in the same animal.
In conclusion, the present study is the first to investigate the rise of interspecific recombinants in a group of related alphaherpesviruses that can naturally encounter each other. It detects, for the first time, interspecific recombinants between BoHV-1 and BoHV-5 after coinfection. It suggests the crucial importance of nucleic acid sequence homologies in the rise of interspecific recombinants and highlights the potential consequences of interspecific recombination in the context of a BoHV-1 eradication scheme.
Acknowledgments
We thank G. Meyer (Toulouse, France), M. Schwyzer, M. Engels, A. Six, and M. Ackermann (Zürich, Switzerland), H. Reid and I. Campbell (Edinburgh, United Kingdom), C. Ek-Kommonen (Helsinki, Finland), C. Ros and S. Belàk (Uppsala, Sweden), and M. Banks (Addlestone, United Kingdom) for helpful comments, advice, or for providing strains. We also thank G. J. Letchworth and M. Engels for BoHV-1 and BoHV-5 reference MAbs. We thank Régine Denaegel, Pierre Gallego, Luc Bauret, and Katalin de Fays for careful reading of the manuscript.
This study was financially supported by the Service Public Fédéral, Santé Publique, Administration Recherche et Développement and by the Fonds National Belge de la Recherche Scientifique (FNRS; FRFC 2.4508.02 and grant 1.5.105.03). F.M. and B.M. are Research Fellows of the FNRS. The confocal microscope was purchased with funds provided by the following grants: FRFC 2.4532.98 from the FNRS, FNRS LOTTO 9.4592.97 from the Belgian National Lottery, and ARC 98/03-220 from the French Community of Belgium.
REFERENCES
- 1.Babiuk, L. A., H. B. Ohmann, G. Gifford, C. W. Czarniecki, V. T. Scialli, and E. B. Hamilton. 1985. Effect of bovine alpha 1 interferon on bovine herpesvirus type 1-induced respiratory disease. J. Gen. Virol. 66:2383-2394. [DOI] [PubMed] [Google Scholar]
- 2.Baranowski, E., J. Dubuisson, P.-P. Pastoret, and E. Thiry. 1993. Identification of 108K, 93K, and 42K glycoproteins of bovine herpesvirus-1 by monoclonal antibodies. Arch. Virol. 133:97-111. [DOI] [PubMed] [Google Scholar]
- 3.Berrios, P. E., D. G. McKercher, and H. D. Knight. 1975. Pathogenicity of a caprine herpesvirus. Am. J. Vet. Res. 36:1763-1769. [PubMed] [Google Scholar]
- 4.Brown, S. M., D. A. Ritchie, and J. H. Subak-Sharpe. 1973. Genetic studies with herpes simplex virus type 1: the isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J. Gen. Virol. 18:329-346. [DOI] [PubMed] [Google Scholar]
- 5.Brown, S. M., J. H. Subak-Sharpe, J. Harland, and A. R. MacLean. 1992. Analysis of intrastrain recombination in herpes simplex virus type 1 strain 17 and herpes simplex virus type 2 strain HG52 using restriction endonuclease sites as unselected markers and temperature-sensitive lesions as selected markers. J. Gen. Virol. 73:293-301. [DOI] [PubMed] [Google Scholar]
- 6.Buonavoglia, C., M. Tempesta, A. Cavalli, V. Voigt, D. Buonavoglia, A. Conserva, and M. Corrente. 1996. Reactivation of caprine herpesvirus 1 in latently infected goats. Comp. Immunol. Microbiol. Infect. Dis. 19:275-281. [DOI] [PubMed] [Google Scholar]
- 7.Campadelli-Fiume, G., F. Cocchi, L. Menotti, and M. Lopez. 2000. The novel receptors that mediate the entry of herpes simplex viruses and animal alphaherpesviruses into cells. Rev. Med. Virol. 10:305-319. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhury, S. I., B. J. Lee, M. Onderci, M. L. Weiss, and D. Mosier. 2000. Neurovirulence of glycoprotein C (gC)-deleted bovine herpesvirus type-5 (BHV-5) and BHV-5 expressing BHV-1 gC in a rabbit seizure model. J. Neurovirol. 6:284-295. [DOI] [PubMed] [Google Scholar]
- 9.Czarniecki, C. W., E. B. Hamilton, C. W. Fennie, and R. L. Wolf. 1986. In vitro biological activities of Escherichia coli-derived bovine interferons alpha, beta, and gamma. J. Interferon Res. 6:29-37. [DOI] [PubMed] [Google Scholar]
- 10.Dangler, C. A., L. M. Henderson, L. A. Bowman, and R. E. Deaver. 1993. Direct isolation and identification of recombinant pseudorabies virus strains from tissues of experimentally coinfected swine. Am. J. Vet. Res. 54:540-545. [PubMed] [Google Scholar]
- 11.Delhon, G., M. P. Moraes, Z. Lu, C. L. Afonso, E. F. Flores, R. Weiblen, G. F. Kutish, and D. L. Rock. 2003. Genome of bovine herpesvirus 5. J. Virol. 77:10339-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dohner, D. E., S. G. Adams, and L. D. Gelb. 1988. Recombination in tissue culture between varicella-zoster virus strains. J. Med. Virol. 24:329-341. [DOI] [PubMed] [Google Scholar]
- 13.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ek-Kommonen, C., P. Veijalainen, M. Rantala, and E. Neuvonen. 1982. Neutralizing antibodies to bovine herpesvirus 1 in reindeer. Acta Vet. Scand. 23:565-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ek-Kommonen, C., S. Pelkonen, and P. F. Nettleton. 1986. Isolation of a herpesvirus serologically related to bovine herpesvirus 1 from a reindeer (Rangifer tarandus). Acta Vet. Scand. 27:299-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Azhary, M. A., R. S. Roy, and J. L. Frechette. 1979. Serological evidence of IBR and BVD infection in caribou (Rangifer tarandus). Vet. Rec. 105:336. [DOI] [PubMed] [Google Scholar]
- 17.Engels, M., C. Giuliani, P. Wild, T. M. Beck, E. Loepfe, and R. Wyler. 1986. The genome of bovine herpesvirus 1 (BHV-1) strains exhibiting a neuropathogenic potential compared to known BHV-1 strains by restriction site mapping and cross-hybridization. Virus Res. 6:57-73. [DOI] [PubMed] [Google Scholar]
- 18.Fehler, F., J. M. Herrmann, A. Saalmüller, T. C. Mettenleiter, and G. M. Keil. 1992. Glycoprotein IV of bovine herpesvirus 1-expressing cell line complements and rescues a conditionally lethal viral mutant. J. Virol. 66:831-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.French, E. L. 1962. A specific virus encephalitis in calves: isolation and characterization of the causal agent. Aust. Vet. J. 38:216-221. [Google Scholar]
- 20.Fujita, K., K. Maeda, N. Yokoyama, T. Miyazawa, C. Kai, and T. Mikami. 1998. In vitro recombination of feline herpesvirus type 1. Arch. Virol. 143:25-34. [DOI] [PubMed] [Google Scholar]
- 21.Glazenburg, K. L., R. J. Moormann, T. G. Kimman, A. L. Gielkens, and B. P. Peeters. 1994. In vivo recombination of pseudorabies virus strains in mice. Virus Res. 34:115-126. [DOI] [PubMed] [Google Scholar]
- 22.Halliburton, I. W. 1980. Intertypic recombinants of herpes simplex viruses. J. Gen. Virol. 48:1-23. [DOI] [PubMed] [Google Scholar]
- 23.Halliburton, I. W., R. E. Randall, R. A. Killington, and D. H. Watson. 1977. Some properties of recombinants between type 1 and type 2 herpes simplex viruses. J. Gen. Virol. 36:471-484. [DOI] [PubMed] [Google Scholar]
- 24.Henderson, L. M., J. B. Katz, G. A. Erickson, and J. E. Mayfield. 1990. In vivo and in vitro genetic recombination between conventional and gene-deleted vaccine strains of pseudorabies virus. Am. J. Vet. Res. 51:1656-1662. [PubMed] [Google Scholar]
- 25.Inglis, D. M., J. M. Bowie, M. J. Allan, and P. F. Nettleton. 1983. Ocular disease in red deer calves associated with a herpesvirus infection. Vet. Rec. 113:182-183. [DOI] [PubMed] [Google Scholar]
- 26.Javier, R. T., F. Sedarati, and J. G. Stevens. 1986. Two avirulent herpes simplex viruses generate lethal recombinants in vivo. Science 234:746-748. [DOI] [PubMed] [Google Scholar]
- 27.Keuser, V., S. Gogev, F. Schynts, and E. Thiry. 2002. Demonstration of generalized infection with caprine herpesvirus 1 diagnosed in an aborted caprine fetus by PCR. Vet. Res. Commun. 26:221-226. [DOI] [PubMed] [Google Scholar]
- 28.Keuser, V., F. Schynts, B. Detry, A. Collard, B. Robert, A. Vanderplasschen, P.-P. Pastoret, and E. Thiry. 2004. Improved antigenic methods for the differential diagnosis of bovine, caprine, and cervine alphaherpesviruses related to bovine herpesvirus 1. J. Clin. Microbiol. 42:1228-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.King, A.M.Q. 1988. Genetic recombination in positive strand RNA viruses, p. 19-165. In E. Domingo, J. J. Holland, and P. Alhlquist (ed.), Retrovirus, viroids, and RNA recombination. CRC Press, Inc., Boca Raton, Fla.
- 30.Kühnle, G., R. A. Collins, J. E. Scott, and G. M. Keil. 1996. Bovine interleukins 2 and 4 expressed in recombinant bovine herpesvirus 1 are biologically active secreted glycoproteins. J. Gen. Virol. 77:2231-2240. [DOI] [PubMed] [Google Scholar]
- 31.Lemaire, M., F. Schynts, G. Meyer, and E. Thiry. 1999. Antibody response to glycoprotein E after bovine herpesvirus type 1 infection in passively immunized, glycoprotein E-negative calves. Vet. Rec. 144:172-176. [DOI] [PubMed] [Google Scholar]
- 32.Letellier, C., A. Delangre, A. De Smet, and P. Kerkhofs. 2001. Characterization of monoclonal antibodies directed against the bovine herpesvirus-1 glycoprotein E and use for the differentiation between vaccinated and infected animals. Vet. Microbiol. 83:301-315. [DOI] [PubMed] [Google Scholar]
- 33.Liang, X. P., L. A. Babiuk, S. van Drunen Littel-van den Hurk, D. R. Fitzpatrick, and T. J. Zamb. 1991. Bovine herpesvirus 1 attachment to permissive cells is mediated by its major glycoproteins gI, gIII, and gIV. J. Virol. 65:1124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liman, A., M. Engels, G. Meyer, and M. Ackermann. 2000. Glycoprotein C of bovine herpesvirus 5 (BHV-5) confers a distinct heparin-binding phenotype to BHV-1. Arch. Virol. 145:2047-2059. [DOI] [PubMed] [Google Scholar]
- 35.Lyaku, J. R., P. F. Nettleton, and H. Marsden. 1992. A comparison of serological relationships among five ruminant alphaherpesviruses by ELISA. Arch. Virol. 124:333-341. [DOI] [PubMed] [Google Scholar]
- 36.Marshall, R. L., L. L. Rodriguez, and G. J. Letchworth. 1986. Characterization of envelope proteins of infectious bovine rhinotracheitis virus (bovine herpesvirus 1) by biochemical and immunological methods. J. Virol. 57:745-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metler, F., M. Engels, P. Wild, and A. Bivetti. 1979. Herpesvirus-Infektion bei Zieklein in der Schweiz. Schweiz. Arch. Tierheilkd. 121:655-662. [PubMed] [Google Scholar]
- 38.Mettenleiter, T. C. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171:623-625. [DOI] [PubMed] [Google Scholar]
- 39.Meurens, F., F. Schynts, G. M. Keil, B. Muylkens, P. Gallego, and E. Thiry.2004. Superinfection prevents recombination of the alphaherpesvirus bovine herpesvirus 1. J. Virol. 78:3872-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meyer, G., M. Lemaire, C. Ros, K. Belak, A. Gabriel, D. Cassart, F. Coignoul, S. Belak, and E. Thiry. 2001. Comparative pathogenesis of acute and latent infections of calves with bovine herpesvirus types 1 and 5. Arch. Virol. 146:633-652. [DOI] [PubMed] [Google Scholar]
- 41.Nettleton, P. F., C. Ek-Kommonen, R. Tanskanen, H. W. Reid, J. A. Sinclair, and J. A. Herring. 1988. Studies in the epidemiology and pathogenesis of alphaherpesviruses from red deer (Cervus elaphus) and reindeer (Rangifer tarandus), p. 143-147. In H. W. Reid (ed.), The management and health of farmed deer. Kluwer Academic Publishers, Boston, Mass.
- 42.Nishiyama, Y., H. Kimura, and T. Daikoku. 1991. Complementary lethal invasion of the central nervous system by nonneuroinvasive herpes simplex virus types 1 and 2. J. Virol. 65:4520-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nixon, P., S. Edwards, and H. White. 1988. Serological comparisons of antigenically related herpesviruses in cattle, red deer, and goats. Vet. Res. Commun. 12:355-362. [DOI] [PubMed] [Google Scholar]
- 44.Pastoret, P.-P., E. Thiry, B. Brochier, G. Derboven, and H. Vindevogel. 1984. The role of latency in the epizootiology of infectious bovine rhinotracheitis, p. 221-227. In G. Wittmann, R. M. Gaskell, and H. J. Rziha (ed.), Latent herpesvirus infections in veterinary medicine. Kluwer Academic Publishers, Boston, Mass.
- 45.Ronsholt, L., L. S. Christensen, and V. Bitsch. 1987. Latent herpesvirus infection in red deer: characterization of a specific deer herpesvirus including comparison of genomic restriction fragment patterns. Acta Vet. Scand. 28:23-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ros, C., and S. Belak. 1999. Studies of genetic relationships between bovine, caprine, cervine, and rangiferine alphaherpesviruses and improved molecular methods for virus detection and identification. J. Clin. Microbiol. 37:1247-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ros, C., and S. Belak. 2002. Characterization of the glycoprotein B gene from ruminant alphaherpesviruses. Virus Genes 24:99-105. [DOI] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T. Maniatis. 2000. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Savan, M., A. B. Angulo, and J. B. Derbyshire. 1979. Interferon, antibody responses and protection induced by an intranasal infectious bovine rhinotracheitis vaccine. Can. Vet. J. 20:207-210. [PMC free article] [PubMed] [Google Scholar]
- 50.Schwyzer, M., and M. Ackermann. 1996. Molecular virology of ruminant herpesviruses. Vet. Microbiol. 53:17-29. [DOI] [PubMed] [Google Scholar]
- 51.Schynts, F., A. Vanderplasschen, E. Hanon, F. A. M. Rijsewijk, J. T. van Oirschot, and E. Thiry. 2001. Use of PCR and immunofluorescence to detect bovine herpesvirus 1 recombinants. J. Virol. Methods 92:99-104. [DOI] [PubMed] [Google Scholar]
- 52.Schynts, F., F. Meurens, B. Detry, A. Vanderplasschen, and E. Thiry. 2003. Rise and survival of bovine herpesvirus 1 recombinants after primary infection and reactivation from latency. J. Virol. 77:12535-12542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schynts, F., M. A. McVoy, F. Meurens, B. Detry, A. L. Epstein, and E. Thiry. 2003. The structures of bovine herpesvirus 1 virion and concatemeric DNA: implications for cleavage and packaging of herpesvirus genomes. Virology 314:326-335. [DOI] [PubMed] [Google Scholar]
- 54.Six, A., M. Banks, M. Engels, C. R. Bascunana, and M. Ackermann. 2001. Latency and reactivation of bovine herpesvirus 1 (BHV-1) in goats and of caprine herpesvirus 1 (CapHV-1) in calves. Arch. Virol. 146:1325-1335. [DOI] [PubMed] [Google Scholar]
- 55.Slobedman, B., X. Zhang, and A. Simmons. 1999. Herpes simplex virus genome isomerization: origins of adjacent long segments in concatemeric viral DNA. J. Virol. 73:810-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Spear, P. G., R. J. Eisenberg, and G. H. Cohen. 2000. Three classes of cell surface receptors for alphaherpesvirus entry. Virology 275:1-8. [DOI] [PubMed] [Google Scholar]
- 57.Tarigan, S., R. F. Webb, and D. Kirkland. 1987. Caprine herpesvirus from balanoposthitis. Aust. Vet. J. 64:321. [DOI] [PubMed] [Google Scholar]
- 58.Theil, D., I. Paripovic, T. Derfuss, S. Herberger, M. Strupp, V. Arbusow, and T. Brandt. 2003. Dually infected (HSV-1/VZV) single neurons in human trigeminal ganglia. Ann. Neurol. 54:678-682. [DOI] [PubMed] [Google Scholar]
- 59.Thiry, E., and M. Lemaire. 2001. Infection de ruminants par des herpèsvirus hétérologues. Point. Vet. 207:20-25. [Google Scholar]
- 60.Tikoo, S. K., M. Campos, and L. A. Babiuk. 1995. Bovine herpesvirus 1 (BHV-1): biology, pathogenesis, and control. Adv. Virus Res. 45:191-223. [DOI] [PubMed] [Google Scholar]
- 61.Timbury, M. C., and J. H. Subak-Sharpe. 1973. Genetic interactions between temperature-sensitive mutants of types 1 and 2 herpes simplex viruses. J. Gen. Virol. 18:347-357. [DOI] [PubMed] [Google Scholar]
- 62.Tolari, F., H. White, and P. Nixon. 1990. Isolation and reactivation of bovine herpesvirus 1 in goats. Microbiologica 13:67-71. [PubMed] [Google Scholar]
- 63.Trueblood, M. S., B. L. Swift, and L. McHolland-Raymond. 1978. A bovine herpesvirus isolated from sheep. Can. J. Comp. Med. 42:97-99. [PMC free article] [PubMed] [Google Scholar]
- 64.Umene, K. 1985. Intermolecular recombination of the herpes simplex virus type 1 genome analysed using two strains differing in restriction enzyme cleavage sites. J. Gen. Virol. 66:2659-2670. [DOI] [PubMed] [Google Scholar]
- 65.Umene, K. 1999. Mechanism and application of genetic recombination in herpesviruses. Rev. Med. Virol. 9:171-182. [DOI] [PubMed] [Google Scholar]
- 66.Vanderplasschen, A., M. Bublot, P.-P. Pastoret, and E. Thiry. 1993. Restriction maps of the DNA of cervid herpesvirus 1 and cervid herpesvirus 2, two viruses related to bovine herpesvirus 1. Arch. Virol. 128:379-388. [DOI] [PubMed] [Google Scholar]
- 67.Van Oirschot, J. T., M. J. Kaashoek, M. A. Maris-Veldhuis, K. Weerdmeester, and F. A. Rijsewijk. 1997. An enzyme-linked immunosorbent assay to detect antibodies against glycoprotein gE of bovine herpesvirus 1 allows differentiation between infected and vaccinated cattle. J. Virol. Methods 67:23-34. [DOI] [PubMed] [Google Scholar]
- 68.Vogel, F. S., L. Caron, E. F. Flores, R. Weiblen, E. R. Winkelmann, S. V. Mayer, and R. G. Bastos. 2003. Distribution of bovine herpesvirus type 5 DNA in the central nervous systems of latently, experimentally infected calves. J. Clin. Microbiol. 41:4512-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Whetstone, C. A., and J. M. Miller. 1989. Two different strains of an alphaherpesvirus can establish latency in the same tissue of the host animal: evidence from bovine herpesvirus 1. Arch. Virol. 107:27-34. [DOI] [PubMed] [Google Scholar]
- 70.Wildy, P. 1955. Recombination with herpes simplex virus. J. Gen. Microbiol. 13:34-46. [DOI] [PubMed] [Google Scholar]
- 71.Worobey, M., and E. C. Holmes. 1999. Evolutionary aspects of recombination in RNA viruses. J. Gen. Virol. 80:2535-2543. [DOI] [PubMed] [Google Scholar]
- 72.Yirrell, D. L., C. E. Rogers, W. A. Blyth, and T. J. Hill. 1992. Experimental in vivo generation of intertypic recombinant strains of HSV in the mouse. Arch. Virol. 125:227-238. [DOI] [PubMed] [Google Scholar]