Abstract
Our previous studies with influenza A viruses indicated that the association of M1 with viral RNA and nucleoprotein (NP) is required for the efficient formation of helical ribonucleoprotein (RNP) and for the nuclear export of RNPs. RNA-binding domains of M1 map to the following two independent regions: a zinc finger motif at amino acid positions 148 to 162 and a series of basic amino acids (RKLKR) at amino acid positions 101 to 105. Altering the zinc finger motif of M1 reduces viral growth slightly. A substitution of Ser for Arg at either position 101 or position 105 of the RKLKR domain partially reduces the nuclear export of RNP and viral replication. To further understand the role of the zinc finger motif and the RKLKR domain in viral assembly and replication, we introduced multiple mutations by using reverse genetics to modify these regions of the M gene of influenza virus A/WSN/33. Of multiple mutants analyzed, a double mutant, R101S-R105S, of RKLKR resulted in a temperature-sensitive phenotype. The R101S-R105S double mutant had a greatly reduced ratio of M1 to NP in viral particles and a weaker binding of M1 to RNPs. These results suggest that mutations can be introduced into the RKLKR domain to control viral replication.
The core of the influenza A virus consists of eight ribonucleoproteins (RNPs). The viral RNA, nucleoprotein (NP), and polymerases are closely associated in RNPs (11, 17, 23). The matrix protein (M1) is associated with the RNP and with the inner surface of the lipid envelope in the intact virion (1, 3, 33). Two major external glycoproteins, hemagglutinin (HA) and neuraminidase (NA), and the small protein M2, which serves as an ion channel, are anchored in the viral envelope (19, 36).
M1 is a major structural component of the virion and has multiple functions during viral replication. The dissociation of M1 from RNP is required for the entry of viral RNP into the cytoplasm of the host cell during initial infection (4, 12, 22). Dissociation is triggered by the transport of H+ ions across the viral membrane by M2 (12, 18, 36). It has also been shown that M1 is transported during early viral replication from the cytoplasm into the nucleus, where M1 associates with newly synthesized RNPs (4, 26, 28). Later in the replication cycle, M1 accumulates in the cytoplasm concomitant with the export of RNP from the nucleus (4, 5, 13, 16, 22, 35). The transport of RNP from the nucleus to the cytoplasm requires the binding of M1 to RNP (15, 22), which also prevents RNP from reentering the nucleus (22). Interactions of M1 with HA, NA, M2, and host cell lipid membranes occur on the cytoplasmic side of the membrane as part of the process of virion maturation and budding at the cell surface (3, 9, 10, 17, 19, 31, 39). The ratio of M1 to NP also affects the morphological features and infectivity of the mature released viruses (21, 29, 30).
The interactions of M1 with RNP have been studied extensively (2, 6, 27, 32, 33, 42). Two RNA-binding domains in M1 have been demonstrated (39, 42). One RNA-binding domain contains a zinc finger motif (148C-C-…-H-H162) (7, 8), and a synthetic peptide containing this motif has been shown to inhibit viral replication (24). The other RNA-binding domain, which resides in a palindromic stretch of basic amino acids, 101RKLKR105, has been shown to bind viral RNA (8, 37, 39), which fulfills a prediction based on X-ray crystallographic studies of M1 (34). The 101RKLKR105 domain also serves as a nuclear translocation signal for M1 (40, 41). Our previous studies demonstrated that viral RNP is not assembled in the absence of M1 (15) and that mutation in the 101RKLKR105 domain of M1 affects viral growth (20). Although mutations in the RKLKR domain have a negative impact on viral growth, the mechanisms are not fully understood (21).
Here we report the impact of multiple mutations of the RNA- and RNP-binding domains of the M gene of influenza virus A/WSN/33. We analyzed the rescued M mutants by comparing their viral replication rates at restrictive and permissive temperatures and studied the RNP-binding strengths of the rescued mutants by inducing the dissociation of M1/RNP complexes. Our studies indicate that the introduction of a double mutation in the RKLKR domain (altered by site-directed mutagenesis to SKLKS) of M1 results in the introduction of temperature sensitivity, a reduced incorporation of M1 into viral particles, and the susceptibility of M1/RNP complexes to salt dissociation.
MATERIALS AND METHODS
Virus and cells.
Influenza virus A/WSN/33 and its M gene mutants were propagated in the allantoic cavities of 9-day-old embryonated eggs at 34°C for 2 days. The viruses were stored in allantoic fluid at −70°C or concentrated and purified by banding in 15 to 60% sucrose gradients (42). Madin-Darby canine kidney (MDCK) and 293T cells were grown and maintained in minimal essential medium (MEM) supplemented with 5 and 10% fetal bovine serum, respectively. Infectivity titers of the viruses were determined by a plaque assay with MDCK cells by use of an agar overlay system containing 1 μg of trypsin/ml.
Plasmids.
Plasmids pHW181-PB2, pHW182-PB1, pHW183-PA, pHW184-HA, pHW185-NP, pHW186-NA, and pHW188-NS, which were used for reverse genetic studies, were obtained from R. Webster, St. Jude Children's Research Hospital, Memphis, Tenn. Plasmids coding for the M protein with mutations to amino acids RKLKR at positions 101 to 105 or to Cys at position 148 of the zinc finger motif region were incorporated into the plasmid pPol I-WSN-M (provided by Y. Kawaoka, University of Wisconsin, Madison) by a PCR using primers (Core Facility, Center for Biologics Evaluation and Research) containing the mutant sequences. An additional control plasmid was constructed to provide an M gene that mimicked the variant of the A/WSN/33 virus known as ts51, which has an amino acid substitution in M1 (F79S) that confers temperature sensitivity. The mutated M genes in these plasmids were confirmed by DNA sequencing.
Production of transfectant influenza viruses carrying M gene mutations.
Transfectant influenza viruses carrying M gene mutations were generated by the reverse genetic techniques described by Hoffmann et al. (14) and Neumann et al. (25), with modifications. Briefly, 1 day before transfection, 293T and MDCK cells from confluent cultures were transferred to 12-well plates (1:1 293T to MDCK cells) in Opti-MEM I medium (Invitrogen, Gaithersburg, Md.). For transfection, a mixture of plasmids was prepared with 1 μl of TransIT LT-1 (Panvera, Madison, Wis.) per 0.5 μg of plasmid DNA. Each transfection mixture included nonmutated influenza A virus genes (contained in pHW181-PB2, pHW182-PB1, pHW183-PA, pHW184-HA, pHW185-NP, pHW186-NA, and pHW188-NS) and one of the plasmids expressing the relevant individual M1 mutation. The plasmid DNA-TransIT LT-1 mixture was incubated at room temperature for 45 min before it was added to the cells. Six hours later, the plasmid DNA transfection mixture was replaced with 0.5 ml of Opti-MEM I medium. Twenty-four hours after transfection, 0.5 ml of Opti-MEM I containing 1 μg of tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin/ml was added to each well. At predetermined time intervals posttransfection, samples were harvested. The titers of viruses were determined in hemagglutination units (HAU) or PFU. The virus particles generated by reverse genetics were purified three times by plaque plating on MDCK cells and were amplified in the allantoic cavities of 9-day-old embryonated eggs. Viral RNAs were isolated from transfectant viruses by use of a QIAamp viral RNA mini kit (Qiagen, Valencia, Calif.). The M genes were amplified by reverse transcription-PCR, gel purified, and used for sequencing analysis.
Temperature sensitivity of mutant viruses.
The temperature sensitivity of the recombinant viruses was determined by a plaque assay with MDCK cells. MDCK cells in 12-well plates were infected with 200 μl of 10-fold serially diluted viruses, which were adsorbed at 37°C for 60 min. The inocula were removed and replaced with 1.5 ml of Dulbecco's modified Eagle's medium containing 0.7% agarose and 1 μg of TPCK-trypsin/ml. The infected cells were incubated at 33, 37, or 39.5°C for 3 days, after which the cells were fixed with 100% methanol and stained with 0.1% crystal violet. Plaque counts obtained at each of the temperatures were compared to assess the ts phenotype of each virus. Each virus was plaque plated in a minimum of three assays. Temperature sensitivity (ts) was defined as the temperature at which the virus yield was reduced 100-fold or more compared to that at 33°C.
Immunofluorescence staining.
MDCK cells grown on glass coverslips were infected with influenza viruses at a multiplicity of infection of 3 PFU per cell and subsequently incubated in MEM containing 2% fetal bovine serum at 33 or 39.5°C for 5 h. Infected MDCK cells were fixed with freshly prepared 4% formaldehyde in phosphate-buffered saline (PBS) for 20 min at room temperature. Background staining was blocked with 3% powdered skim milk in PBS for 1 h. The cells were then incubated at room temperature for 40 min with a monoclonal antibody to M1. The cells were incubated with donkey anti-mouse immunoglobulin G conjugated with fluorescein for 40 min at room temperature. Washed coverslips were mounted in 90% glycerol and 10% PBS in 3,4,5-trihydroxybenzoic acid N-propylester to prevent photobleaching. The cellular distribution of immunofluorescence was visualized with an epifluorescent UV microscope.
Dissociation of M1 protein from M1/RNP complexes.
An assay to induce the dissociation of M1/RNP complexes from the viruses was performed as described previously (21). Purified M1/RNP complexes were obtained by incubating viruses (0.5 mg/ml) for 40 min at room temperature in disruption buffer containing 10 mM Tris-HCl (pH 7.0), 1% Nonidet P-40, 0.05 M NaCl, and 1.25 mM dithiothreitol. M1/RNP complexes released from virions in the reaction mixtures were pelleted by centrifugation through 25% glycerol with a cushion of 50% glycerol in an SW 55Ti rotor at 120,000 × g for 60 min. The supernatant fluid containing lipids and membrane proteins was discarded, and the pellet containing the M1/RNP complexes was resuspended in 10 mM Tris-HCl (pH 7.4).
To study the effect of ionic strength on M1/RNP complex association, we varied the salt concentration by adding NaCl to the reaction buffer. For each salt concentration, a 20-μl aliquot of the RNP-M1-detergent suspension was centrifuged through a 150-μl cushion of 20% sucrose at 8,100 × g for 15 min in a 0.5-ml Eppendorf tube, and the resulting pellet was collected. The protein composition of the RNPs was analyzed by electrophoresis in 12.5% polyacrylamide-sodium dodecyl sulfate (SDS) gels under nonreducing conditions, with the gels being stained with Coomassie blue. The amounts of M1 and NP proteins were determined by densitometry of the protein bands.
RESULTS
Replication and recovery of M1 mutants from transfected cells.
Our previous studies indicated that substituting a single amino acid in the zinc finger motif or replacing an Arg with Ser at position 101 or 105 of the RKLKR domain in M1 did not have a major impact on viral replication. To assess whether alterations of viral replication could be manipulated by alteration of the RKLKR sequence and the zinc finger motif of M1, we altered nucleotide sequences within these two domains by site-directed mutagenesis. Figure 1A shows a schematic diagram of the RNA and RNP-binding domains of M1. The RKLKR sequence is located between amino acids 101 and 105, and the zinc finger motif is located between amino acids 148 and 162 of M1 (40-42). Figure 1B shows that the wild-type (WT) plasmid expressed the WT A/WSN/33 virus M gene, the plasmid DelRKLKR contained an altered M gene coding for a complete deletion of RKLKR, plasmids R101S and R105S contained altered M genes expressing single amino acid substitutions, plasmid C148S contained a DNA sequence coding for an alteration predicted to disrupt the zinc finger motif (7), plasmid R101S-R105S contained an altered M gene designed to express M1 with amino acids RKLKR replaced with SKLKS, and plasmid C148S-R105S contained an M gene designed to express M1 with substituted amino acids in RKLKR (R105S) and an altered zinc finger motif (C148S).
FIG. 1.
(A) Schematic model of RNP- and RNA-binding domains (RKLKR) at amino acids 101 to 105 and a zinc finger RNA-binding domain at amino acids 148 to 162 of WSN M1 protein. (B) Amino acid sequences of WSN WT and mutant M1 constructs.
To determine the effect of multiple substitutions of RKLKR and the zinc finger regions in M1 on viral replication, we recovered M1 mutant viruses generated by reverse genetic techniques 48 to 144 h after transfection from supernatants of mixed cultures of 293T and MDCK cells incubated at 33°C (14). As a control, a temperature-sensitive mutant virus (F79S) mimicking ts51, a previously described temperature-sensitive variant of A/WSN/33, was generated by substituting the amino acid Phe for Ser at position 79 of WT M1. Posttransfection titers, measured in HAU, are shown in Fig. 2. Either the delayed detection of HAU or a lower HAU titer during virus rescue may be interpreted to demonstrate a growth disadvantage compared to the WT virus. The RKLKR deletion mutant (DelRKLKR) had no detectable HAU titer and was a lethal mutation (20), while the single amino acid mutants R101S, R105S, and C148S and the double amino acid mutant C148S-R105S gave results similar to those for the WT, except that the WT had a relatively high HAU titer at 48 h posttransfection. The double mutant R101S-R105S had the lowest HAU titer, which was undetectable until 144 h posttransfection. The ts51 equivalent mutant, F79S, had HAU titers intermediate between those of the WT and the R101S-R105S double mutant.
FIG. 2.
293T and MDCK cells were transfected by use of a modified reverse genetic system to rescue M gene mutants as described in the text. The supernatants of transfected 293T and MDCK cells were harvested at 48 to 144 h posttransfection. Virus titers were analyzed by measuring the HAU.
Double mutation of RKLKR in M introduces ts phenotype.
Because the double amino acid mutations of Arg to Ser at positions 101 and 105 of RKLKR resulted in a growth disadvantage, the viral growth characteristics were studied in more detail by comparing the growth rates of this mutant at different temperatures. The naturally occurring temperature-sensitive mutant ts51 (20, 38) has a cutoff temperature of 39.5°C, which is the result of the substitution of Phe to Ser at position 79 in M1, the same change we introduced to produce mutant F79S (20, 38). We therefore used mutant F79S as a positive control for ts analysis. The temperature sensitivities of the M1 mutants were determined by plaque assays with MDCK cells at 33, 37, and 39.5°C. Figure 3A illustrates the phenotypes of the M1 mutants. Single amino acid alterations in either the zinc finger motif or in RKLKR at amino acid position 101 or 105 had a minimal impact on temperature sensitivity relative to the WT virus. The double mutant R101S-C148S also did not exhibit significant temperature sensitivity. All of the single mutants and the double mutant R101S-C148S yielded 6.3 to 6.5 log10 PFU/ml. However, the double mutant R101S-R105S yielded only 4.8 log10 PFU/ml at 33°C and 3.5 log10 PFU/ml at 37°C. At the restrictive temperature for ts51, 39.5°C (38), the F79S mutant had a >2-log reduction in virus titer, whereas the R101S-R105S mutant was undetectable by a plaque assay. Figure 3B summarizes the observed temperature sensitivity of each M1 mutant by comparing the ratios of PFU yielded at 39.5 and 33°C or at 37 and 33°C. While the reduction in virus yield at 39.5°C relative to 33°C was approximately 100-fold for the F79S mutant, reductions at 39.5°C for the WT and the C148, R101S, R105S, and R101S-C148S mutants were approximately 10-fold or less. Mutant R101S-R105S, however, had a >1,000-fold reduction at 39.5°C and approximately 10-fold at 37°C, while other M1 mutants were reduced <2-fold at 37°C, with the exception of mutant R101S-C148S, which was reduced approximately 4-fold. The plaque size and morphology of mutant R101S-R105S were also influenced by the increased temperature. Figure 4 shows comparison studies of the plaque size and morphology of M1 mutants. Mutant F79S formed small plaques at 39.5°C, while mutant R101S-R105S formed no plaques (at a 1:10 dilution) under the same conditions but formed small plaques at 37°C. Mutant F79S formed turbid plaques at 37°C, while mutant R101S-R105S formed turbid plaques at 33°C. Comparisons of viral titers, plaque sizes, and plaque type in M1 mutants demonstrated, therefore, that the substitution of Ser for Arg at positions 101 and 105 of the M1 protein of mutant R101S-R105S resulted in a temperature-sensitive virus strain, R101S-R105S.
FIG. 3.
Temperature sensitivity of M1 mutants. The temperature sensitivity of the M1 mutants was analyzed by a plaque assay with MDCK cells at 33, 37, or 39.5°C (A), and the reductions in titers at 37 or 39.5°C versus the titers at 33°C were compared (B).
FIG. 4.
Plaque morphology of mutant viruses. MDCK cells were infected with the mutant viruses in 12-well plates at 33, 37, and 39.5°C in the presence of 1 μg of trypsin/ml. After 2 days of incubation, the cells were fixed and stained with crystal violet.
Temperature-sensitive M1 mutants had reduced amounts of M1 in virions and a decreased RNP-binding capacity.
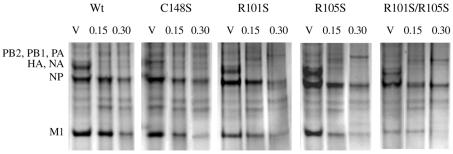
To relate the growth properties of the mutant viruses to the RNP-binding activity of the M1 proteins, we isolated M1/RNP complexes from viral particles and performed a dissociation assay by treating M1/RNP complexes with various salt concentrations to release the M1 protein from the complexes. Complexes containing RNP with an undisrupted M1 protein were separated from dissociated M1 by centrifugation through 20% glycerol. The protein complexes were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), as shown in Fig. 5. An SDS-PAGE analysis of M1/RNP complexes of WT and mutant viruses showed NP migrating at approximately 56 kDa and M1 migrating at approximately 27 kDa. With increasing salt concentrations (0.15 to 0.3 M), the amount of M1 retained in the M1/RNP complexes decreased. Table 1 summarizes the ratios and percentages of M1 to NP based on the density analysis of M1 and NP bands in Fig. 5. The ratio of M1 to NP represents the relative amount of M1 in M1/RNP complexes, and the percentage of M1 represents the binding capacity of M1 to RNP. At a 0.3 M salt concentration, 64% of the M1 protein from the WT virus remained associated with RNP (M1 to NP ratio = 0.93), compared with 47% of M1 from the C148S virus (M1 to NP ratio = 0.62), 40% of M1 from the R101S virus (M1 to NP ratio = 0.30), 39% of M1 from the R105S virus (M1 to NP ratio = 0.28), and only 12% of M1 from the R101S-R105S virus (M1 to NP ratio = 0.05). These results indicated that the M1 protein of mutant R101S-R105S had the lowest binding activity for RNP. In viral particles, the M1/NP ratios for the WT and C148S viruses were similar (1.45 versus 1.33). However, the ratios for the single amino acid mutants of RKLKR (R101S and R105S) were 0.74 and 0.71, respectively. The double mutant R101S-C148S had no further reduction in the ratio of M1 to NP compared with the R101S or C148S virus (data not shown). However, the double mutant R101S-R015S had the lowest M1/NP ratio (0.42) in the viral particle. The results from the analysis of salt dissociation of M1 and RNP demonstrated that the substitution of a Ser for an Arg at either position 101 or 105 of the M1 protein results in a reduced ratio of M1 to NP in viral particles and that the substitution of Ser residues for both Arg residues results in a much more reduced ratio of M1 to NP in viral particles and a weaker association of M1 with RNPs.
FIG. 5.
Effect of salt on M1/RNP association. The dissociation of M1 from RNP was induced by exposing virions or M1/RNP complexes to NaCl at 0.15 or 0.3 M. The protein components of RNPs and virions (V) were analyzed by SDS-12.5% PAGE and stained with 0.1% Coomassie brilliant blue.
TABLE 1.
Effect of salt on dissociation of M1 from RNPa
| Strain | M1/NP ratio in presence of NaCl (M)
|
% of M1 bound to RNP in presence of NaCl (M)
|
||||
|---|---|---|---|---|---|---|
| V | 0.15 | 0.3 | V | 0.15 | 0.3 | |
| WT | 1.45 | 1.25 | 0.93 | 100 | 86 | 64 |
| C148S | 1.33 | 1.21 | 0.62 | 100 | 91 | 47 |
| R101S | 0.74 | 0.52 | 0.30 | 100 | 70 | 40 |
| R105S | 0.71 | 0.47 | 0.28 | 100 | 64 | 39 |
| R101S-R105S | 0.42 | 0.30 | 0.05 | 100 | 71 | 12 |
The RNP-associated M1 and the ratio of M1 to NP were quantified by densitometry. The percentages of binding reactivities of M1s to RNP were calculated by comparisons of M1 protein to NP in virions and the M1/RNP ratio. The data are means of three independent experiments, and the standard deviation of each experimental point was below 10%. V, virion alone.
M1 proteins with double amino acid mutations at positions 101 and 105 prevent nuclear localization at nonpermissive temperatures.
Because the double mutant R101S-R105S resulted in a temperature-sensitive phenotype of influenza virus, the cellular distribution of M1 in the resulting viruses was studied by the infection of MDCK cells with mutant viruses followed by indirect immunofluorescence staining of M1 proteins during early viral infection. The cellular distribution of M1 at a permissive or nonpermissive temperature during early viral infection was visualized by indirect immunofluorescence staining with monoclonal antibodies to the M1 protein. Figure 6 shows the cellular distribution of M1 proteins at a nonpermissive temperature. Under the conditions of the study, WT M1 mostly resided in the nucleus of the infected cells (panel A). A similar cellular distribution was found for the M1 protein with a mutation in the zinc finger motif (C148S) (panel B). A somewhat reduced nuclear localization of M1 in the single amino acid mutants R101S and R105S was observed (panels C and D) compared with the WT virus under the same conditions. However, all of the M1 protein of the double amino acid mutant R101S-R105S was located in the cytoplasm and none was detected in the nuclei of infected cells (panel E). Since the virus was in the early viral replication phase (<5 h postinfection), a similar cellular distribution of NP with M mutant and WT viruses was observed in the same study. The majority of NPs were still located in the nuclei of infected cells (data not shown). These results indicate that substitutions of the Arg residues at both positions 101 and 105 in RKLKR are needed to prevent the nuclear localization of M1 at the restricted temperature, while single amino acid substitutions at either position had a minimal effect on the nuclear localization of M1.
FIG. 6.
Immunofluorescence staining of MDCK cells infected with M1 mutants. MDCK cells were infected for 5 h with M1 mutants at 39.5°C and were incubated with or without (−) a mouse anti-M1 monoclonal antibody and donkey anti-mouse immunoglobulin G conjugated with fluorescein. The cellular distribution of immunofluorescence was determined with a transmission microscope under an epifluorescent UV light source. Magnification, ×472.
DISCUSSION
Our previous studies with the M gene of influenza A viruses (20) demonstrated that a substitution of Ser for either Lys 102 or Lys 104 of the 101RKLKR105 motif results in a lethal mutation, while a substitution of Ser for either Arg 101 or Arg 105 in RKLKR or a mutation of the zinc finger motif permits the recovery of viruses generated by reverse genetic techniques. Since 101RKLKR105 serves as a nuclear localization signal and facilitates M1 binding to RNP, it is possible to hypothesize that a lethal mutation in this region could be due either to alterations in RNP binding or to alterations in the nuclear localization activities of M1. Because the substitution of either Lys 102 or Lys 104 is lethal to the mutated viruses, the binding of the mutant M1 proteins to RNP is not easily examined. However, in this study, we demonstrated that the replacement of both Arg residues in 101RKLKR105 of A/WSN/33 produces a viable but impaired virus with a ts phenotype. With an analysis of the intracellular distribution of M1 at a restricted temperature and the binding avidity of M1 variants to RNPs, we demonstrate here that an alteration in the zinc finger motif or a single replacement of Arg with Ser at position 101 or 105 of 101RKLKR105 in M1 has relatively little effect on the RNP binding of M1, the nuclear localization of M1 proteins, or viral replication. Combining a mutation of one of the Arg residues in 101RKLKR105 with a mutation in the zinc finger motif (mutant R101S-C148S) also did not significantly reduce viral replication at the nonpermissive temperature. In marked contrast, replacing both Arg residues in 101RKLKR105 created a mutant virus (R101S-R105S) that is extremely temperature sensitive, more so than a well-described temperature-sensitive mutant, ts51. Mutant R101S-R105S also exhibited a decreased ratio of M1 to NP in virions, a reduction in RNP-binding activity, and the exclusion of M1 from the nucleus at a nonpermissive temperature. However, there was no significant difference in the morphology of the mutant viruses compared with that of the WT virus by electron microscopy (data not shown).
Although a predicted consensus sequence of the nuclear localization signal of M1 proteins in influenza A viruses is 101XKLKR105 (20), we demonstrate here that the R at position 105 may also be replaced by an alternate amino acid without preventing nuclear localization. However, the lack of nuclear localization of the M1 protein of the double mutant R101S-R105S in infected cells at a nonpermissive temperature is not the result of decreasing amounts of M protein in the infected cells. As determined by immunofluorescence staining of MDCK cells infected with R101S, R105S, C148S, and R101S-R105S mutants, the expression level of the mutated M1 protein in infected cells was not significantly reduced at 39.5°C compared with that at 33°C. Therefore, the failure of the M1 protein of the double mutant R101S-R105S to localize to the nucleus is a functional effect of the mutation.
The M1 protein plays multiple roles in viral replication. Both the dissociation of M1 from RNP in virions entering the infected cell and the association of M1 and RNP in the later phases of infection are required for the viral replication cycle to be completed. The basic amino acid sequence 101RKLKR105 of M1 plays an important role in several steps of replication. The sequence 101RKLKR105 permits the translocation of M1 from the cytoplasm to the nucleus. In the nucleus, M1 and RNP bind, presumably partly because of the RNP-binding activity of 101RKLKR105. The maturation of RNP by folding to the final quaternary helical structure and transport out of the nucleus requires viral RNA (but not cRNA or mRNA) and M1 (15). The RNP-binding activity of the M1 protein, however, varies among viruses, with stronger binding in viruses that replicate more efficiently (21). The results presented in this paper provide further direct evidence of the importance of the 101RKLKR105 motif of the M1 protein in relation to the RNP-binding and growth characteristics of influenza virus. Whether introducing a ts phenotype into the A/WSN/33 virus also results in an attenuation phenotype of the virus, animal models are needed to evaluate the pathogenesis of the M mutant viruses in a future study. Since 101RKLKR105 has many interactions, it is difficulty to attribute the ts phenotype of mutant R101S-R105S to a single M1 function. Further studies of M1 variants may allow us to resolve the basis for the observed effects, with the expectation that such knowledge will enhance our ability to identify mechanisms that can be exploited to understand and prevent influenza virus pathogenesis.
Acknowledgments
We thank Ronald Lundquist and Ketha Mohan for critically reading and improving the manuscript and Michael Klutch at the Center for Biologics Evaluation and Research, Food and Drug Administration, for DNA sequencing. We are especially grateful to Roland Levandowski for a valuable discussion and for improving the manuscript.
REFERENCES
- 1.Allen, H., J. McCauley, M. Waterfield, and M. J. Gething. 1980. Influenza virus RNA segment 7 has the coding for two polypeptides. Virology 107:548-551. [DOI] [PubMed] [Google Scholar]
- 2.Baudin, F., I. Petit, W. Weissenhorn, and R. W. Ruigrok. 2001. In vitro dissection of the membrane and RNP binding activities of influenza virus M1 protein. Virology 281:102-108. [DOI] [PubMed] [Google Scholar]
- 3.Bucher, D. J., I. G. Kharitonenkov, J. A. Zakomirdin, V. B. Grigoriev, S. M. Klimenko, and J. F. Davis. 1980. Incorporation of influenza virus M protein into liposomes. J. Virol. 36:586-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bui, M., G. Whittaker, and A. Helenius. 1996. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J. Virol. 70:8391-8401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bui, M., E. G. Wills, A. Helenius, and G. R. Whittaker. 2000. Role of the influenza virus M1 protein in nuclear export of viral ribonucleoproteins. J. Virol. 74:1781-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Compans, R. W., and H. D. Klenk. 1979. Viral membranes, p. 293-407. In H. Fraenkel-Conrat and R. R. Wagner (ed.), Comprehensive virology, vol. 13. Plenum Publishing Corp., New York, N.Y. [Google Scholar]
- 7.Elster, C., E. Fourest, F. Baudin, K. Larsen, S. Cusack, and R. W. Ruigrok. 1994. A small percentage of influenza virus M1 protein contains zinc but zinc does not influence in vitro M1-RNA interaction. J. Gen. Virol. 75:37-42. [DOI] [PubMed] [Google Scholar]
- 8.Elster, C., K. Larsen, J. Gagnon, R. W. H. Ruigrok, and F. Baudin. 1997. Influenza virus M1 protein binds to RNA through its nuclear localization signal. J. Gen. Virol. 78:1589-1596. [DOI] [PubMed] [Google Scholar]
- 9.Enami, M., and K. Enami. 1996. Influenza virus hemagglutinin and neuraminidase glycoproteins stimulate the membrane association of the matrix protein. J. Virol. 70:6653-6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gregoriades, A. 1980. Interaction of influenza M protein with viral lipid and phosphatidylcholine vesicles. J. Virol. 36:470-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heggeness, M. H., P. R. Smith, L. Ulmanen, R. M. Krug, and P. W. Choppin. 1982. Studies on the helical nucleocapsid of influenza virus. Virology 118:466-470. [DOI] [PubMed] [Google Scholar]
- 12.Helenius, A. 1992. Unpacking of the incoming influenza virus. Cell 69:577-578. [DOI] [PubMed] [Google Scholar]
- 13.Herz, C., E. Stavnezer, and R. M. Krug. 1981. Influenza virus, an RNA virus, synthesizes its messenger RNA in the nucleus of influenza cells. Cell 26:391-400. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann, E., G. Neumann, Y. Kawaoka, G. Hobom, and R. G. Webster. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA 97:6108-6113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, X., T. Liu, J. Muller, R. A. Levandowski, and Z. Ye. 2001. Effect of influenza virus matrix protein and viral RNA on ribonucleoprotein formation and nuclear export. Virology 287:405-416. [DOI] [PubMed] [Google Scholar]
- 16.Krug, R. M., R. V. Alonso-Coplen, L. Julkumen, and M. G. Katze. 1989. Expression and replication of the influenza virus genome, p. 89-162. In R. M. Krug (ed.), The influenza viruses. Plenum Press, New York, N.Y.
- 17.Lamb, R. A., and P. W. Choppin. 1983. The structure and replication of influenza virus. Annu. Rev. Biochem. 52:467-506. [DOI] [PubMed] [Google Scholar]
- 18.Lamb, R. A., L. J. Holsinger, and L. H. Pinto. 1994. The influenza A virus M2 ion channel protein and its role in the influenza virus life cycle, p. 303-321. In E. Wimmer (ed.), Receptor-mediated virus entry into cells. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 19.Lamb, R. A., and S. L. Zebedee. 1985. Influenza virus M2 protein is an integral membrane protein expressed on the infected-cell surface. Cell 40:627-633. [DOI] [PubMed] [Google Scholar]
- 20.Liu, T., and Z. Ye. 2002. Restriction of viral replication by mutation in matrix protein of influenza virus. J. Virol. 76:13055-13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, T., J. Muller, and Z. Ye. 2002. Association of influenza virus matrix protein with ribonucleoproteins may control viral growth and morphology. Virology 304:89-96. [DOI] [PubMed] [Google Scholar]
- 22.Martin, K., and A. Helenius. 1991. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67:117-130. [DOI] [PubMed] [Google Scholar]
- 23.Murti, K. G., R. G Webster, and I. M. Jones. 1988. Localization of RNP polymerases on influenza viral ribonucleoprotein by immunogold labeling. Virology 164:562-566. [DOI] [PubMed] [Google Scholar]
- 24.Nasser, E. H., A. K. Judd, A. Sanchez, D. Anastasiou, and D. J. Bucher. 1996. Antiviral activity of influenza virus M1 zinc finger peptides. J. Virol. 70:8639-8644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neumann, G., T. Watanabe, H. Ito, S. Watanabe, H. Goto, P. Gao, M. Hughes, D. R. Perez, R. Donis, E. Hoffmann, G. Hobom, and Y. Kawaoka. 1999. Generation of influenza A viruses entirely from cloned cDNAs. Proc. Natl. Acad. Sci. USA 96:9345-9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patterson, S., J. Gross, and J. S. Oxford. 1988. The intercellular distribution of influenza virus matrix protein and nucleoprotein in infected cells and their relationship to haemagglutinin in the plasma membrane. J. Gen. Virol. 69:1859-1872. [DOI] [PubMed] [Google Scholar]
- 27.Rees, P. J., and N. J. Dimmock. 1982. Kinetics of synthesis of influenza virus ribonucleoprotein structures. J. Gen. Virol. 59:403-408. [DOI] [PubMed] [Google Scholar]
- 28.Rey, O., and D. P. Nayak. 1992. Nuclear retention of M1 protein in a temperature-sensitive mutant of influenza (A/WSN/33) virus does not affect nuclear export of viral ribonucleoproteins. J. Virol. 66:5815-5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts, P. C., and R. W. Compans. 1998. Host cell dependence of viral morphology. Proc. Natl. Acad. Sci. USA 95:5746-5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roberts, P. C., R. A. Lamb, and R. W. Compans. 1998. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology 240:127-137. [DOI] [PubMed] [Google Scholar]
- 31.Robertson, B. H., J. C. Bennett, and R. W. Compans. 1982. Selective dansylation of M protein within intact influenza virus. J. Virol. 44:871-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruigrok, R. W. H., and F. Baudin. 1995. Structure of influenza virus ribonucleoprotein particles. II. Purified RNA-free influenza virus ribonucleoproteins form structures that are indistinguishable from the intact influenza virus ribonucleoprotein particles. J. Gen. Virol. 76:1009-1014. [DOI] [PubMed] [Google Scholar]
- 33.Schulze, I. T. 1972. The structure of influenza virus. II. A model based on the morphology and composition of subviral particles. Virology 47:181-196. [DOI] [PubMed] [Google Scholar]
- 34.Sha, B., and M. Luo. 1997. Structure of a bifunctional membrane-RNA binding protein, influenza virus matrix protein M1. Nat. Struct. Biol. 4:239-244. [DOI] [PubMed] [Google Scholar]
- 35.Silver, P. A. 1991. How proteins enter the nucleus. Cell 64:489-497. [DOI] [PubMed] [Google Scholar]
- 36.Sugrue, R. J., and A. J. Hay. 1991. Structural characteristics of the M2 protein of the influenza A viruses: evidence that it forms a tetrameric channel. Virology 180:617-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wakefield, L., and G. G. Brownlee. 1989. RNA-binding properties of influenza A virus matrix protein M1. Nucleic Acids Res. 17:8569-8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whittaker, G., I. Kemler, and A. Helenius. 1995. Hyperphosphorylation of mutant influenza virus matrix protein, M1, causes its retention in the nucleus. J. Virol. 69:439-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ye, Z., R. Pal, J. W. Fox, and R. R. Wagner. 1987. Functional and antigenic domains of the matrix (M1) protein of influenza virus. J. Virol. 61:239-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ye, Z., N. W. Baylor, and R. R. Wagner. 1989. Transcription-inhibition and RNA-binding domains of influenza virus matrix protein mapped with anti-idiotype antibodies and synthetic peptides. J. Virol. 63:3586-3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ye, Z., D. Robinson, and R. R. Wagner. 1995. Nucleus-targeting domain of the matrix protein (M1) of influenza virus. J. Virol. 69:1964-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye, Z., T. Liu, D. P. Offringa, J. McInnis, and R. A. Levandowski. 1999. Association of influenza virus matrix protein with ribonucleoproteins. J. Virol. 73:7467-7473. [DOI] [PMC free article] [PubMed] [Google Scholar]