Abstract
Biochemical characterization of hepatitis C virus (HCV) replication using purified, membrane-associated replication complexes is hampered by the presence of endogenous nuclease activity that copurifies with the replication complex. In this study, pulse-chase analyses were used to demonstrate that newly synthesized replicon RNA was protected from nuclease activity by a factor(s) that was sensitive to 0.5% NP-40 or protease treatment. Nuclease susceptibility was not related to disruption of lipid membranes, since NP-40 did not significantly affect the buoyant density of HCV replication complexes or protease susceptibility of HCV NS3 and NS5A proteins. These results suggest that a protease-sensitive factor(s) protects newly synthesized RNA from nuclease degradation.
The hepatitis C virus (HCV) replicase is a membrane-associated nucleoprotein complex composed of viral nonstructural proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B that catalyzes the synthesis of positive- and negative-strand genomic RNA during HCV replication (2). Replicase activity can be enriched by subcellular fractionation of intracellular membranes from replicon-containing cells (1, 4, 5). Replicase activity is measured by incorporation of radiolabeled nucleotides into genomic-length positive-strand RNA, and the products can be resolved from replication intermediates by native or denaturing agarose gel electrophoresis (1, 4, 5). Initiation of RNA synthesis occurs in the absence of added negative-strand template RNA, suggesting that preinitiated template RNA copurifies with the replicase complex (1, 4, 5). The ability to reconstitute HCV replication in a cell-free system will facilitate molecular dissection of the replication process and provide a system to evaluate potential antiviral drugs.
One of the drawbacks of the cell-free HCV replication system is the presence of endogenous RNase activity that copurifies with the replicase complex (1). This nuclease activity degrades exogenously added RNA, limiting the utility of the cell-free replication system for studying processes like replication initiation by using defined full-length HCV RNA templates. While nuclease activity degrades exogenously added RNA, newly synthesized RNA and endogenous template RNA are protected from degradation by a detergent-sensitive factor(s) (6). In this report, we demonstrate that an additional subtilisin-sensitive factor(s) is also necessary for protection of newly synthesized RNA.
As a first step towards understanding how newly replicated RNA is protected from endogenous nuclease activity, a cell-free HCV replication system was established by using membrane fractions from Huh-7 cells containing an HCV replicon derived from the infectious Con1 clone (genotype 1b) (3). Replicon-containing cells (107) were scraped into ice-cold 1× phosphate-buffered saline and concentrated by centrifugation at 1,000 × g for 10 min at 4°C. The cell pellet was resuspended in hypotonic lysis buffer (HLB) containing 10 mM HEPES (pH 8.0), 10 mM KCl, 1 mM dithiothreitol (DTT), and 1× protease inhibitor cocktail (Sigma-Aldrich, St. Louis, Mo.) and incubated for 10 min on ice. The cells were lysed by Dounce homogenization using 40 strokes of a type A pestle, and the nuclei and insoluble debris were removed by centrifugation at 1,000 × g for 5 min at 4°C. The supernatant fluid was transferred to a new tube, and the heavy membrane fraction was collected by centrifugation at 20,000 × g (P20) for 20 min at 4°C.
The P20 fraction was further purified by equilibrium centrifugation on a discontinuous sucrose gradient. The gradient was prepared by resuspending the P20 fraction in 72% sucrose in HLB (3 ml). The 72% sucrose solution was overlaid with 1 ml of 55% sucrose in HLB followed by 1 ml of 10% sucrose in HLB. In some samples, NP-40 was added to the P20 fraction to a final concentration of 0.5% prior to equilibrium centrifugation. The gradients were centrifuged at 100,000 × g for 14 h at 4°C. The material that partitioned in the 55-to-72% sucrose interface was collected, diluted in HLB, and concentrated by centrifugation at 100,000 × g for 2 h at 4°C. The membrane pellet was resuspended in 112 μl of HLB containing 5% glycerol, 1 mM DTT, and 1,920 U of RNasin (Promega Inc., Madison, Wis.). The resuspended membrane fraction was stored in 20-μl aliquots at −80°C.
Reaction mixtures (final volume, 50 μl) contained 50 mM HEPES-KOH (pH 8.0), 0.5 mM MgCl2, 1.5 mM MnCl2, 5 mM DTT, 4 μg of actinomycin D/ml, 3.2 U of RNasin/μl, 50 mM potassium acetate, 0.5 mM ATP, UTP, and GTP, 0.5 μM CTP, 1 mM creatine phosphate, 3.5 U of creatine phosphokinase/μl, 10 μCi of [α-32P]CTP (800 Ci/mmol), and 2.5 μl of membrane fractions prepared from either replicon-containing cells or parental Huh-7 cells. The reactions were initiated by the addition of nucleotide triphosphates, and the mixtures were incubated at 37°C. At 0, 5, 10, 15, 25, and 35 min, reactions were terminated by addition of 50 μl of a solution containing 10 mM HEPES (pH 8.0), 1% sodium dodecyl sulfate (SDS), and 50 mM EDTA. The RNA products were purified by phenol extraction and Sephadex G-50 spin column chromatography and concentrated by ethanol precipitation. The ethanol precipitate was resuspended in 10 μl of diethyl pyrocarbonate-treated water and denatured by treatment with 1.2 M glyoxal for 20 min at 50°C. The RNA was resolved by agarose gel electrophoresis in 1× glyoxal gel running buffer (Ambion, Austin, Tex.). The gel was dried under vacuum onto Whatman 3MM paper, and RNA products were visualized and quantified by phosphorimager analysis.
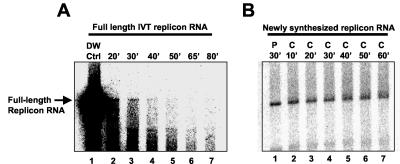
Reaction mixtures supplemented with membranes from replicon-containing cells generated radiolabeled RNA products that comigrated with full-length HCV replicon RNA, as well as products that migrated more slowly than full-length replicon RNA (Fig. 1B, lane 1). The amounts of both RNA species increased linearly as a function of time, suggesting that replicase activity was stable for at least 35 min (data not shown). Upon purification, the full-length replicon RNA products were susceptible to S1 nuclease digestion, while the more slowly migrating RNA species were less susceptible to nuclease treatment (data not shown). In vitro-transcribed (IVT) full-length replicon RNA was also susceptible to S1 nuclease digestion (data not shown). These results suggest that the full-length replicon RNA products were single stranded while the more slowly migrating RNA species contained double-stranded regions, making them less susceptible to S1 nuclease digestion. The more slowly migrating RNA species can be resolved into full-length single-strand replicon RNA and lower-molecular-weight species by methylmercury agarose gel electrophoresis, suggesting that the more slowly migrating RNA species detected on glyoxal gels are likely RNA replication intermediates that are not fully denatured by glyoxal treatment (1).
FIG. 1.
Stability of IVT replicon RNA and newly synthesized replicon RNA in the presence of membrane fractions. Replication reaction mixtures were supplemented with radiolabeled IVT replicon RNA (A) or 10 μCi of [α-32P]CTP (800 Ci/mmol) (B) to generate de novo-synthesized radiolabeled replicon RNA. The reaction mixtures were incubated for 30 min at 37°C followed by addition of 50 μl of 300 μM unlabeled CTP. At 10-min intervals, a portion of the reaction mixture was removed and added to an equal volume of 10 mM Tris (pH 7.5), 1 mM EDTA, 150 mM NaCl, and 0.5% SDS to stop the reaction. Distilled water (DW) instead of membrane fractions was added to the reaction mixture in lane 1 (A). For both panels, the RNA products were purified and analyzed by glyoxal gel electrophoresis.
To determine whether purified membrane fractions contained RNase activity, the stability of IVT HCV replicon RNA was measured in the presence and absence of purified membrane fractions. Radiolabeled HCV replicon RNA synthesized with a MAXIscript SP6/T7 kit (Ambion) was added to reaction mixtures in the presence and absence of membrane fractions from replicon-containing cells. The reaction mixtures were incubated at 37°C, and at 0, 20, 30, 40, 50, 65, and 80 min the RNA was purified from the reaction mixtures and analyzed by glyoxal gel electrophoresis. The stability of the exogenously added IVT RNA was determined by measuring the amount of full-length replicon RNA remaining in the reaction mixtures as a function of time. The results show that radiolabeled IVT replicon RNA was unstable in the presence of membrane fractions with a half-life of less than 20 min (Fig. 1A). In contrast, this same RNA was stable for over 60 min in the absence of membrane fractions (Fig. 1A, lane 1).
The presence of RNase was confirmed using a commercially available test kit (RNaseAlert; Ambion) that quantifies RNase activity by measuring cleavage of an RNA substrate tagged with a fluorescent reporter molecule. Using this assay, RNase activity was readily detected in membrane fractions from replicon containing cells (data not shown).
While exogenously added RNA was rapidly degraded in membrane fractions, newly synthesized RNA products appeared to be resistant to degradation. This observation suggested that newly synthesized RNA was protected from nuclease activity. To test this hypothesis, the stability of newly synthesized RNA was measured by pulse-chase analysis (Fig. 1B). Replication reaction mixtures were incubated for 30 min in the presence of 10 μCi of [α-32P]CTP (800 Ci/mmol) (pulse) to generate radiolabeled RNA. The reaction mixtures were then supplemented with 300 μM unlabeled CTP (chase), and at 10-min intervals, a portion of the reaction mixture was removed and added to an equal volume of 10 mM Tris (pH 7.5), 1 mM EDTA, 150 mM NaCl, and 0.5% SDS (stop buffer) to terminate the reaction. The RNA was purified from the reaction mixture and analyzed by glyoxal agarose gel electrophoresis. Newly synthesized full-length replicon RNA was found to be stable for up to 60 min in the presence of membrane fractions (Fig. 1B). These results suggest that newly synthesized full-length replicon RNA is protected from endogenous nuclease activity.
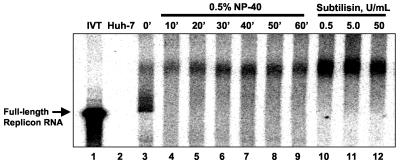
To characterize the nature of the activity that protects full-length HCV replicon RNA from nuclease degradation, the stability of newly synthesized RNA was measured under pulse-chase labeling conditions in the presence and absence of NP-40 or the protease subtilisin. Replicase reaction mixtures were incubated for 30 min in the presence of 10 μCi of [α-32P]CTP (800 Ci/mmol) (pulse) to generate radiolabeled RNA. The reaction mixtures were then supplemented with 300 μM unlabeled CTP (chase) in the presence of either 0.5% NP-40 or 0.5, 5, or 50 U of subtilisin/μl. At 10-min intervals, the reactions were terminated by the addition of stop buffer, and the RNA was purified and analyzed by glyoxal agarose gel electrophoresis. The results show that treatment with NP-40 or subtilisin during the chase period rendered newly synthesized full-length replicon RNA fully susceptible to nuclease digestion (Fig. 2). The more slowly migrating RNA species were resistant to degradation under these conditions. These results are consistent with the observation that purified full-length replicon RNA is susceptible to S1 nuclease digestion while the more slowly migrating RNA species is resistant to S1 nuclease digestion (data not shown). This suggests that the more slowly migrating RNA species may be a replication intermediate containing portions of double-stranded RNA that are resistant to nuclease digestion. The ability of NP-40 to destabilize newly synthesized full-length replicon RNA is consistent with the observation that replicon RNA in permeabilized replicon-containing cells was sensitive to nuclease digestion after addition of detergent (6). These results suggest that the replicon RNA is protected by a component that is sensitive to both detergent and protease treatment.
FIG. 2.
Stability of newly replicated HCV RNA in the presence of NP-40 or subtilisin treatment. Pulse-chase analysis of radiolabeled replicon RNA from replication reaction mixtures supplemented with membrane fractions from Huh-7 cells (lane 2) or replicon containing cells (lanes 3 to 12) was done. The reaction mixtures were incubated for 30 min at 37°C (pulse) followed by the addition of 50 μl of 300 μM unlabeled CTP (chase) in the presence and absence of 0.5% NP-40 or 0.5, 5, and 50 U of subtilisin/ml. At 10-min intervals (NP-40-treated samples) or at 60 min (subtilisin-treated samples), a portion of the reaction mixture was removed and added to an equal volume of 10 mM Tris (pH 7.5), 1 mM EDTA, 150 mM NaCl, and 0.5% SDS to stop the reaction. The RNA products were analyzed by glyoxal gel electrophoresis.
The ability of detergent and protease treatment to render replicon RNA susceptible to nuclease digestion suggests that protein and membranes (lipid-containing material) are required to protect newly synthesized RNA from nuclease activity. A simple explanation for this observation is that NP-40 and subtilisin treatment destabilize protein components of the replication complex that protect newly synthesized RNA from degradation. To determine if NP-40 or subtilisin treatment affect the integrity of the replicase complex and microsomes in the membrane fraction, the protease susceptibilities of viral and cellular proteins in purified membrane fractions were measured in the presence and absence of NP-40.
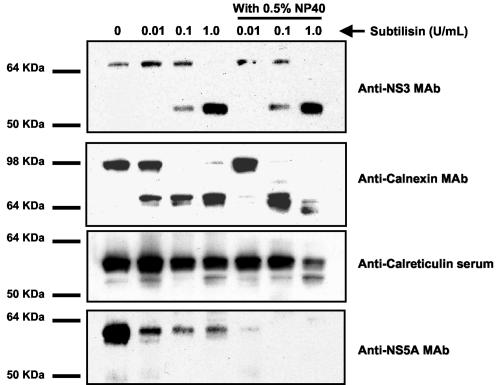
Membrane fractions from replicon-containing cells were treated with 0, 0.01, 0.1, and 1.0 U of subtilisin/ml in the presence and absence of 0.5% NP-40. The protease susceptibilities of HCV proteins were determined by Western blot analysis using monoclonal antibody (MAb) NCL-HCV-NS3 (Novocastra Laboratories Ltd., Newcastle, United Kingdom) and MAb 1877 (ViroStat Inc., Portland, Maine), which are specific for the HCV NS3 and NS5A proteins, respectively (Fig. 3) NP-40 had little effect on the susceptibility of NS3 to subtilisin digestion. Full-length NS3 was cleaved with increasing concentrations of subtilisin. At the highest subtilisin concentration (1.0 U/ml), a protease-resistant domain of NS3 of 53 kDa was observed. This protease-resistant domain was also detected after subtilisin treatment of purified NS3 protein, suggesting that protease resistance is an intrinsic property of NS3 (data not shown). NP-40 had a slight effect on the susceptibility of NS5A to subtilisin digestion. Approximately 90% of NS5A was degraded by subtilisin in the absence of NP-40, while in the presence of NP-40, NS5A was completely digested (Fig. 3).
FIG. 3.
Effects of NP-40 on the subtilisin susceptibilities of HCV NS3, NS5A, and NS5B and the cellular proteins calnexin and calreticulin in purified membrane fractions. Purified membrane fractions (15 μg) were treated with 0.01, 0.1, and 1.0 U of subtilisin/ml for 30 min at 37°C in the presence and absence of 0.5% NP-40. The proteins in the reaction mixtures were resolved by SDS-polyacrylamide gel electrophoresis and transferred to Hybond C+ membranes (Amersham, Inc. Piscataway, N.J.). The membranes were probed with a 1-to-100 dilution of anti-HCV NS3 MAb. The membranes were stripped of primary antibody by incubation in 0.2 N NaOH for 5 min at room temperature and reprobed with a 1-to-100 dilution of anti-HCV NS5A MAb or a 1-to-500 dilution of anticalnexin MAb or anticalreticulin antiserum. The Western blots were developed with an enhanced chemiluminescence detection system (Amersham, Inc.).
To determine if NP-40 treatment disrupted endoplasmic reticulum (ER)-derived microsomal membranes, the effects of NP-40 on protease susceptibility of two ER-resident proteins, calnexin and calreticulin, was examined. Calnexin is a transmembrane protein that contains a C-terminal cytosolic domain and an ER luminal domain, while calreticulin resides completely within the ER lumen (8, 9). The luminal portions of these proteins should be protected from protease digestion in intact ER-derived microsomal membranes.
Purified membrane fractions from replicon containing cells were treated with subtilisin at 0, 0.01, 0.1, and 1 U/ml in the presence and absence of 0.5% NP-40, and the protease susceptibilities of calreticulin and calnexin were measured by Western blot analysis using rabbit polyclonal antiserum PA3-900 (Affinity BioReagents, Golden, Colo.), which is specific for calreticulin, and MAb RDI-CALNEabm (Research Diagnostics Inc., Flanders, N.J.), which is specific for calnexin (Fig. 3). NP-40 treatment of membrane fractions did not significantly change the protease susceptibility of calnexin and calreticulin at 0.01 and 0.1 U of subtilisin/ml (Fig. 4). Importantly, calreticulin, which resides entirely in the ER lumen, was fully protected from protease treatment, while a portion of calnexin was protease resistant. The protease-resistant region of calnexin had the same molecular weight as the luminal domain of calnexin, suggesting that protease resistance was likely due to the presence of intact ER membranes. At the highest concentration of subtilisin (1 U/ml), partial protease susceptibility of both calreticulin and the protease resistant domain of calnexin was observed in the presence of NP-40. These results suggest that 0.5% NP-40 had a slight effect on the integrity of ER-derived microsomes; however, detergent treatment did not completely disrupt ER-derived microsomal membranes.
FIG. 4.
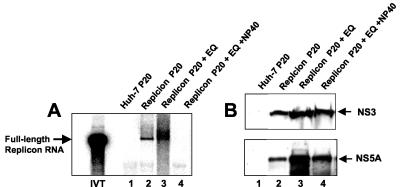
Effects of NP-40 on the integrity of HCV membrane complexes. Membrane fractions were incubated for 15 min at 37°C in the presence and absence of 0.5% NP-40 prior to purification by equilibrium centrifugation. Replicase activity and protein content were measured in purified membrane fractions. The RNA products from the replicase reaction mixtures were resolved by glyoxal gel electrophoresis. (A) Lane IVT, IVT replicon RNA; lane 1, RNA from reaction mixtures supplemented with the 20,000 × g membrane pellet (P20) from Huh-7 cells; lane 2, P20 pellet from replicon containing cells prior to equilibrium sedimentation; lane 3, untreated P20 membrane pellet from replicon containing cells purified by equilibrium centrifugation; lane 4, P20 membrane pellet from replicon containing cells treated with NP-40 prior to equilibrium centrifugation. (B) Western blot analysis of proteins from membranes purified by subcellular fractionation and equilibrium sedimentation. The Western blots were probed with a 1:100 dilution of anti-NS3 MAb and anti-NS5A MAb.
Since HCV susceptibility of HCV proteins to protease treatment was similar in the presence and absence of NP-40, it could not be established if NP-40 disrupted HCV membrane complexes. To determine if HCV membrane complexes were disrupted by NP-40 treatment, membrane fractions from replicon-containing cells were extracted with 0.5% NP-40 during their preparation for equilibrium centrifugation. The levels of viral proteins in resulting purified membrane fractions were determined by Western blot analysis. NP-40 extraction did not significantly affect the level of NS3 and only slightly reduced the level of NS5A in membrane fractions (Fig. 4). These results are consistent with the observation that the HCV replication complex resides on detergent-resistant membranes that cofractionate with lipid raft markers (7). While the levels of NS3 and NS5A were relatively constant following NP-40 treatment, the purified NP-40-treated replicase complex did not support RNA synthesis, suggesting that an NP-40-sensitive factor(s) was required for replicase activity (Fig. 4A).
Taken together, the results presented here suggest that under conditions where viral membrane complexes remain intact, both NP-40 and subtilisin treatment were able to render newly synthesized HCV replicon RNA susceptible to nuclease digestion. These results suggest that newly synthesized HCV replicon RNA is protected by proteins within the replication complex.
Acknowledgments
We thank Linda Barone and Michelle Kimberland for helpful discussions and critical reading of the manuscript.
This work was supported by ViroPharma Incorporated.
REFERENCES
- 1.Ali, N., K. D. Tardif, and A. Siddiqui. 2002. Cell-free replication of the hepatitis C virus subgenomic replicon. J. Virol. 76:12001-12007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 3.Blight, K., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 4.Hardy, R. W., J. Marcotrigiano, K. J. Blight, J. E. Majors, and C. M. Rice. 2003. Hepatitis C virus RNA synthesis in a cell-free system isolated from replicon-containing hepatoma cells. J. Virol. 77:2029-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai, V. C. H., S. Dempsey, J. Y. N. Lau, Z. Hong, and W. Zhong. 2003. In vitro RNA replication directed by replicase complexes isolated from the subgenomic replicon cells of hepatitis C virus. J. Virol. 77:2295-2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyanari, Y., M. Hijikata, M. Yamaji, M. Hosaka, H. Takahashi, and K. Shimotohno. 2003. Hepatitis C virus non-structural proteins in the probable membranous compartment function in viral genome replication. J. Biol. Chem. 278:50301-50308. [DOI] [PubMed] [Google Scholar]
- 7.Shi, S. T., K. J. Lee, H. Aizaki, S. B. Hwang, and M. M. C. Lai. 2003. Hepatitis C virus RNA replication occurs on a detergent-resistant membrane that cofractionates with caveolin-2. J. Virol. 77:4160-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wada, I., D. Rindress, P. H. Cameron, W. J. Ou, J. J. Doherty, D. Louvard, A. W. Bell, D. Dignard, D. Y. Thomas, and J. J. Bergeron. 1991. SSR alpha and associated calnexin are major calcium binding proteins of the endoplasmic reticulum membrane. J. Biol. Chem. 266:19599-19610. [PubMed] [Google Scholar]
- 9.Wada, I., S. i. Imai, M. Kai, F. Sakane, and H. Kanoh. 1995. Chaperone function of calreticulin when expressed in the endoplasmic reticulum as the membrane-anchored and soluble forms. J. Biol. Chem. 270:20298-20304. [DOI] [PubMed] [Google Scholar]