Abstract
The retroviral Gag precursor plays an important role in the assembly of virion particles. The capsid (CA) protein of the Gag molecule makes a major contribution to this process. In the crystal structure of the free CA protein of the human immunodeficiency virus type 1 (HIV-1), 11 residues of the C terminus were found to be unstructured, and to date no information exists on the structure of these residues in the context of the Gag precursor molecule. We performed phylogenetic analysis and demonstrated a high degree of conservation of these 11 amino acids. Deletion of this cluster or introduction of various point mutations into these residues resulted in significant impairment of particle infectivity. In this cluster, two putative structural regions were identified, residues that form a hinge region (353-VGGP-356) and those that contribute to an α-helix (357-GHKARVL-363). Overall, mutations in these regions resulted in inhibition of virion production, but mutations in the hinge region demonstrated the most significant reduction. Although all the Gag mutants appeared to have normal Gag-Gag and Gag-RNA interactions, the hinge mutants were characterized by abnormal formation of cytoplasmic Gag complexes. Gag proteins with mutations in the hinge region demonstrated normal membrane association but aberrant rod-like membrane structures. More detailed analysis of these structures in one of the mutants demonstrated abnormal trapped Gag assemblies. These data suggest that the conserved CA C terminus is important for HIV-1 virion assembly and release and define a putative target for drug design geared to inhibit the HIV-1 assembly process.
The retroviral Gag protein is crucial for late stages in the viral life cycle, as it mediates virion assembly through complex interactions with RNA, protein and lipid molecules (reviewed in references 23, 24, 32, and 81). The Gag protein of type C retroviruses is expressed as a precursor protein, modified by myristylation, and targeted to the plasma membrane. Multimerization of the Gag precursors induces membrane curvature and the formation and release of virion particles through a budding process. It is estimated that 1,000 to 2,000 Gag molecules compose one particle (72). The Gag protein is necessary and sufficient for the production of virus particles. Many mutations in the gag gene block virion formation (2, 17, 48, 62, 70), and expression of the Gag protein alone in mammalian cells is sufficient to direct the assembly and release of virus-like particles (VLPs) from cells (75). In addition, other mutations in the gag gene have been found to block early stages of infection, suggesting a role for some of the Gag domains in the processes of uncoating, reverse transcription, and integration (12, 15, 39, 70, 83).
During and after the assembly process, the Gag protein (Pr55gag) of human immunodeficiency virus type 1 (HIV-1) is cleaved by the viral protease into four products found in the mature virion: matrix (MA), capsid (CA), nucleocapsid (NC), and p6. In addition, two spacer peptides, SP1 and SP2, that flank the NC domain are also cleaved from the Gag precursor. The MA domain is required for membrane binding of Gag and for virion assembly (11, 33, 44, 62, 63, 76, 88). The CA domain is also important for virion assembly, as many point mutations in CA block this process (2, 10, 48, 49, 69, 70, 77, 78). Other mutations in CA block the early stages of infection (69, 83). Interaction between the CA domain and the cellular protein cyclophilin A results in incorporation of the cellular factor into the assembled virion, and this interaction is important for HIV-1 infectivity (22, 26, 52, 84). In the mature virion, CA forms an inner shell (47) that surrounds the RNA genome and core-associated proteins.
The NC domain is highly basic, containing two repeats of a conserved zinc-binding motif. NC contains a sequence, termed the interaction (I) domain (18, 45, 81), that is important for multimerization of Gag; subsequently, deletion of NC results in a drastic reduction in particle production (13, 16). Point mutations in the NC often affect the binding of the Gag protein to the viral RNA (9, 93) and the process of viral RNA encapsidation during virion assembly (9, 21, 93). The ability of the Gag protein to bind RNA through the NC domain is also thought to affect Gag dimerization and to enhance virion assembly (13, 14, 16, 37, 54, 55, 60).
The p6 domain of Gag includes sequences that act as late (L) assembly domains. These are required for late stages of viral assembly and efficient release from the cell (reviewed in references 3, 25, 67, and 73). The L domains serve as binding sites for members of the cellular vacuolar protein sorting (Vps) system, like Tsg101 and AIP1 proteins, which act to release the budding particle from the cell surface (28, 79, 87). The p6 domain also mediates the internalization of the Vpr protein into the assembled virion (36, 40, 50, 65).
The SP1 and SP2 sequences (formerly termed p2 and p1, respectively) also play a role in virion assembly and maturation. The SP1 sequence, which connects the CA domain to the NC domain, is included in minimal Gag constructs that are capable of producing VLPs (2, 89), consistent with the observation that SP1 is important for proper assembly of the Gag molecules and for virion morphogenesis (1, 41, 57, 59, 66, 69). The proteolytic processing of SP1 is important for virion maturation, as removal of SP1 from the C terminus of CA regulates the transition of the particle from an immature spherical appearance to a mature virion harboring a conical capsid (1, 34). In addition, cleavage of SP1 from the N terminus of NC leads to condensation of the ribonucleoprotein core (90) and maturation of the RNA dimer (74). Cleavage of SP2 has also been reported to play a role in the formation of the HIV-1 cone-shaped core (91). Furthermore, the NC-SP2 region is thought to cooperate with the L domain of p6 in Gag ubiquitination (80).
The HIV-1 CA contains two helix-rich domains (26, 27, 30, 58). The first of these is the core domain, which is formed by the N-terminal two thirds of the protein. This domain contains seven α-helices, two β-hairpins, and an exposed loop. The C-terminal one-third of CA forms a globular domain, called the dimerization domain, which is connected to the core domain with a flexible strand. The dimerization domain consists of four α-helices followed by the last 11 amino acids of CA (VGGPGHKARVL). No structure is available for these 11 amino acids, as they are disordered in the dimerization domain crystals (27), yet recent studies have indicated the importance of this region for multimerization and membrane binding of the Gag proteins (48, 49). Overall, mutagenesis and biochemical studies have demonstrated that the core domain is important for core formation in the mature particle, while the dimerization domain plays a role in both particle assembly and core formation (10, 20, 56, 57, 69, 70, 77, 88).
Several residues in the Gag protein are conserved among different retroviruses and are important for the function of the precursor protein and its cleavage products. Some examples of these significant residues include the glycine at the N terminus of the MA domain, which serves as a myristylation site (63, 72, 76, 88), the 20 residues forming the major homology region in CA, which generate a hydrogen-bonding network and contribute to the hydrophobic core of the protein (27), and the zinc finger motifs in the NC that bind RNA (9). We performed a phylogenetic analysis of the HIV-1 CA protein family and found that the 11 residues at the CA C terminus are also conserved. We tested the significance of these residues, and we provide evidence for the importance of this conserved stretch of amino acids for the proper assembly and release of HIV-1 virions.
MATERIALS AND METHODS
Phylogenetic analysis.
The CA protein sequence of strain HXB2 was analyzed with the maximum-likelihood-based algorithm Rate4site (68) and the ConSeq server (http://conseq.bioinfo.tau.ac.il/) (8). This automated analysis created a phylogenetic tree, which was composed of homologous CA protein sequences derived from the following retroviruses (accession numbers appear in parentheses): HIV-1 (P0588, P12494, P03347, Q70622, P03350, P05890, P03349, P20889, P04594, P24736, P18800, P12495, P04592, P05887, P20873, P12493, P35962, and P05889); HIV-2 (P15832, P20874, P04590, P05891, Q74119, P12450, P24106, P18095, P18041, and P17756); simian immunodeficiency virus (P17282, P31634, P05894, P05893, P12496, P19504, Q02843, P27978, P05892, P27972, and P22381); equine infectious anemia virus (P03351); feline immunodeficiency virus (P31821, Q05313, P16087, and P19027); bovine immunodeficiency virus (P19558); caprine arthritis encephalitis virus (P33458); visna-related ovine lentivirus (P23424, P23425, P35955, and P16900); human endogenous retrovirus K-10 (P10264); and Jaagsiekte sheep retrovirus (P31622). The degree of conservation was calculated for each residue according to its preservation along the phylogenetic tree (68). The conservation level (ranging from 0 to 1, where 0 is not conserved and 1 is highly conserved) of each of the last 11 residues of the CA domain was as follows: V353, 0.895 ± 0.015; G354, 0.985 ± 0.015; G355, 0.925 ± 0.015; P356, 0.895 ± 0.015; G357, 0.085 ± 0.085; H358, 0.87 ± 0.01; K359, 0.985 ± 0.015; A360, 0.955 ± 0.015; R361, 0.925 ± 0.015; V362, 0.895 ± 0.015; and L363, 0.925 ± 0.015.
Yeast plasmids.
The Saccharomyces cerevisiae expression vector pGADZX2, generously provided by J. Luban (Columbia University), carries the LEU2 marker and encodes a fusion protein with an N-terminal Gal4 activation domain (Gal4AD) and a C-terminal HIV-1 Gag polyprotein (Gal4AD-HIV Gag), derived from the infectious molecular clone HXBC2 and flanked by BamHI and SalI sites (52). To remove the NC domain from the Gal4AD-HIV Gag fusion protein, the HIV-1 gag open reading frame was excised from plasmid pGADZX2 at the BamHI and SalI sites and cloned into the same sites in plasmid pUC19, creating pUC19-HIV-1gag. An internal PpuMI-BglII fragment (244 bp) containing the C terminus of CA, the entire SP1 and NC domains, and the N terminus of the SP2 domain was deleted from pUC19-HIV-1gag; the termini were blunted with Vent DNA polymerase (New England Biolabs) and ligated to a NotI linker (New England Biolabs), to create pUC19-HIV-1gag-NotI-linker. A BamHI-SalI fragment harboring the deletion was than excised from pUC19-HIV-1gag-NotI-linker and used to exchange the wild-type sequences in pGADZX2, with the same sites, to create pGADZX2-NotI-linker.
pGADZX2-MA-CA is pGADZX2-based plasmid harboring a unique PmeI site that replaced the sequence encoding the last 11 amino acids of CA and introduced a stop codon close to the CA end. This plasmid expresses a Gal4AD-HIV Gag fusion protein lacking the last 10 amino acids of CA as well as the SP1, NC, SP2, and p6 domains. To construct this plasmid, we first PCR amplified two HXBC2 gag sequences with 5′-HIVupPpumI together with MA-CA-R primers and MA-CA-F together with 3′-HIVdownBglII primers (Table 1). The resulting PCR fragments share complementary ends containing the PmeI substitution mutation. These were joined by a subsequent overlapping PCR (5) with the 5′-HIVupPpumI and 3′-HIVdownBglII primers.
TABLE 1.
Oligodeoxynucleotides used in this studya
| Mutation | Oligodeoxynucleotide | Sequence (5′-3′) |
|---|---|---|
| V353A | F-V353A | TGTCAGGGAGCAGGAGGACCC |
| R-V353A | GGGTCCTCCTGCTCCCTGACA | |
| G354A | F-G354A | GTAGCAGGACCCGGCCATAAGGCA |
| R-G354A | ATGGCCGGGTCCTGCTACTCCCTGACATGCTGTC | |
| G355A | F-G355A | GGAGTAGGAGCACCCGGCC |
| R-G355A | GGCCGGGTGCTCCTACTCC | |
| P356A | F-P356A | GGAGCCGGCCATAAGGCAAGAGTT |
| R-P356A | TGCCTTATGGCCGGCTCCTCCTACTCCCTGACAT | |
| G357A | F-G357A | GTAGGAGGACCCGCCCATAAGGC |
| R-G357A | GCCTTATGGGCGGGTCCTCCTAC | |
| H358A | F-H358A | GGACCCGGCGCTAAGGCAAGAG |
| R-H358A | CTCTTGCCTTAGCGCCGGGTCC | |
| K359A | F-K359A | GGACCCGGCCATGCGGCAAGAGTTTTG |
| R-K359A | CAAAACTCTTGCCGCATGGCCGGGTCC | |
| A360L | F-A360L | GGCCATAAGCTAAGAGTTTTGGC |
| R-A360L | GCCAAAACTCTTAGCTTATGGCC | |
| R361A | F-R361A | GGCCATAAGGCAGCAGTTTTGGCTGAAGC |
| R-R361A | GCTTCAGCCAAAACTGCTGCCTTATGGCC | |
| V362A | F-V362A | CCATAAGGCAAGAGCTTTGGCTGAAGC |
| R-V362A | GCTTCAGCCAAAGCTCTTGCCTTATGG | |
| L363A | F-L363A | GGCAAGAGTTGCGGCTGAAGCAATG |
| R-L363A | CATTGCTTCAGCCGCAACTCTTGCC | |
| Δ11 | F-Δ11 | GACAGCATGTCAGGGA- - -GCTGAAGCAATGAGCCAA |
| R-Δ11 | AGC- - -TCCCTGACATGCTGTCATC | |
| MA-CA | F-MA-CA | GGGAGTTTAAAC- - -GCTGAAGCAATGAGCCAAGT |
| R-MA-CA | CTTCAGC- - -GTTTAAACTCCCTGACATGCTGTCATC | |
| G2A | F-G2A | AGGAGAGAGATGGCTGCGAGAGCGTC |
| R-G2A | GACGCTCTCGCAGCCATCTCTCTCC | |
| 5′HIVupPpumI | CAGGAGGTAAAAAATTGGATGACAGAAA | |
| 3′HIVdownBglII | GAGGGGGAGTTGTTGTCTCTACCCCAGA | |
| 2LTR forward | AACTAGGGAACCCACTGCTTAAG | |
| Gag6 | CTTTGGTTCCTAAAATTGCC |
Sequences were derived from HXB2 strain (accession no. K03455). Substitution mutations are underlined, and deletions are represented by dashed lines.
PCRs were carried out with the Expand High Fidelity PCR system according to the manufacturer's instructions (Roche). The resulting PCR product was transformed into S. cerevisiae strain GGY::171 together with pGADZX2-NotI-linker DNA linearized by NotI digestion. Homologous recombination between the ends of the PCR fragment and the linearized plasmid generated pGADZX2-MA-CA DNA, in which the CA edge harboring the PmeI substitution together with the SP1, NC, and SP2 domains were restored in the context of the gag open reading frame. The resulting plasmid was extracted from Leu+ yeast colonies (grown on medium lacking leucine) and recovered by transformation into Leu− Escherichia coli KC8 bacteria (Clontech).
All other mutations in the CA C terminus were generated similarly by overlapping PCR with the appropriate pair of forward (F) and reverse (R) primers (Table 1) and by recombining the resulting PCR fragments with PmeI-linearized pGADZX2-MA-CA. The resulting pGADZX2 plasmids express the full-length Gal4AD-HIV Gag fusion protein with point mutation V353A, G354A, G355A, P356A, G357A, H358A, K359A, A360L, R361A, V362A, or L363A, each of which is named after the mutation that it carries. Similarly, an in-frame deletion mutation (Δ11) of the last 11 amino acids of CA was created, maintaining the downstream SP1, NC, SP2, and p6 domains (Table 1) to generate pGADZX2Δ11.
The yeast expression vector pMA424HIVgag, generously provided by J. Luban, carries the HIS3 marker and encodes a fusion protein with an N-terminal Gal4 DNA binding domain (Gal4DBD) and a C-terminal HIV-1 Gag polyprotein (Gal4DBD-HIV Gag), derived from the HXBC2 clone and flanked by BamHI and SalI sites. To express these CA mutations in the context of Gal4DBD-HIV Gag, BamHI-SalI fragments containing the mutated gag sequences were excised from the pGADZX2 array of plasmids and used to replace the wild-type sequence in pMA424HIVgag at the same sites. This procedure resulted in the creation of a panel of pMA424HIVgag-derived plasmids, each named after the mutation that it carries. The presence of all mutations was confirmed by sequencing. pIIIA/HIVΨ-MS2-2 and pIIIA/HaMSVΨ-MS2-2 encode chimeric RNA molecules that contain the encapsidation signals of HIV-1 and Harvey murine sarcoma virus, respectively (5).
Yeast two- and three-hybrid systems.
S. cerevisiae strains GGY::171 and L40-coat were used in the yeast two- and three-hybrid assays, respectively, as described before (5, 6, 51). To test homodimerization of Gag protein mutants in the yeast-two hybrid system, plasmids pGADZX2 and pMA424HIVgag (see above) containing the same CA mutation were cotransformed into S. cerevisiae GGY::171, and the interaction of the Gag proteins encoded by these plasmids was tested. Similarly, heterodimerization was tested between Gag protein mutants encoded by the pGADZX2 plasmids and the wild-type Gag proteins encoded by the pMA424HIVgag plasmid. Yeast two- and three-hybrid components were generously provided by S. Goff, J. Luban (Columbia University), and M. Wickens (University of Wisconsin-Madison).
Mammalian expression plasmids.
The HIV-1 Gag and Pol proteins were expressed in mammalian cells from plasmid pHIVgptSVPA (53). This plasmid contains the HXB2 provirus except that 1.2 kb of env coding sequence was replaced with the simian virus 40 (SV40) origin of replication and promoter and coding sequence for the gpt gene (64). In addition, the 3′ long terminal repeat (LTR) was replaced by SV40 sequences containing a polyadenylation sequence (53). pHIVgptSVPA also encodes the Rev, Tat, and Vif proteins but not Vpu, Nef, or Vpr.
To introduce the CA mutations into plasmid pHIVgptSVPA, an SpeI-SalI fragment (4,279 bp) was excised from pHIVgptSVPA and cloned into the same sites in pBCSK(+) (Stratagene) to create pBC-SpeI-SalI-wt. An internal SpeI-BglII fragment (589 bp) was excised from pBC-SpeI-SalI-wt and replaced with the corresponding fragment derived from pGADZX2 V353A, G354A, G355A, P356A, G357A, H358A, K359A, A360L, R361A, V362A, L363A, or Δ11. The mutated gag sequences were then exchanged for the wild-type gag sequence in pHIVgptSVPA, with the SpeI-SalI sites, to form pHIVgptSVPA V353A, G354A, G355A, P356A, G357A, H358A, K359A, A360L, R361A, V362A, L363A, and Δ11.
To mutate the Gag myristylation site, overlapping PCR was used to substitute the N-terminal glycine residue of Gag with alanine (G2A mutation), with the HXBC2 gag sequence as a template, with G2A-F and Gag6 as one pair of primers and 2LTRforward and G2A-R as a second pair (Table 1). The resulting PCR fragments with complementary ends containing the G2A mutation were joined by a subsequent overlapping PCR. The resulting fragment was digested with BssHI and SpeI. The mutated Gag sequence was then exchanged for the wild-type gag sequence in pHIVgptSVPA and pHIVgptSVPA G355A with the BssHI and SpeI sites to form pHIVgptSVPA G2A and pHIVgptSVPA G2A/G355A, respectively. The last clone contains both the mutated myristylation site and the CA mutation.
pHR′-CMV-GFP encodes an HIV-1-derived retroviral vector carrying the green fluorescent protein (GFP) marker. pMD.G expresses the vesicular stomatitis virus G envelope protein. Both plasmids were generously provided by I. Verma (Salk Institute). pCDNA1 CD4, generously provided by J. Gershoni (Tel Aviv University), encodes the human CD4 protein. pSV-E-MLV-env (43) expresses the murine leukemia virus ecotropic envelope glycoprotein.
Transformation of 293T and HeLa cells.
293T and HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, l-glutamine, penicillin, streptomycin, and nystatin (Biological Industries, Israel), at 37°C and 5% CO2. Cells were transfected with calcium phosphate (6) with the indicated amount of plasmid DNA, and expression of the encoded proteins was analyzed 2 days after transfection.
Transduction assays.
To quantify the influence of Gag mutations on the virus infectious cycle, we used the single-cycle infectivity assay (6). 293T cells (approximately 80% confluent in 60-mm plates) were cotransfected with wild-type or mutated pHIVgptSVPA clones (7.5 μg), pHR′-CMV-GFP (10 μg), and pMD.G (2.5 μg). After 6 h, the supernatants were replaced with 5 ml of fresh medium. Two days after transfection, the cells were resuspended, fixed with 2% paraformaldehyde, and analyzed by fluorescence-activated cell sorting (FACS) for GFP fluorescence with a FACSort apparatus (Becton Dickinson). Mock-transfected cells (transfection without plasmid DNA) were used to determine the background autofluorescence level of GFP-negative cells.
The total fluorescence of the GFP-positive cell population was calculated by multiplying the percentage of GFP-positive cells by their mean fluorescence value. Supernatants of transfected cells were supplemented with HEPES (pH 7.0; 50 mM final concentration), filtered through a 0.45-μm-pore-size filter, and diluted 1:3 with culture medium; Polybrene (hexadimethrine bromide; Sigma) was added to a final concentration of 8 μg/ml. These viral preparations (2 ml) were used to infect naive 293T cells (2 × 105 cells in 60-mm-diameter dishes) for 2 h, after which 3 ml of fresh medium was added. Two days postinfection, the fluorescence of the infected, paraformaldehyde-fixed, GFP-positive cells was measured by FACS, and total fluorescence was calculated. Normalized infectivity was calculated by dividing the total fluorescence of infected cells by the total fluorescence of transfected cells. The normalized infectivity of the indicated Gag mutants was expressed as a percentage of the normalized infectivity measured for the wild-type protein.
VLP purification and analysis.
293T cells grown in 60-mm plates were transfected with wild-type or mutated pHIVgptSVPA clones (10 μg). Two days posttransfection, culture supernatants were harvested for VLP purification (see below), and the transfected cells were collected. The cells were extracted in 1 ml of ice-cold radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% NP-40, 0.5% deoxycholate, 0.1% sodium dodecyl sulfate, 50 mM Tris [pH 8.0]) supplemented with the protease inhibitors pepstatin (1 μg/ml), leupeptin (1 μg/ml), and aprotinin (1 μg/ml). Extracts were cleared by centrifugation (5 min, 16,000 × g, 4°C) and wre boiled with an equal volume of 2× sample buffer (5% β-mercaptoethanol, 125 mM Tris [pH 6.8], 4% sodium dodecyl sulfate, 0.005% bromophenol blue, 20% glycerol).
To purify the VLPs from the culture medium, VLP-containing supernatants (4.5 ml) were supplemented with 0.5 ml of 0.5 M HEPES (pH 7.0) and 2 ml of TEN buffer (100 mM NaCl, 10 mM Tris, 1 mM EDTA, pH 8.0), and filtered through a 0.45-μm-pore-size filter. VLPs were purified on a 25% (wt/vol) sucrose cushion (2 ml) by centrifugation at 100,000 × g for 2 h at 4°C. VLP pellets were then resuspended and boiled in 60 μl of 2× sample buffer. VLPs (10 μl) and cell extract (20 μl) were further analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blot.
Velocity sedimentation analysis of cellular Gag complexes.
293T cells were transfected with 5 μg of pHIVgptSVPA DNA expressing wild-type or mutant Gag proteins. To inhibit Gag cleavage, 6 h after transfection, culture medium were replaced by fresh medium containing 20 μM saquinavir, an HIV-1 protease inhibitor (Roche; Ro-31-8959; generously provided by M. Kotler, Hebrew University); 48 h posttransfection, culture supernatants were discarded, and cells were washed once with TEN buffer and resuspended in 1 ml of ice-cold TEN buffer containing a protease inhibitor cocktail of pepstatin (1 μg/ml), leupeptin (1 μg/ml), aprotinin (1 μg/ml), and saquinavir (20 μΜ). After 5 min of incubation on ice, the cells were subjected to 10 strokes in a Dounce homogenizer, and the extracts were cleared by centrifugation (30 min, 1,800 × g at 4°C). Postnuclear extracts (1 ml) were placed on top of three successive 1-ml sucrose layers (25, 35, and 45% from top to bottom, all containing Tris-EDTA buffer [pH 8.0] and 20 μM saquinavir), followed by centrifugation (Beckman L-70 ultracentrifuge; 100,000 × g for 1 h at 4°C). Four equal fractions were collected from the top of the gradient, and an aliquot from each fraction was weighed to determine its density. Total proteins were precipitated from each fraction with trichloroacetic acid (10% final concentration) in the presence of 1 μg of the carrier, bovine serum albumin, resuspended in 50 μl of 2× sample buffer, mixed with 50 μl of 1 M Tris (pH 8.0) to adjust the sample pH, and analyzed by SDS-PAGE and Western blotting.
Equilibrium flotation centrifugation assay.
To monitor the membrane binding of wild-type and mutant Gag proteins, we performed the membrane flotation centrifugation assay as detailed previously (62), with minor modifications. 293T cells were transfected with 5 μg of mutant or wild-type pHIVgpt-SVPA DNA; 6 h posttransfection, cell medium were replaced by fresh medium containing 20 μM saquinavir; 48 h posttransfection, the cells were washed once with 1 ml of 10 mM Tris-HCl (pH 7.5) containing 1 mM EDTA and 1 mM EGTA and resuspended in 1 ml of 10 mM Tris-HCl (pH 7.5) containing 1 mM EDTA and 10% (wt/vol) sucrose supplemented with protease inhibitor cocktail containing pepstatin (1 μg/ml), leupeptin (1 μg/ml), aprotinin (1 μg/ml), and 20 μM saquinavir. Following 5 min of incubation on ice, the cell suspensions were subjected to 10 strikes with a Dounce homogenizer and centrifuged at 500 × g for 5 min at 4°C. Then, 250 μl of postnuclear supernatant was mixed with 1.25 ml of 85.5% (wt/vol) sucrose in TE (10 mM Tris, 1 mM EDTA, pH 8.0) containing 20 μM saquinavir and placed on the bottom of a centrifuge tube. On top of this postnuclear supernatant-containing 73% (wt/vol) sucrose mixture was layered 7 ml of 65% (wt/vol) sucrose in TE and 3 ml of 10% (wt/vol) sucrose in TE, all containing 20 μM saquinavir. The gradients were centrifuged at 100,000 × g for 18 h at 4°C (Beckman L-70 ultracentrifuge). Ten 1.2-ml fractions were collected from the top of the gradient, and an aliquot from each fraction was weighed to determine its density. Total proteins were precipitated from each fraction by trichloroacetic acid and analyzed by Western blot as described below.
Western blot analysis and antibodies.
Protein samples were separated on sodium dodecyl sulfate-polyacrylamide (12%) gels, and the proteins were transferred to an Immobilon-P membrane (Millipore) according to the manufacturer's protocol. For Western blot analysis, membranes were blocked with 5% skim milk, incubated with the relevant antibody for 1 h, followed by incubation with a secondary, peroxidase-conjugated antibody. Peroxidase activity was detected by reacting the membrane with chemiluminescence solution containing 150 mM Tris (pH 8.9), 0.22 mg/ml Luminol (Sigma), 0.033 mg/ml paracoumaric acid, and 0.015% H2O2. The intensity of protein bands was measured by densitometry scanning with the TINA2.0 program.
Monoclonal anti-HIV-1 capsid antibody, purified from ascites fluid of hybridoma clone 183-H12-5C (National Institutes of Health AIDS Research and Reference Program) was used at a 1:10,000 dilution. Monoclonal anti-CD4 (7B4) antibody, generously provided by. J. Gershoni, Tel Aviv University, was used at a 1:5,000 dilution. Goat polyclonal anti-Env antiserum raised against Rauscher murine leukemia virus gp69/71 was obtained from the American National Cancer Institute (product no. 81S-127). This antiserum cross-reacts with the Moloney murine leukemia virus ecotropic envelope and was used at a 1:5,000 dilution. Horseradish peroxidase-conjugated polyclonal goat anti-mouse and donkey anti-goat antisera (Jackson Immunoresearch Laboratories) were used at a 1:10,000 dilution.
Immunohistochemistry.
HeLa cells were seeded on glass coverslips in 24-well plates (105 cells/well) in medium lacking phenol red to reduce background fluorescence. The next day, the cells were transfected with 1 μg of the indicated pHIVgptSVPA DNA clone; 48 h posttransfection, glass-attached cells were washed in ice-cold phosphate-buffered saline (PBS), fixed (4% paraformaldehyde in PBS, 20 min), permeabilized (0.1% Triton X-100 in PBS, 3 min), and then blocked (20% normal goat serum and 1% bovine serum albumin in PBS, 30 min). The cells were stained with monoclonal anti-HIV-1 capsid antibody (183-H12-5C, 1:200 dilution, 1 h), followed by the secondary indocarbocyanine-conjugated goat anti-mouse immunoglobulin G antibody (Jackson Immunoresearch Laboratories, product no.115-166-072, 1:500 dilution, 45 min). Extensive PBS washes followed each of the above steps. Stained cells on mounted coverslips were examined with a confocal laser scan microscope (LSM510, Zeiss).
Scanning electron microscopy.
293T cells were seeded on gelatin-coated glass coverslips in 24-well plates (105 cells/well), and the cells were transfected with 1 μg of the indicated pHIVgptSVPA DNA clone the next day; 48 h posttransfection, the cells were fixed (2.5% glutaraldehyde in PBS, 2 h at room temperature), washed with PBS, and dehydrated with graded ethanol concentrations (25, 50, 75, 95, and 100%). Samples coated with sputtered gold were examined with a scanning electron microscope (Jeol 840A).
Transmission electron microscopy.
293T cells in 60-mm plates were transfected with 20 μg of pHIVgptSVPA or pHIVgptSVPA V353A DNA; 48 h posttransfection, the cells were collected in 1 ml of PBS and pelleted in an Eppendorf tube (1,800 × g, 5 min, at 4°C). Cell pellets were overlaid with 100 μl of fixative (2.5% glutaraldehyde), postfixed with 1% osmium tetroxide, and embedded in Epon. Thin sections (60 to 80 nm) were stained with 1% uranyl acetate and 0.3% lead citrate and analyzed with an electron microscope (Jeol JEM 1200EX).
RESULTS
C-terminal region of the HIV-1 capsid domain is evolutionarily conserved.
The retroviral Gag precursor is a multifunctional protein that binds many viral and host factors (23, 67, 81). The Gag protein CA domain is involved in some of these interactions and plays an important role in virion assembly and maturation (1, 10, 48, 49, 56, 69, 78, 83). To look for potentially functionally important residues in the CA protein of HIV-1, we analyzed its amino acid sequence with an advanced algorithm, named Rate4Site (68). This maximum-likelihood-based algorithm provides an estimate of the degree of conservation of each amino acid site among the homologous CA proteins, based on the site's rate of evolution; slowly evolving sites are evolutionarily conserved, while rapidly evolving sites are variable. Retroviruses are excellent candidates for such analysis because the sequences of many members of this family are known.
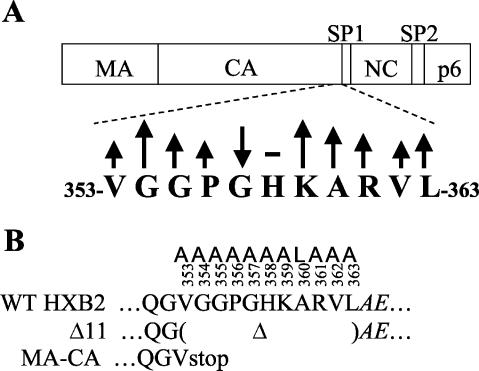
With this method, we analyzed more than 50 capsid sequences, including sequences from human, feline, and bovine immunodeficiency viruses, equine infectious anemia virus, caprine arthritis encephalitis virus, visna-related ovine lentivirus, Jaagsiekte sheep retrovirus, and endogenous retrovirus K-10 (Materials and Methods). This analysis highlighted evolutionarily conserved amino acids along the capsid sequence, such as the first proline residue and the aspartic acid residue with which it interacts upon folding (85), the major homology region (27), and the two cysteines that are bridged by a disulfide bond (27) (data not shown). In addition, we observed that the 11 amino acids at the C terminus of CA protein (positions 353 to 363, numbering beginning at the first methionine of Gag), which are disordered in the crystal structure of the CA protein (27), also scored with a relatively high degree of evolutionary conservation (Fig. 1A). Of these 11 amino acids, only one (glycine 357) was not conserved, while the other 10 residues scored an average to very high degree of conservation.
FIG. 1.
Evolutionary conservation and mutagenesis of the C terminus of the CA domain. (A) Schematic representation of the organization of HIV-1 Gag domains (upper drawing), and an expanded view of the 353-GVGGPGHKARVL-363 sequences located at the C terminus of the CA domain (lower drawing). Arrows indicate a high (↑), average (-), or low (↓) degree of conservation, and the size of the arrow schematically represents the approximate deviation from the average conservation score (Materials and Methods). The positions of the residues relative to the first methionine of Gag are indicated. (B) The structure of the CA C-terminal mutations. Substitution, deletion, and truncation mutations were introduced into the C terminus of the CA domain in Gag. Alanine and leucine substitutions are shown as A and L, respectively, above the numbered wild-type residues of the HXB2 clone (WT HXB2). The first two residues of SP1 domain are in italics. The in-frame deletion mutation (Δ11) consisting of the 11 C-terminal CA residues in Gag is indicated (Δ). A TAA (stop) codon was introduced downstream of the V353 residue to generate a truncated Gag protein (MA-CA).
Mutations of the conserved C-terminal residues of the HIV-1 CA domain impair virion production and infectivity.
To evaluate the role of the evolutionarily conserved CA C terminus in the HIV-1 replication cycle, we either deleted the terminal 11 amino acids of CA from the gag sequence to create the Δ11 mutant or changed each of the amino acids to alanine, while the alanine at position 360 was changed to leucine (Fig. 1B). Mutations were introduced into the gag open reading frame of the pHIVgptSVPA plasmid, generating V353A, G354A, G355A, P356A, G357A, H358A, K359A, A360L, R361A, V362A, and L363A mutants. The pHIVgptSVPA plasmid contains a derivative of the HIV-1 HXB2 clone carrying the SV40 promoter-gpt cassette and the SV40 polyadenylation signal, which replaced the env coding sequences and the 3′ LTR, respectively. The HIVgptSVPA construct produces virion particles from transfected cells (53).
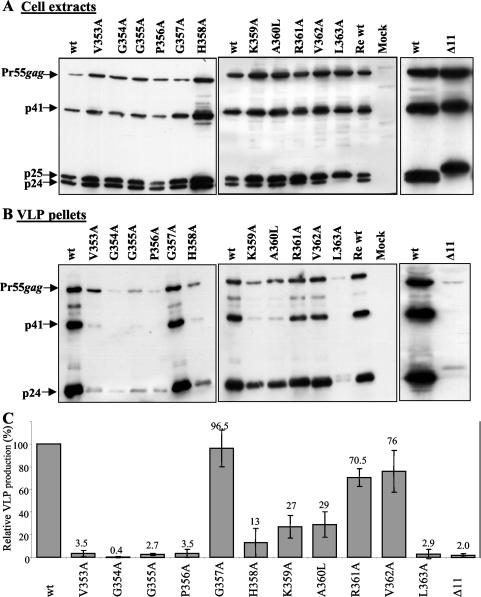
To test the mutants for Gag expression and for their ability to assemble and release VLPs, 293T cells were transfected with the wild-type or mutant proviral DNA. Two days posttransfection, VLPs from the culture supernatants were harvested by centrifugation through a 25% sucrose cushion, and cell lysates were prepared. Gag protein levels were assayed in cell lysates and in VLP pellets by Western blot analysis (Fig. 2). All mutants exhibited levels of Gag proteins similar to those in the wild-type strain in cell lysates. Gag processing was also normal for all of the mutants except the L363A and Δ11 mutants (Fig. 2A), which produced an abnormal ratio of p25 to p24 products, as expected for mutations that disrupt the CA-p2 cleavage site (41, 90).
FIG. 2.
VLP production by wild-type and mutant Gag proteins. Cell extracts from 293T cells transiently transfected with 10 μg of the indicated pHIVgptSVPA-based clone (A) and purified VLPs from the culture medium (B) were analyzed for Gag protein expression and cleavage by Western blot with an anti-CA monoclonal antibody. Mock samples indicate cells transfected without plasmid DNA. The wild type (wt) was the HIVgptSVPA clone expressing wild-type Gag proteins. Re wt designates an HIVgptSVPA clone into which the wild-type Gag sequence was introduced with the same cloning procedure that was used to construct the clones with the mutated gag sequences. This clone was used to ensure the absence of secondary mutations in the HIV sequence and the plasmid backbone that might affect viral protein expression. The migration positions of the Gag polyprotein (Pr55gag) and Gag cleavage products (p41, p25, and p24) are indicated with arrows. (C) VLP production by Gag protein mutants relative to that by the wild-type proteins was calculated after densitometry scanning and analysis of Gag protein bands (B). The data are presented as mean ± standard deviation (n = 2).
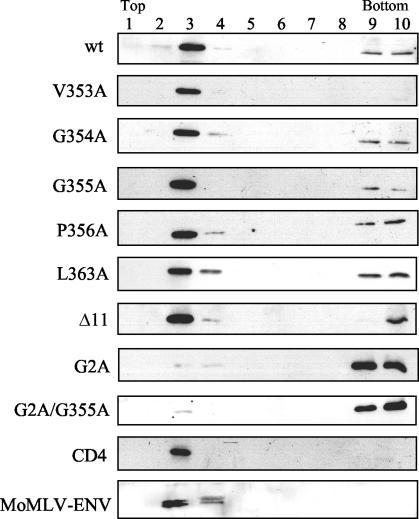
When VLP-associated Gag proteins were analyzed, a strong reduction in Gag levels was observed for the Δ11 mutant (Fig. 2B and C). Strong reduction in pelletable Gag protein levels was also obtained for mutants V353A, G354A, G355A, P356A, H358A, L363A, and Δ11. The level of expression ranged from approximately 0.5 to 15% of the wild-type levels. Mutants K359A and A360L showed a more moderate reduction of about threefold in pelletable Gag levels compared to the wild-type level, whereas mutants R361A and V362A had about 75% of the wild-type level. Mutant G357, which carries a substitution of the least conserved residue of the 11 residues tested, showed wild-type levels of virion-associated Gag proteins. Overall, mutations in the VGGP cluster reduced VLP-associated Gag levels to larger extents than did mutations in the GHKARVL sequence.
In principle, the reduction in the level of Gag proteins that was associated with VLPs may be the result of poor VLP production or instability of VLP mutants. To distinguish between these two possibilities, the total protein content of culture supernatants was precipitated with trichloroacetic acid, and the levels of the Gag proteins in the pellets were examined by Western blot. A reduction in the total Gag protein levels was observed in the trichloroacetic acid-extracted pellets, similar to the reduction that was obtained in the pellets of the sucrose-purified particles (data not shown). This indicates that the reduced level of the Gag protein mutants in the culture supernatants is the result of inefficient VLP production.
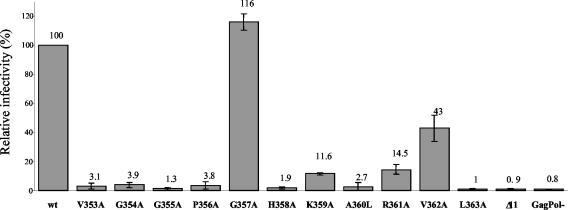
Further evidence that correlates with these findings was obtained from the single-cycle infection assay, measuring the transduction efficiencies of a GFP-containing retroviral vector coexpressed with wild-type or mutated Gag proteins. 293T cells were transfected with wild-type or mutated pHIVgptSVPA clones together with plasmids expressing a GFP retroviral vector and the vesicular stomatitis virus envelope G protein. Pseudotyped particles in equal volumes of culture supernatants were then used to infect naïve 293T cells, and the transduction efficiencies were calculated (Materials and Methods). This analysis revealed a strong reduction in the transduction levels (10% or less of wild-type levels; Fig. 3) for most of the mutants, including mutants V353A, G354A, G355A, P356A, H358A, K359A, A360L, L363A, and Δ11. The transduction levels of mutants R361A and V362A were approximately 15 and 45% of that of the wild type, while mutant G357A was as infectious as the wild type. These data demonstrate that mutations in the conserved C terminus of CA alter HIV-1 virion transduction and overall are in a good agreement with the recent analysis of some identical mutations reported by Liang and coworkers (48, 49).
FIG. 3.
Mutations of the conserved sequence at the CA C terminus reduce virus infectivity. (A) 293T cells were transfected with 7.5 μg of wild-type pHIVgptSVPA DNA or the indicated mutant together with 10 μg of HIV-1-based vector expressing the GFP marker (pHR′-CMV-GFP) and 2.5 μg of plasmid DNA expressing the vesicular stomatitis virus G envelope protein (pMD.G). Two days posttransfection, culture supernatants were used to infect naïve 293T cell. FACS analysis of transfected and infected cells was used to calculate and normalize the infectivity of the pseudotyped particles (Materials and Methods). Values are expressed as a percentage of wild-type infectivity. GagPol- indicates transfection of pHR′-CMV-GFP and pMD.G without pHIVgptSVPA plasmid DNA to control for transduction-independent expression of GFP in infected cells made by carryover of pHR′-CMV-GFP DNA. The data are presented as mean ± standard deviation (n = 3).
Pr55gag mutants show normal RNA binding and normal Gag-Gag interactions in yeast three- and two-hybrid systems.
The reduction in virion production and consequent decreased infectivity caused by the mutations in the C terminus of CA probably reflect impairment of particle assembly. As a series of complex interactions of the Gag proteins is crucial for proper virion assembly (23), we tested Pr55gag mutants for interactions that have been implicated in the assembly process. A high-affinity and specific binding of the NC domain of Gag to the HIV-1 RNA encapsidation signal (HIVΨ) is crucial for genome packaging. In addition, the NC domain is characterized by nonspecific RNA binding. The interaction of Gag with genomic RNA or other RNA molecules is thought to enhance the assembly process, either by supplying a nucleic acid scaffold for the assembled particle or by promoting Gag dimerization (13, 14, 16, 37, 54, 55, 60).
Since the CA mutations are localized in close proximity to the NC domain and might, in principle, alter Gag binding to RNA through indirect conformational changes, we tested the effect of these mutations on RNA recognition. To compare the RNA binding activity of wild-type Gag to that of the mutant Gag proteins, we used the yeast three-hybrid system. In the past, we demonstrated that this system provides a measure of the interaction of HIV-1 Gag with RNA that reflects the interactions seen in vitro and in vivo (5). In this system, wild-type HIV-1 Gag proteins bind with high affinity to RNA molecules that contain the HIV Ψ and show only weak nonspecific interaction with RNA molecules harboring the Harvey murine sarcoma virus encapsidation signal. Likewise, all the Gag protein CA mutants demonstrated strong binding to the HIV Ψ and only weak, nonspecific binding to the Harvey murine sarcoma virus encapsidation signal (data not shown). These results suggest that Gag-RNA interactions were not impaired by the CA mutations.
The CA domain is important for the oligomerization of Pr55gag, and several mutations in CA that damage Gag oligomerization and virion assembly have been described (48, 49, 70, 86). To test whether the Pr55gag mutants were able to participate in Gag-Gag interactions, we used the yeast two-hybrid system to measure the level of interactions between Gag molecules (51). The CA mutants were tested for both homodimerization (interaction between Gag proteins harboring the same mutation), and heterodimerization (interaction between mutant and wild-type Gag proteins). We found that all of the mutant Gag proteins homodimerized, and heterodimerized as efficiently as the wild-type molecules (data not shown). Overall, these results suggest that the CA mutations did not cause gross conformational changes in the Gag proteins and, importantly, did not abolish Gag-Gag interactions.
Intracytoplasmic multimerization of Gag protein mutants in mammalian cells.
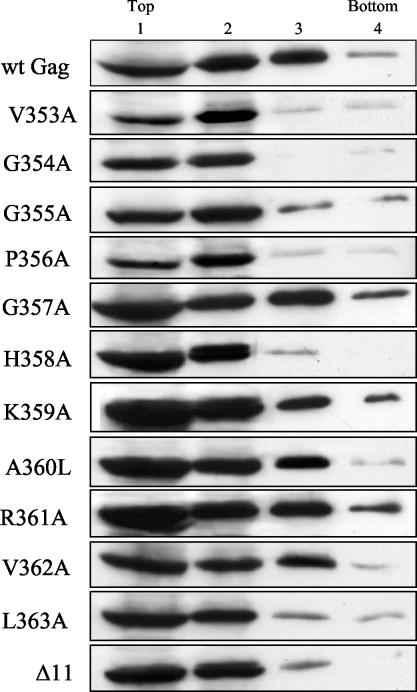
The retroviral assembly process involves multimerization of many Gag molecules to create higher-order structures. Because the analysis of the Gag protein mutants showed that many were defective in virion production, we were interested in assessing their ability to form aggregates in the cytoplasm. To test this, we examined the sedimentation profiles of cellular Gag complexes on nonequilibrium velocity gradients. 293T cells were transfected with pHIVgptSVPA or pHIVgptSVPA derivatives expressing the Gag mutants, and the cultures were grown in the presence of the HIV-1 protease inhibitor saquinavir for 2 days. Under these conditions, 20 μM saquinavir efficiently blocked Pr55gag cleavage (data not shown). The cells were then disrupted by Dounce homogenization, cell debris and nuclei were removed from the homogenate, and the resulting cleared extracts were subjected to velocity sedimentation through 25, 35, and 45% sucrose step gradients.
Western immunoblotting was performed on fractions of the sucrose gradients to detect the migration of the Gag complexes through the gradients (Fig. 4). Wild-type Gag protein complexes were abundant in the first three fractions of the gradient and could also be detected in the fourth-densest fraction. In contrast, the V353A, G354A, G355A, P356A, H358A, L363A, and Δ11 Gag proteins were mainly located at fractions composed of the cell lysate and the 25% sucrose cushion (fractions 1 and 2). For these mutants, only relatively small amounts of Gag proteins could be detected in the 35 and 45% sucrose fractions (fractions 3 and 4). The Gag proteins of mutants G357A, K359A, A360L, R361A, and V362A had migration profiles similar to that of the wild-type protein. This analysis demonstrated that mutants that are severely defective in particle production (Fig. 2C) also produce Gag protein aggregates in the cell cytoplasm that are abnormal in either size, mass, or shape.
FIG. 4.
Velocity sedimentation analysis of cytoplasmic Gag complexes. 293T cells were transfected with 5 μg of pHIVgptSVPA DNA expressing wild-type Gag (wt) or the indicated Gag mutants, and the cells were grown in the presence of saquinavir (20 μΜ). Two days posttransfection, the cells were subjected to Dounce homogenization, and Gag complexes from cytoplasmic extracts were separated by velocity sedimentation in a 25, 35, and 45% sucrose step gradient. Fractions (1 to 4) were collected from the gradient, and the presence of Gag precursors in each fraction was analyzed by Western blotting with an anti-CA monoclonal antibody. Top and Bottom designate the top and bottom gradient fractions, respectively.
Membrane association of Gag protein mutants.
Several of the mutants tested (V353A, G354A, G355A, P356A, L363A, and Δ11) showed both abnormal Gag aggregation and a dramatic reduction in VLP production. Notably, most of these point mutations are found in the predicted hinge region (353-VGGP-356) (49). The aberrant Gag complexes formed by these mutants might be defective in targeting to the cell surface membrane, as has been reported for identical mutants in a previous study (49). To test this, we compared the membrane association of wild-type Gag proteins to that of the mutant Gag proteins with the membrane flotation centrifugation assay as described before (62).
Postnuclear supernatants of cell homogenates made from Gag-expressing cells grown in the presence of saquinavir were subjected to equilibrium flotation centrifugation. Membrane-bound proteins were separated from non-membrane-bound proteins with sucrose step gradients, and unbound and membrane-bound Gag complexes were detected in the gradient fractions by Western blot analysis (Fig. 5). This analysis revealed that the majority of the wild-type Gag proteins were membrane bound and thus had floated to less-dense fractions (mainly to fraction 3), as reported by Ono and Freed (62). A similar migration pattern throughout the gradient was clearly demonstrated by positive controls made of membrane proteins CD4 and Moloney murine leukemia virus Env. Similarly, all of the Gag protein mutants tested floated to the same fractions, demonstrating efficient membrane binding.
FIG. 5.
Membrane binding of Gag protein mutants in the equilibrium flotation centrifugation assay. 293T cells were transfected with 5 μg of pHIVgptSVPA DNA expressing wild-type Gag (wt) or the indicated Gag mutants, and the cells were grown in the presence of saquinavir (20 μΜ). Two days posttransfection the cells were subjected to Dounce homogenization, and Gag proteins from cytoplasmic extracts were allowed to float through 73, 65, and 10% sucrose step gradients during an 18-h ultracentrifugation. Fractions (1 to 10) were collected from the gradient, and the presence of Gag precursors in each fraction was analyzed by Western blotting with an anti-CA monoclonal antibody. Top and Bottom designate the top and bottom gradient fractions, respectively.
We constructed a Gag protein mutant with a glycine-to-alanine substitution (G2A) in its myristylation site to serve as a negative control. The G2A mutation results in myristate-negative Gag proteins that do not associate with the cell surface membrane (33, 62, 63, 72, 76, 88) and do not efficiently float to the membrane-bound protein fraction in this assay (62). As expected, the majority of the G2A Gag protein mutants were retained in the denser portion of the sucrose gradient (fractions 9 and 10) and failed to float to the membrane-bound protein fractions. Likewise, a double mutant harboring both the G2A and the G355A mutations (G2A/G355A) did not migrate efficiently to the membrane-bound protein fraction.
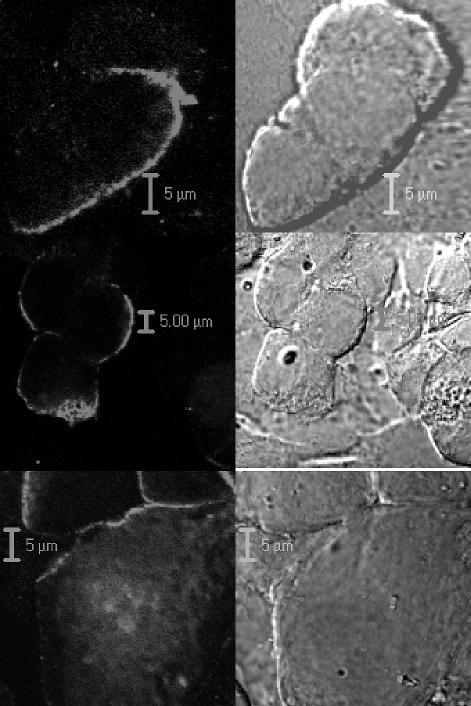
We further compared the ability of wild-type and mutant Gag proteins to target the plasma membrane with a immunohistochemistry procedure. HeLa cells were transfected with pHIVgptSVPA DNA expressing wild-type, V353A, or G355A Gag protein, and 2 days later the cells were fixed, immunostained, and examined with a confocal microscope (Fig. 6). We found that the majority (about 90%) of the cells that expressed the wild-type Gag proteins showed both cytoplasmic and membrane staining with an anti-CA monoclonal antibody (data not shown). About 10% of the Gag-expressing cells showed a clear membrane staining on a background of weak cytoplasmic staining (Fig. 6, upper panel). A similar pattern of plasma membrane staining was also detected for the G355A (Fig. 6, middle panel) and V353A (Fig. 6, lower panel) Gag protein mutants, consistent with previous data obtained with the membrane flotation centrifugation assay. These results suggest that the CA mutations that cause abnormal Gag aggregation and poor virion production did not impair plasma membrane targeting and binding, in contrast to what has been reported before for identical mutations (49).
FIG. 6.
Confocal analysis of Gag protein mutants. HeLa cells were transfected with 1 μg of pHIVgptSVPA DNA expressing wild-type (WT; upper panel), G355A (middle panel), or V355A (lower panel) Gag proteins. Two days later the cells were fixed, permeabilized, and stained with a monoclonal anti-HIV-1 capsid antibody and a secondary indocarbocyanine-conjugated antibody. Cells were examined with a confocal laser scan microscope. Fluorescence is shown on the left, and Nomarski images of the same field are shown on the right. Note the selective staining of only a portion of the cells. Similarly, no membrane staining was observed in control cells that were transfected without plasmid DNA (data not shown). Bars represent the indicated length.
Pr55gag mutants form aberrant structures at the plasma membrane.
The findings that the Gag complexes of the V353A, G354A, G355A, P356A, L363A, and Δ11 mutants targeted the cell surface membrane yet failed to produce significant amounts of VLPs suggest that late stages of the assembly process are impaired in these mutants. One possible explanation is that the aberrant Gag aggregates of these mutants are not efficiently released from the cell surface and are trapped in the plasma membrane. To test this hypothesis, we transfected pHIVgptSVPA DNA encoding wild-type or mutant Gag proteins and examined the cell surface membranes by scanning electronic microscopy (Fig. 7). Cells that expressed wild-type Gag proteins had relatively smooth membranes that resembled those of mock-transfected cells (Fig. 7A and B), with few cells showing spherical bulges with a diameter of about 0.1 μm (Fig. 7B). Similarly, cells that were transfected with pHIVgptSVPA G357A DNA, which encodes mutant Gag proteins that produce wild-type levels of VLPs (Fig. 2C), showed relatively smooth surfaces (Fig. 7C).
FIG. 7.
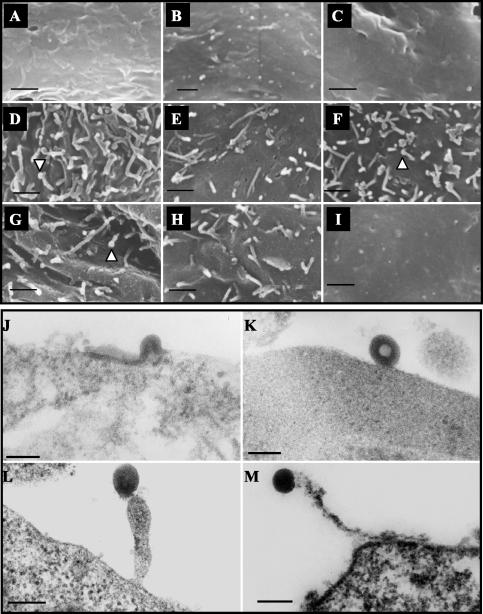
Electron microscopic analysis of assemblies of wild-type and Gag protein mutants. 293T cells were transfected with wild-type or mutant pHIVgptSVPA DNA clones; 48 h posttransfection, the cells were fixed and their plasma membrane was analyzed with a scanning electron microscope (upper panel, A to I), or the cells were pelleted, fixed, and embedded in Epon, and thin sections were stained with uranyl acetate and lead citrate for analysis with a transmission electron microscope (lower panel, J to M). The images represent cells transfected with no DNA (A), wild-type (B, J, and K), G357A (C), V353A (D, L, and M), G354A (E), G355A (F), L363A (G), Δ11 (H), or G2A/G355A (I). Arrowheads point to rod-like structures with swollen rounded ends. Bars: 1 μm (A to I); 100 nm (J and K); and 200 nm (L and M).
In contrast, cells that were transfected with pHIVgptSVPA-V353A, G354A, G355A, L363A, or Δ11 DNA showed excessive rod-like structures protruding from the cell surface (Fig. 7D to H), and these structures were readily detected on the majority of the cultured cells. Similar results were obtained for the P356A mutant (data not shown). We also noted that some of these rod-like structures appeared to have swollen rounded ends (Fig. 7D, F, and G). As a control for this experiment, we transfected the cells with pHIVgptSVPA-G2A/G355A DNA, which expresses myristate-negative Gag proteins. These cells did not show the typical rod-like structures that were present on cells expressing the parental G355A mutant (compare Fig. 7F to 7I), demonstrating that the formation of these shapes was indeed dependent on targeting of the plasma membrane by Gag proteins.
To further analyze these rod-like structures, we expressed wild-type or V353A mutant Gag proteins in 293T cells and examined the plasma membrane by transmission electron microscopy. Whereas wild-type proteins gave rise to immature budding structures that assembled on the cell surface (Fig. 7J and K), the V353A Gag protein mutants formed dense aggregates that appeared to be trapped in structures protruding from the plasma membrane (Fig. 7L and M). These results demonstrate that the mutant Gag proteins are assembled in a way that restricts normal budding.
DISCUSSION
We characterized the phenotypes of mutations localized to the last 11 amino acids of the HIV-1 capsid domain (VGGPGHKARVL). The results presented here demonstrate that these conserved residues define a region that is highly sensitive to the introduction of point mutations, as 10 of 11 mutants were defective in their infectivity.
The results from our experiments define two categories of mutants. One set of mutants were characterized by a drastic reduction in particle production, while the other set of mutants produced significant levels of virions but showed low infectivity levels. Of the 11 amino acid substitutions, those that changed the 353-VGGP-356 cluster or the last leucine residue in the capsid (L363) caused a strong reduction in particle production. For these mutants, the decline in virion production could account for their impaired infectivity. Yet, for mutants that harbored mutations in the 358-HKARV-362 cluster, the reduction in VLP production could not fully explain the reduction in the infectivity levels. For example, compared to the wild type, the H358A, A360L, and R361A mutants produced about 15, 30, and 70% of the wild-type particle levels but were only 2, 3, and 15% as infectious, respectively. These mutants probably possess additional defects, perhaps in the early stages of infection, and it will be of interest to evaluate the contribution of this 11-amino-acid cluster to the function of the capsid protein in this context.
We found a good correlation between normal Gag aggregation in the cytoplasm and virion production. The G357A, K359A, A360L, R361A, and V362A mutants, which were characterized by moderate to normal particle production, generated cytoplasmic Gag aggregates that migrated like wild-type complexes in sucrose gradients. In contrast, the V353A, G354A, G355A, P356, H358A, L363A and Δ11 mutants, which produced low particle levels, generated cytoplasmic Gag complexes that were poor in penetrating the denser fractions of the sucrose gradient in velocity sedimentation analysis. This indicated that some of the CA mutations altered Gag-Gag interactions, resulting in Gag complexes that were abnormal in size, mass, or shape and that could not be released as virions. Yet even Gag mutants that failed to assemble into normal Gag complexes in the cell cytoplasm were all able to form Gag-Gag contacts that resulted in high levels of β-galactosidase activity in the yeast two-hybrid system, similar to the readout obtained by contacts of the wild-type proteins. These data suggest that no overt conformational changes to the Gag molecules have been induced by the CA mutations, as wild-type RNA binding activity was maintained by these mutants.
The V353A, G354A, G355A, P356, L363A and Δ11 Gag protein mutants, which assembled into defective cytoplasmic complexes and produced very low levels of particles, efficiently bound to the cell surface membrane. This was evident biochemically by the flotation of these Pr55gag mutants to the membrane-bound protein fraction in membrane flotation centrifugation assays. Similar flotation profiles were obtained for membrane-associated proteins such as the CD4 and Env molecules as well as for wild-type Gag but not for myristylation-negative Gag proteins. Clear localization of the mutated Gag proteins to the cell surface membrane was also detected by confocal microscopy. These results clearly demonstrate that the defective Gag complexes were still able to traffic and target to the cell surface membrane.
Recently, Liang and coworkers described the phenotypes of a similar set of mutations in the HIV-1 capsid (48, 49). A good correlation exists between the results reported by them and the data presented here regarding the formation of aberrant Gag complexes and low particle production of viruses with mutations in the VGGP sequence. However, these investigators reported that the Gag proteins of these mutants did not target the cell surface membrane efficiently and thus concluded that this defect is the major cause of the severe reduction in virus production. Our results are in clear contrast to these findings and suggest a different explanation for the poor production of particles (see below).
Although we do not know the exact reasons for the different results observed, differences in experimental conditions may have contributed to the variation. For example, the pHIVgptSVPA clone used in this study carries an SV40 origin of replication in the plasmid backbone, which allows high levels of expression in T-antigen-containing 293T cells. This may result in a clearer appearance of Gag assemblies on the plasma membrane. In addition, Liang and coworkers used monkey cells in both the membrane flotation assay and the electron microscopic analysis, in contrast to the human cells that were used in the current study.
Since the movement of cytoplasmic Gag molecules towards the plasma membrane is poorly understood, it is conceivable that factors that exist in human cells assist Gag trafficking more efficiently than factors in monkey cells. While wild-type proteins assemble and bud normally, abnormal Gag aggregates, like the ones formed by the hinge mutants, may not be efficiently targeted to the plasma membrane in simian cells. In line with this hypothesis, recent studies have demonstrated that the different intracellular milieu of monkey and human cells can greatly influence retroviral replication; factors in monkey cells appear to restrict viral replication through unknown interactions with the capsid protein (reviewed in reference 31). Nevertheless, our results are in agreement with the findings of Ono and Freed that showed that a Pr55gag mutant which contained only the MA domain and the N-terminal domain of the capsid exhibited wild-type levels of membrane binding (61).
The finding that the Gag complexes of the V353A, G354A, G355A, P356A, L363A, and Δ11 mutants were targeted to the cell surface membrane yet failed to produce significant amounts of VLPs suggests that the aberrant Gag aggregates accumulated at the plasma membrane. Indeed, scanning electron microscopy analysis of cells expressing the multimerization-defective Gag mutants readily identified the accumulation of rod-like structures protruding from the cell surface. These structures could not be detected on cells expressing the myristylation-negative Gag double mutant G2A/G355A, in contrast to the abundance of such structures on cells that expressed the G355A Gag mutant. Since Gag myristylation is necessary for stable membrane binding and assembly (11, 33, 71), this result indicates that the appearance of the rod-like structures is indeed dependent on the expression of the mutant Gag proteins and their ability to reach the cell surface membrane.
The scanning electron microscopy analysis only rarely detected assembled particle structures on the surface of cells expressing the wild-type Gag proteins or the assembly-competent G357A Gag mutant, perhaps because wild-type or wild-type-like particles bud quickly from the cell surface. In the cases where budding structures of wild-type Gag proteins could be observed, they had a spherical appearance, easily distinguishable from the tubular organization exhibited by the mutant Gag proteins. The appearance of tubular structures rather than spheres has also been reported for other Gag mutants of HIV-1 and other retroviruses (4, 29, 33, 35, 38, 42, 59, 92). In addition, the swollen rounded tips of some of these rod-like structures is reminiscent of structures that were reported for an HIV-1 mutant in which the two residues that flank the CA-SP1 cleavage site were mutated (33).
Transmission electron microscopy analysis of the rod-like structures made by the V353A mutant showed that these structures indeed contained dense assemblies that seemed to be trapped on the inner side of the plasma membrane. Notably, these structures appeared to have denser centers than the wild-type immature particles. Overall, our results suggest that Gag proteins with mutations in the conserved amino acids at the C-terminal edge of the CA domain can form aberrant complexes that are targeted to the cell surface membrane but defective in budding.
What structure might be affected by the mutations described here? As mentioned above, the last 11 amino acids of the free capsid protein are disordered in the crystals (27), and the structure of the CA-p2 region, which is more relevant for the assembly of Gag precursors, has not been determined yet. Nonetheless, calculations based on amino acid composition have predicted two putative structural units which divide the 353-VGGPGHKARVL-363 sequence. The 353-VGGP-356 portion has been suggested to form a flexible hinge (together with an additional glycine residue at position 352) (49), while the 357-GHKARVL-363 sequence has been suggested to be part of a putative α-helix that extends into SP1 (1).
Previous reports showed that mutations in the SP1 section of this α-helix caused defects in particle assembly and budding (1, 33, 41, 48, 59). The L363A mutation caused the most severe reduction in particle production compared to the other mutations that we generated in the CA portion of the putative α-helix. The L363A Gag protein generated aberrant Gag aggregates in the cytoplasm and aberrant assemblies on the cell surface. This result is consistent with the previous suggestion, made by Gottlinger, that the CA-p2 boundary is primarily required for the induction of membrane curvature, as mutations that disrupt the putative α-helix generate grossly aberrant budding structures (32). Our data also correlate well with the results published by Liang and coworkers, demonstrating the importance of the putative α-helix to virus production, infectivity, and Gag multimerization (48). These authors showed that the H358A and L363A mutations damaged the formation of Gag complexes, and our velocity sedimentation analysis revealed similar results. In addition, we found that both mutations did not alter the Gag-membrane association, as determined by the membrane flotation centrifugation assay. However, the L363A Gag mutants appeared to form rod-like structures on the cell surface, while no such aberrant assemblies could be detected for the H358A Gag mutants (data not shown). The reason for this difference is not clear, but it may be explained by the fact that the H358A Gag mutants were released to the culture supernatants in higher quantities (13% of wild-type levels) than the L363A Gag mutants (3% of wild-type levels).
The dramatic reduction in particle formation made by single alanine substitutions in the 353-VGGP-356 putative hinge region highlights the importance of this cluster to the assembly process. The proline-to-alanine substitution in this sequence was also recently shown to drastically reduce virion production and to alter particle morphology (86). Mutations in this sequence might hamper its putative flexibility (49), restricting possible changes in Pr55gag conformation that are important for Gag lattice interactions and virion assembly (86). In addition, this region might interact with cellular factors that are important for efficient assembly and release of virus particles, similar to the function played by L assembly domains in other portions of the retroviral Gag precursors (3, 25, 67, 79).
The assembly of an infectious particle is a step unique to the virus, and hence drugs that operate to interfere with this process are of great interest. Indeed, antiviral compounds that target capsid proteins of picornaviruses (7) and of hepatitis B virus (19) have been developed. Recently, drugs that bind the core domain of HIV-1 CA (82) or inhibit CA-SP1 cleavage (46) have been identified and shown to inhibit viral replication in tissue culture. In addition, a truncated version of the nucleolin protein that binds the CA-NC region in the Moloney murine leukemia virus Gag protein has been found to strongly suppress virion assembly (6).
In this paper, we demonstrated the importance of the C terminus of the HIV-1 CA domain to the assembly and release of infectious particles. The strong suppression of these processes by even subtle changes in the CA C terminus implies that the development of drugs that target this region might be beneficial in suppressing HIV-1 replication.
Acknowledgments
We thank Stephen Goff, Jeremy Luban, Marvin Wickens, Moshe Kotler, and Jonathan Gershoni for generously providing various reagents. We also thank Ruti Herzog for technical assistance, Alexander Barbul and Orit Sagi-Assif for confocal microscopy work and FACS analysis, respectively, and Yacov Delarea for scanning electron microscopy and transmission electron microscopy analysis.
This research was supported by the German-Israeli Foundation for Scientific Research and Development Young Investigator Program, by the Israel Science Foundation, and by the Jakov, Marianna and Jorge Saia Scholarship Fund for HIV and Parkinson Diseases Research.
REFERENCES
- 1.Accola, M. A., S. Hoglund, and H. G. Gottlinger. 1998. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amara, A., and D. R. Littman. 2003. After Hrs with HIV. J. Cell Biol. 162:371-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arvidson, B., J. Seeds, M. Webb, L. Finlay, and E. Barklis. 2003. Analysis of the retrovirus capsid interdomain linker region. Virology 308:166-177. [DOI] [PubMed] [Google Scholar]
- 5.Bacharach, E., and S. P. Goff. 1998. Binding of the human immunodeficiency virus type 1 Gag protein to the viral RNA encapsidation signal in the yeast three-hybrid system. J. Virol. 72:6944-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bacharach, E., J. Gonsky, K. Alin, M. Orlova, and S. P. Goff. 2000. The carboxy-terminal fragment of nucleolin interacts with the nucleocapsid domain of retroviral Gag proteins and inhibits virion assembly. J. Virol. 74:11027-11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Badger, J., I. Minor, M. J. Kremer, M. A. Oliveira, T. J. Smith, J. P. Griffith, D. M. Guerin, S. Krishnaswamy, M. Luo, M. G. Rossmann, et al. 1988. Structural analysis of a series of antiviral agents complexed with human rhinovirus 14. Proc. Natl. Acad. Sci. USA 85:3304-3308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berezin, C., F. Glaser, J. Rosenberg, I. Paz, T. Pupko, P. Fariselli, R. Casadio, and N. Ben-Tal. 2004. ConSeq: the identification of functionally and structurally important residues in protein sequences. Bioinformatics 20:1322-1324. [DOI] [PubMed] [Google Scholar]
- 9.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 10.Borsetti, A., A. Ohagen, and H. G. Gottlinger. 1998. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J. Virol. 72:9313-9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryant, M., and L. Ratner. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc. Natl. Acad. Sci. USA 87:523-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buckman, J. S., W. J. Bosche, and R. J. Gorelick. 2003. Human immunodeficiency virus type 1 nucleocapsid Zn2+ fingers are required for efficient reverse transcription, initial integration processes, and protection of newly synthesized viral DNA. J. Virol. 77:1469-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Hum. immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Callahan, E. M., and J. W. Wills. 2003. Link between genome packaging and rate of budding for Rous sarcoma virus. J. Virol. 77:9388-9398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Casella, C. R., L. J. Raffini, and A. T. Panganiban. 1997. Pleiotropic mutations in the HIV-1 matrix protein that affect diverse steps in replication. Virology 228:294-306. [DOI] [PubMed] [Google Scholar]
- 16.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawson, L., and X. F. Yu. 1998. The role of nucleocapsid of HIV-1 in virus assembly. Virology 251:141-157. [DOI] [PubMed] [Google Scholar]
- 18.Derdowski, A., L. Ding, and P. Spearman. 2004. A novel fluorescence resonance energy transfer assay demonstrates that the human immunodeficiency virus type 1 Pr55Gag I domain mediates Gag-Gag interactions. J. Virol. 78:1230-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deres, K., C. H. Schroder, A. Paessens, S. Goldmann, H. J. Hacker, O. Weber, T. Kramer, U. Niewohner, U. Pleiss, J. Stoltefuss, E. Graef, D. Koletzki, R. N. Masantschek, A. Reimann, R. Jaeger, R. Gross, B. Beckermann, K. H. Schlemmer, D. Haebich, and H. Rubsamen-Waigmann. 2003. Inhibition of hepatitis B virus replication by drug-induced depletion of nucleocapsids. Science 299:893-896. [DOI] [PubMed] [Google Scholar]
- 20.Dorfman, T., A. Bukovsky, A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Functional domains of the capsid protein of human immunodeficiency virus type 1. J. Virol. 68:8180-8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorfman, T., J. Luban, S. P. Goff, W. A. Haseltine, and H. G. Gottlinger. 1993. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J. Virol. 67:6159-6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 23.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 24.Freed, E. O. 2001. HIV-1 replication. Somat. Cell Mol. Genet. 26:13-33. [DOI] [PubMed] [Google Scholar]
- 25.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 27.Gamble, T. R., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 28.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 29.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 30.Gitti, R. K., B. M. Lee, J. Walker, M. F. Summers, S. Yoo, and W. I. Sundquist. 1996. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science 273:231-235. [DOI] [PubMed] [Google Scholar]
- 31.Goff, S. P. 2004. HIV: replication trimmed back. Nature 427:791-793. [DOI] [PubMed] [Google Scholar]
- 32.Gottlinger, H. G. 2001. The HIV-1 assembly machine. AIDS 15(Suppl. 5):S13-S20. [DOI] [PubMed] [Google Scholar]
- 33.Gottlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gross, I., H. Hohenberg, T. Wilk, K. Wiegers, M. Grattinger, B. Muller, S. Fuller, and H. G. Krausslich. 2000. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 19:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hockley, D. J., M. V. Nermut, C. Grief, J. B. Jowett, and I. M. Jones. 1994. Comparative morphology of Gag protein structures produced by mutants of the gag gene of human immunodeficiency virus type 1. J. Gen. Virol. 75:2985-2997. [DOI] [PubMed] [Google Scholar]
- 36.Jenkins, Y., O. Pornillos, R. L. Rich, D. G. Myszka, W. I. Sundquist, and M. H. Malim. 2001. Biochemical analyses of the interactions between human immunodeficiency virus type 1 Vpr and p6Gag. J. Virol. 75:10537-10542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson, M. C., H. M. Scobie, Y. M. Ma, and V. M. Vogt. 2002. Nucleic acid-independent retrovirus assembly can be driven by dimerization. J. Virol. 76:11177-11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joshi, S. M., and V. M. Vogt. 2000. Role of the Rous sarcoma virus p10 domain in shape determination of gag virus-like particles assembled in vitro and within Escherichia coli. J. Virol. 74:10260-10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kiernan, R. E., A. Ono, G. Englund, and E. O. Freed. 1998. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J. Virol. 72:4116-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kondo, E., F. Mammano, E. A. Cohen, and H. G. Gottlinger. 1995. The p6gag domain of human immunodeficiency virus type 1 is sufficient for the incorporation of Vpr into heterologous viral particles. J. Virol. 69:2759-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krausslich, H. G., M. Facke, A. M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuznetsov, Y. G., S. Datta, N. H. Kothari, A. Greenwood, H. Fan, and A. McPherson. 2002. Atomic force microscopy investigation of fibroblasts infected with wild-type and mutant murine leukemia virus (MuLV). Biophys. J. 83:3665-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Landau, N. R., K. A. Page, and D. R. Littman. 1991. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J. Virol. 65:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, P. P., and M. L. Linial. 1994. Efficient particle formation can occur if the matrix domain of human immunodeficiency virus type 1 Gag is substituted by a myristylation signal. J. Virol. 68:6644-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee, Y. M., B. Liu, and X. F. Yu. 1999. Formation of virus assembly intermediate complexes in the cytoplasm by wild-type and assembly-defective mutant human immunodeficiency virus type 1 and their association with membranes. J. Virol. 73:5654-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li, F., R. Goila-Gaur, K. Salzwedel, N. R. Kilgore, M. Reddick, C. Matallana, A. Castillo, D. Zoumplis, D. E. Martin, J. M. Orenstein, G. P. Allaway, E. O. Freed, and C. T. Wild. 2003. PA-457: a potent HIV inhibitor that disrupts core condensation by targeting a late step in Gag processing. Proc. Natl. Acad. Sci. USA 100:13555-13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 48.Liang, C., J. Hu, R. S. Russell, A. Roldan, L. Kleiman, and M. A. Wainberg. 2002. Characterization of a putative alpha-helix across the capsid-SP1 boundary that is critical for the multimerization of human immunodeficiency virus type 1 Gag. J. Virol. 76:11729-11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liang, C., J. Hu, J. B. Whitney, L. Kleiman, and M. A. Wainberg. 2003. A structurally disordered region at the C terminus of capsid plays essential roles in multimerization and membrane binding of the gag protein of human immunodeficiency virus type 1. J. Virol. 77:1772-1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu, Y. L., P. Spearman, and L. Ratner. 1993. Hum. immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J. Virol. 67:6542-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luban, J., K. B. Alin, K. L. Bossolt, T. Humaran, and S. P. Goff. 1992. Genetic assay for multimerization of retroviral Gag polyproteins. J. Virol. 66:5157-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Hum. immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 53.Luban, J., and S. P. Goff. 1994. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J. Virol. 68:3784-3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma, Y. M., and V. M. Vogt. 2004. Nucleic acid binding-induced Gag dimerization in the assembly of Rous sarcoma virus particles in vitro. J. Virol. 78:52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ma, Y. M., and V. M. Vogt. 2002. Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J. Virol. 76:5452-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mammano, F., A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J. Virol. 68:4927-4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McDermott, J., L. Farrell, R. Ross, and E. Barklis. 1996. Structural analysis of human immunodeficiency virus type 1 Gag protein interactions, using cysteine-specific reagents. J. Virol. 70:5106-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Momany, C., L. C. Kovari, A. J. Prongay, W. Keller, R. K. Gitti, B. M. Lee, A. E. Gorbalenya, L. Tong, J. McClure, L. S. Ehrlich, M. F. Summers, C. Carter, and M. G. Rossmann. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 59.Morikawa, Y., D. J. Hockley, M. V. Nermut, and I. M. Jones. 2000. Roles of matrix, p2, and N-terminal myristoylation in human immunodeficiency virus type 1 Gag assembly. J. Virol. 74:16-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ono, A., D. Demirov, and E. O. Freed. 2000. Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding. J. Virol. 74:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ono, A., and E. O. Freed. 1999. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J. Virol. 73:4136-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ono, A., J. M. Orenstein, and E. O. Freed. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J. Virol. 74:2855-2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Page, K. A., N. R. Landau, and D. R. Littman. 1990. Construction and use of a human immunodeficiency virus vector for analysis of virus infectivity. J. Virol. 64:5270-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Paxton, W., R. I. Connor, and N. R. Landau. 1993. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J. Virol. 67:7229-7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pettit, S. C., M. D. Moody, R. S. Wehbie, A. H. Kaplan, P. V. Nantermet, C. A. Klein, and R. Swanstrom. 1994. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 68:8017-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pornillos, O., J. E. Garrus, and W. I. Sundquist. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12:569-579. [DOI] [PubMed] [Google Scholar]
- 68.Pupko, T., R. E. Bell, I. Mayrose, F. Glaser, and N. Ben-Tal. 2002. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics 18(Suppl. 1):S71-S77. [DOI] [PubMed] [Google Scholar]
- 69.Reicin, A. S., A. Ohagen, L. Yin, S. Hoglund, and S. P. Goff. 1996. The role of Gag in human immunodeficiency virus type 1 virion morphogenesis and early steps of the viral life cycle. J. Virol. 70:8645-8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reicin, A. S., S. Paik, R. D. Berkowitz, J. Luban, I. Lowy, and S. P. Goff. 1995. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J. Virol. 69:642-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rein, A., M. R. McClure, N. R. Rice, R. B. Luftig, and A. M. Schultz. 1986. Myristylation site in Pr65gag is essential for virus particle formation by Moloney murine leukemia virus. Proc. Natl. Acad. Sci. USA 83:7246-7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Royer, M., S. S. Hong, B. Gay, M. Cerutti, and P. Boulanger. 1992. Expression and extracellular release of human immunodeficiency virus type 1 Gag precursors by recombinant baculovirus-infected cells. J. Virol. 66:3230-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scarlata, S., and C. Carter. 2003. Role of HIV-1 Gag domains in viral assembly. Biochim. Biophys. Acta 1614:62-72. [DOI] [PubMed] [Google Scholar]
- 74.Shehu-Xhilaga, M., H. G. Kraeusslich, S. Pettit, R. Swanstrom, J. Y. Lee, J. A. Marshall, S. M. Crowe, and J. Mak. 2001. Proteolytic processing of the p2/nucleocapsid cleavage site is critical for human immunodeficiency virus type 1 RNA dimer maturation. J. Virol. 75:9156-9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shioda, T., and H. Shibuta. 1990. Production of human immunodeficiency virus (HIV)-like particles from cells infected with recombinant vaccinia viruses carrying the gag gene of HIV. Virology 175:139-148. [DOI] [PubMed] [Google Scholar]
- 76.Spearman, P., J. J. Wang, N. Vander Heyden, and L. Ratner. 1994. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J. Virol. 68:3232-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Srinivasakumar, N., M. L. Hammarskjold, and D. Rekosh. 1995. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J. Virol. 69:6106-6114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Srivastava, M., M. Cartas, T. A. Rizvi, S. P. Singh, D. Serio, V. S. Kalyanaraman, H. B. Pollard, and A. Srinivasan. 1999. HIV-1 Gag shares a signature motif with annexin (Anx7), which is required for virus replication. Proc. Natl. Acad. Sci. USA 96:2704-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Strack, B., A. Calistri, S. Craig, E. Popova, and H. G. Gottlinger. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689-699. [DOI] [PubMed] [Google Scholar]
- 80.Strack, B., A. Calistri, and H. G. Gottlinger. 2002. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J. Virol. 76:5472-5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 82.Tang, C., E. Loeliger, I. Kinde, S. Kyere, K. Mayo, E. Barklis, Y. Sun, M. Huang, and M. F. Summers. 2003. Antiviral inhibition of the HIV-1 capsid protein. J. Mol. Biol. 327:1013-1020. [DOI] [PubMed] [Google Scholar]
- 83.Tang, S., T. Murakami, N. Cheng, A. C. Steven, E. O. Freed, and J. G. Levin. 2003. Hum. immunodeficiency virus type 1 N-terminal capsid mutants containing cores with abnormally high levels of capsid protein and virtually no reverse transcriptase. J. Virol. 77:12592-12602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Thali, M., A. Bukovsky, E. Kondo, B. Rosenwirth, C. T. Walsh, J. Sodroski, and H. G. Gottlinger. 1994. Functional association of cyclophilin A with HIV-1 virions. Nature 372:363-365. [DOI] [PubMed] [Google Scholar]
- 85.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.von Schwedler, U. K., K. M. Stray, J. E. Garrus, and W. I. Sundquist. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 77:5439-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.von Schwedler, U. K., M. Stuchell, B. Muller, D. M. Ward, H. Y. Chung, E. Morita, H. E. Wang, T. Davis, G. P. He, D. M. Cimbora, A. Scott, H. G. Krausslich, J. Kaplan, S. G. Morham, and W. I. Sundquist. 2003. The protein network of HIV budding. Cell 114:701-713. [DOI] [PubMed] [Google Scholar]
- 88.Wang, C. T., and E. Barklis. 1993. Assembly, processing, and infectivity of human immunodeficiency virus type 1 gag mutants. J. Virol. 67:4264-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang, C. T., H. Y. Lai, and J. J. Li. 1998. Analysis of minimal human immunodeficiency virus type 1 gag coding sequences capable of virus-like particle assembly and release. J. Virol. 72:7950-7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, and H. G. Krausslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yu, X. F., Z. Matsuda, Q. C. Yu, T. H. Lee, and M. Essex. 1995. Role of the C terminus Gag protein in human immunodeficiency virus type 1 virion assembly and maturation. J. Gen. Virol. 76:3171-3179. [DOI] [PubMed] [Google Scholar]
- 92.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang, Y., and E. Barklis. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 71:6765-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]