Abstract
Rift Valley fever virus (RVFV) is an important cause of epizootics and epidemics in Africa and a potential agent of bioterrorism. A better understanding of the factors that govern RVFV virulence and pathogenicity is required, given the urgent need for antiviral therapies and safe vaccines. We have previously shown that RVFV strains with mutations in the NSs gene are excellent inducers of α/β interferon (IFN-α/β) and are highly attenuated in mice. Here, we demonstrate that NSs is sufficient to block IFN-β gene expression at the transcriptional level. In cells transiently expressing NSs, IFN-β transcripts were not inducible by viral infection or by transfection of poly(I:C). NSs with anti-IFN activity accumulated in the nucleus. In contrast, mutant forms of NSs that had lost their IFN-inhibiting activity remained in the cytoplasm, indicating that nuclear localization plays a role. IFN synthesis is regulated by specific transcription factors, including interferon regulatory factor (IRF-3), NF-κB, and AP-1. In the presence of NSs, IRF-3 was still activated and moved to the nucleus. Likewise, NF-κB and AP-1 were activated normally, as shown in electrophoretic mobility shift assays. Moreover, NSs was found to inhibit transcriptional activity of a constitutive promoter, in agreement with recent findings showing that NSs targets the basal cellular transcription factor TFIIH. The present results suggest that NSs, unlike other viral IFN antagonists, does not inhibit IFN-specific transcription factors but blocks IFN gene expression at a subsequent step.
Interferons (IFNs) constitute a first line of host defense against viral infections. IFN-α/β is induced by virus or by double-stranded RNA (dsRNA) in many cell types, whereas IFN-γ is produced by activated T cells and natural killer cells. To overcome the antiviral response, viruses have evolved various strategies to inhibit either IFN production, IFN signaling, or IFN action (13, 16). The efficiency by which a virus antagonizes the IFN system is critical for its pathogenicity and its ability to infect and spread in the host organism.
Rift Valley fever virus (RVFV), a phlebovirus, belongs to the family Bunyaviridae. It is transmitted by mosquitoes (11) and infects a wide range of vertebrate hosts. In humans, infection can lead to fatal hepatitis with hemorrhagic fever and encephalitis. In cattle, infection generally causes death in young animals and abortion and teratogenesis in pregnant females (14, 30). The disease is endemic in many countries of sub-Saharan Africa and in Egypt, where it repeatedly provokes serious epizootics and concomitant epidemics. Recently, simultaneous outbreaks occurred in Yemen and Saudi Arabia (9, 10). This was the first time that RVFV manifested itself outside of Africa, raising fears that the virus will emerge in new areas.
RVFV has a tripartite single-stranded RNA genome consisting of L, M, and S segments. The L and M segments are of negative polarity and code for the RNA-dependent RNA polymerase L, the precursor of the glycoproteins Gn and Gc, and the nonstructural protein NSm, respectively. The S segment of RVFV uses an ambisense strategy to code for the nucleoprotein N in the antigenomic sense and the nonstructural protein NSs in the genomic sense (15). NSs of RVFV is a 31-kDa protein which is phosphorylated by casein kinase II at two serine residues located in the carboxy terminus (20). It accumulates in the nuclei of infected cells, where it forms filamentous structures (38). A carboxy-terminal domain mediates oligomerization and is responsible for filament formation (46). The nuclear localization of NSs is intriguing because all steps of the viral life cycle are known to occur in the cytoplasm.
Clone 13 is a clonal isolate of RVFV with a large in-frame deletion in the S segment which leads to a truncated NSs protein (32). This virus is highly attenuated and immunogenic in IFN-competent mice (8, 32, 41). However, clone 13 was found to be highly virulent in IFN-α/β-nonresponsive mice, pointing to a crucial role of IFN-α/β in the attenuation phenotype (8). In addition, clone 13 proved to be an excellent inducer of early IFN-α/β production in vivo. In contrast, the virulent strain ZH548 failed to induce detectable amounts of IFN-α/β and replicated extensively in both IFN-competent and IFN-defective mice. Analyses of reassortant viruses with the S segment of either clone 13 or ZH548 provided strong evidence that NSs is an IFN-α/β antagonist in vivo (8). Comparisons of the S segment sequence of clone 13 with that of ZH548 revealed minor differences in addition to the large deletion in the NSs gene. These correspond to a single amino acid change (glycine to glutamic acid) at position 159 in the N protein sequence and six nucleotide changes in the intergenic region (8).
Here we show that NSs is sufficient for suppression of IFN induction but does not interfere with the activation of IFN-specific transcription factors. Furthermore, our data demonstrate that the inhibitory effect of NSs is not restricted to IFN gene expression but affects constitutive promoters as well, in agreement with recent findings demonstrating that NSs targets a basal cellular transcription factor (23).
MATERIALS AND METHODS
Cells and viruses.
Vero cells, murine fibroblasts derived from BALB/c mice (BF cells), kindly provided by Stephen Goodbourn, St. George's Hospital Medical School, London, United Kingdom, and 293 cells were grown as described (41). Stocks of the RVFV strain ZH548 and clone 13 were produced by infecting Vero cells with 0.01 PFU per cell. The reassortants were obtained after coinfection of cells with ZH548 and clone 13 as described previously (40).
Plasmids for NSs expression.
Plasmids were constructed by standard procedures, and the correct sequences were confirmed by restriction enzyme analysis and sequencing (2). The sequence coding for NSs (positions 34 to 836 of the genomic sense RNA) was reverse transcribed and amplified by reverse transcription (RT)-PCR from RNAs extracted from Vero cells infected with ZH548 or clone 13 by using primers already described (46). The cDNA fragment was ligated to plasmid pCI (Promega) downstream of the human cytomegalovirus (CMV) immediate-early enhancer/promoter at the unique NheI and KpnI sites in the polylinker. This resulted in plasmids pCI-NSsZH548 and pCI-NSsC13, respectively. The NSs mutated sequences inserted into plasmids pCI-NSs(S252A), pCI-NSs(S256A), and pCI-NSs(S252A/S256A) were generated from template plasmids described previously (20). pCI-NSsPP1, pCI-NSsPP2, and pCI-NSsPP3/4 containing substitutions of alanine for proline were generated by directed mutagenesis using the ExSite kit (Stratagene). The mutated sequences are indicated below (see Fig. 4).
FIG. 4.
Generation of mutant NSs proteins. Proline residues in PXXP motifs (bold letters) were replaced by alanines (arrowheads) leading to the mutant NSs proteins designated PP1, PP2, and PP3/4, respectively. Exchange of the serine residues (bold, italic letters) by alanines resulted in the single mutants S252A and S256A and the double mutant S252A/S256A. These mutations removed one or both casein kinase II phosphorylation sites in the NSs protein. The sequence of the clone 13 NSs protein is shown in the gray boxes. In addition to the large in-frame deletion, a V-to-A substitution at position 216 is present.
IFN titrations.
A culture medium of RVFV- or mock-infected BF cells was exposed to overnight inactivation at pH 2, and IFN-α/β activity was determined in a standard bioassay using vesicular stomatitis virus infection of L cells (36).
Transfections and reporter assays.
The plasmid pIFΔ(−125)lucter (pIF-luc) containing the IFN-β promoter (positions −125 to 72) fused to the firefly luciferase gene was kindly provided by Stephen Goodbourn (19). The NF-κB promoter-reporter plasmid p55A2-luc and the IRF-3-responsive promoter-reporter plasmid p55C1B-luc were both kindly provided by Takashi Fujita, The Tokyo Metropolitan Institute of Medical Sciences, Tokyo, Japan (48), and pAP1-luc and pCF-MEKK were purchased from Stratagene, La Jolla, Calif. For transfection experiments, the reporter plasmids expressing the firefly luciferase gene were cotransfected with a control plasmid expressing either β-galactosidase under the transcriptional control of the CMV promoter (pCMV-βGal) or Renilla luciferase under the transcriptional control of the SV40 early promoter (pRL-SV40; Promega, Mannheim, Germany). Cellular extracts for the measurement of luciferase activity were prepared, and assays were performed as recommended by the manufacturer (Promega). Beta-galactosidase was assayed by the standard method. Plasmid IRF-3(5D) was kindly provided by John Hiscott, McGill University, Montreal, Canada (26).
RT-PCR.
Total RNA was extracted from BF cells infected by RVFV at a multiplicity of infection of 5 PFU per cell by using Trizol reagent (Invitrogen, Cergy Pontoise, France), digested with DNase I, and subjected to RT-PCR, using primers specific for murine IFN-β mRNA and the GAPDH mRNA. The amplified DNAs were examined after migration in an agarose gel and staining with ethidium bromide. To confirm the absence of genomic DNA in the RNA preparation, a PCR was carried out without the preceding RT step.
Northern blot analysis.
After denaturation in the presence of 50% formamide, total RNA was separated by electrophoresis in a 1% agarose gel containing 6% formaldehyde and transferred onto a Hybond N membrane (Amersham Biosciences, Orsay, France). Hybridization with the riboprobes was performed as described elsewhere (5). IFN-β- and β-actin-specific riboprobes were generated with SP6 RNA polymerase in the presence of [α-32P]ATP (Amersham Biosciences) using partial mouse IFN-β and β-actin cDNA inserted into plasmid pGEM-4Z (Promega) as a template.
Western blotting.
Cellular extracts were obtained by lysis with 50 mM Tris (pH 8), 1% NP-40, and 2 mM EDTA in the presence of protease inhibitors (Roche, Mannheim, Germany), 50 mM NaF, and 1 mM sodium vanadate. The proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto a Hybond C Extra membrane (Amersham Biosciences) which was incubated overnight with primary antibodies directed against RVFV nucleoprotein, IκB-β (a gift from Robert Weil, Institut Pasteur, Paris, France), or β-tubulin (Sigma, Lyon, France). Following incubation with horseradish peroxidase-labeled anti-mouse immunoglobulin G, detection was performed by using Supersignal (Pierce, Rockford, Ill.) according to the manufacturer's instructions.
Indirect immunofluorescence assay.
Vero cells cultured on coverslips were transfected with wild-type or mutant NSs expression plasmids using Effectene (QIAGEN, Hilden, Germany) as the transfection reagent. At 24 h posttransfection, the cells were fixed with 3.2% paraformaldehyde and permeabilized with 0.5% Triton X-100 (Sigma). The permeabilized cells were then incubated with a mouse anti-NSs antibody diluted 1:200 (46), followed by incubation with fluorescein-labeled goat anti-mouse immunoglobulin G (Sigma) diluted 1:100 and counterstaining with Evans blue (Sigma).
IRF-3 translocation assay.
Vero cells were infected with RVFV with a multiplicity of infection of 5. Four hours postinfection, the cells were fixed with 3% paraformaldehyde and permeabilized with 0.5% Triton X-100. Endogenous IRF-3 and RVFV N protein was detected by using a polyclonal antibody against IRF-3 (FL-425; Santa Cruz Biotechnology, Santa Cruz, Calif.) and a mouse anti-N antibody, respectively (46). IRF-3-specific signals were amplified using the Tyramide signal amplification system (Perkin-Elmer, Rodgau, Germany).
IRF-3 dimerization assay.
The IRF-3 dimerization assay was carried out as described elsewhere (17). Briefly, at 12 h postinfection, cells were resuspended in lysis buffer containing 50 mM Tris HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, protease inhibitors (complete protease inhibitor; Roche) and phosphatase inhibitors (phosphatase inhibitor cocktail II; Calbiochem, Bad Soden, Germany). The lysates were mixed thoroughly, incubated on ice for 10 min, and then centrifuged at 4°C for 5 min at 10,000 × g. Cleared cell lysates were separated by nondenaturating gel electrophoresis in a 7.5% native gel. IRF-3 monomers and dimers were detected by Western blot analysis using a polyclonal antibody against IRF-3 (FL-425; Santa Cruz).
Electrophoretic mobility shift assay.
Nuclear extracts from RVFV-infected and mock-infected BF cells were prepared 5 h after infection as described previously (47) and assayed with a double-stranded oligonucleotide κB probe derived from the H2Kb promoter (H2Kb, 5′ TGGGGATTCCCCAT 3′; H2Kb anti, 5′ ATGGGGAATCCCCA 3′), or an ATF/cJun binding domain derived from the interleukin 8 promoter (AP1, 5′ GAAGTGTGATGACTCAGGTTTGCCTGA 3′; AP1 anti, 5′ TCAGGCAAACCTGAGTCATCACACTTC 3′) (22). The probe labeled with [32P]ATP in the presence of 10 U of polynucleotide kinase and [γ-32P]ATP (3,000 Ci per mmol) (Amersham Biosciences) was incubated with the cellular extract and analyzed in a 6% polyacrylamide gel.
RESULTS
The S segment of RVFV determines IFN-α/β synthesis in infected cells.
First, we determined the ability of different RVFV strains to induce IFN-α/β in infected cells. IFN-competent BF cells were infected with either the virulent wild-type virus ZH548, the attenuated clone 13 virus, or specific reassortant viruses of the two strains. Reassortant R406 contains the truncated S segment of clone 13 and the L and M segments of ZH548 and is designated Z/Z/C, whereas reassortant R414 contains the intact S segment of ZH548 on a clone 13 genetic background and is designated C/C/Z (8, 40). High levels of IFN were detected in cells infected with clone 13 or Z/Z/C, whereas reduced amounts were found in cells infected with ZH548 or C/C/Z (Table 1). These results demonstrate that the ability to suppress IFN production cosegregates with an intact NSs gene and confirmed our previous in vivo data (8).
TABLE 1.
The S segment of RVFV determines IFN production
| RVFV strain | IFN-α/β titera
|
|
|---|---|---|
| 24 h p.i. | 48 h p.i. | |
| Mock | <2 | <2 |
| ZH548 | 4 | <2 |
| Clone 13 | 128 | 128 |
| Z/Z/C | 128 | 128 |
| C/C/Z | 16 | <2 |
Reciprocal values of the IFN-α/β titers were determined in a bioassay as described in Materials and Methods. p.i., postinfection.
NSs-deficient clone 13, but not wild-type virus, induces IFN-β gene expression.
To monitor IFN-β gene expression in infected cells, accumulation of specific transcripts was analyzed by RT-PCR using total RNA isolated from BF cells infected with either wild-type virus or NSs-deficient clone 13. As shown in Fig. 1A, IFN-β transcripts were detected as early as 3 h after infection with clone 13 and became more prominent thereafter. In contrast, no signal was detectable at any time following infection with wild-type virus. When IFN-β transcripts were analyzed by Northern blotting, similar results were obtained (Fig. 1B). To exclude the possibility that the absence of IFN-β gene transcription was simply caused by an inability of the wild-type virus to replicate in BF cells, viral protein synthesis was assessed by Western blotting. Equivalent amounts of N protein were found in cells infected with wild-type or mutant virus (Fig. 1C).
FIG. 1.
Induction of IFN-β gene expression in RVFV-infected cells. (A and B) Detection of IFN-β transcripts. Total RNA was extracted from BF cells infected with either strain ZH548 or clone 13 at the indicated time points of infection. p.i., postinfection. (A) RT-PCR. Following RT, cDNAs were amplified using primers specific for mouse IFN-β or GAPDH. (B) Northern blotting. RNA samples were separated on a 1% gel, transferred to a nylon membrane, and hybridized with probes specific for mouse IFN-β mRNA or β-actin mRNA. (C) Viral protein synthesis. Lysates of cells either mock-infected or infected with the indicated RVFV strains were monitored for expression of the viral N protein and β-tubulin (as a loading control) by Western blotting. (D) Activation of the IFN-β promoter. 293 cells were transfected with the reporter plasmid pIF-luc. At 5 h after transfection, cells were infected with the wild-type or mutant RVFV strain. Cell lysates were assayed for firefly luciferase activities 16 h after infection.
To assess whether the IFN-β promoter is transactivated in RVFV-infected cells, we determined the IFN-β promoter activity using a luciferase reporter construct. Human 293 cells were transfected with the reporter plasmid pIF-luc and infected with RVFV at 6 h posttransfection. Cell lysates were prepared at 24 h after infection and assayed for luciferase activity. Infection with ZH548 stimulated luciferase activity approximately 2-fold, whereas clone 13 infection resulted in 10-fold stimulation (Fig. 1D). Experiments with BF cells led to similar results (data not shown).
Taken together, these results indicated that clone 13, but not wild-type virus, is a strong inducer of IFN-α/β gene expression and suggested that NSs is the inhibitory factor.
NSs protein suppresses IFN-β promoter activation.
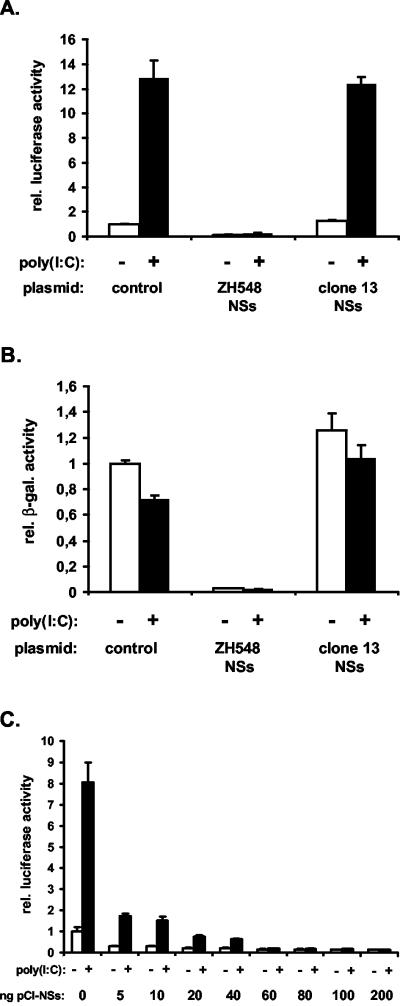
To explore the possibility that NSs itself was suppressing IFN-β gene expression, we expressed NSs from appropriate plasmids in the absence of other viral components and analyzed its effect on IFN-β promoter activity. Recombinant NSs of wild-type strain ZH548 formed filamentous structures in the nuclei of transiently transfected cells; these structures were identical to those observed in ZH548-infected cells (Fig. 2A). In contrast, the truncated NSs of clone 13 was barely detectable and located mainly in the cytoplasm (Fig. 2B), in agreement with previous findings (40). To test the activity of recombinant NSs, a firefly luciferase reporter assay was used in which activation of the IFN-β promoter by synthetic dsRNA [poly(I:C)] is measured. Poly(I:C) efficiently activated the IFN-β promoter in cells that were transfected with a control plasmid. Expression of the truncated NSs of clone 13 had no negative effect on the activation of the IFN-β promoter (Fig. 3A). In contrast, expression of wild-type NSs almost completely blocked IFN-β promoter activation (Fig. 3A). To evaluate the efficiency with which NSs was able to block IFN-β promoter activation, decreasing amounts of NSs expression plasmids were used. While 60 ng of plasmids resulted in over 95% inhibition of reporter gene expression, a 12-fold lower dose of 5 ng still led to a reduction of approximately 75% (Fig. 3C), indicating that NSs is a potent inhibitor of promoter activation. Interestingly, in untreated cells expressing wild-type NSs, the background level of luciferase activity was always lower than in control cells not expressing NSs (Fig. 3A and C). Likewise, the β-galactosidase activity driven by a constitutive promoter was also lower (Fig. 3B), indicating that NSs was a general inhibitor of transcription.
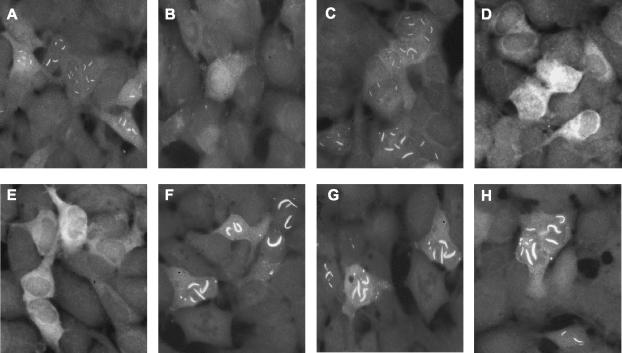
FIG. 2.
Localization and filament formation of wild-type and mutant NSs proteins. ZH548 NSs (A), clone 13 NSs (B), NSs-PP3/4 (C), NSs-PP1 (D), NSs-PP2 (E), NSs-S252A (F), NSs-S256A (G), NSs-S252A/S256A (H). For an explanation of the introduced mutations, see the legend to Fig. 4.
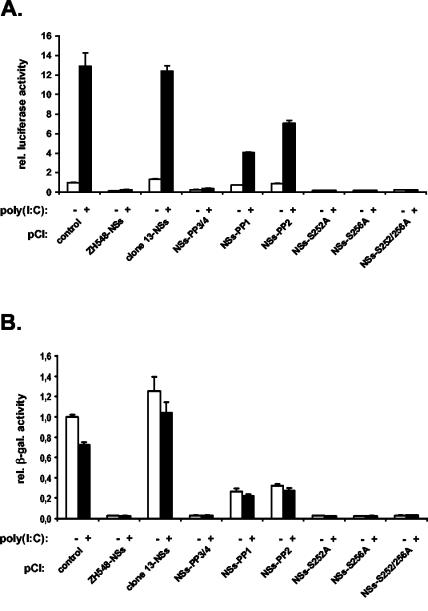
FIG. 3.
NSs protein suppresses IFN-β promoter activation. 293 cells were cotransfected with the indicated NSs expression plasmids, the reporter construct pIF-luc, and the control plasmid pβGal. At 24 h posttransfection, cells were mock treated or stimulated with poly(I:C) for 16 h. Cell lysates were assayed for firefly luciferase and β-galactosidase (β-gal.) activity. rel., relative. (A and C) Effect of NSs on the activation of the IFN-β promoter. (B) Effect of NSs on the constitutively active CMV promoter.
Nuclear accumulation is required for NSs activity.
Sequence analysis revealed that wild-type NSs contained four PXXP motifs (where P is proline and X is any amino acid) (6, 29), namely, motif 1 at positions 29 to 32, motif 2 at positions 82 to 85, and motifs 3 and 4 at positions 240 to 246 (Fig. 4). It should be noted that motifs 1 and 2 are absent in clone 13 NSs. In an attempt to analyze the role of these proline-rich motifs, mutant NSs proteins were generated in which proline residues were replaced by alanine. Substitutions at motifs 3 and 4 were well tolerated, and the resulting NSsPP3/4 mutant protein behaved like the wild type, forming nuclear filaments (Fig. 2C). In contrast, substitutions at motifs 1 and 2 changed the properties of the molecules. Both mutant forms did not produce nuclear filaments and remained mainly in the cytoplasm (Fig. 2D and E). When analyzed for their suppressive effect on IFN-β promoter activation, the two mutants residing in the cytoplasm had lost most of their inhibitory capacity, whereas the nuclear NSsPP3/4 mutant protein still inhibited luciferase activity similarly to wild-type NSs (Fig. 5A).
FIG. 5.
Suppression of IFN-β promoter activation by mutant NSs proteins. rel., relative; β-gal., β-galactosidase. (A) Effect of NSs on the activation of the IFN-β promoter. (B) Effect of NSs on the constitutively active CMV promoter. The experiment was performed as described in the legend to Fig. 3.
Next, we investigated mutant forms of NSs in which the major phosphorylation sites were changed (20). NSs is phosphorylated by casein kinase II at serine residues 252 and 256. Both serines were replaced by alanine, resulting in the single mutants NSs(S252A) and NSs(S256A) as well as the double mutant (S252A/S256A) (Fig. 4). The three mutants behaved like wild-type NSs (Fig. 2F to H) and blocked IFN-β promoter activity (Fig. 5A). Again, they also affected the basal levels of β-galactosidase expression driven by the CMV promoter (Fig. 5B). Taken together, these results suggested that NSs or its mutant forms need to be in the nucleus for efficient suppression of transcription, whereas serine phosphorylation does not seem to matter.
NSs inhibits promoter transactivation without affecting IRF3, NF-κB, or AP1.
Virus-induced activation of the IFN-β gene is an immediate early event which involves the three key transcription factors interferon regulatory factor 3 (IRF3), NF-κB, and ATF2/cJun (AP-1). These are known to cooperate in transactivating the IFN-β promoter (43). One of the characteristic steps during activation of IRF3 or NF-κB is nuclear translocation. In the case of IRF3, phosphorylation and dimerization events trigger nuclear translocation (25, 37). NF-κB is normally retained in the cytoplasm by its inhibitor, IκB, which upon activation is phosphorylated and degraded by the proteasome, thereby releasing NF-κB (18).
To investigate whether activation of IRF-3 is inhibited by NSs, we assessed the fate of IRF-3 in cells infected with either wild-type RVFV or clone 13. IRF-3 dimerization and nuclear translocation were both induced in cells infected with the NSs-deficient clone 13 virus, as expected (Fig. 6A and B). Surprisingly, however, both events also occurred unhindered in cells infected with the NSs-expressing wild-type virus (Fig. 6A and B). In uninfected control cells, IRF-3 remained in the cytoplasm in its monomeric form (Fig. 6A and B). Obviously, NSs did not inhibit activation of IRF-3 but rather blocked a different or subsequent step in promoter activation. If this is the case, NSs should also be able to block IFN-β promoter activation by IRF-3(5D), a constitutively active phosphomimetic form of IRF-3 (26). We therefore examined whether NSs would inhibit IRF-3(5D)-mediated gene activation in an appropriate reporter assay. Indeed, wild-type NSs completely abolished the activation of an IRF-3-dependent promoter, whereas clone 13 NSs was inactive (Fig. 7A). Next, we investigated whether NSs would interfere with NF-κB signaling. Figure 6C shows that infection with clone 13 as well as wild-type virus led to the degradation of IκB-β, as revealed by Western blotting. Compared to that in uninfected cells, the amount of IκB-β was clearly decreased in cells infected with either virus. This effect was not observed in cells treated with the proteasome inhibitor MG132, indicating true degradation of IκB-β by the proteasome (Fig. 6C). Translocation of NF-κB into the nucleus was confirmed by gel shift assays using a probe which contained the NF-κB binding site. Incubation with nuclear extracts from both clone 13- and ZH548-infected cells led to retardation in the electrophoretic mobility of the probe (Fig. 6D). Similar results were obtained with a probe specific for ATF2/cJun (AP-1) (Fig. 6E), indicating that NSs did not interfere with the NF-κB and AP-1 pathways. Nevertheless, wild-type NSs but not clone 13 NSs was able to block activation of an NF-κB-responsive promoter by tumor necrosis factor alpha (Fig. 7B). Likewise, activation of an AP1-responsive promoter mediated by MEK kinase overexpression was inhibited in the presence of wild-type NSs, whereas clone 13 NSs had no effect (Fig. 7C).
FIG. 6.
RVFV infection activates IRF-3, NF-κB, and AP-1. (A) Dimerization of IRF-3. 293 cells were infected with the indicated RVFV strains for 12 h, and formation of IRF-3 dimers was detected as described in Materials and Methods. (B) Nuclear translocation of IRF-3. Vero cells were assayed for localization of IRF-3 and expression of the RVFV N protein by immunofluorescence 4 h after infection. (C) Degradation of IκB-β. RVFV-infected or mock-infected BF cells were incubated in the absence or presence of the proteasome inhibitor MG132, beginning at 1 h postinfection and ending at 5 h postinfection. Cell lysates were analyzed for degradation of IκB-β by Western blotting. Detection of β-tubulin was performed as a loading control. (D and E) Activation of NF-κB and AP-1. Nuclear extracts from RVFV-infected and mock-infected BF cells were prepared 5 h after infection and incubated with [32P]ATP-labeled probes specific for NF-κB or AP-1, and binding of the transcription factors was detected in gel shift assays.
FIG. 7.
(A to C) NSs protein blocks promoter activation mediated by IRF-3, NF-κB, and AP-1. 293 cells were cotransfected with expression plasmids for NSs or GFP (as a control) and reporter constructs containing the indicated transcription factor binding sites of the IFN-β promoter. Promoter activation was achieved by coexpression of the constitutively active IRF-3 mutant IRF-3(5D) (A), incubation with tumor necrosis factor alpha (TNF-α) (B), or coexpression of MEK kinase (MEKK) (C). (D) NSs inhibits transcriptional activation of a constitutive promoter. Cells were cotransfected with expression plasmids for NSs or GFP (as a control) and a reporter construct containing a constitutively active SV40 promoter. Promoter-independent activation of transcription was achieved by incubation with TSA. rel., relative.
NSs inhibits nonspecific activation of transcription.
Since inhibition of IFN-β promoter inducibility by NSs did not rely on inactivation of specific transcription factors, we analyzed the effect of NSs on nonspecific activation of transcription in the presence of trichostatin A (TSA). TSA inhibits the deacetylation of histones, leading to a less condensed state of the chromatin (31, 45). As a consequence, the RNA polymerase II can access the DNA more easily, resulting in nonspecific activation of transcription. Thus, various NSs expression plasmids were cotransfected into 293 cells together with a reporter plasmid expressing the Renilla luciferase under the control of the SV40 early promoter. Twenty-four hours later, transcription was stimulated with TSA. Treatment with TSA resulted in activation of the SV40 early promoter, irrespective of whether the cells expressed NSs of clone 13 or green fluorescent protein (GFP) as a control (Fig. 7D). In contrast, expression of wild-type NSs completely suppressed the TSA-mediated promoter activation (Fig. 7D). These results indicate that NSs is capable of inhibiting inducible and constitutive transcription alike.
DISCUSSION
The nonstructural NSs protein of RVFV is a major virulence factor subverting the innate immune defenses of the host (8, 40). Here we demonstrate that the NSs protein blocks IFN-α/β production in virus-infected cells at the transcriptional level. Induction of IFN mRNA synthesis depends critically on key cellular transcription factors such as IRF-3, NF-κB, and AP-1, which are known to be targeted by IFN antagonists of various viruses. For example, the influenza A virus NS1 protein, which is a multifunctional protein (21), prevents IRF-3 activation and subsequent IFN production by binding to and sequestering dsRNA (13, 39). It interferes also with the activation of NF-κB and AP-1 (28, 42). Several other viruses express proteins that inhibit the function of IRF-3 by additional mechanisms (3, 4, 7, 12, 24, 33, 34). In contrast, as shown here, RVFV NSs does not affect IRF-3 activation. Likewise, NSs does not interfere with the activation of NF-κB or AP-1. We therefore conclude that NSs does not prevent the activation of IFN-specific transcription factors but acts at a subsequent step. Therefore, the IFN-antagonistic activity of NSs seems to stem from a general negative effect on the host cell transcriptional machinery. This is supported by the fact that NSs (i) was capable of blocking the IFN-inducing activity of a constitutively activated form of IRF-3, (ii) inhibited transcriptional activity of constitutive promoters, and (iii) prevented nonspecific upregulation by the general transcriptional activator TSA. Our results are in good agreement with recent findings demonstrating that NSs targets the p44 component of the multisubunit TFIIH basal transcription factor complex, leading to a general suppression of host cellular RNA synthesis (23). Therefore, it is quite likely that NSs inhibits IFN-β gene transcription through the same mechanism. Interestingly, the NSs protein of Bunyamwera virus (an orthobunyavirus) has no sequence similarity to NSs of RVFV (a phlebovirus) and is expressed by a different coding strategy. Yet, it has a comparable effect on host gene expression (39a) and, in particular, IFN-β gene transcription (44). Indeed, recent evidence suggests that Bunyamwera NSs targets an important process in cellular transcription, namely, serine phosphorylation of the C-terminal domain of RNA polymerase II (39a). Since TFIIH is involved in RNA polymerase II phosphorylation (27, 35), it would be interesting to know if the two unrelated NSs proteins use the same strategy.
To further elucidate the mechanism of NSs action, we introduced several mutations destroying known phosphorylation sites (20) or proline-rich motifs of NSs. Interestingly, substitutions at the major phosphorylation sites were well tolerated, indicating that serine phosphorylations by casein kinase II are not required for the effector function of NSs. In contrast, nuclear accumulation of NSs is indispensable for this function. We observed a clear correlation between filament formation in the nucleus and suppression of reporter gene transcription. Disruption of either of two proline-rich PXXP motifs prevented nuclear import, filament formation, and, concomitantly, inhibition of transcription. Proline-rich sequences are known to mediate protein-protein interactions (6). Since NSs has no nuclear localization signal, it probably gains access to the nucleus by binding to a nuclear protein of the host cell. The p44 subunit of the TFIIH transcription factor interacts with NSs (23) and is likely to perform this function. It remains to be demonstrated whether the inactive NSs mutants have lost the capacity to associate with p44. It should be noted, however, that clone 13 NSs which lacks PXXP motifs both 1 and 2 can no longer interact with p44 (23).
The inactive NSs mutants were defective in suppressing both induced expression driven by the IFN-β promoter and constitutive expression from the SV40 promoter, suggesting that a common mechanism is at work. Additional mutations should help to resolve this question. An interesting example in this context is the matrix (M) protein of vesicular stomatitis virus. It inhibits transcription by all three host RNA polymerases and is also a strong inhibitor of IFN-β promoter activity (1, 49). When mutant M proteins were tested for their transcription inhibitory activity, the ability of specific mutants to suppress IFN-β gene expression always correlated with their ability to inhibit host RNA synthesis in general. The conclusion was that wild-type M protein functions as a suppressor of IFN gene expression through its general inhibitory effect on host gene transcription (1).
NSs synthesis is a comparatively late event in the life cycle of RVFV, occurring after secondary transcription of the ambisense S segment. To be effective early on, NSs has to be a very potent repressor of IFN-β gene expression, as clearly indicated by the present results (Fig. 3C). While IFN-β mRNA synthesis was induced by the NSs-deficient clone 13 virus as early as 3 h after infection, no stimulation was detectable in wild-type ZH548-infected cells. Apparently, no IFN-β gene transcription occurred even at early time points when NSs was barely visible in the nucleus and overall mRNA synthesis was still unaffected (23). Thus, the NSs-mediated block in IFN-β gene expression seems to precede the general decline in mRNA synthesis which occurs much later in infection (23). It is conceivable that, for transcriptional activation, inducible genes are generally much more sensitive than housekeeping genes to quantitative changes in the availability of basal transcription factors. If so, TFIIH may be a critical limiting factor for stimulated IFN-β gene transcription. Alternatively, NSs may target additional, and as yet unknown, cellular factors involved in IFN-β gene induction.
It has previously been proposed that NSs is a pathogenicity factor because of its IFN-antagonistic function in vivo (8). Here we demonstrate that NSs indeed suppresses IFN-α/β synthesis. The inhibitory mechanism most likely involves the basal transcriptional machinery of the host cell. It might therefore be argued that the high pathogenicity of wild-type RVFV is caused by the cytotoxic effect of NSs rather than its IFN-antagonistic activity. This, however, is certainly not the case, as demonstrated in experiments with IFN-nonresponsive animals (8). The NSs-deficient clone 13 virus was more pathogenic than the wild-type virus in these animals. The lack of NSs allowed the virus to grow faster and to higher titers, causing an increase in viremia and early death of the animals. This was equally true for a reassortant wild-type virus carrying the NSs deletion of clone 13, indicating that NSs was the decisive factor rather than some other genetic property. Thus, NSs seems to have a slowing effect on virus growth, at least in the mammalian host. This might be explained by the fact that RVFV requires capped RNA primers for its transcription which are derived from the cellular mRNA pool in the cytoplasm. In wild-type but not clone 13 virus-infected cells, the cytoplasmic mRNA levels drop dramatically, thereby limiting viral transcription. Concomitantly, the cytotoxic effect of NSs presumably has additional consequences that are not favorable for virus growth. The growth disadvantage can be envisaged as the price paid by the virus for the tremendous benefit that results from shutting down the IFN system. Recent work with Bunyamwera virus indicates that NSs has no effect on cellular transcription in the insect vector (39a). If applicable to other arboviruses of the bunyavirus family, NSs might well represent a gain in function adaptation to the mammalian host, with the prime task to subvert the IFN defense system without causing much loss in general fitness.
Acknowledgments
We thank Peter Staeheli, Georg Kochs, and Nicolas Le May for valuable comments, Robert Weil for antibodies against IκB-β, Stephen Goodbourn for BF cells, and Takashi Fujita and John Hiscott for plasmids.
This work was supported by grant HA 1582/4-1 of the Deutsche Forschungsgemeinschaft to O.H. and by a grant from the Institut Pasteur to M.B.
REFERENCES
- 1.Ahmed, M., M. O. McKenzie, S. Puckett, M. Hojnacki, L. Poliquin, and D. S. Lyles. 2003. Ability of the matrix protein of vesicular stomatitis virus to suppress beta interferon gene expression is genetically correlated with the inhibition of host RNA and protein synthesis. J. Virol. 77:4646-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidmann, J. A. Smith, and K. Struhl. 1994. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 3.Baigent, S. J., G. Zhang, M. D. Fray, H. Flick-Smith, S. Goodbourn, and J. W. McCauley. 2002. Inhibition of beta interferon transcription by noncytopathogenic bovine viral diarrhea virus is through an interferon regulatory factor 3-dependent mechanism. J. Virol. 76:8979-8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler, C. F., A. Mikulasova, L. Martinez-Sobrido, J. Paragas, E. Muhlberger, M. Bray, H. D. Klenk, P. Palese, and A. Garcia-Sastre. 2003. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J. Virol. 77:7945-7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billecocq, A., P. Vialat, and M. Bouloy. 1996. Persistent infection of mammalian cells by Rift Valley fever virus. J. Gen. Virol. 77:3053-3062. [DOI] [PubMed] [Google Scholar]
- 6.Bliska, J. 1996. How pathogens exploit interactions mediated by SH3 domains. Chem. Biol. 3:7-11. [DOI] [PubMed] [Google Scholar]
- 7.Bossert, B., S. Marozin, and K. K. Conzelmann. 2003. Nonstructural proteins NS1 and NS2 of bovine respiratory syncytial virus block activation of interferon regulatory factor 3. J. Virol. 77:8661-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouloy, M., C. Janzen, P. Vialat, H. Khun, J. Pavlovic, M. Huerre, and O. Haller. 2001. Genetic evidence for an interferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J. Virol. 75:1371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Centers for Disease Control and Prevention. 2000. Outbreak of Rift Valley fever—Saudi Arabia, August-October, 2000. Morb. Mortal. Wkly. Rep. 49:905-908. [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention. 2000. Outbreak of Rift Valley fever—Yemen, August-October, 2000. Morb. Mortal. Wkly. Rep. 49:1065-1066. [PubMed] [Google Scholar]
- 11.Elliott, R. M. 1997. Emerging viruses: the Bunyaviridae. Mol. Med. 3:572-577. [PMC free article] [PubMed] [Google Scholar]
- 12.Foy, E., K. Li, C. Wang, R. Sumpter, Jr., M. Ikeda, S. M. Lemon, and M. Gale, Jr. 2003. Regulation of interferon regulatory factor-3 by the hepatitis C virus serine protease. Science 300:1145-1148. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Sastre, A. 2001. Inhibition of interferon-mediated antiviral responses by influenza A viruses and other negative-strand RNA viruses. Virology 279:375-384. [DOI] [PubMed] [Google Scholar]
- 14.Gerdes, G. H. 2002. Rift Valley fever. Vet. Clin. N. Am. Food Anim. Pract. 18:549-555. [DOI] [PubMed] [Google Scholar]
- 15.Giorgi, C., L. Accardi, L. Nicoletti, M. C. Gro, K. Takehara, C. Hilditch, S. Morikawa, and D. H. Bishop. 1991. Sequences and coding strategies of the S RNAs of Toscana and Rift Valley fever viruses compared to those of Punta Toro, Sicilian sandfly fever, and Uukuniemi viruses. Virology 180:738-753. [DOI] [PubMed] [Google Scholar]
- 16.Goodbourn, S., L. Didcock, and R. E. Randall. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341-2364. [DOI] [PubMed] [Google Scholar]
- 17.Iwamura, T., M. Yoneyama, K. Yamaguchi, W. Suhara, W. Mori, K. Shiota, Y. Okabe, H. Namiki, and T. Fujita. 2001. Induction of IRF-3/-7 kinase and NF-κB in response to double-stranded RNA and virus infection: common and unique pathways. Genes Cells 6:375-388. [DOI] [PubMed] [Google Scholar]
- 18.Karin, M., and Y. Ben-Neriah. 2000. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18:621-663. [DOI] [PubMed] [Google Scholar]
- 19.King, P., and S. Goodbourn. 1994. The beta-interferon promoter responds to priming through multiple independent regulatory elements. J. Biol. Chem. 269:30609-30615. [PubMed] [Google Scholar]
- 20.Kohl, A., V. di Bartolo, and M. Bouloy. 1999. The Rift Valley fever virus nonstructural protein NSs is phosphorylated at serine residues located in casein kinase II consensus motifs in the carboxy-terminus. Virology 263:517-525. [DOI] [PubMed] [Google Scholar]
- 21.Krug, R. M., W. Yuan, D. L. Noah, and A. G. Latham. 2003. Intracellular warfare between human influenza viruses and human cells: the roles of the viral NS1 protein. Virology 309:181-189. [DOI] [PubMed] [Google Scholar]
- 22.Kunz, M., A. Hartmann, E. Flory, A. Toksoy, D. Koczan, H. J. Thiesen, N. Mukaida, M. Neumann, U. R. Rapp, E. B. Brocker, and R. Gillitzer. 1999. Anoxia-induced up-regulation of interleukin-8 in human malignant melanoma. A potential mechanism for high tumor aggressiveness. Am. J. Pathol. 155:753-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le May, N., S. Dubaele, L. P. De Santis, A. Billecocq, M. Bouloy, and J. M. Egly. 2004. TFIIH transcription factor, a target for the Rift Valley hemorrhagic fever virus. Cell 116:541-550. [DOI] [PubMed] [Google Scholar]
- 24.Lin, R., P. Genin, Y. Mamane, M. Sgarbanti, A. Battistini, W. J. Harrington, Jr., G. N. Barber, and J. Hiscott. 2001. HHV-8 encoded vIRF-1 represses the interferon antiviral response by blocking IRF-3 recruitment of the CBP/p300 coactivators. Oncogene 20:800-811. [DOI] [PubMed] [Google Scholar]
- 25.Lin, R., C. Heylbroeck, P. M. Pitha, and J. Hiscott. 1998. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol. Cell. Biol. 18:2986-2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, R., Y. Mamane, and J. Hiscott. 1999. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol. Cell. Biol. 19:2465-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, H., L. Zawel, L. Fisher, J.-M. Egly, and D. Reinberg. 1992. Human general transcription factor IIH phosphorylates the C-terminal domain of RNA polymerase II. Nature 358:641-645. [DOI] [PubMed] [Google Scholar]
- 28.Ludwig, S., X. Wang, C. Ehrhardt, H. Zheng, N. Donelan, O. Planz, S. Pleschka, A. Garcia-Sastre, G. Heins, and T. Wolff. 2002. The influenza A virus NS1 protein inhibits activation of Jun N-terminal kinase and AP-1 transcription factors. J. Virol. 76:11166-11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer, B. J., and R. Gupta. 1998. Functions of SH2 and SH3 domains. Curr. Top. Microbiol. Immunol. 228:1-22. [DOI] [PubMed] [Google Scholar]
- 30.Meegan, J. M., H. Hoogstraal, and M. I. Moussa. 1979. An epizootic of Rift Valley fever in Egypt in 1977. Vet. Rec. 105:124-125. [DOI] [PubMed] [Google Scholar]
- 31.Minucci, S., V. Horn, N. Bhattacharyya, V. Russanova, V. V. Ogryzko, L. Gabriele, B. H. Howard, and K. Ozato. 1997. A histone deacetylase inhibitor potentiates retinoid receptor action in embryonal carcinoma cells. Proc. Natl. Acad. Sci. USA 94:11295-11300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muller, R., J. F. Saluzzo, N. Lopez, T. Dreier, M. Turell, J. Smith, and M. Bouloy. 1995. Characterization of clone 13, a naturally attenuated avirulent isolate of Rift Valley fever virus, which is altered in the small segment. Am. J. Trop. Med. Hyg. 53:405-411. [DOI] [PubMed] [Google Scholar]
- 33.Poole, E., B. He, R. A. Lamb, R. E. Randall, and S. Goodbourn. 2002. The V proteins of simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33-46. [DOI] [PubMed] [Google Scholar]
- 34.Ronco, L. V., A. Y. Karpova, M. Vidal, and P. M. Howley. 1998. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 12:2061-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy, R., J. P. Adamczewski, T. Seroz, W. Vermeulen, J. P. Tassan, L. Schaeffer, E. A. Nigg, J. H. Hoeijmakers, and J. M. Egly. 1994. The MO15 cell cycle kinase is associated with the TFIIH transcription-DNA repair factor. Cell 79:1093-1101. [DOI] [PubMed] [Google Scholar]
- 36.Rubinstein, S., P. C. Familletti, and S. Pestka. 1981. Convenient assay for interferons. J. Virol. 37:755-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schafer, S. L., R. Lin, P. A. Moore, J. Hiscott, and P. M. Pitha. 1998. Regulation of type I interferon gene expression by interferon regulatory factor-3. J. Biol. Chem. 273:2714-2720. [DOI] [PubMed] [Google Scholar]
- 38.Struthers, J. K., and R. Swanepoel. 1982. Identification of a major non-structural protein in the nuclei of Rift Valley fever virus-infected cells. J. Gen. Virol. 60:381-384. [DOI] [PubMed] [Google Scholar]
- 39.Talon, J., C. M. Horvath, R. Polley, C. F. Basler, T. Muster, P. Palese, and A. Garcia-Sastre. 2000. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J. Virol. 74:7989-7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39a.Thomas, D., G. Blakqori, V. Wagner, M. Banholzer, N. Kessler, R. M. Elliott, O. Haller, and F. Weber. 2004. Inhibition of RNA polymerase phosphorylation by a viral interferon antagonist. J. Bio. Chem. 279:31471-31477. [DOI] [PubMed] [Google Scholar]
- 40.Vialat, P., A. Billecocq, A. Kohl, and M. Bouloy. 2000. The S segment of Rift Valley fever phlebovirus (Bunyaviridae) carries determinants for attenuation and virulence in mice. J. Virol. 74:1538-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vialat, P., R. Muller, T. H. Vu, C. Prehaud, and M. Bouloy. 1997. Mapping of the mutations present in the genome of the Rift Valley fever virus attenuated MP12 strain and their putative role in attenuation. Virus Res. 52:43-50. [DOI] [PubMed] [Google Scholar]
- 42.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. García-Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wathelet, M. G., C. H. Lin, B. S. Parekh, L. V. Ronco, P. M. Howley, and T. Maniatis. 1998. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-beta enhancer in vivo. Mol. Cell 1:507-518. [DOI] [PubMed] [Google Scholar]
- 44.Weber, F., A. Bridgen, J. K. Fazakerley, H. Streitenfeld, N. Kessler, R. E. Randall, and R. M. Elliott. 2002. Bunyamwera bunyavirus nonstructural protein NSs counteracts the induction of alpha/beta interferon. J. Virol. 76:7949-7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Workman, J. L., and R. E. Kingston. 1998. Alteration of nucleosome structure as a mechanism of transcriptional regulation. Annu. Rev. Biochem. 67:545-579. [DOI] [PubMed] [Google Scholar]
- 46.Yadani, F. Z., A. Kohl, C. Prehaud, A. Billecocq, and M. Bouloy. 1999. The carboxy-terminal acidic domain of Rift Valley fever virus NSs protein is essential for the formation of filamentous structures but not for the nuclear localization of the protein. J. Virol. 73:5018-5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yamaoka, S., G. Courtois, C. Bessia, S. T. Whiteside, R. Weil, F. Agou, H. E. Kirk, R. J. Kay, and A. Israel. 1998. Complementation cloning of NEMO, a component of the IkappaB kinase complex essential for NF-kappaB activation. Cell 93:1231-1240. [DOI] [PubMed] [Google Scholar]
- 48.Yoneyama, M., W. Suhara, Y. Fukuhara, M. Fukuda, E. Nishida, and T. Fujita. 1998. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 17:1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan, H., S. Puckett, and D. S. Lyles. 2001. Inhibition of host transcription by vesicular stomatitis virus involves a novel mechanism that is independent of phosphorylation of TATA-binding protein (TBP) or association of TBP with TBP-associated factor subunits. J. Virol. 75:4453-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]