Abstract
Genetic relationships among flaviviruses within the yellow fever (YF) virus genetic group were investigated by comparing nucleotide sequences of the 3′ noncoding region (3′NCR). Size heterogeneity was observed between members and even among strains of the same viral species. Size variation between YF strains was due to duplications and/or deletions of repeated nucleotide sequence elements (RYF). West African genotypes had three copies of the RYF (RYF1, RYF2, and RYF3); the Angola and the East and Central African genotypes had two copies (RYF1 and RYF3); and South American genotypes had only a single copy (RYF3). Nucleotide sequence analyses suggest a deletion within the 3′NCR of South American genotypes, including RYF1 and RYF2. Based on studies with the French neurotropic vaccine strain, passage of a YF virus strain in cell culture can result in deletion of RYF1 and RYF2. Taken together, these observations suggest that South American genotypes of YF virus evolved from West African genotypes and that the South American genotypes lost RYF1 and RYF2, possibly in a single event. Repeated sequence elements were found within the 3′NCR of other members of the YF virus genetic group, suggesting that it is probably characteristic for members of the YF virus genetic group. A core sequence of 15 nucleotides, containing two stem-loops, was found within the 3′NCR of all members of the YF genetic group and may represent the progenitor repeat sequence. Secondary structure predictions of the 3′NCR showed very similar structures for viruses that were closely related phylogenetically.
Members of the genus Flavivirus, family Flaviviridae, are small positive-sense, single-stranded RNA viruses. The genome is arranged into a short 5′ noncoding region (5′NCR), a single open reading frame that encodes the structural proteins (C, prM, and E) and nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, 2K, NS4B, and NS5), and a 3′NCR without a poly(A) tail (6). The genus Flavivirus consists of approximately 70 viruses, many of which are pathogens of public health and/or veterinary importance. Fifty-seven of these (80%) are arthropod borne and can be divided into two major groups: 42 mosquito-borne and 15 tick-borne viruses (17).
By using serological methods, the genus Flavivirus has been divided into eight antigenic complexes: tick-borne encephalitis (TBE), Tyuleniy, Japanese encephalitis, Ntaya, Uganda S, dengue, Rio Bravo, and Modoc (5). However, 17 viruses, including yellow fever (YF) virus, the prototype member of the genus, were not sufficiently related to any of the eight antigenic groups and were therefore ungrouped. Subsequently, several studies (3, 8, 12, 17) have taken a phylogenetic approach to analyze the nucleotide sequences of specific regions of Flavivirus genomes. These studies have confirmed the serologic relationships and established relationships between the flaviviruses beyond that possible using serologic testing. On the basis of phylogenetic analyses, a YF genetic group was proposed that included YF, Saboya, Potiskum, Wesselsbron, and Sepik (SEP) viruses plus the Uganda S serogroup, which includes Uganda S virus (UGS), Jugra, Banzi (BAN), Bouboui, and Edge Hill viruses (12, 13, 17). Furthermore, Gaunt et al. (12) proposed phylogenetic differences between viruses vectored by mosquitoes in either the Aedes or Culex genera, suggesting a genetic basis for the adaptation of these viruses to particular mosquito vectors. It was suggested that viruses in the clade transmitted by Culex spp. were associated with encephalitic disease, whereas viruses associated with hemorrhagic disease were transmitted by Aedes spp. (12).
Previous studies on the 3′NCR of flaviviruses have shown extensive differences in size and sequence among species and between strains of the same viral species in the genus Flavivirus (25, 27, 31, 36). The divergence is mostly concentrated at the beginning of the 3′NCR directly following the NS5 gene stop codon (2, 27, 36). The distal part of the 3′NCR usually shows high sequence homology among strains of the same viral species and members of the same serogroup (27). Proutski et al. (28) suggested that the conserved distal regions for the 3′NCR may represent a functional core where most of the important elements in viral translation, replication, and assembly are located.
Compared to the detailed studies on the genetic relationships and evolution of tick-borne flaviviruses (reviewed by Gould et al. [13]), genetic relationships among mosquito-borne flaviviruses are not well established. Gould et al. (13) observed that phylogenetic topologies of mosquito-borne flaviviruses show a two-phase pattern of evolution, in which a slow phase is followed by rapid growth and apparent phylogenetic gaps in which lineages have died out. In contrast, phylogenetic topologies of tick-borne flaviviruses show that they have diverged continuously (10, 13). In the present study, we investigated genetic relatedness between members of the YF virus group by genetic and phylogenetic analysis of the 3′NCR. To date, repeat regions at the beginning of the 3′NCR directly following the NS5 gene stop codon have only been found for one mosquito-borne virus: YF virus. Here we report that all members of the YF virus genetic group studied in this report contain repeat sequences in the 3′NCR. We speculate that the repeat sequences in the 3′NCR of flaviviruses may play a role in regulation of viral replication.
MATERIALS AND METHODS
Viruses.
Low-passage strains of BAN, SEP, UGS, and Zika viruses (Table 1) were obtained from the World Arbovirus Reference Collection at the University of Texas Medical Branch at Galveston. In addition, examples of 15 strains of YF virus were included in the study: 4 strains of YF virus, 14FA (Angola71), A 709-4-A2 (Uganda 48b), M90-5 (Sudan40a), and Asibi (Ghana27) were sequenced, and 11 sequences, ArB9005 (Car77b; GenBank accession no. U52396), 69056 (Nigeria46; GenBank accession no. U52401), and JSS (Brazil35; GenBank accession no. U52390), 85-82H (Ivory Coast82; GenBank accession no. U54798), B4.1 (Peru81; GenBank accession no. U52411), 153 (Peru95c; GenBank accession no. U52407), French viscerotropic virus (FVV, or Senegal27; GenBank accession no. U21056), Dak 1279 (Senegal65; GenBank accession no. U52414), ArB883 (Car77a; GenBank accession no.U52393), MR896 (Uganda48a; GenBank accession no. U52423), and 788379 (Trinidad79A; GenBank accession no. U52420) were obtained from GenBank and included in the nucleotide sequence analysis. A homologous sequence of dengue type 1 virus strain FGA/98 (GenBank accession no. AF226687) (9) was obtained from GenBank and included in the analysis as an outgroup.
TABLE 1.
Virus strains
| Virus | Strain | Yr of isolation | Abbre- viation | Source (country) | Principal mosquito vector | Geographic distribution |
|---|---|---|---|---|---|---|
| Banzi | SA H336 | 1956 | BAN | Human (South Africa) | Culex rubinotus | Africa |
| Sepik | MK71448 | 1966 | SEP | Mansonia septempunctata (New Guinea) | Mansonia septempunctata | New Guinea |
| Uganda S | Prototype | 1947 | UGS | Mixed Aedes spp. (Uganda) | Aedes longipalpis | Africa |
| Uganda S | Makonde | 1957 | UGS | Unknown (Tanzania) | Aedes longipalpis | Africa |
| Yellow fever | Asibi | 1927 | Ghana27 | Human (Ghana) | Aedes africanus | West Africa |
| Yellow fever | 69056 | 1946 | Nigeria46 | Human (Nigeria) | Aedes africanus | West Africa |
| Yellow fever | A 709-4-A2 | 1948 | Uganda48b | Unknown (Uganda) | Aedes africanus | East Africa |
| Yellow fever | ArB9005 | 1977 | Car77b | Aedes africanus (Central African Republic) | Aedes africanus | East/Central Africa |
| Yellow fever | 14FA | 1971 | Angola71 | Human (Angola) | Aedes africanus | Angola |
| Yellow fever | JSS | 1935 | Brazil35 | Human (Brazil) | Heamagogus janthinomys | South America |
| Zika | MR 766 | 1947 | Zika | Rhesus Monkey (Uganda) | Aedes africanus | Africa |
| Dengue type 1 | FGA/89 | 1989 | DEN1 | Human | Aedes aegypti | South America |
Viral RNA extraction and RT-PCR.
Methods used to grow virus and to extract viral RNA have been described in detail elsewhere (37). Reverse transcription-PCR (RT-PCR) was performed on purified viral RNA using the procedures described by Wang et al. (38). Flavivirus universal primers VD8 (antisense, 5′-GGGTCTCCTCTAACCTCTAG-3′) and EMF1 (sense, 5′-TGGATGAC[C,G]AC[G,T]GA[C,T]ATG-3′) (25) were used in the RT-PCR. VD8 is complementary to an element region in the 3′NCR, conserved sequence 2 (CS2), which is conserved in all mosquito-borne flaviviruses (6, 15, 39). EMF1 is complementary to a conserved region in the NS5 gene. These primers amplified regions 300 to 600 nucleotides long. PCR products were screened using agarose gels and ethidium bromide staining. The PCR products were then extracted from the gels and either cloned into pGEM Easy vectors (Promega) or directly sequenced depending on the staining intensity. For cloned isolates, three clones were sequenced in both directions to provide a representative consensus sequence for the isolate. Sequencing was done using an ABI automatic sequencer.
Data analysis.
Each nucleotide sequence was translated into the amino acid sequence to determine the end of the NS5 gene and the beginning of the 3′NCR, using the Vector NTI sequence analysis program (Informax). The beginning of the 3′NCR was identified by the first stop codon in the sequence. The portion of the NS5 gene was eliminated, and further analysis was undertaken using the region of the 3′NCR from the beginning (the stop codon) to the end of the VD8 primer. Nucleotide sequences were processed to identify repeated sequences using the computer program REPEAT, a GCG program available as part of the allele package. The sequences were aligned using the Vector NTI sequence analysis program (Informax). Nucleotide percent similarities and differences were calculated using the MegAlign program (DNASTAR, Lasergene). Phylogenetic analysis of the aligned sequences was performed using PAUP (35) and NEIGHBOR, a neighbor-joining program in the PHYLIP package (11). Parsimony analysis was implemented using the heuristic algorithm. The one-parameter formula was used to generate the distance matrix for neighbor-joining analysis (16). Bootstrap analysis with 1,000 resamplings was used to determine confidence values for groupings within the phylogenetic tree.
Secondary structure predictions.
RNA folding predictions were performed online using the mfold software (http://bioinfo.math.rpi.edu/∼zukerm/) (19, 40). Variable overlapping segments of the RNA sequence for the different 3′NCRs (50 to 377 nucleotides) were folded using mfold, and the predicted RNA structure and several suboptimal foldings were analyzed against the sequence alignments of the different viruses. The presence of a similar structure in the other viruses or isolates and covariant substitutions were taken as strong evidence for the presence of an observed RNA structure. The observed structures were then introduced in the mfold program using the forced folding option, and the RNA segments were folded once more. The recurrence of specific structures in variable-sized fragments of RNA and in multiple viruses or isolates was taken to be indicative for the presence of these structures. Therefore, the observed structures do not necessarily represent the lowest-energy folding.
Nucleotide sequence accession number.
The nucleotide sequences determined in this study have been deposited in GenBank with accession numbers AY326407 to AY326418.
RESULTS
Variation of nucleotide sequence for 3′NCRs of different genotypes of YF virus: repeated sequences in the 3′NCR of YF virus.
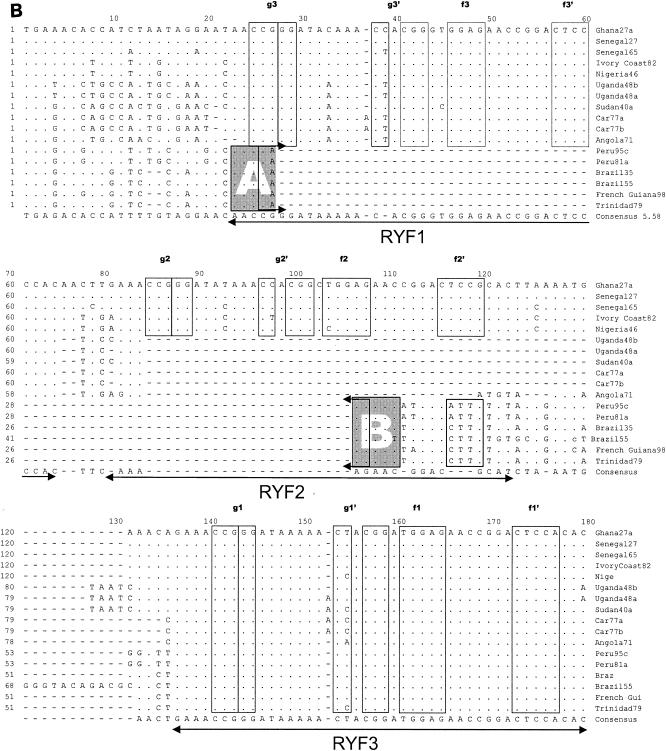
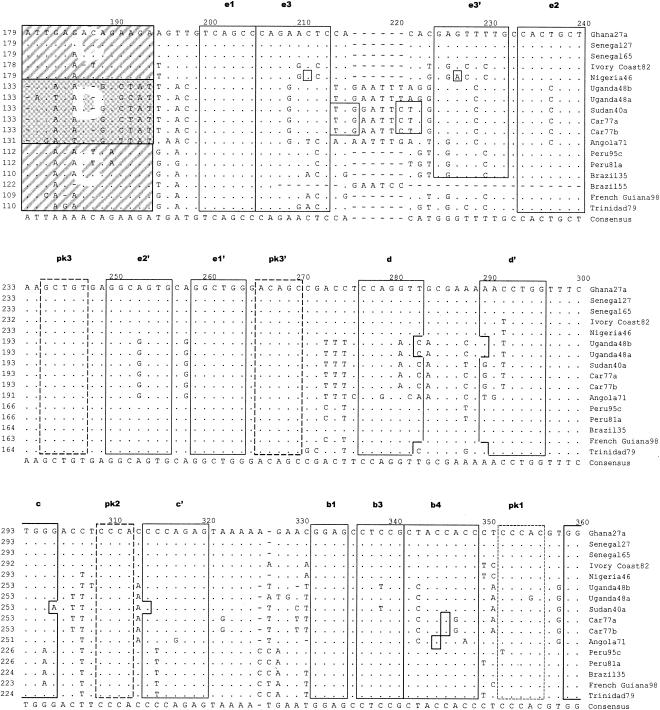
Table 2 shows sizes of the proximal portion of the 3′NCR of the different YF virus strains sequenced and containing nucleotides from the beginning of the 3′NCR (beginning with the stop codon) to the 3′ terminus of the conserved sequence (CS2) element. Based on prM sequences, our investigators previously identified five genotypes of YF virus in Africa: two in West Africa (West Africa I and II) and three in East and Central Africa (East Africa, East/Central Africa, and Angola) (21). In addition, we observed imperfectly repeated sequences in the 5′-proximal region of the 3′NCR of YF virus strains (38). The size of the repeated sequences (RYFs) was 41 to 44 nucleotides, and the number of copies of the repeat sequences varied from one to three depending on the genotype of the YF virus. Two strains of YF virus representing genotypes West Africa I (Ghana27) and West Africa II (Nigeria46) each had three copies (RYF 1, RYF2, and RYF3) of the repeated nucleotide sequence 5′-GAAACCGGGATAAAAACTACGGATGGAGAACCGGACTCCACAC-3′ (Fig. 1B; see also Fig. 6, below). In contrast, YF virus strains from the three East and Central African genotypes (Angola71, Car77b, and Uganda48b) had two copies (RYF1 and RYF3) of the repeated sequence, while strain Brazil35 from South America had only one copy (RYF3) of the repeated sequence (Fig. 1B). The nucleotide sequence between RYF1 and RYF3 in Car77a, Car77b, Uganda48a, Uganda48b, and Angola71 showed no homology to RYF2, which suggested that RYF2 was probably absent in the progenitor virus for these strains. Furthermore, nucleotide sequence homology between West African and South American strains of YF virus in an 18-nucleotide sequence (Fig. 1B, box C) immediately downstream of RYF3 showed a closer genetic relationship between these strains than to strains of the East and Central African genotypes (Fig. 1B, boxes C and D). Nucleotide homology in the same region (Fig. 1B, box D) between Car77b, Uganda48b, and Angola71 suggested that these three YF virus strains are genetically close; however, Angola71 was different from strains Uganda48b and Car77b at five nucleotide positions (Fig. 1B, box D), suggesting genetic differentiation between the Angola genotype and the East and East/Central African genotypes of YF virus.
TABLE 2.
Characteristics of the 3′NCR
| Virus | Strain | Abbre- viation | Length of PCR frag- ment (nt) | No. of repeats | Stop codon |
|---|---|---|---|---|---|
| Banzi | SA H336 | BAN | 342 | 3 | TAA |
| Sepik | MK71448 | SEP | 331 | 2 | TAG |
| Uganda S | Prototype | UGS | 236 | 1 | TAA |
| Uganda S | Makonde | UGS | 236 | 1 | TAA |
| Yellow fever | Asibi | Ghana27 | 377 | 3 | TGA |
| Yellow fever | 69056 | Nigeria46 | 377 | 3 | TGA |
| Yellow fever | A 709-4-A2 | Uganda48b | 336 | 2 | TGA |
| Yellow fever | ArB9005 | Car77b | 337 | 2 | TGA |
| Yellow fever | 14FA | Angola71 | 334 | 2 | TGA |
| Yellow fever | JSS | Brazil35 | 309 | 1 | TGA |
| Zika | MR 766 | Zika | 314 | 0 | TAA |
| Dengue type 1 | FGA/89 | DEN1 | 343 | 0 | TAA |
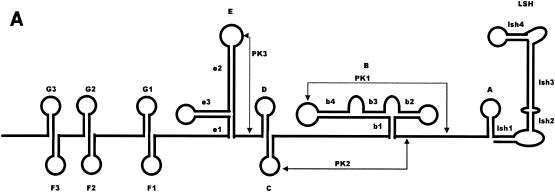
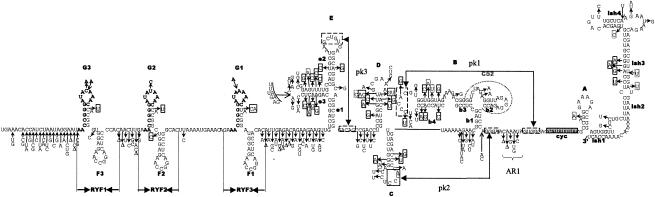
FIG. 1.
Alignment of nucleotide sequence for the 3′NCR of YF virus strains showing structural regions for the complete 3′NCR. (A) Predicted secondary structure for the entire 3′NCR of YF virus. (B) Nucleotide sequence and identity of sequence elements in the predicted secondary structure shown in panel A. The bottom line indicates the consensus sequence for all strains. Regions involved in base pairing are boxed, and the letters above the boxes mark specific stem regions. Regions involved in formation of pseudoknot structures are marked by hatched boxes. The cyclization signal is marked by a grey box. The RYF repeat regions are marked as RYF1, RYF2, and RYF3. Boxes A and B show pentanucleotide sequences in Brazil35, which are very similar to the sequences at the 5′ end of RYF1 and the 3′ end of RYF2, respectively. Box C highlights nucleotide sequence similarity in the 3′-flanking region of RYF3 between West African and South American genotypes. Box D highlights nucleotide sequence similarity in the 3′ flanking region of RYF3 among East and Central African genotypes. A dash indicates spacing introduced to optimize alignment, while a dot indicates nucleotides identical with the Ghana27 sequence.
FIG. 6.
Predicted stem-loop structure for the repeats and flanking nucleotide sequences in the 3′NCR of flaviviruses in the YF virus genetic group. A dash indicates a missing nucleotide.
Secondary structure predictions for the 3′NCR of YF virus.
RNA structure predictions for segments of the YF virus 3′NCR have previously been described (15, 23, 28, 31, 32). The 3′NCR for flaviviruses, with the exception of TBE virus, can be divided into three domains (Fig. 2). Domain I contains a highly conserved 3′-terminal RNA structure and the cyclization sequence (15, 28-31, 39). Domain II contains a conserved element that includes the CS2 and RCS2 regions and partially forms similar structures in YF, dengue, and Japanese encephalitis viruses, including two to three RNA pseudoknot structures (23). Domain III encompasses the region most proximal to the NS5 stop codon, for which very few RNA structures have been proposed.
FIG. 2.
Predicted secondary structure for the 3′NCR of Ghana27 virus. The folding for stem-loops LSH, E, and F1 is based on that of Proutski et al. (28). The terminal base pairing has been modified based on our sequence comparison. Stem-loops B, C, and D are based on the model proposed by Olsthoorn and Bol (23). The location of RYF1, RYF2, and RYF3 repeat elements is indicated, and the core elements are highlighted in bold print. The repeated stem-loop structures present in the RYF repeats are indicated as F1, F2, F3, G1, G2, and G3. Hatched boxes mark nucleotides involved in pseudoknot formation (pk1 to pk3). Arrows indicate the locations of positions of nucleotide variation. Nucleotides in solid boxes mark changes that leave stem formation unchanged. Dotted boxes indicate nucleotide changes that are expected to disrupt base pairing. Nucleotide insertions are marked by open arrows. The cyclization element is indicated as a shaded box. The locations of the conserved CS2 sequences are marked by gray lines. AR1, A-rich region 1 (28).
Since previous predicted RNA structures have focused on domains I and II located in the terminal 150 to 200 nucleotides of the 3′NCR, we have used the mfold program to predict RNA structures in domain III. The identified structures correlated directly with the observed repeat region. The structure of the entire 3′NCR is shown in Fig. 1A and includes the newly proposed structures described in this study and the structures reported previously for the more-3′-located nucleotide sequence. Each RYF repeat element was able to form two stem-loop structures. These structures were termed F and G to keep the nomenclature consistent with that proposed by Olsthoorn and Bol (23). Although the stem regions were in general well preserved between the three RYF repeats, some polymorphism was observed. However, the nucleotide variation was limited to one single position in the stem, which still allowed base pairing to occur (Fig. 1 and 2). Interestingly, all three copies of stem G contained a single un-base-paired adenosine residue in the 3′ section of the stem. The loop region was well conserved but showed some variation in the middle of the loop by either nucleotide substitution and/or insertions, giving an AUA C/U/A AAA loop motif. Stem-loop F was completely conserved in the loop region and apical part of the stem GAG AACCGGA CUC sequence. The base of the stem showed less conservation. The longest stem, 5 bp, was found in F2. Nucleotide changes either disrupted the bottom base pair (F3 and F1) or changed the base-pairing nucleotides (U-G to C-G in F2, G-C to U-A) (Fig. 1A; see also Fig. 4, below).
FIG. 4.
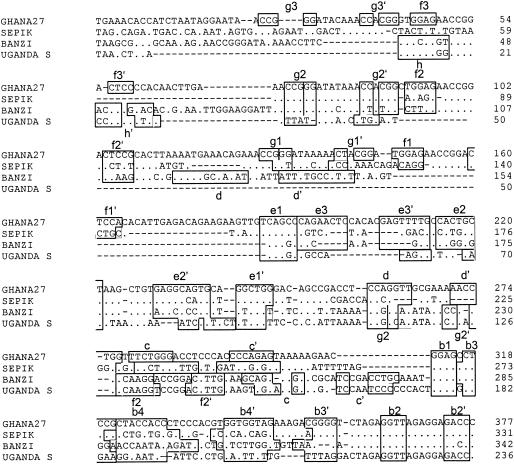
Alignment of nucleotide sequences for the 3′NCR of YF (Ghana27), BAN, SEP, and UGS viruses, showing the nucleotide sequence and identity of sequence elements in the predicted secondary structure. Regions involved in base paring are boxed, and the letters above the boxes mark specific stem regions. For further details, see the legend for Fig. 2.
Most notable was the location of three hot spots for nucleotide variation in this section of the 3′NCR. The region in between the stop codon and G3, a 7-nucleotide region just upstream of G2, and the region in between G1 and E1 had an extraordinarily high rate of nucleotide conversion and single nucleotide deletions. Significantly, this is in agreement with the RNA structure predictions indicating that these regions do not appear to be involved in the well-conserved repeat structures that are preserved between the different YF virus strains.
Comparison of the 15 YF virus strains in this study revealed that the variations observed in the two other structural domains (E to A and LSH) were, with few exceptions, in agreement with the model proposed by Olsthoorn and Bol (23). When we compared nucleotide variation in the region between stem-loops D and E and in the A-rich region 1 (AR1), the sequence elements proposed to be involved in the PK2 and PK3 base pairing were conserved (Fig. 2). In contrast, substantial nucleotide variation was found in the regions flanking these conserved sequences (Fig. 2). Large amounts of covariance were observed for stems e3, b4, and slr3. Covariance was also present to a lesser extent for stems C and e2. Some of the nucleotide variance was specific for particular YF virus genotypes, e.g., the A insertion in the G1 and G3 loops in the Central African strains, suggesting that this insertion was acquired before these viruses started to diverge.
Nucleotide variation among YF virus FNV strains.
We examined nucleotide sequence variation of the 3′NCR in four samples of the French neurotropic vaccine (FNV) virus, termed FNV-IP, FNV-FC, FNV-YALE, and FNV-NT, which were obtained from the Insitut Pasteur (Paris, France), the Division of Vector-borne Infectious Diseases, Centers for Disease Control and Prevention (Fort Collins, Colo.), Yale University (New Haven, Conn.), and the Microbiological Research Establishment, Porton Down, Salisbury, United Kingdom, respectively (37). The passage history of these viruses following derivation of the original FNV after 128 mouse brain passages is not known but has involved multiple mouse brain passages, as the commercial vaccine was used after at least 260 mouse brain passages, and there are no samples of the commercial vaccine available, as it was discontinued in 1971. Sequence alignment of the 3′NCRs showed that FNV-NT had lost two of the three RYFs totaling 95 nucleotides (from YF virus genome nucleotides 10373 to 10467), which included RYF1 and RYF2, compared to other FNV vaccine viruses examined and Senegal27 (also known as the French viscerotropic virus), the wild-type parent to FNV (Fig. 3, which shows the 5′-terminal 200 nucleotides only, as the remaining 216 nucleotides were identical for all four FNV samples). Repeating the RT-PCR and sequencing procedures twice using independent RNA preparations of the four FNV viruses confirmed the deletion. Other than the deletion, the sequences of the four FNV viruses were very similar, with only three nucleotide differences (Fig. 3). Thus, passaging of a YF virus strain under laboratory conditions in mice and cell culture can result in deletion of RYF1 and RYF2.
FIG. 3.
Nucleotide sequence alignment for 3′NCR of four strains of FNV and the parent FVV (or Senegal27). For further details, see the legend for Fig. 2.
Variation of the nucleotide sequence for the 3′NCR of the different viruses in the YF virus group.
Following analysis of different genotypes of YF virus, other viruses in the YF virus genetic group were studied. Table 2 shows nucleotide sequence sizes for the 5′-proximal portions of the 3′NCRs of the different flaviviruses sequenced. The numbers represent nucleotides from the beginning of the 3′NCR (beginning with the NS5 stop codon, nucleotide 10352) to the end of the CS2 element. The sizes varied from 236 nucleotides (UGS virus) to 377 (YF virus strain Ghana27) (Table 2); BAN (342 nucleotides) and SEP (331 nucleotides) were intermediate in length.
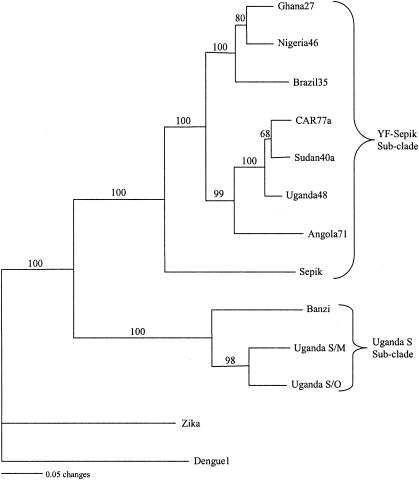
Alignment of the 3′NCR nucleotide sequences for the YF virus group revealed extreme heterogeneity in the 5′-proximal portion of the sequences following the NS5 stop codon (Fig. 4). Most of the variation was in the form of deletions, as demonstrated by the gaps in the alignment. No two viral sequences were identical, even though two strains of UGS virus and representatives of six genotypes of YF virus were included. Nucleotide sequence variations between the two strains of UGS virus sequenced in this study suggest that they are probably representatives of different genotypes. Due to extensive size variation in the region, it was not possible to obtain percent similarity between sequences, but sequence similarity was observed between BAN and UGS 3′NCR sequences, both in the 5′-proximal and the 3′-distal regions. Also, sequence similarity was observed between SEP and YF viruses. A neighbor-joining phylogenetic tree for all the 3′NCR sequences in this study (Fig. 5) showed very close relationships among YF virus strains despite size heterogeneity and variations in numbers of copies of repeated sequences among YF virus strains. BAN, strains of UGS, SEP, and the six genotypes of YF virus clustered together into a single large clade (Fig. 5), suggesting a close phylogenetic relationship among these viruses. This clade was further subdivided into two subclades, one consisting of members of the UGS antigenic group (BAN and UGS viruses) and the other consisting of SEP and YF viruses. Zika virus, a member of the Spondweni virus group, and dengue type 1 virus, a member of the dengue virus group, were included for comparison and were found to be phylogenetically distinct from each other and from all the other viruses in this study, confirming the relatedness of the YF group of viruses.
FIG. 5.
A neighbor-joining tree generated from the 3′NCR nucleotide sequences of 13 strains of flaviviruses. The numbers at the nodes are bootstrap values for 1,000 replicates.
Repeat sequences are present in the 3′NCRs of other flaviviruses in the YF virus group.
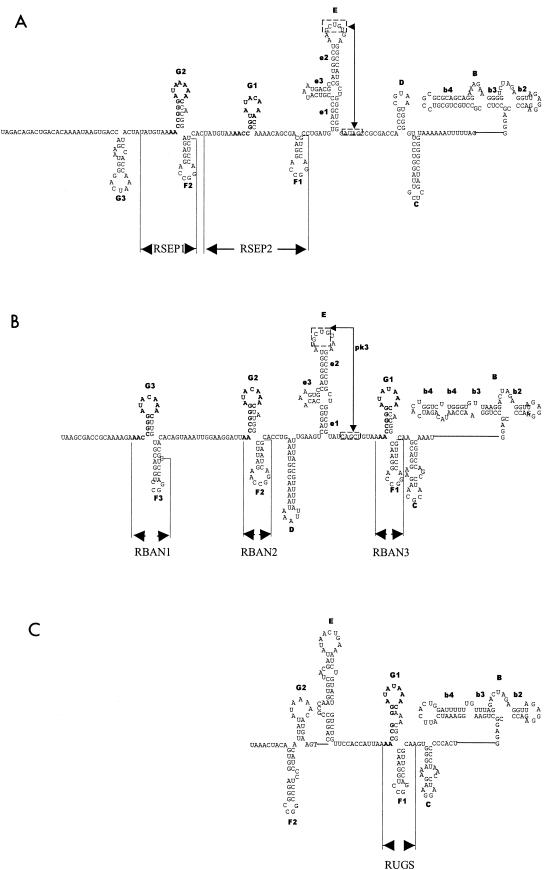
Like YF virus, imperfect repeated sequences were found in the 3′NCR of BAN, SEP, UGS (original strain), and UGS (Makonde strain) viruses (see Table 2 and Fig. 6 for a summary). The number of copies of the repeated sequences varied from one to three, depending on the virus. Three copies of a repeat sequence were identified in the 3′NCR of BAN virus (Fig. 6 and 7B). The repeat sequences (RBAN1 to RBAN3) were 1 to 4 nucleotides shorter than those of YF virus (RYF1 to RYF3), and RBAN3 was further downstream than RYF3. Nonetheless, the nucleotide sequences of RBAN1 to RBAN3 had greater than 90% similarity to RYF1 to RYF3. SEP virus had two repeat sequences (RSEP) (Fig. 6 and 7A). Each RSEP was 42 to 45 nucleotides long and shared 80% homology and 32 to 48% similarity to RYFs. Compared to RYF1, RSEP1 was 10 nucleotides downstream, while RSEP2 was 20 nucleotides downstream of RYF2. UGS virus had one copy, RUGS, which was approximately 40 nucleotides (Fig. 6 and 7C). RUGS was approximately in the same position as RBAN3 (Fig. 7B and C). In comparison, Zika virus, a member of the Spondweni virus group, and dengue type 1 virus, a member of the dengue virus group, had no repeated sequences of 20 nucleotides or longer in the region of the 3′NCR 3′ from the stop codon to the CS2 element.
FIG. 7.
Predicted RNA structures for the 3′NCR of members of the YF virus subgroup. The predicted structures based on computer-based folding and phylogenetic analysis for the 3′NCR upstream of the CS2 sequence for SEP(A), BAN (B), and UGS (C) viruses are shown. The repeat regions for each virus are indicated. The nomenclature of the RNA structures follows that used for YF virus.
Identification of a core sequence in the repeats.
Comparison of the repeat sequences in the members of the YF virus genetic group revealed a 15-nucleotide core sequence, 5′-AACCGGGATACAAAC-3′ in the 3′NCRs of YF, BAN, SEP, and UGS viruses (Fig. 6). Since the core sequence was found within RYF, RSEP, RUGS, and RBAN, this would suggest that the core is probably the progenitor sequence. The core sequence was found at the 5′ end of RYF1, RYF2, RYF3, RBAN1, RBAN2, RBAN3, and RUGS (Fig. 1, 6, and 7B and C), but in the middle of RSEP1 and RSEP2 (Fig. 7A). Examination of the entire YF virus genome in positive- or negative-sense orientation revealed no other copy of the core sequence.
When the RYF sequence was compared against the GenBank database, the most remarkable alignment was a complete match found between stem-loop G and a sequence in the Drosophila melanogaster genome (GenBank accession numbers AC010661 and AY094944). The sequence upstream of stem-loop G showed no similarity, while the two nucleotides downstream of the Drosophila element were identical to the two nucleotides downstream of G1, but no further similarity was present. This could suggest that this element was acquired during replication in an insect host, possibly by recombination with an RNA segment of an insect host, or that the element is essential for replication in certain insect hosts.
Secondary structure for the 3′NCR of other flaviviruses within the YF virus group. (i) SEP virus.
When the sequences of the SEP, BAN, and UGS viruses were compared with that of YF virus, it was clear that the elements required for the formation of the dumbbell structure were found as a single copy (Fig. 4 and 7B). SEP virus appeared to adapt a structure most closely related to YF virus (Fig. 7A). It was able to form a structure E similar to that formed in YF virus. Furthermore, the loop, apical, and base sections of the e2 stem were identical to those found in the YF virus structure E, while for stem-loop e3 only the closing CG base pair was preserved. In the pk3 element a single C-to-U change was present. This still allowed the G in the pk3 element to base pair. In addition, structural equivalents to the YF virus stem-loops C and D can be folded in the SEP virus sequence. In stem-loop D, four out of the five loop nucleotides were identical to the YF virus stem-loop D. However, in the stem region only the two closing base pairs were identical. Stem-loop C in SEP virus showed no sequence similarity with the equivalent stem-loop in YF virus. As we had no nucleotide sequences 3′ of structure B, it was impossible to determine if the loop nucleotides for structure C could form pk2.
The structural similarity between SEP and YF viruses was substantiated by the presence of repeat elements (RSEPs), which are very similar to the RYFs and can form the two stem-loop structures (Fig. 7A). In SEP virus the sequence of the F2/G2 tandem was closer to that present in YF virus compared to all other flaviviruses in this study. The only differences were the closing base pair in F2 and the deletion of the nucleotide position that was also variable in the YF virus F loops. The F2 loop in SEP virus appeared to have lost a single A residue compared to the YF virus F loops. There were two incomplete repeats of stem-loop G (Fig. 7A, G1 and G3) and a single incomplete repeat of stem-loop F (Fig. 7A, F1). The apical section of stem-loop F1 was still identical to that of F2, but the stem contained base substitutions and had only 4 bp instead of the 5 present in F2. Stem-loop G1 only consisted of 3 bp and had an A residue in the loop converted to C. In G3, only the apical base pairs and most of the loop nucleotides showed similarity to the YF virus G and SEP virus G2 stem-loops.
(ii) BAN virus.
BAN virus shares many of the main structural features present in both YF and SEP viruses, such as the dumbbell structure B, structure E; however, e3 was much smaller in size than in YF or SEP viruses (compare Fig. 2, 4, and 7B). In the pk3, the U at the 3′ side of the pk3 element had lost base-pairing capability; however, the A at the 5′ side of the sequence had acquired a base-pairing partner (Fig. 7B). Two repeat elements, RBAN1 and RBAN2, were located upstream of structure E, but stem-loop D was located in between the E and F structures. Surprisingly, element RBAN3 was located just upstream of the stem-loop C equivalent (Fig. 7B). The loop of G2 had a nucleotide change compared to G1 at the variable position identified in the YF virus stem-loop G. The stem contained some nucleotide changes, but these still allowed base pairing to occur. Stem-loop F2 had lost a single A from the loop, and the stem region had accumulated several non-base-pair-disrupting nucleotide changes.
(iii) UGS virus.
The predicted UGS virus structure (Fig. 7C) was different than that of YF, SEP, and BAN viruses. Although UGS virus can form a structure similar to domain E in YF, SEP, and BAN viruses, structure e3 was absent (Fig. 7C). Although UGS virus can form a structural equivalent to stem-loop C, it was impossible to evaluate if the loop region could form an RNA pseudoknot. Like RBAN3, RUGS was immediately upstream of stem-loop C (Fig. 7A and C). The sequence of the UGS F1/G1 tandem was almost identical in sequence to that of BAN virus. The exceptions were the presence of a double-A bulge-loop in the UGS virus stem-loop G and an A-to-U conversion in the loop of stem-loop F resulting in an additional base pairing in UGS virus. A hairpin structure with a loop reminiscent of the YF virus G structures was present close to the 5′ terminus of the fragment (G2) UGS virus shares with BAN virus, and stem-loop F2 in RUGS shares high sequence identity with the RBAN stem-loop. An additional structure 3′ of f2 can be folded in UGS virus, but there was no phylogenetic evidence to provide support for this structure.
DISCUSSION
Many phylogenetic analyses of the flaviviruses have focused on protein-coding gene sequences, such as the envelope protein gene (12) and nonstructural protein genes NS3 (3) and NS5 (2, 12, 17). Other studies (1, 2, 25, 27, 28, 33, 34, 36) have used the 3′NCR to study relationships between flaviviruses. The heterogeneity in size, due to repeat sequence insertion or deletion at the beginning of the 3′NCR directly following the NS5 gene stop codon, enables the 3′NCR to be used as a tool to investigate the evolution and genetic relationships of flaviviruses. Prior to this study, repeat sequences had only been demonstrated in the 3′NCR of YF (15) and TBE (14) viruses, and no repeat elements had been identified in the 3′NCR of dengue, Japanese encephalitis, Modoc, or Rio Bravo genetic groups in the domain immediately 3′ of the NS5 stop codon (3, 7, 18, 22, 24, 34). Furthermore, studies with other flaviviruses have shown that strain genotypic differences are associated with insertions or deletions, rather than repeat sequences, in the 5′ region proximal to the NS5 stop codon (27, 34). The repeated sequences found in members of the YF virus group (i.e., BAN, SEP, and UGS viruses) were restricted to the 5′-proximal region of the 3′NCR, upstream of CS2. Deletions or duplications of these repeated sequences contributed to the observed nucleotide sequence heterogeneity.
Despite the nucleotide sequence heterogeneity in the 3′NCR of the flaviviruses in the present study, phylogenetic and structural analyses in this study grouped these viruses into one clade, as has been reported previously by analysis of other regions of the genome. Kuno et al. (17) and Gould et al. (13) grouped 10 flaviviruses, BAN, UGS, Jugra, Potiskum, Saboya, Bouboui, Edge Hill, YF, Wesselsbron, and SEP, together in the YF virus genetic group based primarily on sequence data. These findings suggest that studies of phylogenetic analysis of the 3′NCR are representative of the genome despite the size heterogeneity in this region.
Since there was little evidence of deletions in the 3′NCR of East and Central African and Angola genotypes of YF virus, it was hypothesized that RYF2 did not exist in the progenitor to East and Central African and Angola YF virus. We speculate that the duplication events that resulted in RYF2 (i.e., structures g2 and f2) probably occurred in West Africa. An alternative pathway is through a stepwise deletion process of the RYF2 from West Africa, resulting in the Central and East African strains, followed by deletion of RYF1, leading to the South American strains. However, sequence identity in the regions flanking the RYF repeats clearly indicated that South American strains are more closely related to West African isolates, suggesting that either South American and Central and East African strains developed independently from the progenitor strain(s) or, more likely, that South American strains were derived from West Africa, as is supported by phylogenetic analysis of both the 3′NCR and other genomic sequences. Furthermore, two short pentanucleotide sequences, AACCA (Fig. 1B, box A) and AGAAC (Fig. 1B, box B), in the 3′NCR of Brazil35 were homologous to sequences at the 5′ end of RYF1 and near the 3′ end of RYF2, respectively, which suggests that RYF1 and RYF2 could have been lost due to deletion from the 3′NCR of South American YF virus, probably as a single event. Nonetheless, as the sequence 3′ of box B is only present in YF virus isolates from South America, the sequence similarity in box B could be coincidental. Another alternative is that an ancestral YF virus strain that had accumulated three copies of the RYF repeat region could, while migrating into Central and Eastern Africa, have lost a single copy of the repeats. In a separate event, perhaps while migrating to South America, two copies of the repeats became lost due to a changed selection pressure. However, since South American YF virus strains are phylogenetically closely related to West African strains (38), we speculate that RYF1 and RYF2 were deleted from South American strains subsequent to the introduction of West African strains into South America, probably during the process of adaptation to a new host-vector system in South America (i.e., Aedes spp. mosquitoes transmit YF virus in Africa, while Hemagogus and Sabethes spp. mosquitoes transmit YF virus in South America). Deletion of RYF1 and RYF2 from South American genotypes of YF virus appears to have been a single event, suggesting that introduction of YF virus into South America from West Africa took place during one period of time, presumably during the slave trade. The fact that all South American strains of YF virus that have been sequenced to date have only RYF3 suggests that genetic adaptation to vectors and/or vertebrate hosts occurs rapidly. Pisano et al. (26) proposed that YF virus strain Trinidad79a may have been a recent introduction from West Africa, but this strain like all the others in South America has only one RYF3 in its 3′NCR.
Comparison of the four FNV viruses with FVV indicated that the deletion seen in the 3′NCR of FNV-NT did not arise during the attenuation process of FVV through mouse brain to derive FNV. Consequently, the deletion must have taken place during the subsequent passage history of FNV-NT in the laboratory. Furthermore, the deletion of RYF1 and RYF2 in FNV (nucleotides 20 to 114) and South American strains (nucleotides 28 to 95) took place at similar nucleotide positions (Fig. 1 and 3) and corresponds to deletion of structures g3, f2, and g2 (Fig. 1). Although the passage history of FNV-NT is unknown, this result shows that deletions may occur in the 3′NCR of YF virus by passaging the virus in the laboratory. We speculate that a similar process of passaging YF virus in nature, or the adaptation of YF virus to South American host-vector systems, occurred when West Africa YF virus strains were introduced into South America, leading to deletions of RYF1 and RYF2 in the South American genotypes of YF virus. It should be noted that the wild-type strains used in these studies are all low-passage isolates (i.e., less than five passages) from the World Arbovirus Reference Collection, and so we believe that the RYFs identified in the genotypes studied are representative of the situation in nature.
The 3′NCR is thought to play a critical role in virus replication. However, it was recently shown that a recombinant YF virus lacking all RYFs was still able to support RNA synthesis and formed plaques with wild-type efficiency, indicating that the repeats are not required for replication in vertebrate cells (4). Since host-specific factors interact with regions of the 3′NCR, then one would expect a relationship between 3′NCRs of viruses transmitted by the same or similar vector species. Flaviviruses replicate within the arthropod vector for considerable periods of time, while only replicating for brief periods of time in the mammalian host before being taken up by a new vector during blood feeding. This would suggest that the major selection pressure to maintain the RYFs is most likely present in the mosquito host.
However, the above hypothesis was not well supported by our results. We did not see close genetic relatedness between viruses transmitted by the same vector. For example, YF virus in tropical Africa is maintained in the sylvatic cycle by the mosquito Aedes africanus. This same species is the only natural vector known for Zika virus. However, there were extensive nucleotide sequence differences between YF and Zika viruses despite their sharing the same mosquito vector (Fig. 5). Similarly, dengue viruses are transmitted by the mosquito Aedes aegypti, the same urban vector for YF virus. However, phylogenetic analysis using nucleotide sequences for the 3′NCR showed that YF and dengue viruses are evolutionally more distant compared to viruses in the YF virus genetic group, which are transmitted by mosquitoes in different species (Fig. 5). Interestingly, the mosquito vector of SEP virus, the closest genetic relative to YF virus, is Mansonia septempuncatata, which is not only a different species but also a different genus from Aedes mosquitoes. Although higher taxonomic units, such as mosquitoes (family Culicidae) and ticks (order Acari), correlate well with the phylogenetic grouping of flaviviruses (17), the relationship breaks down at the species level. Gaunt et al. (12) suggested some correlation between mosquito genera and phylogenetic clades of mosquito-borne flaviviruses, but there were some exceptions to their groupings. For example, the principal vector for BAN virus is Culex rubinotus, but Banzi virus was included in the Aedes clade. Based on the 3′NCR, we have shown that the primary nucleotide sequence does not show any correlation to vector species. We speculate that adaptation of a flavivirus to a mosquito species is probably based on the structural aspect of the sequence rather than sequence homology. Significantly, this hypothesis is supported by the finding that each repeat element can form two RNA stem-loop structures (Fig. 6). Even if the nucleotide sequence were not completely conserved in the different inter- and intraspecies repeats, the capacity for base pairing suggests that the sequence of the base-paired elements is of lesser importance, and the loop sequences are well conserved between the different repeats. This hypothesis is consistent with the findings of Proutski et al. (28), who reported that structural elements at the 3′ terminus were preserved among the flavivirus 3′NCR, despite the high degree of sequence divergence among them.
We did not find any correlation between number, length, and similarity of repeated nucleotide sequences and phylogenetic relationship. Surprisingly, the nucleotide sequences for RBANs were very similar to that of RYFs (88.4 to 90.9%), but BAN virus was phylogenetically closer to UGS virus than to YF virus (Fig. 5). It was possible that the similarity between RYFs and RBANs was due to BAN virus being a recombinant between 3′NCRs of YF virus and another flavivirus. However, when RYFs were swapped with RBANs using computer programs, the resultant sequences clustered in the UGS-BAN subclade and not in the YF-SEP subclade (data not shown), which suggests that BAN virus was not a recombinant. In comparison, RSEP was only 32.3 to 48.4% similar to RYF, but the two viruses were very close phylogenetically (Fig. 5). The lack of correlation between sequence repeats and phylogenetic relationships is intriguing and may be related to RNA structures, but the function of these repeats is unknown. We speculate that repeat sequences probably have a regulatory role in viral replication and translation and arose during adaptation to arthropod vectors. The similarity between the core sequence in the repeat sequence of the YF virus group and a Drosophila sequence element may support this hypothesis. Also, studies by Men et al. (20) showed that deletion of the 3′NCR of dengue type 4 virus, 5′ to the CS2 element, resulted in reduced infectivity and less-efficient replication of the virus. Since dengue and YF viruses are both flaviviruses, it is possible that homologous regions of the 3′NCR have similar functions in both viruses.
Overall, our results show close phylogenetic relationships between flaviviruses in the YF virus genetic group. The closest relative to YF virus is SEP virus. The geographical distribution of YF virus is limited to tropical Africa and South America and does not overlap with that of SEP virus, which is found in New Guinea; this probably indicates that the two viruses diverged a long time ago and have evolved independently of each other. We did not find a correlation between the nucleotide sequence of the 3′NCR and mosquito vector species. However, despite the extensive sequence heterogeneity between viruses, phylogenetic analysis grouped viruses according to already established groups but showed closer relationships between the YF-SEP group and the UGS group.
Acknowledgments
We thank Robert Tesh and Robert Shope for the virus strains analyzed in this study. We also thank Mike Holbrook, David Beasley, Tom Solomon, Jacqueline Scherret, and Amy Shurtleff, and Monica McArthur and Juliet Bryant, for helpful discussions, comments, and suggestions during the course of this work. We thank the UTMB Sequence Core Lab (S. Smith and E. Surriga) for their help in this project. Last, we thank N. Barrett and Li Li for technical assistance.
This work was supported in part by grant AI10986 and by the 2000-2001 Colin Powell Minority Postdoctoral Fellowship in Tropical Disease Research to J.-P.M.
REFERENCES
- 1.Batista, W. C., S. Kashima, C. Marques, and L. T. M. Figueiredo. 2001. Phylogenetic analysis of Brazilian flaviviruses using nucleotide sequences of parts of NS5 gene and 3′ non-coding regions. Virus Res. 75:35-42. [DOI] [PubMed] [Google Scholar]
- 2.Beasley, D. W. C., M. T. Suderman, M. R. Holbrook, and A. D. T. Barrett. 2001. Nucleotide sequencing and serological evidence that the recently recognized deer tick virus is a genotype of Powassan virus. Virus Res. 79:81-89. [DOI] [PubMed] [Google Scholar]
- 3.Billoir, F., R. de Chesse, H. Tolou de Micco, E. A. Gould, and X. de Lamballerie. 2000. Phylogeny of the genus Flavivirus using complete coding sequences of arthropod-borne viruses and viruses with no known vector. J. Gen. Virol. 81:781-790. [DOI] [PubMed] [Google Scholar]
- 4.Bredenbeek, P. J., E. A. Kooi, B. Lindenbach, N. Huikman, C. M. Rice, and W. J. Spaan. 2003. A stable full-length yellow fever virus cDNA clone and the role of conserved RNA elements in flavivirus replication. J. Gen. Virol. 84:1261-1268. [DOI] [PubMed] [Google Scholar]
- 5.Calisher, C. H., N. Karabatsos, J. M. Dalrymple, R. E. Shope, J. S. Porterfield, E. G. Westway, and W. E. Brandt. 1989. Antigenic relationships between flaviviruses as determined by cross-neutralization tests with polyclonal antisera. J. Gen. Virol. 70:37-43. [DOI] [PubMed] [Google Scholar]
- 6.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 7.Deubel, V., R. M. Kinney, and D. W. Trent. 1988. Nucleotide sequence and deduced amino acid sequence of nonstructural proteins of dengue type 2 virus, Jamaica genotype: comparative analysis of the full-length genome. Virology 165:234-244. [DOI] [PubMed] [Google Scholar]
- 8.de Zanotto, P. M., E. A. Gould, G. F. Gao, P. H. Harvey, and E. C. Holmes. 1996. Population dynamics of flaviviruses revealed by molecular phylogenies. Proc. Natl. Acad. Sci. USA 93:548-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duarte dos Santos, C. N., M.-P. Frenkiel, M.-P. Courageot, C. F. S. Rocha, M.-C. Vazeille-Falcoz, M. W. Wien, F. A. Rey, V. Deubel, and P. Despres. 2000. Determinants in the envelope E protein and viral RNA helicase NS3 that influences the induction of apoptosis in response to infection with dengue type 1 virus. Virology 274:292-308. [DOI] [PubMed] [Google Scholar]
- 10.Ebel, G. D., A. Speilman, and S. R. Telford III. 2001. Phylogeny of North American powassan virus. J. Gen. Virol. 82:1657-1665. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1993. PHYLIP (phylogeny inference package), version 3.5. Department of Genetics, University of Washington, Seattle.
- 12.Gaunt, M. W., A. A. Sall, X. de Lamballerie, A. K. I. Falconar, T. I. Dzhivanian, and E. A. Gould. 2001. Phylogenetic relationships of flaviviruses correlate with their epidemiology, disease association and biogeography. J. Gen. Virol. 82:1867-1876. [DOI] [PubMed] [Google Scholar]
- 13.Gould, E. A., X. de Lamballerie, P. M. A. Zanotto, and E. C. Holmes. 2001. Evolution, epidemiology and dispersal of flaviviruses revealed by molecular phylogenies. Adv. Virus Res. 57:71-104. [DOI] [PubMed] [Google Scholar]
- 14.Gritsun, T. S., K. Venugopal, P. M., Zanotto, M. V. Mikhailov, A. A. Sall, E. C. Holmes, I. Polkinghorne, T. V. Frolova, V. V. Pogodina, V. A. Lashkevich, and E. A. Gould. 1997. Complete sequences of two tick-borne flaviviruses isolated from Siberia and the UK: analysis and significance of the 5′ and 3′-UTRs. Virus Res. 49:27-39. [DOI] [PubMed] [Google Scholar]
- 15.Hahn, C. S., Y. S. Hahn, C. M. Rice, E. Lee, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198:33-41. [DOI] [PubMed] [Google Scholar]
- 16.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munros (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 17.Kuno, G., G. J. Chang, R. Tsuchiya, N. Karabastos, and C. B. Cropp. 1998. Phylogeny of the genus Flavivirus. J. Virol. 72:73-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leyssen, P., N. Charlier, P. Lemey, F. Biloir, A.-M. Vandamme, E. De Clercq, X. de Lamballarie, and J. Neyts. 2002. Complete genome sequence, taxonomic assignment, and comparative analysis of the untranslated regions of Modoc virus, a flavivirus with no known vector. Virology 293:125-140. [DOI] [PubMed] [Google Scholar]
- 19.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 20.Men, R., M. Bray, D. Clark, R. M. Chanock, and C.-J. Lai. 1996. Dengue type 4 mutants containing deletions in the 3′ noncoding region of the RNA genome: analysis of growth restriction in cell culture and altered viremia pattern and immunogenicity in rhesus monkeys. J. Virol. 70:3930-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mutebi, J.-P., H. Wang, L. Li, J. E. Bryant, and A. D. T. Barrett. 2001. Phylogenetic and evolutionary relationships among yellow fever virus isolates in Africa. J. Virol. 75:6999-7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nam, J. H., S. L. Chae, S. Y. Won, E. J. Kim, K. S. Yoon, B. I. Kim, and H. W. Cho. 2001. Short report: genetic heterogeneity of Japanese encephalitis virus assessed via analysis of the full-length genome sequence of Korean isolate. Am. J. Trop. Med. 65:388-392. [DOI] [PubMed] [Google Scholar]
- 23.Olsthoorn, R. C. L., and J. F. Bol. 2001. Sequence comparison and secondary structure analysis of the 3′ noncoding region of flavivirus genomes reveals multiple pseudoknots. RNA 7:1370-1377. [PMC free article] [PubMed] [Google Scholar]
- 24.Osatomi, K., and H. Sumiyoshi. 1990. Complete nucleotide sequence of dengue type 3 virus genome RNA. Virology 176:643-647. [DOI] [PubMed] [Google Scholar]
- 25.Pierre, V., M.-T. Drouet, and V. Deubel. 1994. Identification of mosquito-borne flavivirus sequences using universal primers and reverse transcription/polymerase chain reaction. Res. Virol. 145:93-104. [DOI] [PubMed] [Google Scholar]
- 26.Pisano, M. R., V. Mercier, V. Deubel, and H. Tolou. 1999. Complete nucleotide sequence and phylogeny of an American strain of yellow fever virus, Trinidad79A. Arch. Virol. 144:1837-1843. [DOI] [PubMed] [Google Scholar]
- 27.Poidinger, M., R. A. Hall, and J. S. Mackenzie. 1996. Molecular characterization of the Japanese encephalitis serocomplex of the Flavivirus genus. Virology 218:417-421. [DOI] [PubMed] [Google Scholar]
- 28.Proutski, V., E. A. Gould, and E. C. Holmes. 1997. Secondary structure of the 3′ untranslated region of flaviviruses: similarities and differences. Nucleic Acids Res. 25:1194-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Proutski, V., M. W. Gaunt, E. A. Gould, and E. C. Holmes. 1997. Secondary structure of the 3′ untranslated region of yellow fever virus: implications for virulence, attenuation and vaccine development. J. Gen. Virol. 78:1543-1549. [DOI] [PubMed] [Google Scholar]
- 30.Proutski, V., T. S. Gritsun, E. A. Gould, and E. C. Holmes. 1999. Biological consequences of deletions within the 3′-untranslated region of flaviviruses may be due to rearrangements of RNA secondary structure. Virus Res. 64:107-123. [DOI] [PubMed] [Google Scholar]
- 31.Rauscher, S., C. Flamm, C. W. Mandl, F. X. Heinz, and P. F. Stadler. 1997. Secondary structure of the 3′-noncoding region of flavivirus genomes: comparative analysis of base pairing probabilities. RNA 3:779-791. [PMC free article] [PubMed] [Google Scholar]
- 32.Rice, C. M., E. G. Strauss, and J. H. Strauss. 1996. Structure of the flavivirus genome, p. 279-326. In S. Schlesinger and M. J. Schlesinger (ed.), The Togaviridae and Flaviviridae. Plenum, New York, N.Y.
- 33.Scherret, J. H., M. Poidinger, J. S. Mackenzie, A. K. Broom, V. Deubel, W. I. Lipkin, T. Briese, E. A. Gould, and R. A. Hall. 2001. The relationships between West Nile and Kunjin viruses. Emerg. Infect. Dis. 4:697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shurtleff, A. C., D. W. C. Beasley, J. J. Y. Chen, H. Ni, M. T. Sudermann, R. X. Wang, E. Wang, S. C. Weaver, D. M. Watts, K. L. Russell, and A. D. T. Barrett. 2001. Genetic variation in the 3′ non-coding region of dengue viruses. Virology 281:75-87. [DOI] [PubMed] [Google Scholar]
- 35.Swofford, D. L. 1991. PAUP: phylogenetic analysis using parsimony, version 3.0. Illinois Natural History Survey, Champaign, Ill.
- 36.Wallner, G., C. W. Mandl, C. Kunz, and F. X. Heinz. 1995. The flavivirus 3′-non-coding region: extensive size heterogeneity independent of evolutionary relationships among strains of tick-borne encephalitis virus. Virology 213:169-178. [DOI] [PubMed] [Google Scholar]
- 37.Wang, E., K. D. Ryman, A. D. Jennings, D. J. Wood, F. Taffs, P. D. Minor, P. G. Sanders, and A. D. T. Barrett. 1995. Comparison of the genomes of the wild-type French viscerotropic strain of yellow fever virus with its vaccine derivative French neurotropic vaccine. J. Gen. Virol. 76:2749-2755. [DOI] [PubMed] [Google Scholar]
- 38.Wang, E., S. C. Weaver, R. E. Shope, R. S. Tesh, D. M. Watts, and A. D. T. Barrett. 1996. Genetic variation in yellow fever virus: duplication in the 3′ noncoding region of strains in Africa. Virology 225:274-281. [DOI] [PubMed] [Google Scholar]
- 39.Wengler, G., and E. Castle. 1986. Analysis of structural properties, which possibly are characteristic for 3′-terminal sequence of the genome RNA of flaviviruses. J. Gen. Virol. 67:1183-1188. [DOI] [PubMed] [Google Scholar]
- 40.Zuker, M., D. H. Mathews, J. Sabina, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. In J. J. Barciszewski and B. F. C. Clark (ed.), RNA biochemistry and biotechnology. Kluwer Academic Publishers, Boston, Mass.