Abstract
Retroviral vectors produced from packaging cells are invariably contaminated by protein, nucleic acid, and other substances introduced in the manufacturing process. Elimination of these contaminants from retroviral vector preparations is helpful to reduce unwanted side effects, and purified vector preparations are desirable to improve reproducibility of therapeutic effect. Here we report a novel approach to engineer a metal binding peptide (MBP)-tagged murine leukemia virus (MuLV), allowing for one-step purification of retroviral vectors by immobilized metal affinity chromatography (IMAC). We inserted a His6 peptide into an ecotropic envelope protein (Env) by replacing part of its hypervariable region sequence with a sequence encoding the His6 peptide. Display of the His6 tag on the surface of Env endowed the vectors with a high affinity for immobilized metal ions, such as nickel. We demonstrated that the His6-tagged MuLV could be produced to high titers and could be highly purified by one-step IMAC. The protein and DNA contaminants in the purified vector supernatants were below 7 μg/ml and 25 pg/ml, respectively, indicating a 1,229-fold reduction in protein contaminant level and a 6,800-fold reduction in DNA contaminant level. About 56% of the viral vectors were recovered in the IMAC purification. The purified vectors retained their functionality and infectivity. These results establish that an MBP can be functionally displayed on the surface of ecotropic retroviruses without interfering with their integrity, and MBP-tagged retroviral vectors can be highly purified by one-step IMAC.
Recombinant retroviral vectors are used in the majority of gene therapy trials to accomplish life-long cures of inherited diseases. Retroviral vectors produced from packaging cell lines are invariably contaminated by proteins and nucleic acids, as well as other substances introduced in the manufacturing process (7, 22). Elimination of these contaminants from retroviral vector preparations is helpful to reduce unwanted side effects, and purified vector preparations are desirable to improve reproducibility of therapeutic effect.
At the laboratory scale, the processing of retroviral vectors is relatively straightforward. The viral vector supernatants are usually prepared by separation of viral particles from particulates and cell debris by filtration through 0.45-μm-pore-size membranes. Nevertheless, the manufacture of retroviral vectors for use in human gene therapy is quite complicated. It requires not only large volumes but also high purity of the viral supernatants (2). Although large-scale production of retroviral vectors can be readily achieved with existing technologies, purification of the retroviral vectors remains a difficult technical challenge. Considerable efforts have been made to develop a variety of processes for purification of retroviral vectors. Both ultracentrifugation and low-speed centrifugation have been employed to prepare highly concentrated retroviral vectors (3, 8, 10, 36). Cosedimentation of small cell-derived vesicles, as well as serum proteins, with the viral particles led to even more serious contamination in those concentrated retroviral vectors (4, 19). The retroviral vectors are 80 to 100 nm in diameter (6) and have a density of 1.16 to 1.18 g/ml, which is similar to the density of cell culture medium (21). Consequently, it is impossible to remove all of the contaminants from viral supernatants by centrifugation. The removal of serum protein contaminants may be achieved by a size-exclusion membrane filtration using a 100-kDa molecular mass cutoff membrane (5, 18, 29, 34). Large-molecule contaminants (i.e., mass of >100 kDa), such as bovine immunoglobulins and proteoglycans, however, cannot be eliminated from the vector supernatants with this method (2, 18, 20). Other methods, including polyethylene glycerol precipitation (1), calcium phosphate precipitation (26), and two-phase extraction (13, 14), have also been examined for purification of retroviral vectors. Cosedimentation of impurities along with retroviral particles limits the use of any of these methods. Nevertheless, all of these methods are time- and cost-consuming, difficult to scale up and, most importantly, they can significantly reduce the transduction ability of retroviral vectors (2, 34).
More recently, several groups have developed a number of rapid purification techniques with affinity chromatography by utilizing some specific ligands on a virus surface. A good example is the alphaherpesvirus, which attaches to cells by binding to the negatively charged sulfate groups of the cell surface heparin sulfate. Since sulfonic acid contains an SO3H group which is chemically similar to the heparin sulfate, a sulfonic acid-modified cation membrane has been tested to purify the virus by chromatography (16). However, this methodology requires a specific glycoprotein called gC for the virus' binding to the cation exchange membrane. Size-exclusion chromatography has also been explored for virus purification (24). As with the size-exclusion membrane filtration, the removal of high-molecular-weight contaminants is difficult to achieve (2). While chemically stable and inexpensive ligands with a relatively high specificity for the viruses are attractive for large-scale purification of viral vectors, the requirement of specific acceptors on the viral surface to mediate their binding to the ligands used in the chromatography has hindered the use of this technology for retroviral vector preparation (27).
Immobilized metal affinity chromatography (IMAC) has the potential to become a new methodology for preparation of highly purified retroviral vectors. In IMAC, the composition and conformation of a protein dictate the binding affinity of the protein for immobilized metal ions (32). The difference in binding affinities of proteins for immobilized metal ions is the basis for protein fractionation in IMAC (9). Typically, nitrilotriacetic acid (NTA) or iminodiacetic acid is linked to an agarose or silica gel support to provide chelation sites for a metal ion (Zn2+, Cu2+, or Ni2+) via the NTA or iminodiacetic acid functional groups. Factors such as the accessibility, microenvironment of the binding residue (i.e., histidine, cysteine, and tryptophan), cooperation between neighboring amino acid side groups, and local conformations play important roles in the strength of binding and, thus, protein retention and specificity. Subsequent destruction of the bond by lowering the pH or addition of a competing ligand such as imidazole in the mobile phase displaces the bound protein and results in the elution of the metal binding protein. Because the binding of the protein to the immobilized metal ions is specific, impurities which do not possess the metal binding site will not bind and can be purified away from the bound material. When the prerequisites for retention on IMAC resin are absent, genetic modifications may endow a protein with increased metal binding affinity by insertion of the binding moiety into its amino acid sequence. The amino acid sequences of the metal affinity tags are selected based on either (i) a previously noticed capacity of a given peptide sequence to interact with metal ions, or (ii) the premise that multiplicity of histidine residues will increase metal binding. To adopt IMAC for the purification of retroviral vector supernatants, we proposed to display a metal binding peptide (MBP) on the surface of a retrovirus by inserting an MBP into the surface unit protein (SU) of the retrovirus, thereby endowing the retrovirus with a high affinity for immobilized metal ions. The MBP-tagged viral vectors can then be highly purified by IMAC.
MATERIALS AND METHODS
Cell lines.
293T cells and the packaging cell line GP293 (BD Clontech, Palo Alto, Calif.) were grown in Dulbecco's modified Eagle's medium (DMEM) with 10% heat-inactivated fetal bovine serum (FBS; American Type Culture Collection, Manassas, Va.). The retrovirus producer cell line RVPC-1 was grown in DMEM with 10% non-heat-inactivated FBS (40).
Mutagenesis.
E/A-PRR was kindly provided by Paula M. Cannon (39). This plasmid encodes an ecotropic Env protein with the PRR region replaced by that of an amphotropic virus. pRVHis_e encodes a His6-tagged ecotropic Env, which was constructed by inserting the His6 peptide sequence into the PRR region encoded in E/A-PRR. The oligonucleotides used for His6 peptide insertion were His-1 (5′-ACAATCCTGCAGCATCACCATCACCATCACTCAACCTCCCCTACAAGTCCAAGTGTCC-3′) and His-1R (5′-AGGGAACAAAAGCTGGAGCT-3′). The PstI site is underlined, and the His6 peptide sequence is italicized. The PCR products amplified from E/A-PRR using His-1 and His-1R were digested with restriction enzymes and ligated into E/A-PRR through the restriction enzyme sites, PstI and NotI, resulting in pRVHis_e. As a result, the sequence between PstI and StuI in the PRR region was replaced by the His6 peptide sequence.
Western blotting.
In order to analyze the form of the His6-tagged Env present in cell lysates, 293T cells were transfected with envelope protein expression plasmids alone (8 μg of DNA per 100-mm plate). Cell lysates were prepared by incubating the cells in 500 μl of M-PER lysis buffer (Pierce, Rockford, Ill.) for 10 min at room temperature, followed by centrifugation at 10,000 × g for 10 min at 4°C to pellet nuclei. Envelope proteins were denatured by treating the lysates with 0.5% sodium dodecyl sulfate (SDS) and 1% β-mercaptoethanol for 5 min at 100°C and then deglycosylated in a deglycosylation buffer (New England BioLabs, Beverly, Mass.) for 1 h at 37°C. Proteins were separated on a 4-to-20% gradient polyacrylamide gel and transferred onto a nitrocellulose membrane (0.4 μm) by using a semidry electroblotter (Fisher Scientifics, Pittsburgh, Pa.). The blot was blocked overnight at 4°C in 5% nonfat milk in Tris-buffered saline (TBS)-Tween buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, and 0.1% Tween 20), followed by incubation with primary antibodies, goat anti-Rauscher murine leukemia virus (MuLV) gp69/71 in a 1:1,000 dilution ratio (American Type Culture Collection). After washing with TBS-Tween buffer three times each for 10 min, the blot was incubated with the secondary antibodies, rabbit anti-goat immunoglobulin G (IgG) horseradish peroxidase (HRP) conjugates (Sigma, St. Louis, Mo.), in a 1:10,000 dilution ratio for 1 h at room temperature and thereafter given a further wash three times, each for 10 min in TBS-Tween buffer. Proteins were visualized with the super-chemiluminescence reagent (Pierce).
Immunolocalization of the His6-tagged Env.
293T cells were transiently transfected with an envelope protein expression plasmid alone (8 μg of DNA per 100-mm plate) and harvested 48 h posttransfection. After washing once with phosphate-buffered saline (PBS) buffer, 106 cells were transferred into 50 μl of ice-cold PBS-5% bovine serum albumin (BSA) buffer and incubated for 10 min at 4°C, followed by washing twice with ice-cold PBS buffer. The cells were thereafter incubated with mouse anti-Penta-His antibodies at a 1:50 dilution ratio in the PBS-1% BSA buffer for 1 h at 4°C. After washing three times each with 200 μl of ice-cold PBS buffer, the cells were incubated with anti-mouse IgG fluorescein isothiocyanate (FITC) conjugate (Sigma), with a 1:100 dilution ratio in the PBS-1% BSA buffer for 1 h at 4°C, followed by washing four times each with 200 μl of ice-cold PBS buffer. The expression of the His6-tagged Env was detected by FACScan (Becton Dickinson, San Diego, Calif.). Fluorescence microscopy was also performed to image the His6-tagged Env on the cell surface. The cells were incubated with mouse anti-Penta-His antibodies against the His6 tag. Rabbit anti-mouse IgG FITC conjugate (Sigma) was used as secondary antibody. After staining, the cells were examined under an inverted fluorescence microscope (IX 70; Olympus, Tokyo, Japan).
Viral vector production.
Retroviral vectors were produced by cotransfection of a viral vector, pCMVEGFP_neo (40), into GP293 cells with an Env expression plasmid by using the PolyFect transfection reagent (QIAGEN, Valencia, Calif.) as described elsewhere (41). The viral supernatants were collected 48 h posttransfection, filtered through a 0.45-μm membrane filter, and stored at −70°C until use. The viral titer was determined with NIH 3T3 cells. In brief, the cells were plated in six-well plates at a density of 105 cells per well 1 day prior to infection. One milliliter of 10-fold-serially diluted virus supernatants was used to infect the cells in each well in the presence of 8 μg of Polybrene/ml. The medium was exchanged with fresh medium 24 h postinfection, and 1.2 mg of G418/ml was added to the medium 48 h postinfection. The medium was then exchanged every 4 days until 12 days. The resistant colonies were scored by methylene blue staining. Ecotropic MuLV vectors, which served as a control for the experiments described below, were produced from RVPC-1 cells (40).
IMAC purification of the His6-tagged retroviral vectors.
A column filled with 1 ml of Ni-NTA agarose (QIAGEN) was used to purify the His6-tagged retroviral vectors. The column was preequilibrated with a five-bed volume of PBS buffer (pH 7.5). The vector supernatants were buffered to pH 7.5 with 10× PBS buffer. A 3-ml aliquot of the viral supernatants was gravity fed into the column at a rate of 0.6 ml/min, followed by washing with 10 ml of PBS buffer containing 10 mM imidazole (pH 7.5). The retroviral vectors were eluted with PBS buffers containing high concentrations of imidazole ranging from 40 to 250 mM at a rate of 0.6 to 0.7 ml/min.
SDS-PAGE.
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was performed to determine the removal of protein contaminants from the vector supernatants by IMAC. The proteins were resolved onto a 4-to-20% gradient precast gel (Bio-Rad Laboratories, Hercules, Calif.), visualized with CBB-R250 dye, and detected with a Kodak 1-D gel analysis system (Kodak, Tokyo, Japan).
Protein and DNA assays.
Protein contaminants in the vector supernatant, flowthrough, washing, and elution fractions were determined by protein microassay using the reagents from Bio-Rad Laboratories. The lowest protein concentration that can be detected in the microassay is 1 μg/ml. DNA contaminants in the vector supernatant were measured with PicoGreen dye (Molecular Probes) according to the manufacturer's instructions. A standard curve was prepared using λ DNA concentrations between 25 pg/ml and 50 ng/ml.
RESULTS
Construction of the His6-tagged Env.
The retention of ecotropic MuLV vectors on the Ni-NTA chromatography column was first examined. As presented in Table 1, almost 100% of the MuLV vectors passed through the column without binding. It was postulated that fusion of a short MBP to a retroviral surface protein might endow the retrovirus with sufficient affinity for immobilized metal ions, e.g., Ni2+. An important question was whether an insertion site could be identified such that the MBP would be surface exposed and accessible for binding to the immobilized nickel. Another issue of concern was the infectivity of the engineered vectors. To be effective as an affinity peptide for retroviral vector purification, the peptide should not lead to any significant decrease in viral titer.
TABLE 1.
Binding affinity of retrovirus vectors for Ni-NTA agarose
| Fraction | Viral titer (105 CFU/ml)a
|
||
|---|---|---|---|
| Ecotropic MuLV | E/A-PRR | RVHis_e | |
| Feed | 4.5 ± 0.3 | 4.3 ± 0.3 | 3.5 ± 0.5 |
| Flowthrough | 4.0 ± 0.5 | 4.4 ± 0.1 | 0.042 ± 0.004 |
Each titer is the average of three independent experiments ± the standard error. Each fraction contained 3 ml of the vector supernatant. E/A-PRR, the viruses enveloped with an ecotropic MuLV Env protein with the PRR region replaced by that of an amphotropic virus. RVHis_e: the virus enveloped with His6-tagged ecotropic MuLV Env.
MuLV encodes an Env precursor, Pr85, that is initially glycosylated in the endoplasmic reticulum, further processed in the Golgi apparatus, and proteolytically cleaved to generate two subunits, the surface protein unit (SU), gp70, and the transmembrane protein (TM), p15E (25, 30, 38). The signal sequence directs Env to enter a secretory pathway and reach the cell surface, where they are incorporated into budding virions (15) and form the surface proteins. The N-terminal portion of SU is responsible for receptor recognition, which determines virus host range (23, 25), whereas the C-terminal portion of SU is more conserved and is believed to be associated with TM (31, 35). A hypervariable PRR links the N and C termini of SU (37). The N terminus of PRR is conserved among five subtypes of MuLVs with a consensus sequence of GPR(I/V)PIGPNP (12) and is essential for viral infection (33, 37), whereas the C terminus of PRR is variable in length and in sequence among different viruses (17, 28, 39). X-ray structural analyses of feline leukemia virus revealed that PRR forms an exposed loop between the N- and C-terminal domains of SU (11). Accordingly, the insertion of a small peptide in the hypervariable region of PRR would most likely lead to surface exposure of the peptide. Mutagenesis studies demonstrated that truncation of up to 29 amino acids in the hypervariable region of PRR had very little effect on the viral titer (37). Taken together, we decided to insert a His6 peptide in the hypervariable region of PRR. The His6 peptide was selected because of its ability to interact strongly with immobilized nickel and other divalent metal ions, such as copper and zinc. The choice of the His6 peptide, therefore, enabled us to test the availability of the peptide for binding to the immobilized nickel and the peptide's effect on virus infectivity. The His6-tagged Env was engineered by inserting a His6 peptide sequence into the hypervariable region of PRR by using site-directed mutagenesis (Fig. 1). The sequence between PstI and StuI in the amphotropic PRR encoded by E/A-PRR was deleted and replaced by a sequence (CATCACCATCACCATCAC) encoding the His6 peptide (HHHHHH). To facilitate screening for positive mutants in the site-directed mutagenesis, we replaced the StuI site with a sequence that was originally used in the amphotropic PRR. As a result, the chimeric PRR consisted of 55 amino acid residues, including 15 highly conserved amino acid residues at the N terminus of PRR. The viruses bearing the chimeric PRR on their surface were produced and titers were determined for examining the effect of the His6 peptide insertion on the viral integrity. We found no significant reduction in viral titer for the His6-tagged viruses. As shown in Table 1, the titer of the His6-tagged RVHis_e vectors was about (3.5 ± 0.5) ×105 CFU/ml (mean ± standard error), whereas the titer of the ecotropic MuLV vectors was (4.5 ± 0.3) ×105 CFU/ml. Accordingly, we concluded that the insertion of the His6 peptide in PRR did not interfere with the viral integrity.
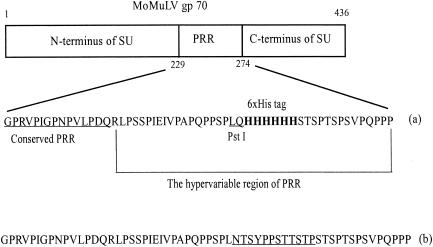
FIG. 1.
Construction of the His6-tagged Env. (a) The His6 peptide was inserted in the hypervariable region of PRR downstream of the PstI site. The sequence between PstI and StuI in the PRR region encoded by E/A-PRR was deleted and replaced by a sequence encoding the His6 peptide. The StuI site was removed as well, to facilitate screening for positive mutants in the site-directed mutagenesis. (b) Sequence of the PRR region encoded by E/A-PRR. The underlined sequence was replaced by the His6 peptide sequence shown in panel a.
Immunolocalization of His6-tagged Env.
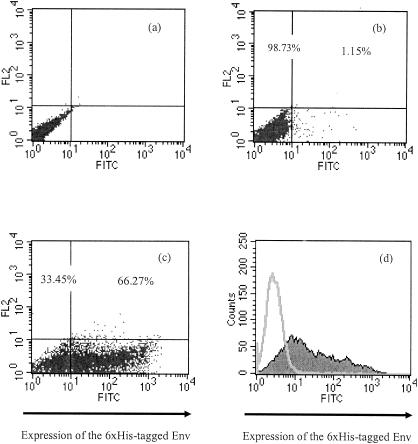
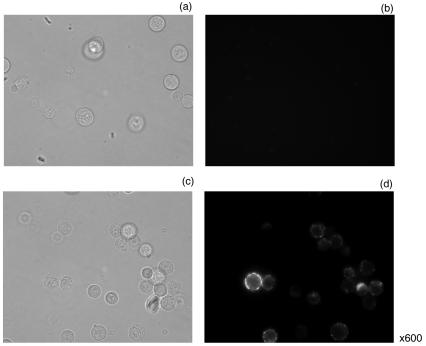
To determine the accessibility of the His6 site, we detected His6-tagged Env by immunolocalization assay. The chimeric Env proteins were expressed in 293T cells, and the cell surface-localized Env was examined by flow cytometry with an anti-Penta-His antibody. Anti-Penta-His antibody specifically recognizes the 5-adjacent histidine peptide (xHHHHHx; x signifies any amino acid other than histidine). The nontransfected 293T cells served as a negative control (Fig. 2a), and E/A-PRR-transfected 293T cells were used as a mock for the assays. Compared to the mock and control, the indirect immunostaining of pRVHis_e-transfected cells gave rise to a shift of the mean fluorescence peak to 8 arbitrary units (relative fluorescence), resulting in a 25.96-fold enhanced signal (Fig. 2d). About 66.3% of pRVHis_e-transfected 293T cells were labeled by anti-Penta-His antibody (Fig. 2c), indicating a good accessibility of the peptide ligands. The other 33.5% of cells did not show fluorescence, partly due to low transfection efficiency. Indeed, the transfection efficiency that we tested (using enhanced green fluorescent protein as a reporter) was around 70%. Besides, a histogram of the antibody-labeled cells showed a wide spread of fluorescence intensity. This may be attributable to a variable expression level of Env proteins on the cell surface. Fluorescence microscopy confirmed the display of the His6-tagged Env on the surface of pRVHis_e-transfected 293T cells. To detect the display of the His6-tagged Env on the cell surface of pRVHis_e-transfected 293T cells, the cells were stained with anti-Penta-His antibody as primary antibody and anti-mouse IgG FITC conjugate as secondary antibody. We observed a green fluorescence ring around the pRVHis_e-transfected cells under a fluorescence microscope, indicating the surface exposure of the His6-tagged Env to the antibodies (Fig. 3d).
FIG. 2.
Immunolocalization assay of cell surface-displayed His6-tagged ecotropic Env. (a) 293T cells. (b) Mock transfection of 293T cells transfected with E/A-PRR. (c) 293T cells transfected with pRVHis_e encoding the His6-tagged Env. (d) Histogram of the cells expressing the His6-tagged Env on their surface. Cells were collected 48 h posttransfection and stained with mouse anti-Penta-His antibodies as the primary antibody (1:50 dilution ratio) and goat anti-mouse IgG FITC conjugate as the second antibody (1:100 dilution ratio). After staining, the samples were applied to flow cytometry immediately. All the samples contained 106 cells. The FL2 channel measured autofluorescence emitted from the cells, which corrected the measurements from the FL1 channel (labeled as FITC channel) by adjusting the color compensation in the flow cytometry.
FIG. 3.
Expression of His6-tagged Env on the cell surface. Cells were collected 48 h posttransfection and stained with mouse anti-Penta-His antibodies as primary antibody and goat anti-mouse IgG FITC conjugate as secondary antibody. The cells were mounted onto a glass slide and examined under an inverted fluorescence microscope with a 60× oil objective. (a and b) E/A-PRR-transfected cells; (c and d) pRVHis_e-transfected cells.
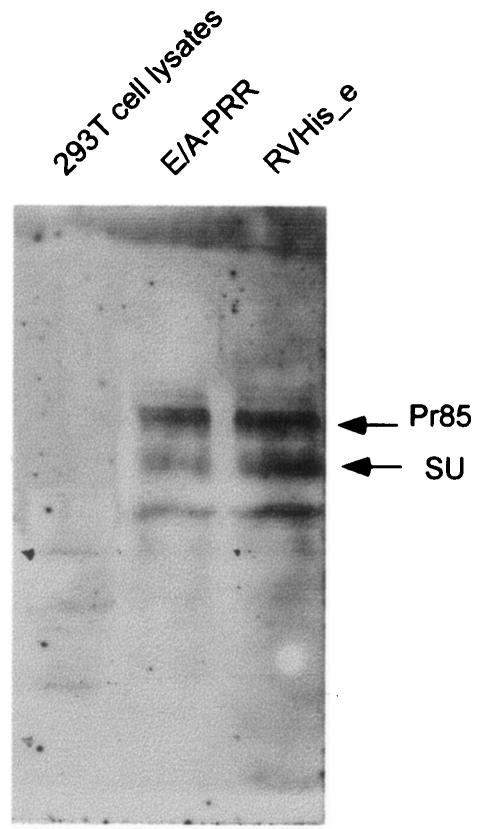
To examine the effect of the His6 peptide on the processing of Pr85 to SU and TM subunits, we analyzed the form of Env proteins present in the lysates of 293T cells. We employed N-glycosidase to treat the lysates in order to clearly discriminate Pr85, as well as SU, from various processing intermediates of Pr85. Probing a Western blot of Pr85 and SU with an anti-SU antibody revealed approximately equal amounts of Pr85 and SU in the E/A-PRR- and pRVHis_e-transfected 293T cell lysates, indicating a similar level of Pr85 processing of SU for both constructs (Fig. 4).
FIG. 4.
Western blot of the His6-tagged envelope proteins in cell lysates. The proteins were deglycoslated and probed with an anti-SU antibody, goat anti-Rauscher MuLV gp69/71 at a 1:1,000 dilution as primary antibody, and a rabbit anti-goat IgG HRP conjugate as second antibody at a 1:10,000 dilution. Pr85 is the precursor form of the envelope proteins, and SU is the mature form of the envelope proteins.
One-step purification of the His6-tagged retroviral vectors using one-step IMAC.
In the next experiment, we investigated the purification of the His6-tagged retroviral vectors with a Ni-NTA column. The experiments were conducted under conditions identical to those reported in Table 1. Unlike the nontagged retroviral vectors, the His6-tagged retroviral vectors were almost completely retained in the column. Only about 1% of the viruses were detected in the flowthrough fraction (Table 1). These experiments provided fundamental evidence that the His6 peptide inserted in the PRR region was not only accessible to its antibody, but also offered a high binding affinity for the immobilized nickel. Subsequently, we determined the optimal conditions for IMAC purification of the His6-tagged retroviral vectors. We used 10× PBS buffer to adjust the vector supernatants to pH 7.3. To wash away nonspecifically or weakly bound proteins and other contaminants from the Ni-NTA column, we included 10 mM imidazole in the washing buffer. It was postulated that the His6-tagged viral vectors could be eluted from the Ni-NTA column by either reducing the pH in the elution buffer or by increasing the concentration of imidazole in the elution buffer. Based on the data accumulated in the protein purification, we expected that the His6-tagged viral vectors would completely dissociate from the Ni-NTA by raising the imidazole concentration to 100 to ∼250 mM in the elution buffer.
We first employed 150 mM imidazole to elute the viral vectors from the Ni-NTA column. Approximately 35% of the viral vectors were recovered (Table 2). To determine what caused the low recovery rate, we assessed the effect of imidazole on the viral titer. We found that the viral titers were reduced nearly 75% when the vectors were exposed to 150 mM imidazole for 20 min at room temperature (Table 3). Exposure of the vectors to higher concentrations of imidazole led to a further decline in the viral titers. The vectors retained high titers when exposed to a lower concentration of imidazole. Exposure of the vectors to 75 mM imidazole for 20 min at room temperature resulted in only a 40% reduction in the viral titers. No decline in titers was observed when the vectors were exposed to 40 mM imidazole for 20 min at room temperature. Accordingly, recovery of the vectors in the IMAC purification could be improved by eluting the vectors with lower concentrations of imidazole. In the next experiment, we employed 75 mM imidazole to elute the vectors from the Ni-NTA column. About 56% of the vectors were recovered in an elution buffer containing 75 mM imidazole (Table 2). However, a further decrease in the concentration of imidazole in the elution buffer did not result in an increase in the recovery rate. As shown in Table 2, no vectors were recovered when 40 mM imidazole was employed to elute the vectors. We speculated that the low recovery rate when 40 mM imidazole was used for elution was caused by the retention of the vectors onto the column in a lower concentration of imidazole because of the high affinity of the His6-tagged retroviral vectors. To determine this effect, a 50 mM EDTA solution was passed through the column after the elution of the vectors with 40 mM imidazole. EDTA chelated the nickel ions and removed them from the NTA groups together with the His6-tagged viral vectors. About 36% of the vectors were recovered in the EDTA elution, indicating retention of the His6-tagged retroviral vectors onto the column when 40 mM imidazole was used for elution. It was felt that the lack of complete recovery of the vectors in the EDTA elution might have been the result of inactivation of the vectors in the presence of EDTA. The effect of EDTA on vector infectivity was investigated by incubating the vectors with 50 mM EDTA. We found that the viral titer decreased nearly 79% after exposure of the vectors to EDTA for 20 min at room temperature (data not shown). Thus, the discrepancy between the loaded and recovered viral vectors could be entirely explained by the inhibitory effect of EDTA.
TABLE 2.
Purification of His6-tagged retrovirus vectors by one-step IMAC
| Imidazolea (mM) | Viral titer (105 CFU/ml)b
|
Recovery rate (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| F (3 ml) | E1 (1 ml) | E2 (1 ml) | E3 (1 ml) | E4 (1 ml) | ED 1 (1 ml) | ED 2 (1 ml) | ||
| 40 | 5.1 | 0 | 0 | 0 | 0 | 0.36 | 5.1 | 0 |
| 75 | 7.2 | 3.6 | 8.4 | 0.018 | 0 | 56 | ||
| 150 | 5.0 | 2.1 | 3.1 | 0.12 | 0 | 35 | ||
Concentration of imidazole in the elution buffer.
Each titer is the average of three independent determinations. F, feed fraction; E1 to E4, elution fraction; ED1 and ED2, vectors eluted with 50 mM EDTA after elution with 40 mM imidazole.
TABLE 3.
Effect of imidazole on the infectivity of retroviruses
| Imidazole concn (mM) | Viral titer (105 CFU/ml)a |
|---|---|
| 0 | 3.0 ± 0.2 |
| 10 | 2.3 ± 0.5 |
| 40 | 3.2 ± 0.4 |
| 75 | 1.8 ± 0.3 |
| 100 | 1.2 ± 0.5 |
| 150 | 0.75 ± 0.06 |
| 250 | 0.39 ± 0.07 |
Viral titers were determined after exposure of the RVHis_e vectors to imidazole for 20 min at room temperature. Each titer is the average of three independent experiments ± the standard error.
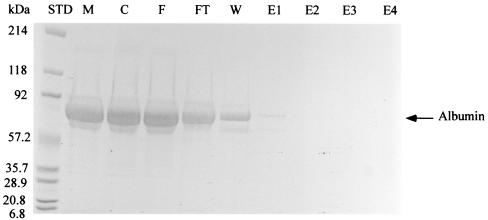
The purity of the retroviral vectors eluted with 75 mM imidazole was determined by SDS-PAGE assay, protein microassay, and DNA PicoGreen assay. As shown in Fig. 5, the majority of the protein contaminants in the vector supernatants were bovine serum proteins that were introduced in the cell culture medium to improve vector production. Most of the serum protein contaminants flowed through the column without any binding to the Ni-NTA matrix (Fig. 5, lane FT). The weakly bound serum proteins could be efficiently purified away by washing the column with 10 mM imidazole (Fig. 5, lane W). No protein contaminants were found in the purified viral supernatants (Fig. 5, lane E2), indicating a high purity of the IMAC-purified vector supernatants. Protein assay revealed that the concentration of protein contaminants in the vector supernatants before purification was 8.4 mg/ml in total (Table 4). It dropped down to 0.23 mg/ml in the first elution fraction and 7 μg/ml in the second elution fraction, indicating a 1,229-fold reduction in protein contamination in the purified vector supernatants. The second elution fraction contained most of the recovered viral vectors. The DNA assay showed that the concentration of DNA contaminants in the vector supernatants before purification was 0.17 μg/ml. About 59% of DNA contaminants flowed through the column without any binding to the Ni-NTA matrix. Nearly all of the weakly bound DNA contaminants were purified away in the washing step, because we could not detect any DNA contaminants in the purified vector supernatants in the PicoGreen DNA assay. The lowest concentration of DNA that can be determined by the PicoGreen DNA assay is 25 pg/ml. We, therefore, estimated that a 6,800-fold reduction in the DNA contamination level was achieved in the one-step IMAC purification.
FIG. 5.
Detection of protein contaminants in purified vector supernatant. The viral vectors were eluted with 75 mM imidazole. Lanes: STD, protein standards; M, DMEM-10% FBS; C, cell culture supernatant; F, feed fraction; FT, flowthrough fraction; W, washing fraction; E1 to E4, elution fractions.
TABLE 4.
Removal of contaminants from retrovirus vector supernatants by IMACa
| Amt in fraction
|
||||||||
|---|---|---|---|---|---|---|---|---|
| DMEM | F | FT | W | E1 | E2 | E3 | E4 | |
| DNAb (μg/ml) | 0 | 0.17 | 0.1 | 0.02 | <2.5 × 10−5 | <2.5 × 10−5 | <2.5 × 10−5 | <2.5 × 10−5 |
| Proteinc (mg/ml) | 8.4 | 8.6 | 3.07 | 1.11 | 0.23 | 0.007 | <0.001 | <0.001 |
The viral vectors were eluted with 75 mM imidazole.
Total double-stranded nucleic acids.
Total protein. DMEM, cell culture medium; F, feed fraction (3 ml); FT: flowthrough fraction (3 ml); W, washing fraction (10 ml); E1 to E4, elution fraction (1 ml).
We also explored the possibility of elution of the His6-tagged retroviral vector by reducing the pH of the elution buffer. It has been shown that histidine residues in the His6 peptide have a pKa of approximately 6.0. The histidine residues become protonated if the pH is reduced to 4.5 to 5.3. Our investigators have demonstrated that MuLV vectors remain infectious in the pH range from 5.5 to 8.0 (40). Accordingly, we investigated the elution of the viral vectors under those pH values. As shown in Table 5, no vectors were eluted from the Ni-NTA column even at low pH. Although most of the DNA contaminants were removed from the vector supernatants, protein contaminants were still present in all elution fractions, indicating the inability to remove protein contaminants under the pH range that we have examined. The SDS-PAGE assay confirmed this result (data not shown). Consequently, changing the pH of the elution buffer is not a good option for eluting His6-tagged vectors from a Ni-NTA column, because of the difficulty in removing all contaminated proteins from the vector supernatants without inactivating the viral particles.
TABLE 5.
Elution of His6-tagged retrovirus vectors under different pHs
| Fraction (3 ml) | Viral titer (105 CFU/ml) | Proteins (mg/ml) | DNA (μg/ml) |
|---|---|---|---|
| Feed | 4.2 | 7.72 | 0.16 |
| Flowthrough | 0.03 | 2.13 | 0.11 |
| pH 7.3a | 0 | 0.89 | 0.059 |
| pH 6.5 | 0 | 1.11 | <2.5 × 10−5 |
| pH 6.0 | 0 | 1.02 | <2.5 × 10−5 |
| pH 5.5 | 0 | 0.57 | <2.5 × 10−5 |
| pH 5.0 | 0 | 0.15 | <2.5 × 10−5 |
The pH value of the elution buffer.
DISCUSSION
The comparison of different methods for preparation of purified retroviral vector supernatants is presented in Table 6. Although the retroviral vectors can be highly concentrated by treating the vector supernatants with centrifugation or hollow fiber filtration, the presence of large molecular contaminants or cosedimentation of contaminants leads to a concern over the safety of the vector supernatants for in vivo administration. Chromatography could be an excellent technique for purification of retroviral vectors, since most protein and DNA contaminants could be eliminated from the purified vector supernatants. Compared to these methods, IMAC purification provides a titer higher than 1.2 × 106 CFU/ml and a 56% recovery rate. Of significance, this method costs less and can easily be scaled up.
TABLE 6.
Comparison of different methods used for purification of retrovirus vectors
| Method (Reference[s]) | Yield (%) | Contaminants in purified vector supernatants
|
Viral titer (CFU/ml) | Large-scale processing | Cost | |
|---|---|---|---|---|---|---|
| Proteins (mg/ml) | DNA (μg/ml) | |||||
| This work | 56 | <0.007 | <2.5 × 10−5 | 1.2 × 106 | Easy | Low |
| Ultra-filtration (12) | 65 | 3.2 | 42.2 | 1.0 × 105 | Difficult | High |
| Hollow fiber filtration (13, 15) | 50-80 | Large-molecular contaminants are co-concentrated with viral vectors | ∼5.5 × 107 | Difficult | Medium | |
| Chromatography (12) | 10 | 0.09 | 3.78 | 1.6 × 104 | Easy | Low |
| Centrifugation (4, 6, 7) | 50-97 | Significant cosedimentation of contaminants, especially those corresponding in size to known viral proteins, including CA, integrase, and matrix proteins | 106-1.1 × 108 | Difficult | Medium | |
Serum proteins with a smaller number of accessible histidine residues as well as amino acids that possess electron-donating groups can compete with the His6-tagged viral vectors for binding sites, resulting in a weak binding of these components to the Ni-NTA matrix. However, these interactions are much weaker than the binding of the His6-tagged retroviral vectors to the Ni-NTA matrix. Indeed, we demonstrated that both protein and DNA contaminants could be efficiently purified away from the Ni-NTA column by inclusion of 10 mM imidazole in the washing buffer. The presence of imidazole and the possibility of leakage of nickel in the purified vector supernatants could be of some concern. Nevertheless, they can be readily removed by dialysis, as these molecules are very small. Coupling a dialysis process to IMAC purification could provide another advantage to allow for the delivery of the purified viral vectors to a preferred buffer for in vivo administration. We have tested the effect of dialysis on the infectivity of the purified viruses. The viruses were dialyzed against PBS buffer with the volume ratio of 1:1,000 (volume of virus in the dialysis membrane/volume of PBS buffer used for dialysis). The dialysis was carried out for 3 h at 4°C with two exchanges of dialysis buffer. We found no reduction in the viral titer after dialysis (data not shown).
In summary, we established that retroviral vectors can be purified by IMAC with a Ni-NTA column through the surface-displayed His6 tag, which endows the viral vectors with an increased affinity for the immobilized nickel. These results serve as a proof of principle that the impurities and/or contaminants introduced in the manufacture of retroviral vectors can be efficiently eliminated from the vector supernatants by IMAC. To apply this technology for preparation of clinical-grade retroviral vectors for human gene therapy applications, it may be necessary to engineer a His6-tagged amphotropic or a His6-tagged GalV Env. Obviously, the elimination of contaminants from the retroviral vector supernatants would significantly improve the safety and reproducibility of therapeutic effect of retroviral vectors in human gene therapy.
Acknowledgments
We thank Paula M. Cannon (Norris Cancer and Department of Biochemistry and Molecular Biology, University of Southern California School of Medicine) for providing the plasmid E/A-PRR. We thank Ali Qzuer (Molecular Genetics and Biochemistry Department, University of Pittsburgh), who helped with the nucleic acid assay.
REFERENCES
- 1.Aboud, M., M. Wolfson, Y. Hassan, and M. Huleihel. 1982. Rapid purification of extracellular and intracellular Moloney murine leukemia virus. Arch. Virol. 71:185-195. [DOI] [PubMed] [Google Scholar]
- 2.Andreadis, S. T., C. M. Roth, J. M. Le Doux, J. R. Morgan, and M. L. Yarmush. 1999. Large-scale processing of recombinant retroviruses for gene therapy. Biotechnol. Prog. 15:1-11. [DOI] [PubMed] [Google Scholar]
- 3.Balliet, J. W., and P. Bates. 1998. Efficient infection mediated by viral receptors incorporated into retroviral particles. J. Virol. 72:671-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Battini, J. L., P. Rodrigues, R. Muller, O. Danos, and J. M. Heard. 1996. Receptor-binding properties of a purified fragment of the 4070A amphotropic murine leukemia virus envelope glycoprotein. J. Virol. 70:4387-4393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braas, G., P. F. Searle, N. K. Slater, and A. Lyddiatt. 1996. Strategies for the isolation and purification of retroviral vectors for gene therapy. Bioseparation 6:211-228. [PubMed] [Google Scholar]
- 6.Burns, J. C., T. Friedmann, W. Driever, M. Burrascano, and J. K. Yee. 1993. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. USA 90:8033-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J., L. Reeves, N. Sanburn, J. Croop, D. A. Williams, and K. Cornetta. 2001. Packaging cell line DNA contamination of vector supernatants: implication for laboratory and clinical research. Virology 282:186-197. [DOI] [PubMed] [Google Scholar]
- 8.Chu, T. H., and R. Dornburg. 1997. Toward highly efficient cell-type-specific gene transfer with retroviral vectors displaying single-chain antibodies. J. Virol. 71:720-725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crowe, J., H. Dobeli, R. Gentz, E. Hochuli, D. Stuber, and K. Henco. 1994. 6xHis-Ni-NTA chromatography as a superior technique in recombinant protein expression/purification. Methods Mol. Biol. 31:371-387. [DOI] [PubMed] [Google Scholar]
- 10.Darling, D., C. Hughes, J. Galea-Lauri, J. Gaken, I. D. Trayner, M. Kuiper, and F. Farzaneh. 2000. Low-speed centrifugation of retroviral vectors absorbed to a particulate substrate: a highly effective means of enhancing retroviral titer. Gene Ther. 7:914-923. [DOI] [PubMed] [Google Scholar]
- 11.Fontenot, J. D., N. Tjandra, C. Ho, P. C. Andrews, and R. C. Montelaro. 1994. Structure and self assembly of a retrovirus (FeLV) proline rich neutralization domain. J. Biomol. Struct. Dyn. 11:821-836. [DOI] [PubMed] [Google Scholar]
- 12.Gray, K. D., and M. J. Roth. 1993. Mutational analysis of the envelope gene of Moloney murine leukemia virus. J. Virol. 67:3489-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hammar, L., and G. Gilljam. 1990. Extraction of HIV-1 in aqueous two-phase systems to obtain a high yield of gp120. AIDS Res. Hum. Retrovir. 6:1379-1388. [DOI] [PubMed] [Google Scholar]
- 14.Hammar, L. 1994. Concentration of biomaterials: virus concentration and viral protein isolation. Methods Enzymol. 228:640-658. [DOI] [PubMed] [Google Scholar]
- 15.Izard, J. W., and D. A. Kendall. 1994. Signal peptides: exquisitely designed transport promoters. Mol. Microbiol. 13:765-773. [DOI] [PubMed] [Google Scholar]
- 16.Karger, A., B. Bettin, H. Granzow, and T. C. Mettenleiter. 1998. Simple and rapid purification of alphaherpesviruses by chromatography on a cation exchange membrane. J. Virol. Methods 70:219-224. [DOI] [PubMed] [Google Scholar]
- 17.Koch, W., G. Hunsmann, and R. Friedrich. 1983. Nucleotide sequence of the envelope gene of Friend murine leukemia virus. J. Virol. 45:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuiper, M., R. M. Sanches, J. A. Walford, and N. K. Slater. 2002. Purification of a functional gene therapy vector derived from Moloney murine leukemia virus using membrane filtration and ceramic hydroxyapatite chromatography. Biotechnol. Bioeng. 80:445-453. [DOI] [PubMed] [Google Scholar]
- 19.Le Doux, J. M., J. R. Morgan, R. G. Snow, and M. L. Yarmush. 1996. Proteoglycans secreted by packaging cell lines inhibit retrovirus infection. J. Virol. 70:6468-6473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Doux, J. M., J. R. Morgan, and M. L. Yarmush. 1998. Removal of proteoglycans increases efficiency of retroviral gene transfer. Biotechnol. Bioeng. 58:23-34. [DOI] [PubMed] [Google Scholar]
- 21.Lowy, D. R. 1985. Transformation and oncogenesis: retroviruses, p. 237. In B. N. Field, J. L. Knipe, R. M. Melnick, and B. Roizman (ed.), Virology. Raven Press, New York, N.Y.
- 22.Lyddiatt, A., and D. A. O'Sullivan. 1998. Biochemical recovery and purification of gene therapy vectors. Curr. Opin. Biotechnol. 9:177-185. [DOI] [PubMed] [Google Scholar]
- 23.MacKrell, A. J., N. W. Soong, C. M. Curtis, and W. F. Anderson. 1996. Identification of a subdomain in the Moloney murine leukemia virus envelope protein involved in receptor binding. J. Virol. 70:1768-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McGrath, M., O. Witte, T. Pincus, and I. L. Weissman. 1978. Retrovirus purification: method that conserves envelope glycoprotein and maximizes infectivity. J. Virol. 25:923-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan, R. A., O. Nussbaum, D. D. Muenchau, L. Shu, L. Couture, and W. F. Anderson. 1993. Analysis of the functional and host range-determining regions of the murine ecotropic and amphotropic retrovirus envelope proteins. J. Virol. 67:4712-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morling, F. J., and S. J. Russell. 1995. Enhanced transduction efficiency of retroviral vectors coprecipitated with calcium phosphate. Gene Ther. 2:504-508. [PubMed] [Google Scholar]
- 27.O'Neil, P. F., and E. S. Balkovic. 1993. Virus harvesting and affinity-based liquid chromatography. A method for virus concentration and purification. Biotechnology 11:173-178. [DOI] [PubMed] [Google Scholar]
- 28.Ott, D., R. Friedrich, and A. Rein. 1990. Sequence analysis of amphotropic and 10A1 murine leukemia viruses: close relationship to mink cell focus-inducing viruses. J. Virol. 64:757-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paul, R. W., D. Morris, B. W. Hess, J. Dunn, and R. W. Overell. 1993. Increased viral titer through concentration of viral harvests from retroviral packaging lines. Hum. Gene Ther. 4:609-615. [DOI] [PubMed] [Google Scholar]
- 30.Pinter, A., and W. J. Honnen. 1988. O-linked glycosylation of retroviral envelope gene products. J. Virol. 62:1016-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinter, A., R. Kopelman, Z. Li, S. C. Kayman, and D. A. Sanders. 1997. Localization of the labile disulfide bond between SU and TM of the murine leukemia virus envelope protein complex to a highly conserved CWLC motif in SU that resembles the active-site sequence of thiol-disulfide exchange enzymes. J. Virol. 71:8073-8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Porath, J., J. Carlsson, I. Olsson, and G. Belfrage. 1975. Metal chelate affinity chromatography, a new approach to protein fractionation. Nature 258:598-599. [DOI] [PubMed] [Google Scholar]
- 33.Rothenberg, S. M., M. N. Olsen, L. C. Laurent, R. A. Crowley, and P. O. Brown. 2001. Comprehensive mutational analysis of the Moloney murine leukemia virus envelope protein. J. Virol. 75:11851-11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saha, K., Y. C. Lin, and P. K. Wong. 1994. A simple method for obtaining highly viable virus from culture supernatant. J. Virol. Methods 46:349-352. [DOI] [PubMed] [Google Scholar]
- 35.Schulz, T. F., B. A. Jameson, L. Lopalco, A. G. Siccardi, R. A. Weiss, and J. P. Moore. 1992. Conserved structural features in the interaction between retroviral surface and transmembrane glycoproteins? AIDS Res. Hum. Retrovir. 8:1571-1580. [DOI] [PubMed] [Google Scholar]
- 36.Spear, G. T., N. S. Lurain, C. J. Parker, M. Ghassemi, G. H. Payne, and M. Saifuddin. 1995. Host cell-derived complement control proteins CD55 and CD59 are incorporated into the virions of two unrelated enveloped viruses. Human T cell leukemia/lymphoma virus type I (HTLV-I) and human cytomegalovirus (HCMV). J. Immunol. 155:4376-4381. [PubMed] [Google Scholar]
- 37.Weimin Wu, B., P. M. Cannon, E. M. Gordon, F. L. Hall, and W. F. Anderson. 1998. Characterization of the proline-rich region of murine leukemia virus envelope protein. J. Virol. 72:5383-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Witte, O. N., and D. F. Wirth. 1979. Structure of the murine leukemia virus envelope glycoprotein precursor. J. Virol. 29:735-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu, B. W., J. Lu, T. K. Gallaher, W. F. Anderson, and P. M. Cannon. 2000. Identification of regions in the Moloney murine leukemia virus SU protein that tolerate the insertion of an integrin-binding peptide. Virology 269:7-17. [DOI] [PubMed] [Google Scholar]
- 40.Ye, K., H. K. Dhiman, J. Suhan, and J. S. Schultz. 2003. Effect of pH on infectivity and morphology of ecotropic Moloney murine leukemia virus. Biotechnol. Prog. 19:538-543. [DOI] [PubMed] [Google Scholar]
- 41.Ye, K., S. Jin, and S. J. Schultz. 2004. Genetically engineered fluorescent cell marker for labeling CD34+ hematopoietic stem cells. Biotechnol. Prog. 20:561-565. [DOI] [PubMed] [Google Scholar]