Abstract
Human cytomegalovirus (HCMV), a ubiquitous human pathogen, is the leading cause of birth defects and morbidity in immunocompromised patients and a potential trigger for vascular disease. HCMV replicates in vascular endothelial cells and drives leukocyte-mediated viral dissemination through close endothelium- leukocyte interaction. However, the genetic basis of HCMV growth in endothelial cells and transfer to leukocytes is unknown. We show here that the UL131-128 gene locus of HCMV is indispensable for both productive infection of endothelial cells and transmission to leukocytes. The experimental evidence for this is based on both the loss-of-function phenotype in knockout mutants and natural variants and the gain-of-function phenotype by trans-complementation with individual UL131, UL130, and UL128 genes. Our findings suggest that a common mechanism of virus transfer may be involved in both endothelial cell tropism and leukocyte transfer and shed light on a crucial step in the pathogenesis of HCMV infection.
Human cytomegalovirus (HCMV) is the leading infectious agent of birth defects and a major cause of morbidity in immunocompromised patients. A crucial step in all clinical conditions related to HCMV infection is virus dissemination mediated by leukocytes and associated with viral replication in endothelial cells (ECs). ECs are natural sites of HCMV infection (33, 37, 38), and the virus may be transmitted bidirectionally between ECs and leukocytes during both primary and reactivated HCMV infections (12, 13). Transfer of HCMV from ECs to leukocytes appears to be mediated by transitory microfusion events of plasma membranes driven by viral infection (12).
The ability to infect ECs and leukocytes is a nonessential virus-encoded function and is characteristic of clinical HCMV isolates, but it is missing in reference laboratory strains such as AD169, Towne, and Davis (11, 13, 28, 29, 36). EC tropism represents the property of a virus strain to undergo productive infection in ECs. Virus transfer to leukocytes is the property of HCMV strains to be transferred during coculture from infected fibroblasts or ECs to leukocytes via microfusion events. This results in abortive infection of either polymorphonuclear leukocytes (PMN) or monocytes (12). Presumably, laboratory strains have lost both characteristics during extensive propagation in fibroblasts (tropism-deficient variants), as reported previously (17, 30, 36). In contrast, EC-tropic and leukocyte-transmissible variants could be obtained by adaptation of tropism-deficient strains to growth in ECs (9, 10). However, the gene(s) responsible for EC tropism and virus transfer to leukocytes are elusive.
The cloning of an EC-tropic clinical HCMV isolate (VR1814) as a bacterial artificial chromosome (BAC) in Escherichia coli (FIX-BAC) by adapting a previously reported method (23) provided standard genetic material with preserved wild-type characteristics for rapid mutagenesis in E. coli (14, 15, 24). Site-directed mutagenesis of FIX-BAC has been used to show that, unlike the murine cytomegalovirus (MCMV) (4), the HCMV-encoded ribonucleotide reductase homolog (UL45) is not the genetic determinant of EC tropism of HCMV (14) and is dispensable for growth in vitro (14, 26).
In searching for the genetic determinants of EC tropism and transfer of HCMV to leukocytes, the ULb′ region of the HCMV genome appeared to be a prime candidate, due to the following observations. First, in AD169 and Towne the loss of EC tropism and leukocyte transmissibility is associated with the loss of the ULb′ region (5, 28). Second, the low-passage strain Toledo, recently shown to be tropism deficient (11), displays an inversion of ULb′ compared to HCMV clinical isolates (28). Third, the extensive fibroblast propagation of an EC-tropic clinical isolate (VR6110) was associated with the selection of a tropism-deficient variant showing a possible deletion of UL132-130 within the ULb′ region (30).
We show here that the UL131-128 locus of HCMV genome is indispensable for both EC tropism and leukocyte transfer, as demonstrated by both loss-of-function phenotypes of deletion or knockout (KO) mutants and natural variants and gain-of-function phenotypes by cis- and trans-complementation.
MATERIALS AND METHODS
Cell cultures.
Two types of cell cultures were used to perform all of the experiments reported: human embryonic lung fibroblast (HELF) and human umbilical vein EC (HUVEC) cultures. HELFs were derived from a cell strain developed in the laboratory in 1980 and were used at passages 20 to 30. HUVECs were obtained by trypsin treatment of umbilical cord veins and used at passages 2 to 5. All HUVEC preparations were tested for HCMV DNA by nested PCR to rule out asymptomatic congenital HCMV infection.
HCMV clinical isolates, variants, and laboratory strains.
Four HCMV clinical isolates were tested for EC tropism and leukocyte (both PMN and monocyte) transfer: VR6110 and VR3480, both originally recovered from blood of AIDS patients (3, 29); VR1814, originally recovered from cervical secretions of an healthy pregnant woman (30); and VR6340, originally isolated from the milk of a breast-feeding mother (30). All four HCMV isolates were extensively propagated in HELFs and tested periodically (every 10 passages on the average) for EC tropism and leukocyte transfer (see below), both as viral isolates and as plaque-purified variants. As a general rule, the loss of leukocyte transmissibility in HELFs was consistently found to be associated with a loss of EC tropism. A recently reported clinical strain (Merlin) recovered from the urine sample of a congenitally infected infant (1) and kindly provided by A. J. Davison (Glasgow, Scotland) and G. W. G. Wilkinson (Cardiff, Wales) was included in the study. Extensive propagation in HUVECs was consistently associated with maintenance of both properties, which, however, were lost after extensive propagation of clinical isolates in HELFs. Strains competent for growth in HUVECs and leukocyte transfer were referred to as Huv+ Leuk+, and their variants deficient for both properties as Huv− Leuk−. The HCMV laboratory reference strains AD169 (originally obtained from the American Type Culture Collection) and Towne, both Towne-Wistar (obtained from E. Gonczol, Wistar Institute, Philadelphia, Pa.) and Towne-RIT (received from RIT, Genval, Belgium) were used as reference high-passage HCMV strains grown in HELFs and lacking both biological properties (29, 30). In addition, both laboratory strains, currently considered to be Huv− Leuk−, were shown to be able to reacquire both properties, and thus to become Huv+ Leuk+ after adaptation to growth in HUVECs, as recently reported (9, 10). In parallel, the Toledo strain (originally obtained from E. Gonczol, Wistar Institute), which is considered a low-passage wild-type HCMV strain, could never be adapted to growth in ECs despite repeated attempts and was thus considered Huv− Leuk−, whereas the reported EC-tropic variant (18) of Toledo was shown to be a different virus strain that possibly originated from a recombination event (2).
Construction of RVFIX deletion mutants by site-directed mutagenesis.
A clinical isolate of HCMV, VR1814, previously shown to be EC- and leukocyte-tropic (30), was cloned as a BAC (fusion-inducing factor X [FIX]-BAC), as recently reported (14). The FIX-BAC reconstituted virus (RVFIX) was shown to preserve the wild-type characteristics of the parental strain VR1814. Deletion mutants of FIX-BAC (14) were obtained by site-directed mutagenesis with PCR-generated linear recombination fragments electroporated into a recombination-proficient E. coli strain containing FIX-BAC and expressing bacteriophage λ functions redαβγ from plasmid pBADαβγ (14). Briefly, primers with 3′ homology to sequences flanking a kanamycin resistance (Kanr) marker and 5′ homology to sequences flanking the viral region to be deleted were used to PCR amplify pAcyc 177 (New England Biolabs) DNA and generate DNA fragments containing the Kanr marker tailed onto short regions of HCMV homology. The primers used to generate the linear PCR recombination fragments for the construction of deletion mutants RVFIXΔULb′, RVFIXΔUL130-132, RVFIXΔUL132-128, RVFIXΔUL132K, RVFIXΔUL131K, RVFIXΔUL130, RVFIXΔUL128K, RVFIXΔUL133-148, RVFIXΔUL148, RVFIXΔUL146-147, and RVFIXΔUL127 are reported elsewhere (http://www.sanmatteo.org/virologia/hahn_et_al.doc). Colonies containing BAC (cmr) with the Kanr marker inserted were selected on plates containing both kanamycin and chloramphenicol (14). The correct Kanr cassette insertion into the virus genome was verified by Southern blot hybridization of FIX-BAC DNA and confirmed by sequencing of all individual reconstituted RVFIX virus mutants. Reconstituted virus RVFIX and deletion mutants were obtained by transfection of the respective BAC DNAs into MRC-5 cells. After three passages in MRC-5 cells, all mutants were cultured in HELFs or HUVECs.
Assays for leukocyte transfer.
PMN preparations from healthy blood donors were cocultured for 18 h with infected HELFs or HUVECs. To separate PMN from infected cells, cell suspensions were placed for 3 h at 37°C in the upper compartment of a cell culture device separated by a Transwell filter (5-μm pore size; Costar) from the lower compartment containing 10−8 M N-formyl-Met-Leu-Phe-Ala (FMLP; Sigma), as reported earlier (29). Ficoll separated peripheral blood mononuclear cell suspensions were enriched in monocytes by purification through a Percoll gradient. After overnight coculture with infected cells, monocytes were purified by chemotaxis (10−7 M FMLP) as described above. Both PMN and monocytes suspensions were then tested for the presence of pp65 by immunofluorescence in cytospin preparations of 2 × 105 cells by using a pool of monoclonal antibodies (9). In each experiment, the mean number of pp65-positive leukocytes per 2 × 105 leukocytes in two cytospin preparations (read under code) was determined, and the mean number of positive leukocytes was calculated for at least five replicate experiments. When required, infectious virus of both leukocyte preparations was also tested (12). The degree of purity of the recovered leukocytes was >95% (29). Leukocyte transfer was measured by counting the number of pp65-positive leukocytes and, when required, the number of leukocytes carrying infectious virus by the shell vial assay. As a general rule, the ratio of the former to the latter parameter was 300:1.
Assays for HUVEC tropism.
For assays for HUVEC tropism, published procedures were used (8). Infected HELFs were mixed with uninfected HUVECs at a ratio of 1:2 and cultured for 7 days. After five passages, infected cultures were sonicated, and cell-free virus was inoculated onto HUVECs. Virus was then propagated until passage 10 by infecting uninfected HUVEC monolayers weekly with infected HUVECs. When EC-tropic viruses were tested at passage 10, >50% of HUVECs stained for viral antigens, whereas no viral antigen was detected after infection with non-EC-tropic viruses. However, at passage 10, HUVEC-adapted viruses still remained cell associated. At about passages 30 to 40, HUVECs could be directly infected with supernatants from infected cultures after centrifugation at 600 × g for 45 min. Accordingly, at first passages, virus growth was monitored by immunofluorescence only, with monoclonal antibodies to the immediate-early (IE) protein p72, the early protein p52, and the late protein gB, whereas at later passages virus growth was monitored by cytopathic effect (CPE).
Transcript analyses by Northern blot, rapid amplification of cDNA ends (RACE), and RT-PCR.
mRNA was extracted from RVFIX-infected HELF 4 days postinfection (p.i.). For Northern blotting, 1 μg of RNA was electrophoresed on agarose gel according to the MOPS-formaldehyde protocol and blotted onto Hybond N+ membranes (Amersham Pharmacia). Blots were hybridized with RNA probes generated with the Riboprobe kit (Promega) by antisense transcription of either the unspliced UL131-128 region or individual UL131, UL130, and UL128 genes (all fully spliced open reading frame [ORFs]) amplified by reverse transcription-PCR (RT-PCR) and cloned into pBluescript SK(−) (Stratagene).
First-strand cDNA synthesis and RACE were performed by using the SMART RACE cDNA amplification kit (Clontech) according to the manufacturer's instructions. RACE products were cloned into the pT-Adv vector (Clontech) by using the AdvanTAge PCR cloning kit for analysis and sequencing. Primers used for RACE and RT-PCR analysis are described online (http://www.sanmatteo.org/virologia/hahn_et_al.doc).
trans-Complementation.
The ORF UL130 was amplified from genomic DNA of RVFIX. The amplimer was cloned into the retroviral vector pLNCX (Clontech). The construct (pLNUL130) expresses the HCMV insert from an internal promoter-enhancer. The ORFs UL131 and UL128 of the same strain (fully spliced versions) were amplified from cDNAs cloned in pBluescript SK(−) (see above) and inserted into pLNCX (constructs pLNUL131 and pLNUL128). The retrovirus constructs were transfected into the 293-Gag-Pol packaging cell line, together with plasmid pVG, expressing the vesicular stomatitis virus G protein for envelope pseudotyping. Stably transduced HUVECs were obtained by selection with 400 μg of G418/ml. Primers used for construction of the retroviral vectors expressing UL131-128 genes are reported online (see above).
Rescue of EC tropism.
Rescue of EC tropism was determined by counting the number of plaques of CPE 7 or 10 days after infection of HUVECs individually expressing UL131, UL130, or UL128 ORFs with EC tropism-deficient FIX-based KO mutants and natural variants by using cell-free virus at a multiplicity of infection of 5 (as determined in HELFs). Experiments were performed in replicates of 24-well microtiter plates. After immunostaining for both IE and gB was done, the plaques were counted. Each plaque consisted of 10 to 20 cytopathic contiguous HUVECs (IE and gB positive) around the parental cytomegalic cell. In parallel, CPE plaques were counted in replicate experiments. The absence of rescue of EC tropism was shown by the absence of plaques in transduced HUVEC monolayers infected with an EC tropism-deficient virus. In addition, in each trans-complementation experiment, HUVECs were transduced with the void retrovirus vector. The assay was based on previous experience showing that all EC-tropic viruses or mutants susceptible to growth in EC were able to induce plaque formation in HUVECs when inoculated for the first time at a multiplicity of infection of 5 (as determined in HELFs). Viruses deficient for EC tropism did not induce plaque formation in HUVECs upon first inoculation.
In addition, HUVEC monolayers transduced with UL131, UL130, UL128, or the void retrovirus vector and infected with the KO or natural variant mutant viruses were cocultured 7 to 10 days p.i. with leukocytes to study leukocyte transfer kinetics.
RESULTS
Mutagenesis of a BAC-cloned EC-tropic and leukocyte-transmissible HCMV clinical isolate.
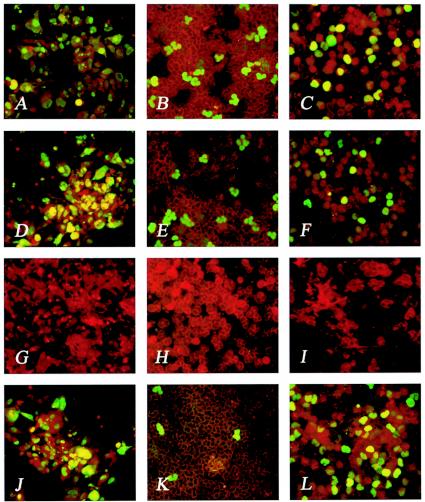
As already reported (14), the BAC-reconstituted virus RVFIX maintained the ability of the parental strain VR1814 (Fig. 1A) to grow in HUVECs (Fig. 1D). Here, transmission to PMN and monocytes of VR1814 (Fig. 1B and C) was also shown to be retained by RVFIX (Fig. 1E and F, respectively, and Table 1). On this basis, to identify the genes responsible for EC tropism and leukocyte transfer, the ULb′ region of RVFIX (Fig. 2A) was systematically mutagenized by deleting large fragments as well as individual ORFs (Fig. 2B). In addition, KO mutants of ORFs adjacent to (UL127) or distant from (UL45) ULb′ were generated. The growth kinetics in HELFs excluded a major growth deficit for each virus mutant, with the exception of RVΔULb′, in which ca. 6% of the genome length is missing (Fig. 3A).
FIG. 1.
EC and leukocyte tropism of VR1814, RVFIX, and deletion mutants. (A to L) EC (A, D, G, and J), PMN (B, E, H, and K), and monocyte (C, F, I, and L) tropism of VR1814 (A, B, and C), RVFIX (D, E, and F), RVFIXΔUL132-128 (G, H, and I) and RVFIXΔ133-148 (J, K, and L). Panels G through I are negative. Panels K and L show the predominant monocyte tropism of RVFIXΔ133-148. Panels A, D, G, and J show immunostaining of EC with monoclonal antibodies to both p72 and gB. Panels B, E, H, and K (PMN) and panels C, F, I, and L (monocytes) show immunostaining with a pool of three pp65-specific monoclonal antibodies.
TABLE 1.
Leukocyte transfer and HUVEC tropism of RVFIX mutants, as well as tropism-deficient (Huv− Leuk−) and tropism-competent (Huv+ Leuk+) clinical isolates and laboratory (Huv− Leuk−) HCMV strains
| HELF-cultured HCMV strain | HCMV leukocyte transfera (no. of leukocytes [range])
|
HCMV HUVEC tropismb (%) | |
|---|---|---|---|
| PMN | Monocytes | ||
| RVFIX mutants | |||
| RVFIX | 8,000 (5,000-13,000) | 6,500 (5,100-9,600) | 80 |
| RVFIXΔULb′ | 0 | 0 | 0 |
| RVFIXΔUL132-128 | 0 | 0 | 0 |
| RVFIXΔUL133-148 | 50 (30-80) | 6,500 (5,000-8,500) | 70 |
| RVFIXΔUL132-130 | 0 | 0 | 0 |
| RVFIXΔUL131K | 0 | 0 | 0 |
| RVFIXΔUL130 | 0 | 0 | 0 |
| RVFIXΔUL128K | 0 | 0 | 0 |
| RVFIXΔUL132K | 7,300 (6,000-9,000) | 7,500 (6,300-9,500) | 70 |
| RVFIXΔUL148 | 7,800 (5,900-11,000) | 7,550 (5,550-10,500) | 75 |
| RVFIXΔUL146-147 | 40 (30-60) | 6,200 (4,200-9,000) | 80 |
| RVFIXΔUL127 | 6,500 (4,200-9,200) | 6,850 (5,000-8,300) | 65 |
| RVFIXΔUL45 | 7,500 (5,500-10,500) | 7,200 (5,100-9,800) | 90 |
| Clinical isolatesc | |||
| VR1814 Huv+ Leuk+ | 9,000 (6,300-14,000) | 9,500 (6,500-13,000) | 90 |
| VR1814 Huv− Leuk− | 0 | 0 | 0 |
| VR6110 Huv+ Leuk+ | 9,500 (5,100-13,200) | 9,000 (5,600-12,000) | 75 |
| VR6110 Huv− Leuk− | 0 | 0 | 0 |
| VR6340 Huv+ Leuk+ | 8,100 (6,300-12,300) | 7,800 (6,100-11,300) | 70 |
| VR6340 Huv− Leuk− | 0 | 0 | 0 |
| VR3480 Huv+ Leuk+ | 8,500 (6,000-9,800) | 9,500 (6,100-9,500) | 75 |
| VR3480 Huv− Leuk− | 0 | 0 | 0 |
| Merlin Huv− Leuk− | 0 | 0 | 0 |
| Laboratory strainsd | |||
| AD169 Huv− Leuk− | 0 | 0 | 0 |
| Towne-Wistar Huv− Leuk− | 0 | 0 | 0 |
| Towne-RIT Huv− Leuk− | 0 | 0 | 0 |
| Toledo Huv− Leuk− | 0 | 0 | 0 |
That is, the median number (range) of pp65-positive leukocytes/2 × 105 leukocytes examined in five independent experiments.
That is, the percentage of infected cells in the cell monolayer after more than 10 passages.
Huv+ Leuk+, low passage (<20) in HELFs; Huv− Leuk−, high passage (>50) in HELFs.
Reference strains were passaged a number of times (>100) in fibroblast cultures.
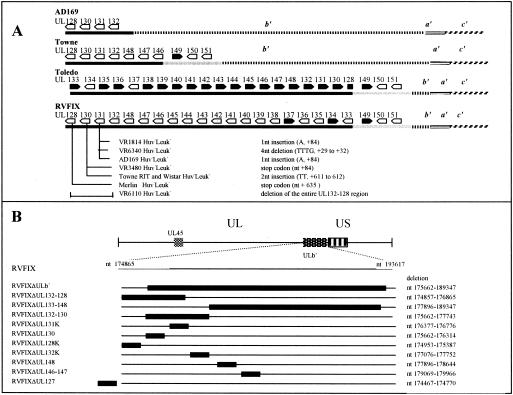
FIG. 2.
Mutagenesis of ULb′ genome region of RVFIX. (A) Diagram of the HCMV ULb′ genome region of RVFIX in comparison with the reference strains AD169, Towne, and Toledo. Mutations in UL131-128 locus of five Huv− Leuk− clinical isolate variants (VR1814, VR6340, VR3480, VR6110, and Merlin) and three reference strains (AD169, Towne and Toledo) are shown. Black bars indicate the ULb′ region variably deleted in AD169 and Towne and inverted in Toledo. Light gray bars indicate the ULb′ region deleted in AD169 only. The ULb′ numbering is as follows: black bar, nt 174865 to 178221 in AD169 (GenBank no. X17403) and nt 175068 to 189515 in Toledo (GenBank no. U33331); gray bar, nt 189515 to 193617 in Toledo (GenBank no. U33331). The numbering of corresponding ULb′ fragments in Towne and RVFIX was as follows: black bar, nt 174865 to 180068 in Towne and nt 174865 to 189515 in RVFIX; and light gray bar, nt 180068 to 184170 in Towne and nt 189515 to 193617 in RVFIX. ULb′ inversion site in Toledo is in UL128 at nt 175082. Positions of nucleotide mutations in UL131, UL130, and UL128 genes are indicated with respect to the relevant ATGs. (B) The position of ULb′ within HCMV genome is indicated according to reference 6. The positions of the Kanr insertion and the deleted region in each RVFIX mutant are indicated by solid black bars.
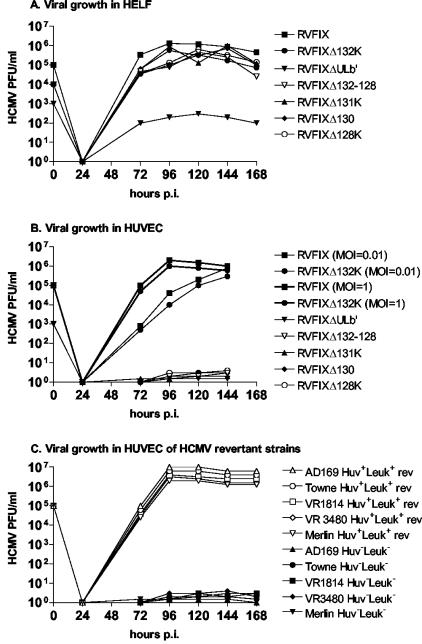
FIG. 3.
Growth kinetics of RVFIX and virus mutants at passages 30 to 40 in HELFs (A) and HUVECs (B). (C) Growth kinetics in HUVECs of Huv− Leuk− clinical isolates and laboratory strains and their relevant revertants.
EC tropism and leukocyte transfer of ULb′ KO mutants.
As shown in Table 1, KO mutants of genes UL131, UL130, and UL128 lost the ability to grow in HUVEC and to be transmitted to leukocytes (PMN and monocytes). In fact, the mutants RVFIXΔULb′, RVFIXΔUL132-128 [lacking the UL131-128 region including a predicted poly(A) signal of UL132, thus knocking out UL132-128], RVFIXΔUL131K, RVFIXΔUL130, and RVFIXΔUL128K showed the complete loss of both EC tropism and transmission to leukocytes (both monocytes and PMN) (Fig. 1G to I). Deletion of any ORF within ULb′ other than UL131, UL130, and UL128, as well as genes adjacent to (UL127) or distant from (UL45) ULb′ (Fig. 2A and B), did not interfere with the ability of virus mutants to productively infect EC or be transmitted to leukocytes (Table 1). The parental RVFIX and the EC-tropic mutants remained cell associated for about the first 20 passages in HUVECs. In subsequent passages, cell-associated virus started to be released in the supernatant, reaching titers of up to 106 to 107 PFU/ml between passages 30 and 40 (Fig. 3B, showing both one-step and multiple-step growth in ECs). At earlier passages, EC infection was therefore quantified by determining the number of antigen-positive plaques, whereas from passage 20 on, virus titers in infected cells supernatant were determined additionally. In general, EC tropism was an all-or-none phenomenon, and mutants could be classified as either EC tropic or non-EC tropic. The same held true for transfer to leukocytes, with the exception of mutants RVFIXΔUL133-148 and RVFIXΔUL146-147, both of which lack the genes UL146 and UL147 (27) and show a substantial reduction (∼2 log10) in transfer to PMN but not to monocytes (Table 1 and Fig. 1J to L).
EC tropism- and leukocyte transfer-deficient HCMV variants have mutations in the UL131-128 locus.
The lack of EC tropism and transfer to leukocytes was also observed in five clinical isolates extensively propagated in HELFs and in three laboratory strains (AD169, Towne, and Toledo), hereafter referred to as Huv− Leuk− (Table 1). Sequencing of the UL131-128 genes of the five EC tropism-deficient and leukocyte transfer-deficient virus variants (VR1814 Huv− Leuk−, VR3480 Huv− Leuk−, VR6110 Huv− Leuk−, VR6340 Huv− Leuk−, and Merlin Huv− Leuk−) selected in vitro (1, 30) showed a consistent pattern of mutations (Fig. 2A; see also the supplemental online data [http://www.sanmatteo.org/virologia/hahn_et_al.doc]). These mutations affected the UL131-128 region in VR6110 Huv− Leuk−, UL131 in VR1814 Huv− Leuk−, and VR6340 Huv− Leuk−, UL130 in VR3480 Huv− Leuk−, and UL128 in the Merlin Huv− Leuk− strain (1). Similar mutations were observed in tropism-deficient laboratory strains and were relevant to UL131 in AD169 Huv− Leuk−, UL130 in Towne Huv− Leuk−, and the inversion of the ULb′ region in Toledo Huv− Leuk−, causing the concomitant truncation of UL128 and dislocation of the downstream poly(A) signal (Fig. 2A and online data).
Rescue of EC tropism and leukocyte transfer after reversal of UL131-128 mutations in virus variants.
RVFIX mutants with deletion of UL131, UL130, and UL128 genes could not be adapted to growth in HUVECs (Table 2). In contrast, the adaptation of two different preparations of Towne (Towne-Wistar Huv− Leuk− and Towne-RIT Huv− Leuk−) (9) as well as of AD169 Huv− Leuk− (10), VR1814 Huv− Leuk−, VR3480 Huv− Leuk−, and Merlin Huv− Leuk− led to rescue of the Huv+ Leuk+ phenotype (rev). Levels of EC tropism and leukocyte transfer of VR1814 Huv+ Leuk+ rev, VR3480 Huv+ Leuk+ rev, and Merlin Huv+ Leuk+ rev were comparable to those of the relevant tropism- and transfer-competent viruses. The growth kinetics in HUVECs of HCMV Huv+ Leuk+ revertant strains in comparison with their relevant tropism- and transfer-deficient variants are shown in Fig. 3C. Sequencing of the UL131-128 locus of the virus revertants revealed that reconstitution of EC tropism was consistently associated with reversal of UL131-128 mutations either by reversal to the original coding sequence or, as in the case of AD169, by acquisition of a compensatory mutation (see supplemental Fig. 1 [http://www.sanmatteo.org/virologia/hahn_et_al.doc]). However, in AD169 Huv+ Leuk+ rev only monocyte tropism could be fully rescued (Table 2). The finding of a very partial (∼1%) rescue of PMN versus monocyte transfer in AD169 Huv+ Leuk+ rev is consistent with the impaired PMN transfer observed in RVFIXΔUL133-148 and RVFIXΔUL146-147 mutants.
TABLE 2.
Rescue in cis of both EC tropism and leukocyte transfer of Huv− Leuk− viral variants showing reversal of UL131-128 mutations after adaptation to growth in HUVECs
| HCMV strain (mutated gene) | Rescue in cis of HUVEC tropism and leukocyte transfer after attempts at adaptation to growth in HUVECs (Huv+ Leuk+ rev) in:
|
|
|---|---|---|
| HUVECa | PMN/monocytesb | |
| RVFIX mutants | ||
| RVFIXΔUL132-128 | 0 | 0/0 |
| RVFIXΔUL131K | 0 | 0/0 |
| RVFIXΔUL130 | 0 | 0/0 |
| RVFIXΔUL128K | 0 | 0/0 |
| Clinical isolates | ||
| VR1814 Huv− Leuk− (UL131) | 80 | 8,800/8,200 |
| VR3480 Huv− Leuk− (UL130) | 80 | 7,000/7,300 |
| Merlin Huv− Leuk− (UL128) | 80 | 7,500/8,000 |
| Laboratory strains | ||
| AD169 Huv− Leuk− (UL131) | 85 | 70/7,500 |
| Towne Huv− Leuk− (UL130) | 85 | 8,500/9,200 |
| Toledo Huv− Leuk− (UL131-128) | 0 | 0/0 |
That is, the percentage of infected cells in the HUVEC monolayer after more than 10 passages.
That is, the median number of pp65-positive leukocytes/2 × 105 leukocytes examined in five independent experiments. Individual figures were within a 30% range.
UL131-128 transcription analysis and gene products prediction.
RACE analysis identified transcripts running through the entire UL131-128 region, and showing a splicing event between UL128x2 and UL128x3 (nucleotides [nt] 175201 to 175081), either exclusively or in conjunction with splicing between UL131x1 and UL131x2 (nt 176589 to 176480). An additional splicing event between UL128x1 and UL128x2 (nt 175459 to 175335) was observed in several clones. All transcripts terminated ∼14 nt after an AAUAAA signal immediately downstream of the UL128 stop codon (nt 174865 to 174863 according to Cha et al. [6]; see Fig. 4A and B and supplemental Fig. 2 [http://www.sanmatteo.org/virologia/hahn_et_al.doc]).
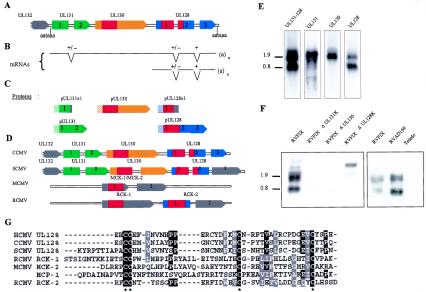
FIG. 4.
Diagram of the UL131-128 locus. The structures of the predicted ORFs (A), the newly identified UL131-128 spanning transcripts (B), and the predicted protein products (C) are shown. In panel C, dashed boxes indicate predicted signal peptides, and red boxes indicate putative chemokine domains. (D) Comparison of UL131-128 locus of CCMV, SCMV, MCMV, and RCMV; color codes are as defined for panels A to C. (E) Northern blots of RVFIX-infected fibroblasts (4 days p.i.). Each lane was separately hybridized with the antisense RNA probe covering the indicated ORF. (F) Northern blots of fibroblasts infected with RVFIX, RVFIXΔUL131K, RVFIXΔUL130, RVFIXΔUL128K, RVAD169, and Toledo. mRNAs from the indicated virus strains and virus mutants were hybridized with a UL131-128-specific probe. (G) Amino acid alignment of UL128 homologs of HCMV, CCMV, and SCMV with RCK-1 and RCK-2 of RCMV, MCK-2 of MCMV, and the prototype cellular CC-chemokine MCP-1. Sequences are ordered on the basis of decreasing homology from top to bottom. Asterisks indicate conserved cysteines.
Northern blots from fibroblasts infected with RVFIX (Fig. 4E), when hybridized with a UL131-128 antisense RNA probe showed an upper (1.8 to 2.0 kb) and a lower (0.7 to 0.8 kb) signal. The 0.7- to 0.8-kb band can be safely interpreted as an UL128-specific transcript, since it hybridized with an UL128-specific probe and not with UL131 or UL130 probes. In contrast, it appears that the upper signal may contain at least two bands. These results are in agreement with a recent report (1). Probably, an upstream promoter drives the long transcripts, which may express either UL131 or UL130, depending on the location of the mRNA 5′ end, upstream or downstream from the UL131 AUG (nt 176825 to 176823 according to Cha et al. [6]). Alternatively, a UL130-specific promoter overlapping UL131 could drive transcription of UL130. Since 5′ RACE analyses have not yet defined the bona fide initiation site for any of the UL131-128 specific mRNAs, the 1.8- to 2.0-kb mRNA 5′-untranslated region might be created via the junction with an upstream exon. On the other hand, the UL128-specific promoter most probably overlaps the UL131-130 region, as further suggested by the analysis of KO mutant transcription (see below).
A summary of the predicted UL131-128 gene products is shown in Fig. 4C. Confidence in these predictions is strengthened by the conservation of proteins in chimpanzee cytomegalovirus (CCMV), green monkey cytomegalovirus (SCMV) and, limited to UL128, rat cytomegalovirus (RCMV) (Fig. 4D). The consensus amino acid sequence of pUL130, and pUL128 from mammal cytomegaloviruses suggests that both proteins bear an N-terminal domain homologous to chemokines (Fig. 4D). HCMV UL130 amino acids 45 to 120 were indeed predicted to have a monocyte chemoattractant protein fold (PDB; 1dok) based on a threading algorithm (25), and a putative CC chemokine domain was independently noticed (1) in the UL128 ORF (Fig. 4G).
Rescue of EC tropism and leukocyte transfer by trans-complementation.
Northern analysis of fibroblasts infected with RVFIX KO mutants showed that targeted insertion of a Kanr marker in UL131 or UL130 genes affects the expression or the stability of transcripts of the entire locus (Fig. 4F, left). Similarly, all UL131-128 transcripts were absent in cells infected with Toledo, suggesting that truncation of UL128 and dislocation of the common poly(A) signal destabilize all transcripts (Fig. 4F, right). In contrast, RVFIXΔUL128K did express the expected upshifted mRNAs (Fig. 4F, left). Finally, the UL131 point mutation in AD169 (Huv− Leuk−) affected neither mRNA mobility nor stability (Fig. 4F, right). The same holds true for the Huv− Leuk− Towne strains, as well as for all of the described UL131-128 point mutants of clinical strains (data not shown).
The transcription data from KO mutants and Huv− Leuk− variants strongly indicated that the UL131-128 locus is indispensable for both HCMV growth in ECs and transmission to leukocytes. In order to define the contribution of the individual genes of the locus, we devised a trans-complementation assay to prove that pUL131, pUL130, and pUL128 are individually essential for HCMV EC tropism and transmission to leukocytes (Fig. 5). Indeed, the spliced ORFs UL131, UL130, and UL128 from RVFIX individually expressed in HUVECs were able to restore EC tropism of the relevant Huv− Leuk− viral variants of either clinical isolates or laboratory strains but not of variants with mutations in the other genes (Fig. 5A). In agreement with the transcription data, EC tropism of RVFIXΔUL128K could be restored by HUVECs expressing UL128, whereas HUVECs individually expressing UL131 or UL130 could not restore the EC tropism of RVFIXΔUL131K and RVFIXΔUL130 (Fig. 5A). Rescue of EC tropism was shown by detection of plaques of CPE (Fig. 5Cb and e), as well as plaques positive for both the IE and gB proteins (Fig. 5Ca and d) in HUVECs transduced with individual genes. In contrast, no plaque was observed in HUVECs transduced with the void or the heterologous retroviral vector. The number of CPE plaques detected after trans-complementation was lower than that measured after infection of HUVECs with EC-tropic viruses (Fig. 5A).
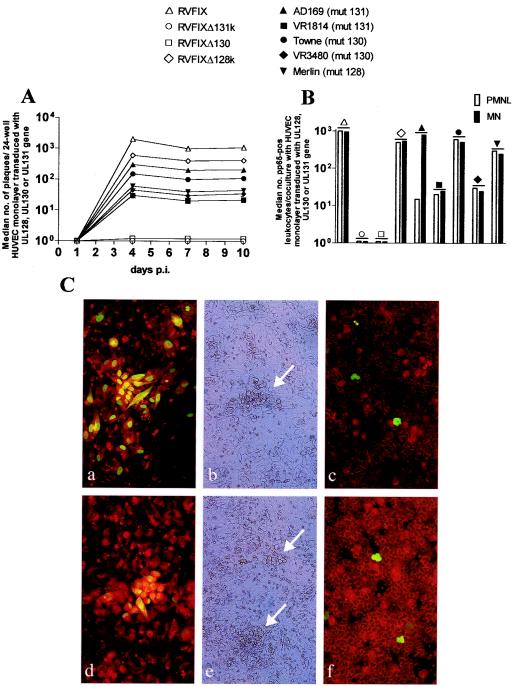
FIG. 5.
Rescue in trans of HUVEC tropism and subsequent leukocyte transfer. (A) Analysis of viral growth showing the median number of CPE plaques per 24-well HUVEC culture (three wells examined) either expressing the void vector and infected with RVFIX or expressing the individual UL131, UL130, or UL128 genes and infected with the relevant KO (Δ) or natural virus mutant (mut). No plaque was observed after infection of HUVECs expressing the void or the heterologous vector with deletion or natural mutants. (B) Median number of pp65-positive PMN (□) and monocytes (MN, ▪) after coculture with 24-well HUVEC monolayers (three wells examined) expressing individual UL131, UL130, or UL128 genes and infected with the homologous KO or natural virus mutant at 10 days p.i. Transfer of pp65 to leukocytes occurs only at the late stage of productive virus replication (12). (C) Virus growth in HUVECs transduced with the UL128 gene and infected with RVFIXΔUL128K (a and b) or Merlin strain (UL128 mutant) (d and e). Panels a and d show gB+IE antigen-positive plaques; panels b and e show CPE plaques (arrows). In panels c and f, pp65-positive leukocytes after coculture with HUVECs transduced with UL128 and infected with RVFIXΔUL128K (c) or Merlin strain (f) (10 days p.i) are shown.
Furthermore, rescue of EC tropism was consistently associated with the rescue of transfer to leukocytes—both PMN and monocytes (Fig. 5B). Coculture of leukocytes with HUVEC monolayers transduced with UL131, UL130, or UL128 and infected with the relevant natural virus mutant (Fig. 5Cf) or transduced with UL128 and infected with RVFIXΔUL128 (Fig. 5Cc) showed rescue of leukocyte (both PMN and monocyte) transfer, whereas HUVECs individually expressing UL131 or UL130 could not restore leukocyte transfer of RVFIXΔUL131K and RVFIXΔUL130. In addition, AD169 (Huv− Leuk−), when supplied with UL131, selectively regained full monocyte, but incomplete PMN transfer (Fig. 5B) No transfer to leukocytes occurred with heterologous complementation of mutant viruses.
DISCUSSION
In this study, we provide evidence that the UL131-128 locus of the HCMV genome is indispensable for HCMV to productively replicate in HUVECs and to be transmitted to PMN and monocytes. In addition, our data suggest that each of the genes of the locus is individually requested. This is not to imply that UL131-128 are the only virus-encoded proteins specifically required for the HCMV growth in ECs and transfer to leukocytes. Indeed, our study implicates an additional locus, UL146-UL147, as necessary for the efficient transmission to PMN (see below).
These conclusions are supported by experimental conditions leading to either a gain or a loss of function. Loss of function, i.e., the loss of both EC tropism and leukocyte transfer was documented by two experimental findings: (i) the experimental introduction of targeted deletions into the UL131-128 locus and (ii) the identification of spontaneous mutations within the UL131-128 locus of natural viral variants. As for the first finding, generation of mutants lacking any ORF within ULb′ other than UL131, UL130, and UL128 did not affect their ability to grow in EC or be transmitted to leukocytes—either PMN or monocytes. Partial exceptions were mutants RVFIXΔUL133-148 and RVFIXΔUL146-147, the transmission of which to PMN, but not to monocytes, was drastically reduced. As for the natural variants, as many as eight independent HCMV strains, deficient for both EC tropism and leukocyte transfer after extensive propagation in HELFs, exhibited a variety of spontaneous loss-of-function mutations, all affecting the coding sequence of one or the other of UL131, UL130, and UL128 genes.
Gain of function was documented by two different and independent procedures: (i) phenotypic reversion of five natural variant strains to EC tropism and leukocyte transfer, associated with reversal of mutations within UL131-128, and (ii) partial restoration of EC tropism and leukocyte transfer by trans-complementation with individual UL131, UL130, or UL128 genes. Rescue of EC tropism and leukocyte transfer of three clinical and one laboratory strains was consistently associated with the selection of a progeny showing the original coding sequence, whereas in the laboratory strain AD169 the correct coding frame of UL131 was associated with the acquisition of a compensatory mutation. In contrast, the irreversible nature of targeted (KO) mutations (replacement of the entire coding sequence of each gene with a Kanr cassette) expectedly prevented the reacquisition of wild-type sequences. None of the KO mutants could be readapted to growth in ECs, which is the conclusive demonstration of the indispensability of the UL131-128 locus, whereas no possible mutation elsewhere in the HCMV genome could compensate for a primary lesion in UL131-128. This observation extends to the Huv− Leuk− Toledo strain: the repeated inability to adapt this strain to growth in ECs is likely to be linked to the reported peculiar mutation of the UL131-128 region (28), which would require recombination for reversion (18).
In trans-complementation experiments, the individual UL131-128 genes were supplied in trans by retrovirus-mediated transduction of HUVECs. The results were fully consistent, since complementation with the single genes could rescue the growth and/or transfer properties only when the complementing gene(s) matched the affected gene of each virus. An apparent exception to this picture was represented by RVFIXΔUL131 and RVFIXΔUL130 mutants, which failed to rescue growth or transmissibility to leukocytes in HUVECs individually expressing UL131 and UL130, respectively. An explanation for this discrepancy came from Northern blot analysis of the UL131-128 region. In fact, it was observed that insertion of the Kanr cassette within the coding sequence of UL131 or UL130 strongly downregulated all transcripts of the entire UL131-128 locus, possibly by affecting the promoter sequences, transcript stability, or both. Thus, of three single-gene KO mutants of the UL131-128 region, only RVFIXΔUL128K exhibited a selective inactivation of UL128, and indeed this mutant could be rescued by the autologous gene in HUVEC trans-complementation assays. RVFIXΔUL131 and RVFIXΔUL130 are functionally UL131-128 knockdown mutants.
The quantitative aspects of trans-complementations need additional consideration. Most of the trans-complementations with UL131-128 genes produced infection levels (number of virus plaques in HUVEC monolayers and number of infected leukocytes) which were 1 to 2 logs lower compared to the reference EC-tropic, leukocyte transfer proficient strains. Thus, although trans-complementation of the UL131-128 genes restores growth in EC and transfer of the virus to leukocytes, this effect is partially efficient. The low efficiency of trans-complementation may reflect the inappropriate levels and/or timing of transduced gene expression compared to natural infection. In fact, a low efficiency of complementation was observed also with the KO derivatives, which were raised through the targeted inactivation of defined genes in a prokaryotic host, a process that is highly unlikely to inadvertently affect other genes related by chance to growth in ECs and transfer to leukocytes.
At present, we cannot formally rule out that part of the quantitative differences in the growth or transfer capacity between parent viruses and complemented mutants may be due to the contribution of other viral genes. In fact, in one subset of mutants (all mutants lacking the UL146-147 locus) we provide evidence that additional virus-encoded proteins are indeed required for virus passage to PMN. The role of leukocyte attraction in HCMV infection was recently underscored by the finding that, in MCMV, two differentially spliced CC chemokine homologs encoded by m131-129 (MCK-1 and MCK-2) are important determinants of virus dissemination (7, 19, 20, 34, 35). MCK-2 acts as a proinflammatory signal that recruits leukocytes to the site of infection to increase virus spread (34, 35). The relevance of chemoattraction is highlighted in our study by the phenotypes of mutants RVΔUL146-147 and RVΔUL133-148, as well as of the laboratory strain revertant AD169 (Huv+ Leuk+), all bearing a functional UL131-128 locus but lacking the viral CXC chemokine genes UL146-147. UL146 product is a potent attractor and activator of human PMN in vitro (27). These strains fully maintain EC tropism and transmissibility to monocyte but are only inefficiently transmitted to PMN. Similarly, the parental AD169 (Huv− Leuk−) strain, when transiently supplied with UL131, selectively regains full monocyte but incomplete PMN transmissibility. Thus, whereas the genes within UL131-128 appear to be indispensable for both local endothelial spread and virus passage to leukocytes, the chemotactic factors coded for by UL146-147 strongly enhance virus passage to PMN in our in vitro assay.
An inference from our data is that a common mechanism of virus transfer, involving UL131-128 protein function, acts in both EC infection and transfer to leukocytes. The importance of cell-to-cell transmission as a mechanism of virus spread has been emphasized for a number of viruses, including lymphotropic human retroviruses such as human immunodeficiency virus type 1 (16, 22). Human immunodeficiency virus type 1 is transmitted between infected T cells, dendritic cells, and monocytes/macrophages and from these to endothelial and epithelial cells through virus-induced structures reminiscent of the immunological synapse, the polarized intercellular adhesion formed between leukocytes and antigen-presenting cells (16, 22). This structure entails integrin-ligand (e.g., LFA-1 and ICAM-1) and receptor-ligand (TCR-MHC) interactions and may permit virus passage through direct intercellular transport. Bidirectional transmission of HCMV between ECs and leukocytes seems in turn to entail a viral usurpation of normal leukocyte-EC adhesion mechanisms, since it is inhibited by anti-LFA-1 and anti-ICAM-1 antibodies (12). Using an in vitro model, we have shown that viral and cellular products are bidirectionally transferred between infected ECs and PMN through cytoplasmic bridges formed by transitory fusion events of the cell membranes, induced by viral infection (11, 12, 29, 30). Leukocytes transporting both infectious virus and virus products are detected in vivo in the blood of patients with disseminated infection (12, 29, 31), thus underscoring their importance for virus dissemination in the host during productive infection. Therefore, HCMV can exploit leukocytes as a transfer vehicle for dissemination to tissues through the vascular tree (12, 13, 21).
The amino acid sequence of the UL131, UL130, and UL128 products qualifies them as lumenal molecules of the secretory pathway. They could be involved in the final stages of virus morphogenesis and maturation at membranes. Alternatively, they could be exported independently from other viral proteins as soluble secreted proteins. In either case, the fact that mutations within any of the three UL131-128 genes result in a common phenotype strongly suggests that they can either physically or functionally interact with each other to aim at a common cellular target. This target seems to include virus spreading from infected to uninfected EC, as well as to uninfected leukocytes. Both events assure virus dissemination and entail microfusion between adhering membranes of contiguous cells. Thus, induction of microfusion events could be a major function of the UL131-128 encoded gene products.
In addition, UL131-128 gene products may be involved in attraction-adhesion of leukocytes to ECs. The presence of chemokine-like domains in UL130 and UL128 proteins suggests a role in leukocyte-EC interaction, e.g., by upregulating adhesion molecules and activating the secondary production of cell-encoded attractants (32), such as IL-8 and GROα, previously shown to be released by HCMV-infected cells (13). The chemotactic activity of each individual gene product of the UL131-128 locus, as well as the potential cooperation with other viral or cellular gene signaling molecules, remains to be further elucidated.
In conclusion, the identification of a genetic locus governing HCMV exchange between ECs and effector cells of the blood appears to be an important step forward in the understanding of the enigmatic pathogenesis of HCMV infection.
Acknowledgments
We thank D. Rose, S. Rhiel, I. Skuratovska, D. Lilleri, and F. Rovida for excellent technical assistance and W. Hell (Medigenomix, Martinsried, Germany) for sequencing of the RACE clones. We are indebted to A. J. Davison for generously sharing sequence information.
This study was supported by a grant of the Wilhelm Sander-Stiftung (to G.H.); grants from the Deutsche Forschungsgemeinschaft (to U.K.); grants from the Italian Ministero della Salute, Ricerca Finalizzata ICS120.5/RF00.124, ICS030.4/RF99.104, and 08920401 (convenzione 126) (to G.G.); grant 80206 from Ricerca Corrente (to E.P.); Programma Nazionale AIDS (grants 40B.66 to G.M. and 50D.12 to G.G.); the Italian MURST Cofinanziamento 2001 (G.M.); and the CNR-Biotechnology Target Project (to A.G. and F.B.).
REFERENCES
- 1.Akter, P., C. Cunningham, B. P. MacSharry, A. Dolan, C. Addison, D. J. Dargan, A. F. Hassan-Walker, V. C. Emery, P. D. Griffiths, G. W. Wilkinson, and A. J. Davison. 2003. Two novel spliced genes in human cytomegalovirus. J. Gen. Virol. 84:1117-1122. [DOI] [PubMed] [Google Scholar]
- 2.Baldanti, F., M. G. Revello, E. Percivalle, N. Labò, and G. Gerna. 2003. Genomes of the endothelial cell-tropic variant and the parental Toledo strain of human cytomegalovirus are highly divergent. J. Med. Virol. 69:76-81. [DOI] [PubMed] [Google Scholar]
- 3.Baldanti, F., E. Silini, A. Sarasini, C. L. Talarico, S. C. Stanat, K. K. Biron, M. Furione, F. Bono, G. Palù, and G. Gerna. 1995. A three-nucleotide deletion in the UL97 open reading frame is responsible for the ganciclovir resistance of human cytomegalovirus clinical isolates. J. Virol. 69:796-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brune, W., C. Menard, J. Heeseman, and U. H. Koszinowski. 2001. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science 291:303-305. [DOI] [PubMed] [Google Scholar]
- 5.Cha, T.-H., E. Tom, G. W. Kemble, G. M. Duke, E. S. Mocarski, and R. S. Spaete. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 7.Fleming, P., N. Davis-Poynter, M. degli Espositi, E. Densley, J. Papadimitriou, G. Shellam, and H. Farrell. 1999. The murine cytomegalovirus chemokine homolog, m 131/129, is a determinant of viral pathogenicity. J. Virol. 73:6800-6809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerna, G., E. Percivalle, A. Sarasini, and M. G. Revello. 2002. Human cytomegalovirus and human umbilical vein endothelial cells: restriction of primary isolation to blood samples and susceptibilities of clinical isolates from other sources to adaptation. J. Clin. Microbiol. 40:233-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerna, G., E. Percivalle, A. Sarasini, F. Baldanti, and M. G. Revello. 2002. The attenuated Towne strain of human cytomegalovirus may revert to both endothelial cell tropism and leuko-(neutrophil- and monocyte-) tropism in vitro. J. Gen. Virol. 83:1993-2000. [DOI] [PubMed] [Google Scholar]
- 10.Gerna, G., E. Percivalle, A. Sarasini, F. Baldanti, G. Campanini, and M. G. Revello. 2003. Rescue of human cytomegalovirus strain AD169 tropism for both leukocytes and human endothelial cells. J. Gen. Virol. 84:1431-1436. [DOI] [PubMed] [Google Scholar]
- 11.Gerna, G., E. Percivalle, F. Baldanti, and M. G. Revello. 2002. Lack of transmission to polymorphonuclear leukocytes and human umbilical vein endothelial cells as a marker of attenuation of human cytomegalovirus. J. Med. Virol. 66:335-339. [DOI] [PubMed] [Google Scholar]
- 12.Gerna, G., E. Percivalle, F. Baldanti, S. Sozzani, P. Lanzarini, E. Genini, D. Lilleri, and M. G. Revello. 2000. Human cytomegalovirus replicates abortively in polymorphonuclear leukocytes after transfer from infected endothelial cells via transient microfusion events. J. Virol. 74:5629-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundy, J. E., K. M. Lawson, L. P. MacCormack, J. M. Fletcher, and K. L. Yong. 1998. Cytomegalovirus-infected endothelial cells recruit neutrophils by the secretion of C-X-C chemokines and transmit virus by direct neutrophil-endothelial cell contact and during neutrophil transendothelial migration. J. Infect. Dis. 177:1465-1474. [DOI] [PubMed] [Google Scholar]
- 14.Hahn, G., H. Khan, F. Baldanti, U. H. Koszinowski, M. G. Revello, and G. Gerna. 2002. The human cytomegalovirus ribonucleotide reductase homolog UL45 is dispensable for growth in endothelial cells, as determined by a BAC-cloned clinical isolate of human cytomegalovirus with preserved wild-type characteristics. J. Virol. 76:9551-9555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hahn, G., D. Rose, M. Wagner, S. Rhiel, and M. McVoy. 2003. Cloning of the genomes of human cytomegalovirus strains Toledo, TownevarRIT3, and Townelong as BACs and directed mutagenesis using a PCR-based technique. Virology 307:164-177. [DOI] [PubMed] [Google Scholar]
- 16.Johnson, D. C., and M. T. Huber. 2002. Directed egress of animal viruses promotes cell-to-cell spread. J. Virol. 76:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kahl, M., D. Siegel-Axel, S. Stenglein, G. Jahn, and C. Sinzger. 2000. Efficient lytic infection of human arterial endothelial cells by human cytomegalovirus strains. J. Virol. 74:7628-7635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacCormac, L. P., and J. L. Grundy. 1999. Two clinical isolates and the Toledo strain of cytomegalovirus contain endothelial cell tropic variants that are not present in the AD169, Towne, or Davis strain. J. Med. Virol. 74:7720-7729. [DOI] [PubMed] [Google Scholar]
- 19.MacDonald, M. R., M. W. Burney, S. B. Resnick, and H. W. Virgin. 1999. Spliced mRNA encoding the murine cytomegalovirus chemokine homolog predicts a beta chemokine of novel structure. J. Virol. 73:3682-3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.MacDonald, M. R., X. Y. Li, and H. W. Virgin. 1997. Late expression of a beta chemokine homolog by murine cytomegalovirus. J. Virol. 71:1671-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maidji, E., E. Percivalle, G. Gerna, S. Fisher, and L. Pereira. 2002. Transmission of human cytomegalovirus from infected uterine microvascular endothelial cells to differentiating/invasive placental cytotrophoblasts. Virology 304:53-69. [DOI] [PubMed] [Google Scholar]
- 22.McDonald, D., L. Wu, S. M. Bohks, V. N. Keval Ramani, D. Unutmaz, and T. J. Hope. 2003. Recruitment of HIV and its receptors to dendritic cell-T cell functions. Science 300:1285-1297. [DOI] [PubMed] [Google Scholar]
- 23.Messerle, M., I. Crnkovic, W. Hammaerschmidt, H. Ziegler, and U. H. Koszinowski. 1997. Cloning and mutagenesis of a herpesvirus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 94:14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murphy, E., Y. Dong, J. Grimwood, J. Schmutz, M. Dickson, M. A. Jarvis, G. Hahn, J. A. Nelson, R. M. Myers, and T. E. Shenk. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. USA 100:14976-14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Novotny, J., I. Rigoutsos, D. Coleman, and T. Shenk. 2001. In silico structural and functional analysis of the human cytomegalovirus (HHV5) genome. J. Mol. Biol. 310:1151-1166. [DOI] [PubMed] [Google Scholar]
- 26.Patrone, M., E. Percivalle, M. Secchi, L. Fiorina, G. Pedrali-Noy, M. Zoppe, F. Baldanti, G. Hahn, U. H. Koszinowski, G. Milanesi, and A. Gallina. 2003. The human cytomegalovirus UL45 gene product is a late, virion-associated protein influencing virus growth at low multiplicities of infection. J. Gen. Virol. 84:3359-3370. [DOI] [PubMed] [Google Scholar]
- 27.Penfold, M. E., D. J. Dairaghi, G. M. Duke, N. Saedrup, E. S. Mocarski, G. W. Kemble, T. J. Schall. 1999. Cytomegalovirus encodes a potent alpha chemokine. Proc. Natl. Acad. Sci. USA 96:9839-9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prichard, M. N., M. E. Penfold, G. M. Duke, R. R. Spaete, and G. W. Kemble. 2001. A review of genetic differences between limited and extensively passaged human cytomegalovirus strains. Rev. Med. Virol. 11:191-200. [DOI] [PubMed] [Google Scholar]
- 29.Revello, M. G., E. Percivalle, E. Arbustini, R. Pardi, S. Sozzani, and G. Gerna. 1998. In vitro generation of human cytomegalovirus pp65 antigenemia, viremia, and leukoDNAemia. J. Clin. Investig. 101:2686-2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Revello, M. G., F. Baldanti, E. Percivalle, A. Sarasini, L. De Giuli, E. Genini, D. Lilleri, N. Labò, and G. Gerna. 2001. In vitro selection of human cytomegalovirus variants unable to transfer virus and virus products from infected cells to polymorphonuclear leukocytes and to grow in endothelial cells. J. Gen. Virol. 82:1429-1438. [DOI] [PubMed] [Google Scholar]
- 31.Revello, M. G., M. Zavattoni, A. Sarasini, E. Percivalle, L. Simoncini, and G. Gerna. 1998. Human cytomegalovirus in blood of immunocompetent persons during primary infection: prognostic implications for pregnancy. J. Infect. Dis. 177:1170-1175. [DOI] [PubMed] [Google Scholar]
- 32.Rott, D., J. Zhu, M. S. Burnett, Y. F. Zhou, A. Wasserman, J. Walker, and S. E. Epstein. 2001. Serum of cytomegalovirus-infected mice induces monocyte chemoattractant protein-1 expression by endothelial cells. J. Infect. Dis. 184:1109-1113. [DOI] [PubMed] [Google Scholar]
- 33.Saederup, N., and E. S. Mocarski, Jr. 2002. Fatal attraction: cytomegalovirus encoded chemokine homologs. Curr. Top. Microbiol. Immunol. 269:235-256. [DOI] [PubMed] [Google Scholar]
- 34.Saederup, N., S. A. Aguirre, T. E. Sparer, D. M. Bouley, and E. S. Mocarski. 2001. Murine cytomegalovirus CC chemokine homolog MCK-2 (m131-129) is a determinant of dissemination that increases inflammation at initial sites of infection. J. Virol. 75:9966-9976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saederup, N., Y. C. Lin, D. J. Dairaghi, T. J. Schall, and E. S. Mocarski. 1999. Cytomegalovirus-encoded beta chemokine promotes monocyte-associated viremia in the host. Proc. Natl. Acad. Sci. USA 96:10881-10886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sinzger, C., M. Kahl, K. Laib, K. Klingel, P. Rieger, B. Plachter, and G. Jahn. 2000. Tropism of human cytomegalovirus for endothelial cells is determined by a post-entry step dependent on efficient translocation to the nucleus. J. Gen. Virol. 81:3021-3035. [DOI] [PubMed] [Google Scholar]
- 37.Streblow, D. N., C. Soderberg-Naucler, J. Vieira, P. Smith, E. Wakabayashi, F. Ruchti, K. Mattison, Y. Altschuler, and J. A. Nelson. 1999. The human cytomegalovirus chemokine receptor US28 mediates vascular smooth muscle cell migration. Cell 99:511-520. [DOI] [PubMed] [Google Scholar]
- 38.Streblow, D. N., S. L. Orloff, and J. A. Nelson. 2001. Do pathogens accelerate atherosclerosis? J. Nutr. 131:2798S-2804S. [DOI] [PubMed] [Google Scholar]