Abstract
Species C human adenovirus mutants that fail to express open reading frame 3 of early region 4 (E4orf3) are phenotypically indistinguishable from the wild-type virus when evaluated in cells cultured in vitro. However, E4orf3 gene function has been productively studied in the context of additional viral mutations. This study identifies diverse roles for the E4orf3 protein that are evident in the absence of early region 1B 55-kDa protein (E1B-55K) function. In an E1B-55K-deficient background, the E4orf3 protein promotes viral replication by increasing both the burst size and the probability that an infected cell will produce virus. Early viral gene expression is not impaired in E1B-55K/E4orf3 double mutant virus-infected cells. Cells infected with the double mutant virus accumulated concatemers of viral DNA. However, the E1B-55K/E4orf3 double mutant virus did not replicate any better in MO59J cells, in which viral DNA concatemers did not accumulate, than in MO59K cells, in which viral DNA concatemers were produced, suggesting that viral DNA concatenation is not the primary growth defect of the E1B-55K/E4orf3 double mutant virus. Accumulation of viral mRNA in the nucleus and cytoplasm of E1B-55K/E4orf3 double mutant virus-infected cells was severely reduced compared to that on wild-type virus-infected cells. Thus, in an E1B-55K mutant background, the E4orf3 protein promotes the accumulation of late viral RNA and enhances late gene expression. Finally, within the context of an E1B-55K mutant virus, the E4orf3 protein acts to suppress host cell translation and preserve the viability of cells at moderately late times of infection.
Diverse cellular activities are targeted by the adenovirus oncoproteins encoded by early regions 1A (E1A), 1B (E1B), and 4 (E4). Key among these activities is the control of normal cell cycle progression. Presumably, the action of these viral oncoproteins on cell cycle progression establishes a favorable environment for virus replication (7, 66). At late times in a productive infection, some of these oncoproteins serve the additional role of overcoming a cell cycle-dependent restriction to virus growth. The cell cycle restriction imposed on virus growth is apparent in the replication of viruses with mutations in either the E1B-55K or E4orf6 gene. Synchronously growing HeLa cells infected during S phase with these mutant viruses are more likely to produce progeny virus than HeLa cells infected during G1 with these mutant viruses (20, 22). It has been suggested that ability of the E1B-55K mutant virus to replicate selectively in human tumors may be linked to its ability to replicate selectively in S-phase-infected cells (21, 37).
Another early gene, E4orf3, was shown to be required for enhanced replication of the E4orf6 mutant virus in S-phase-infected cells. Viruses with mutations in both E4orf6 and E4orf3 no longer showed enhanced replication in S-phase-infected cells (22). Additionally, viruses with multiple E4 mutations were severely defective for replication compared to mutant viruses with defects in only a single open reading frame of E4 (10, 22, 29). With a recently created E1B-55K/E4orf3 double mutant virus, we have shown that E4orf3 is also necessary for enhanced S-phase replication of the E1B-55K mutant virus (62). This double mutant virus was also similar to the E4orf6/E4orf3 double mutant viruses in that it was severely defective for replication. Thus, E4orf3 exerts both cell cycle-dependent and -independent effects to promote virus replication.
At late times of a productive infection, the E1B-55K and E4orf6 proteins maintain several activities in the infected cell, some of which can be affected in a compensatory manner by the E4orf3 protein. First, the E1B-55K and E4orf6 proteins promote the degradation of Mre11, leading to degradation of the Mre11-Rad50-NBS1 protein complex (64), which is important for double-stranded DNA break rejoining (11, 24, 49, 72). By contrast, the E4orf3 protein directs these proteins away from sites of viral DNA replication (64). In addition, the E4orf3 protein binds the catalytic subunit of DNA-dependent protein kinase and may inhibit its activity during the repair of double-strand DNA breaks (8). Thus, both the E4orf3 protein and the E1B-55K-E4orf6 protein complex act to prevent the formation of viral DNA concatemers during an infection (64, 68).
Second, the E1B-55K-E4orf6 protein complex promotes the accumulation of viral mRNA in the cytoplasm while preventing newly synthesized cellular mRNA from reaching the cytoplasm (9, 25, 36, 50). By promoting the nuclear stability of late viral mRNA, the E4orf3 protein also promotes the cytoplasmic accumulation of late viral mRNA although by a mechanism distinct from that of the E1B-55K-E4orf6 protein complex (10).
Third, the E1B-55K-E4orf6 protein complex and the E4orf3 protein alter host cell gene expression late during an infection, although possibly in opposite directions. The E1B-55K-E4orf6 protein complex blocks the translation of most cellular mRNAs, in part by promoting late viral gene expression (4, 5). Also, the E1B-55K protein is tethered to p53-responsive promoters by directly binding p53 and thereby repressing transcription of p53-responsive cellular genes. The E1B-55K-E4orf6 protein complex also destabilizes p53 (53, 63) as part of a ubiquitin ligase composed of Cul5, elongins B and C, and Rbx1 (26, 52). By contrast, the E4orf3 protein may prevent E1B-55K from repressing p53-mediated transcription at early times of infection (33). Finally, the E4orf3 protein may promote the expression of some cellular genes such as glucocorticoid-responsive genes (69).
The focus of this work was to elucidate the E4orf3 protein activity, which promotes virus replication in an E1B-55K mutant background. This work demonstrates that the E4orf3 protein maintains diverse roles at late times of infection. Among these roles are the ability to promote efficient late viral RNA accumulation and late viral gene expression and to block the formation of viral DNA concatemers. Surprisingly, the severely defective nature of the E1B-55K-E4orf3 double mutant virus is not due to the accumulation of concatemeric viral DNA. A novel role for the E4orf3 protein revealed in the context of an E1B-55K deletion was its ability to suppress host gene expression and possibly sustain the viability of the late infected cell.
MATERIALS AND METHODS
Cell culture.
Cell culture media, cell culture supplements, and serum were obtained from Invitrogen (Carlsbad, Calif.) through the Tissue Culture Core Laboratory of the Comprehensive Cancer Center of Wake Forest University. HeLa cells (ATCC CCL 2; American Type Culture Collection, Manassas, Va.) were maintained as monolayer cultures in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% newborn calf serum, 100 U of penicillin, and 100 μg of streptomycin per ml. 293 (ATCC CRL 1573) and A549 (ATCC CCL 185) cells were maintained in DMEM supplemented with 10% fetal bovine serum, 100 U of penicillin, and 100 μg of streptomycin per ml. MO59J and MO59K cells (ATCC CRL-2366 and CRL-2365, respectively) were maintained in 1:1 DMEM-Ham's F12 medium supplemented with 2.5 mM l-glutamine, 1.5 g of sodium bicarbonate per liter, 0.5 mM sodium pyruvate, 0.05 mM nonessential amino acids, and 10% fetal bovine serum. Cells were maintained in subconfluent adherent cultures in a 5% CO2 atmosphere at 37°C by passaging them twice weekly at approximately a 1:10 dilution.
E4orf3-expressing HeLa cells.
Quasistable HeLa cells expressing the E4orf3 gene were established by selecting for a linked drug resistance gene. The E4orf3 gene was subcloned into a plasmid under control of the immediate-early cytomegalovirus promoter. The plasmid also expressed the aminoglycoside 3′-phosphotransferase gene. This E4orf3 expression plasmid, pRT1, was transfected into HeLa cells, and cells expressing the plasmid were selected in the presence of G418 (Invitrogen). After 14 days, G418-resistant colonies were expanded in the presence of selective medium and tested for the presence of E4orf3 by immunofluorescence.
Recombinant virus creation.
An E1B-55K/E4orf3 double mutant recombinant virus was created with viral DNA purified from the E1B-55K mutant virus dl1520 (6) and E4orf3 mutant virus dl341 (59). Viral DNA containing E1B-55K mutant sequences was cleaved to completion with the restriction enzymes EcoRI, SpeI, and Bsu36I. These restriction enzymes recognize sequences limited to the rightmost 43% of the adenovirus type 5 genome. Similarly, viral DNA bearing the E4orf3 mutation was cleaved with NsiI, which recognizes sequences in the leftmost 32% of the adenovirus type 5 genome. To prevent subsequent ligation, the DNA was dephosphorylated and purified before being cleaved with PmeI, a restriction enzyme that cleaves the adenovirus type 5 genome once near the center of the genome (at approximately 37 map units).
The restriction fragments were ligated without further purification, and the products were transfected into 293 cells with Lipofectin (Invitrogen) according to the supplier's recommendations to recover infectious virus by plaque purification. The parental E4orf3 mutant virus shows no growth defect in 293 cells, allowing the E1B-55K/E4orf3 double mutant virus to exhibit the same growth properties as the E4orf3 single mutant virus in these cells. The newly created recombinant virus was purified by three sequential plaque isolations. Mutant viruses were identified by PCR with primers to amplify selected regions of interest, followed by restriction mapping to determine the status of the diagnostic restriction site. Candidate recombinants were partially sequenced to confirm the E1B-55K and E4orf3 mutations and to verify the integrity of the junction.
Viruses.
The phenotypically wild-type adenovirus type 5 parent virus dl309 used in these studies lacks a portion of the E3 region that has been shown to be dispensable for growth in tissue culture (30). The E1B mutant virus dl1520 contains an 827-bp deletion in the 55-kDa protein coding region in combination with a nonsense mutation at the third codon to ensure that a truncated 55-kDa product cannot be expressed (6). The E4 mutant virus dl341 contains a frameshift mutation resulting from a single-base-pair deletion of a cytosine residue at nucleotide position 7143 as measured from the right end (59). This mutation disrupts the E4orf3 coding region and prevents expression of any detectable E4orf3 protein. The E1B-55K/E4orf1/E4orf2/E4orf3 mutant virus dl1016 was generated by Bridge and Ketner (9) from the E1B-55K mutant virus dl110 (4) and the E4 triple mutant virus dl1006 (9).
The propagation of these viruses has been described elsewhere (9, 30). In brief, virus stocks were prepared by infecting 293 cells at a low multiplicity of infection. Virus was harvested 3 to 5 days postinfection from a concentrated freeze-thaw lysate by sequential centrifugation in discontinuous and equilibrium cesium chloride gradients (31). The gradient-purified virus was supplemented with 5 volumes of 0.012 M HEPES (pH 7.4)-0.12 M NaCl-0.1 mg of bovine serum albumin per ml-50% (vol/vol) glycerol and stored at −20°C. The titer of each virus stock was determined by plaque assays with 293 cells.
For infection with adenovirus, cells were passaged 16 to 24 h prior to infection to a density of 1 × 104 to 2 × 104 cells per cm2. The cells were washed once with phosphate-buffered saline (PBS), and the final wash was replaced with virus (10 to 25 PFU per cell) in adenovirus infection medium (PBS supplemented with 0.2 mM CaCl2, 2 mM MgCl2, and 2% calf serum). The virus was added at one-fourth the normal culture volume, and the cells were gently rocked for 60 min at 37°C. The virus suspension was then replaced with normal growth medium, and the infected cells were returned to 37°C.
Plaque assay for viral yields.
Detailed methods for adenovirus plaque assays have been described elsewhere (31). In brief, virus was harvested from infected cells in culture medium at 48 h postinfection by three cycles of freezing and thawing. The cell lysates were clarified by centrifugation and serially diluted in adenovirus infection medium for infection of 293 cells for plaque assays. The diluted lysates were applied to the cells for 1 h and removed, and the 293 cells were overlaid with 0.65% SeaKem ME agarose (Cambrex Bio Science, Rockland, Maine) in DMEM supplemented with 0.75% sodium bicarbonate and 4% fetal bovine serum. The infected cells were supplemented with additional agarose and medium on the fourth and seventh days after infection. The plaques were visualized by staining with neutral red in an agarose overlay. Data were typically collected from three dilutions in each series of dilutions. The virus yield (PFU per milliliter) was determined by linear regression and expressed as PFU per infected cell.
Antibodies.
The primary antibodies specific for adenovirus proteins used in this study include mouse monoclonal antibodies against the E1A (M73) (27), E1B-55K (2A6) (60), E2A DNA-binding protein (DBP) (B6-8) (56), E4orf6 and E4orf6/7 (MAb#3) (39), and the L4-100K (Db5 5C5) (12) proteins. Rat monoclonal antibodies specific for adenovirus proteins recognized the E1B-55K (9C10, Calbiochem, San Diego, Calif.) (70) and E4orf3 (6A11) (41) proteins. Polyclonal rabbit serum raised against adenovirus type 5 was purchased from the American Type Culture Collection. Antibodies specific for cellular proteins included mouse monoclonal antibodies specific for the PML protein (5E10) (65), β-actin (AC-15, A5441; Sigma Chemical Co., St. Louis, Mo.) (19), and β-tubulin (Tub 2.1, T4026; Sigma Chemical) (23).
Primary antibodies were used as hybridoma tissue culture supernatant fluid (undiluted or diluted 1:1) or at a concentration of 1 to 4 μg per ml. Horseradish peroxidase-conjugated secondary antibodies were raised in goats and used at 0.02 to 0.1 μg per ml (Jackson ImmunoResearch, West Grove, Pa.). Fluorescent secondary antibodies, specific for either rat or mouse immunoglobulin G, were also raised in goats and coupled to Alexa Fluor 568 or Alexa Fluor 488 (Molecular Probes, Eugene, Oreg.) or fluorescein (Jackson ImmunoResearch). Fluorescent secondary antibodies were used at 2 μg per ml. Antibodies were diluted in Tris-buffered saline (0.137 M NaCl, 0.003 M KCl, 0.025 M Tris-Cl [pH 8.0], 0.0015 M MgCl2, 0.5% bovine serum albumin, 0.1% glycine, 0.05% Tween 20, 0.02% sodium azide). The antibodies used for immunofluorescence included 10% normal goat serum (Invitrogen). Sodium azide was omitted from solutions used with horseradish peroxidase-conjugated antibodies.
Indirect immunofluorescence.
Indirect immunofluorescence was conducted as previously described (46) with the modification that cells were fixed in freshly prepared 2% formaldehyde and permeabilized in 0.5% Triton X-100 in PBS. Images were acquired as eight-bit TIFF files with a Zeiss LSM 510 (Carl Zeiss, Inc., Thornwood, N.Y.) confocal laser scanning device fitted to a Zeiss Axioplan 2 microscope with a 60× NA 1.4 oil immersion objective. Alexa Fluor 488 and Alexa Fluor 568 dyes were excited by krypton ion and helium-neon laser excitation, respectively. Single optical sections of approximately 1.5-μm depth at the level of the cytoplasm were recorded as TIFF files with the LSM 510 software.
Early viral gene expression.
For immunoblot analysis, at 9 h postinfection, adherent and detached HeLa cells were harvested and washed with ice-cold PBS, and the cells were lysed by the addition of freshly prepared sodium dodecyl sulfate (SDS) protein sample buffer (0.06 M Tris-Cl [pH 6.8], 1% SDS, 0.005 M EDTA, 0.05 M dithiothreitol, 10% [vol/vol] glycerol) at 107 cells per ml. The cells were disrupted by sonication, and the lysates were clarified by centrifugation before separating the proteins from 5 × 104 infected cells per lane by polyacrylamide gel electrophoresis (PAGE) through a 12.5% polyacrylamide gel with a 36:1 ratio of acrylamide to N,N′-methylenebisacrylamide containing 0.1% SDS. The separated proteins were electrophoretically transferred to a nitrocellulose support and visualized by blotting with adenovirus-specific antibodies and chemiluminescent detection with the PicoWest kit as per the manufacturer's recommendations (Pierce, Rockford, Ill.).
Infectivity.
The level of infection in HeLa, MO59J, and MO59K cells was determined by infecting each cell line with dl309, dl1520, or the 55K−/orf3− mutant virus at multiplicities of 40, 100, 200, 2,000, and 6,000 virions per cell. At 24 h postinfection, cells were analyzed by indirect immunofluorescence for the E2A DBP with the B6-8 monoclonal antibody (56). The fraction of DBP-positive cells among approximately 100 to 200 cells per sample was determined with a Nikon TE300 inverted microscope fitted with filters appropriate for 4′,6′-diamidino-2-phenylindole to enumerate total cells and Alexa Fluor 488 excitation to enumerate infected (DBP-positive) cells. The fraction of infected cells was expressed as a function of the multiplicity, and the data were fit to an exponential curve with MacCurvefit (Kevin Raner Software, Waverley, Australia) to determine the effective infectivity for each virus and cell line.
DNA slot blotting.
Total viral DNA present in the infected cell culture was measured by slot blot analysis essentially as described previously (20, 32). Briefly, DNA was isolated from infected HeLa, MO59J, and MO59K cells by detergent lysis and proteinase K treatment followed by extraction with phenol and chloroform. Serial dilutions of infected-cell DNA were immobilized on nylon membranes (Nytran; Schleicher and Schuell, Keene, N.H.) and then hybridized to radioactively labeled adenovirus DNA fragments generated by random-primed extension in the presence of α-32P-labeled dATP with total adenovirus DNA (3). The amount of bound radioactive DNA was quantified by phosphorescence imaging (ImageQuant software; Molecular Dynamics, Sunnyvale, Calif.), and the amount of adenovirus DNA per cell was expressed relative to the amount measured in infected HeLa cells.
Pulsed-field gel electrophoresis.
To detect the formation of viral DNA concatemers, total DNA from infected cells was analyzed by pulsed-field gel electrophoresis with a Bio-Rad CHEF DR II instrument (Bio-Rad, Hercules, Calif.) as described elsewhere (67, 68). In brief, 106 infected cells were detached with trypsin 24 or 48 h after infection and suspended in 0.25 ml of PBS, which was immediately mixed with 0.25 ml of liquid 1.5% low-melting-point agarose in PBS. This solution was cast into blocks, allowed to solidify, and then incubated at 55°C for 24 h in 1% SDS-0.1 M EDTA-0.010 M Tris-Cl (pH 8.0), with 100 μg of proteinase K per ml. The DNA in portions of the agarose blocks representing 2.5 × 105 infected cells was analyzed by electrophoresis through 1.5% agarose at 150 V with ramped pulse times from 10 to 90 s for 45 h at 15°C in 0.5× TBE (0.045 M Tris base, 0.045 M sodium borate, 0.005 M EDTA). The gel was stained with ethidium bromide for 30 min to visualize viral DNA. The gel was washed twice for 30 min each in denaturing buffer (3 M NaCl and 0.4 M NaOH), followed by a 15-min wash in transfer buffer (3 M NaCl and 8 mM NaOH) before Southern transfer (3).
Southern transfer.
Infected-cell DNA was immobilized on a neutral nylon membrane (Nytran; Schleicher and Schuell) by downward capillary transfer under alkaline conditions and then hybridized to radioactively labeled adenovirus DNA fragments. [α-32P]dATP radioactivity was incorporated into probes derived from total adenovirus DNA by random priming.
Cell fractionation and RNA isolation.
All solutions used for RNA purification and in vitro transcription were prepared from diethyl pyrocarbonate-treated water as described by Ausubel et al. (3). RNA was isolated from the nucleus and cytoplasm of adenovirus-infected HeLa cells at 24 h postinfection. Infected cells were suspended in an isotonic buffer (0.140 M NaCl, 0.01 M Tris-Cl [pH 7.4], 0.0015 M MgCl2) containing 0.01 mM vanadyl-adenosine complex and an equal volume of isotonic buffer with 1% (vol/vol) Nonidet P-40 (Calbiochem, San Diego, Calif.) with mixing. The nuclei were collected by centrifugation and washed, and RNA was isolated and purified as previously described (3, 22).
RNase protection assays.
The levels of L3 and L5 RNAs in the cytoplasm and nucleus were determined by RNase protection assays as previously described (22). Briefly, L3 and L5 transcript levels were determined with radioactive RNA probes that span the polyadenylation site of the L3 and L5 families of transcripts. Protected fragments were resolved by denaturing urea-PAGE and quantified by phosphorescence imaging (Molecular Dynamics). The amount of nuclear contamination in the cytoplasmic fractions was estimated from the amount of unprocessed viral mRNA in the cytoplasm as described previously (3, 22). By this measure, no more than 5% of the nuclear material was found in the cytoplasm. The amount of cytoplasmic contamination in the nuclear fractions was not determined in these experiments.
Late viral protein synthesis.
Late viral protein synthesis was analyzed by pulse labeling infected HeLa cells for 1 h with 0.1 mCi of 35S-labeled amino acids (Tran-S-label; ICN Biochemicals, Irvine, Calif.) per ml in cysteine- and methionine-free DMEM supplemented with 2% fetal bovine serum at 36 h postinfection. The cells were then scraped into PBS, pelleted, and lysed in SDS protein sample buffer. Protein from 105 cell equivalents was separated by SDS-PAGE through an 8% polyacrylamide gel. SDS-PAGE gels were fixed in 15% glacial acetic acid-7.5% methanol, and the radioactive proteins were visualized by phosphorescence imaging (Molecular Dynamics). Late adenovirus proteins were identified by comparison to gradient-purified, radioactive virion proteins obtained from 293 cells infected with the wild-type adenovirus dl309 that had been labeled with [14C]amino acids as described previously (21).
Cell killing assay.
Virus-induced cell killing was measured by trypan blue dye exclusion. HeLa cells were plated in 12-well dishes and infected with the indicated viruses at a multiplicity of 10 PFU per cell. At 72 h after infection, the cells were detached with trypsin and collected along with the nonadherent cells in the growth medium. The cells were collected by centrifugation for 5 min at 200 × g and suspended in 0.1 ml of PBS, to which an equal volume of 0.4% trypan blue in PBS was added. The percentage of nonviable (trypan blue positive) cells was determined by bright-field microscopy with the use of a hemacytometer.
RESULTS
E4orf3 enhances virus replication in the absence of E1B-55K.
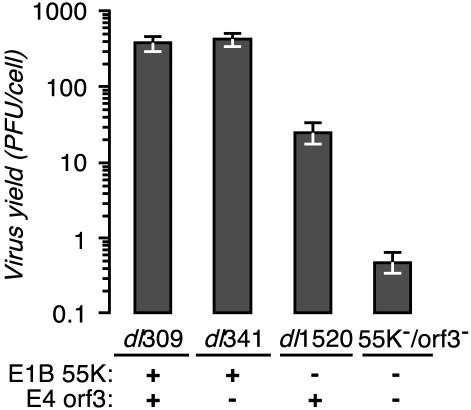
The virus yield from infected HeLa cells reveals that deletion of E4orf3 has a significant effect on replication in an E1B-55K mutant background but not in an otherwise wild-type background (Fig. 1). HeLa cells were infected with the wild-type virus dl309, the E4orf3 mutant virus dl341, the E1B-55K mutant virus dl1520, or the E1B-55K/E4orf3 double mutant virus 55K−/orf3− at a multiplicity of 10 PFU per cell. At 48 h after infection, the total virus present in the culture was measured by plaque assay. The yield of E1B-55K/E4orf3 double mutant virus was approximately 1,000-fold less than the yield of wild-type virus and approximately 10-fold less than that of the E1B-55K single mutant virus. As noted previously, replication of the E4orf3 mutant virus was nearly indistinguishable from that of wild-type virus (10, 25, 29). Thus, although the E4orf3 protein appears to contribute little to the replication of an otherwise wild-type virus, these results show that the E4orf3 protein enhances the replication of the E1B-55K mutant virus.
FIG. 1.
E1B-55K/E4orf3 double mutant virus yield is severely reduced. HeLa cells were infected with the indicated viruses at a multiplicity of 10 PFU per cell. The status of the relevant viral genes (E1B-55K or E4orf3) is indicated below the histogram. The virus yield at 48 h postinfection was measured by plaque assay with 293 cells. The results shown are the averages of three independent infections performed in three independent experiments. Error bars indicate the standard error of the mean.
To confirm that the severe defect of the E1B-55K/E4orf3 double mutant virus was due to loss of the E4orf3 protein and not to the presence of unintended mutations elsewhere in the genome, the growth of the virus was measured in the presence of exogenously supplied E4orf3 (Table 1). Quasistable HeLa cell lines expressing the E4orf3 gene under control of the cytomegalovirus immediate-early promoter were established by selecting for a linked drug resistance gene. By indirect immunofluorescence microscopy, greater than 90% of the G418-resistant cells contained the E4orf3 protein at levels comparable to those produced during a virus infection. However, enforced expression of E4orf3 in these cells slowed the growth of the cells, and E4orf3 expression could not be sustained for more than several weeks (data not shown).
TABLE 1.
Virus production in E4orf3-expressing HeLa cellsa
| Virus | Virus yield, PFU/cell (% of dl309 value)
|
|
|---|---|---|
| Control cells | E4orf3-expressing cells | |
| dl309 | 380 (100) | 13.2 (100) |
| dl341 | 429 (113) | 39 (230) |
| dl1520 | 25 (6.6) | 1.3 (10) |
| 55K−/orf3− | 0.47 (0.1) | 1.1 (8.4) |
The virus yield is expressed as PFU per cell and as a percentage of the corresponding yield of the wild-type virus (dl309).
Curiously, the yield of the wild-type, E4orf3 mutant, and E1B-55K mutant viruses from the E4orf3-expressing cell line was approximately 10- to 30-fold less than that from vector-transfected HeLa cells (Table 1). However, the exogenously expressed E4orf3 compensated for the E4orf3 mutation in the E1B-55K/E4orf3 double mutant virus (Table 1). The yield of double mutant virus from the E4orf3-expressing cells was nearly equivalent to the yield of E1B-55K single mutant virus from the same cells. These results suggest that the additional defect seen in the E1B-55K/E4orf3 double mutant virus is a consequence of the failure to express the E4orf3 protein. This raises the question of where in the infectious cycle of this double mutant virus the function of E4orf3 is required.
Early protein synthesis is not affected by the absence of the E1B-55K and E4orf3 proteins.
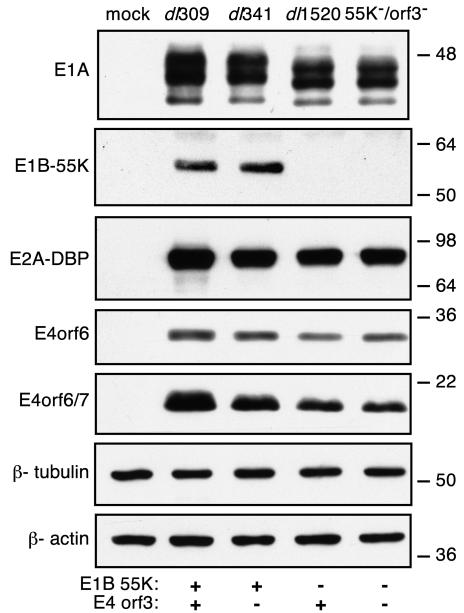
The accumulation of early viral proteins in HeLa cells that were mock infected or infected with the viruses indicated in Fig. 2 was measured by immunoblot. As controls, the levels of the representative host cell proteins β-tubulin and β-actin were measured; these remained unchanged from the levels in mock-infected cells 9 h after infection. As anticipated, both wild-type virus- and E4orf3 mutant virus-infected cells expressed comparable levels of all four early viral proteins measured. The level of E1A protein produced by the E1B-55K mutant virus- and the E1B-55K/E4orf3 double mutant virus-infected cells was slightly less than that detected in the wild-type virus-infected cells. These two mutant viruses encode an E1A protein that is a hybrid of the adenovirus type 2 and type 5 E1A proteins. The different levels measured may reflect an intrinsic difference between type 2 and type 5 E1As. As expected, dl1520- and the 55K−/orf3−-infected cells failed to express any E1B-55K protein. Additionally, these two viruses appeared to direct the accumulation of about half as much E4orf6/7 protein as the wild-type virus. However, all of the viruses analyzed here expressed similar levels of the E4orf6 protein as well as the E2A DBP. Although small differences in the levels of the E1A and E4orf6/7 proteins were noted, other early viral proteins accumulated to wild-type levels. Therefore, the severe defect of the E1B-55K/E4orf3 double mutant virus is not likely due to an inability to synthesize early viral proteins.
FIG. 2.
E1B-55K/E4orf3 double mutant virus directs early protein synthesis to nearly wild-type levels. HeLa cells were mock infected or infected with the indicated viruses at a multiplicity of 20 PFU per cell. The status of the relevant viral genes (E1B-55K or E4orf3) is indicated below the blots. The levels of E1A, E1B-55K, E2A-72K, E4orf6, and E4orf6/7 proteins from equivalent numbers of cells at 9 h postinfection were determined by Western blotting with primary mouse monoclonal antibodies M73, 2A6, B6-8, and MAb#3, respectively. As controls, the levels of the cellular proteins β-actin and β-tubulin were determined with primary mouse monoclonal antibody clones AC-15 and Tub 2.1, respectively. Sizes are shown to the right (in kilodaltons).
Cells infected with the E1B-55K/E4orf3 double mutant virus synthesize wild-type levels of viral DNA that accumulate as concatemers.
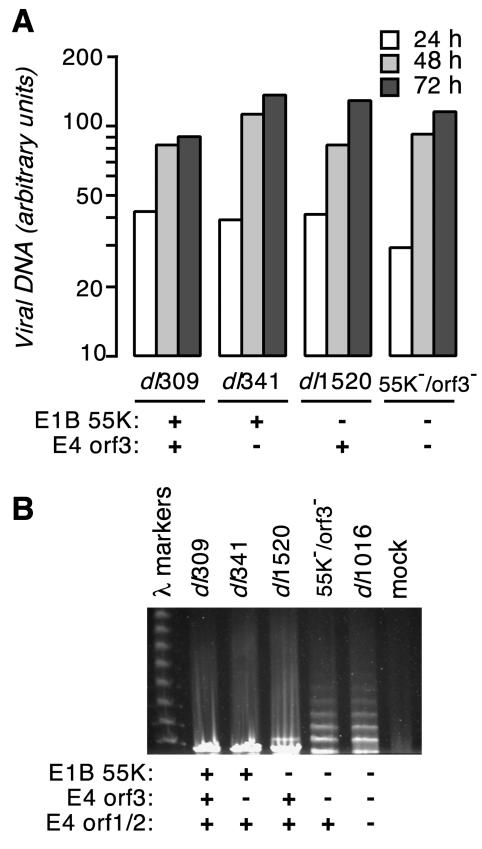
We performed a quantitative and qualitative analysis of the newly synthesized viral DNA in E1B-55K/E4orf3 double mutant virus-infected cells. HeLa cells were infected with each of the viruses identified in Fig. 3 at a multiplicity of 25 PFU per cell. Total DNA was isolated from the infected cells at 24, 48, and 72 h postinfection, and the amount of viral DNA was quantified. Although the double mutant virus-infected cells contained slightly less viral DNA than the other infected cells at 24 h postinfection, cells infected with this virus contained the same amount of viral DNA as the wild-type virus- and single mutant virus-infected cells at 48 and 72 h postinfection (Fig. 3A). These data indicate that, in HeLa cells, loss of the E4orf3 protein in an E1B-55K mutant virus background does not greatly affect viral DNA accumulation.
FIG. 3.
(A) E1B-55K/E4orf3 double mutant virus-infected cells synthesize viral DNA to the same level as in a wild-type virus infection. HeLa cells were infected with the indicated viruses at a multiplicity of 25 PFU per cell. The status of the E1B-55K and E4orf3 genes in each of the viruses is indicated below the histogram. Total cellular DNA was isolated from equal numbers of adenovirus-infected HeLa cells at 24, 48, and 72 h postinfection, and the amount of viral DNA was measured by hybridization with radioactively labeled adenovirus DNA. The results shown are representative of two to three independent experiments. (B) Viral genomes of E1B-55K/E4orf3 double mutant virus-infected cells form concatemers. HeLa cells were mock infected or infected with the indicated viruses at a multiplicity of 25 PFU per cell. The status of the E1B-55K and E4 genes in each of the viruses is indicated below the panel. The total DNA present in the cells at 24 h postinfection was visualized by ethidium bromide staining after pulsed-field gel electrophoresis. The sizes of concatemers of 40-kb bacteriophage lambda DNA provided size standards.
The E1B-55K/E4orf3 double mutant virus produced concatemeric viral DNA during replication. HeLa cells were infected with the viruses indicated in Fig. 3B at a multiplicity of 25 PFU per cell. After 24 h, the total DNA present in the cells was analyzed by pulsed-field gel electrophoresis and Southern blotting to detect viral DNA concatemers. The wild-type virus and the E4orf3 mutant virus produced exclusively monomeric viral DNA. Although the E1B-55K mutant virus produced primarily monomeric viral DNA, we repeatedly observed viral DNA with the mobility of a dimer (Fig. 3B). However, we have not determined if this species is a bona fide dimer of viral DNA or an aberrantly migrating form of monomeric viral DNA (57, 58). The E1B-55K/E4orf3 double mutant virus produced concatemers of viral DNA in excess of five genomes in length. The amount of viral DNA present in the concatemers was quantified by Southern blotting and found to correlate with the levels of viral DNA measured by slot blot analysis (data not shown). These results, which are in agreement with those reported by Weitzman and associates in studies involving dl1016, a virus defective in the E1B-55K E4orf1, E4orf2, and E4orf3 genes (64), support the notion that E4orf3 prevents concatenation of viral genomes in E1B-55K mutant virus-infected cells.
Severe growth defect of the E1B-55K/E4orf3 double mutant virus is not alleviated in MO59J cells, in which viral DNA concatemers no longer accumulate.
It has been suggested that the poor yield of the E1B-55K/E4orf3 double mutant virus could be due to the accumulation of viral DNA concatemers, which are inefficiently packaged into virions. This suggestion was tested with MO59J cells, which are derived from a human glioblastoma and fail to express the catalytic subunit of DNA-dependent protein kinase (DNA-PKcs) (34). This lesion renders MO59J cells defective in the nonhomologous end-joining repair pathway. These cells inefficiently repair double-strand DNA breaks and fail to concatenate linear adenovirus genomes. As a control, MO59K cells, derived from the same tumor as MO59J cells (1), express normal levels of DNA-PKcs and thus have an intact DNA repair program. The ability of the MO59J and MO59K cell lines to be infected was the same. However, approximately 10-fold more viral particles were required to infect a comparable fraction of MO59J or MO59K cells as HeLa cells (data not shown).
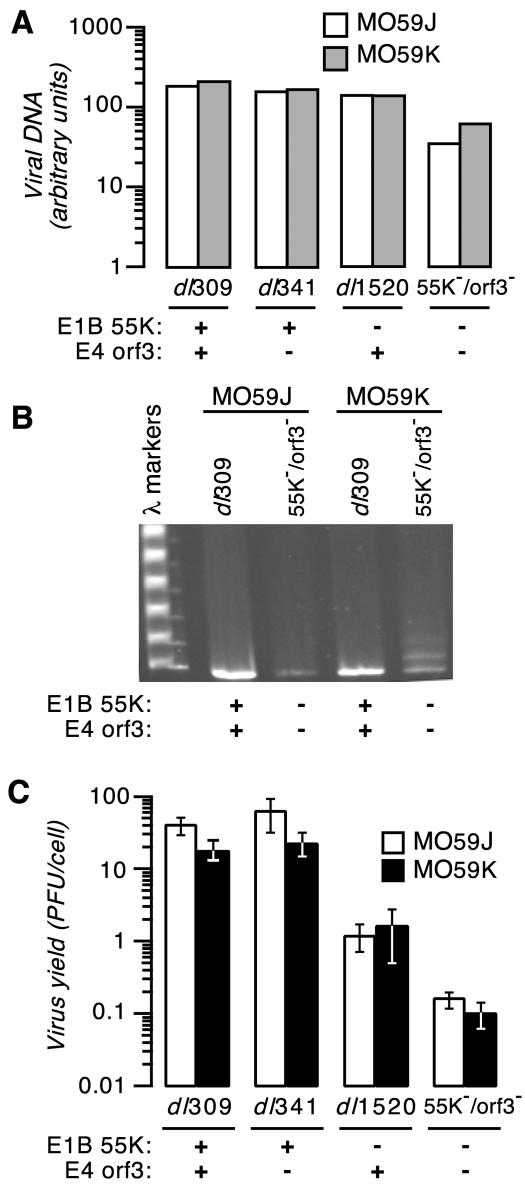
Viral DNA accumulation was compared between MO59J and MO59K cells infected with the viruses indicated in Fig. 4 at an effective multiplicity of one infectious unit per cell. Levels of viral DNA were measured by slot blot analysis at 24 and 48 h postinfection. The amount of viral DNA in cells infected with the E1B-55K/E4orf3 double mutant virus was reduced at 48 h after infection (three- to fivefold) compared to cells infected with the wild-type, E4orf3 mutant, or E1B-55K mutant virus (Fig. 4A). However, each virus directed the synthesis of viral DNA amounts equivalent to or slightly more than those in DNA repair-proficient MO59K cells than in DNA-PKcs mutant MO59J cells.
FIG. 4.
(A) E1B-55K/E4orf3 double mutant virus-infected cells synthesize reduced levels of viral DNA in MO59J and MO59K cells. MO59J and MO59K cells were infected with the indicated viruses at an effective multiplicity of 1 infectious unit per cell. The status of the E1B-55K and E4orf3 genes in each of the viruses is indicated below the histogram. The amount of viral DNA present in equal numbers of adenovirus-infected cells at 48 h postinfection was measured by slot blotting. Representative results from two independent experiments are shown. (B) Viral genomes in E1B-55K/E4orf3 double mutant virus-infected MO59J cells fail to undergo concatemer formation. MO59J and MO59K cells were infected with the wild-type virus and the E1B-55K/E4orf3 double mutant virus at an effective multiplicity of 1 infectious unit per cell, and total infected-cell DNA at 48 h postinfection was analyzed by pulsed-field gel electrophoresis and ethidium bromide staining. (C) The E1B-55K/E4orf3 double mutant virus exhibits a decrease in virus yield in both MO59J and MO59K cells. MO59J and MO59K cells were infected with the indicated viruses, and the virus yield at 48 h postinfection was measured by plaque assay with 293 cells. The status of the relevant viral genes is indicated below the histogram. The results shown are the averages of three independent infections performed in three independent experiments. Error bars indicate the standard error of the mean.
Total DNA present in the infected cells at 48 h after infection was separated by pulsed-field gel electrophoresis and visualized by ethidium bromide staining (Fig. 4B). As anticipated, the E1B-55K/E4orf3 double mutant virus produced monomeric viral DNA in the MO59J cells and concatemeric viral DNA up to four genomes in length in the MO59K cells. As observed in HeLa cells, neither the wild type (Fig. 4B) nor any of the single mutant viruses (data not shown) produced concatemeric viral DNA in either MO59J or MO59K cells. These results confirm that, in the absence of E1B-55K function, the E4orf3 protein is sufficient to prevent viral DNA concatenation mediated by a DNA-PKcs-dependent pathway.
To determine if the poor virus yield of the E1B-55K/E4orf3 double mutant virus was indeed a result of concatemer formation, MO59J and MO59K cells were infected with each of the viruses of interest, and the amount of progeny virus present after 48 h was measured by plaque assay (Fig. 4C). Two observations can be made. First, the yield of virus from these glioblastoma-derived cells lines was approximately 10-fold less than that from HeLa cells (Fig. 1). Nevertheless, the wild-type and E4orf3 mutant viruses replicated to comparable levels in both MO59J and MO59K cells. As observed in HeLa cells, the E1B-55K mutant virus grew to levels approximately 1 to 2 log units less than the wild-type virus. If the failure of the E1B-55K/E4orf3 double mutant virus to replicate was strictly due to the accumulation of viral DNA concatemers, then this defect should have been relieved in cells without an intact DNA repair program, and the yield of the double mutant virus should have been comparable to the yield of the E1B-55K single mutant virus. Surprisingly, however, the E1B-55K/E4orf3 double mutant virus replicated poorly in both MO59J and MO59K cells, and there was no significant difference in growth of the double mutant virus between the two cell lines. From this we conclude that the failure of this virus to replicate is not due to the accumulation of concatemeric viral DNA but must result from defects in other processes subsequent to viral DNA synthesis.
Accumulation of late viral mRNA in the nucleus and cytoplasm is significantly reduced in cells infected with the E1B-55K/E4orf3 double mutant virus.
The E1B-55K/E4orf3 double mutant virus is not defective in its ability to synthesize viral DNA, and the formation of concatemeric viral genomes does not appear to correlate with the poor growth of this virus. To determine if poor growth resulted from a defect after viral DNA synthesis, the levels of polyadenylated L3 and L5 late viral transcripts in the cytoplasm and nucleus were measured by an RNase protection assay.
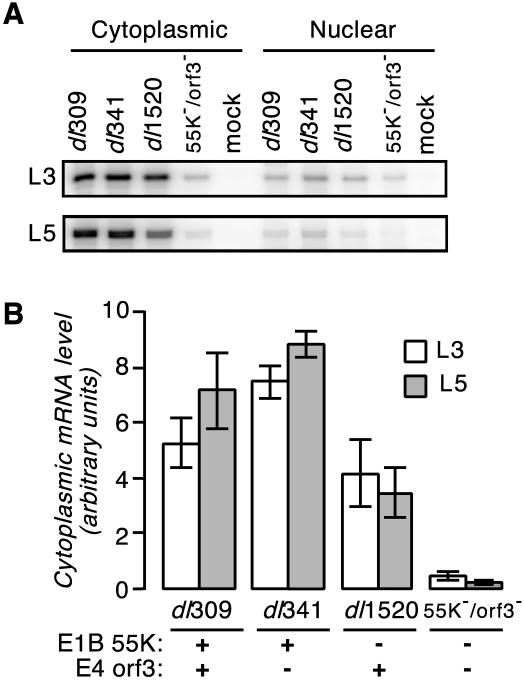
The results representing averages of three independent experiments with cells infected with the wild-type virus, the E4orf3 mutant virus, the E1B-55K mutant virus, or the E1B-55K/E4orf3 double mutant virus are shown in Fig. 5. Cells infected with the double mutant virus contained substantially less cytoplasmic and nuclear L3 and L5 RNA than the other viruses analyzed (Fig. 5A). E4orf3 mutant virus-infected cells had slightly more (20 to 40%) cytoplasmic viral mRNA than wild-type virus-infected cells (Fig. 5B). Cells infected with the E1B-55K mutant virus contained a slight (50 to 70%) but, with respect to L5, significant (P < 0.05) reduction in cytoplasmic viral mRNA compared to wild-type virus-infected cells, consistent with previously published results (22). Significantly, accumulation of viral mRNA in the cytoplasm of E1B-55K/E4orf3 double mutant virus-infected cells was reduced by as much as 30-fold compared to that in wild-type virus-infected cells. Although somewhat variable between experiments, a corresponding decrease in the amount of mature mRNA recovered in the nucleus of cells infected with the double mutant virus was observed as well (Fig. 5A and data not shown).
FIG. 5.
(A) Relative levels of cytoplasmic and nuclear L3 and L5 late viral transcripts determined by RNase protection assays. HeLa cells were mock infected or infected with the indicated viruses at a multiplicity of 20 PFU per cell. At 16 h postinfection, RNA was isolated from both the cytoplasmic and nuclear fractions. L3 and L5 transcript levels were determined by RNase protection. The results of a representative protection assay are shown. (B) E1B-55K/E4orf3 double mutant virus-infected cells contain less late cytoplasmic viral mRNA than cells infected with the wild-type or parent mutant virus. The cytoplasmic levels of L3 and L5 transcripts in HeLa cells infected with the indicated viruses were measured by RNase protection and are expressed relative to the levels in dl309-infected cells, which were set to 100. The status of the relevant genes is indicated below the histogram. The results are averages obtained from four to six experiments. Error bars indicate the standard error of the mean.
The cytoplasmic-to-nuclear ratio of the L3 and L5 mRNAs provides an estimate of the efficiency of nuclear mRNA export (22). The greatest cytoplasmic-to-nuclear ratios (between 4 and 6) were measured for wild-type and E4orf3 mutant virus-infected cells in three independent experiments. Consistent with a defect in the export of late viral mRNA, the cytoplasmic-to-nuclear ratio measured for both E1B-55K mutant and E1B-55K/E4orf3 double mutant virus-infected cells was less, between 2 and 3, measured in three independent experiments. Because the cytoplasmic-to-nuclear ratio of mature mRNA for the double mutant virus was not less than that measured for the E1B-55K single mutant virus, we suggest that mRNA transport is not the major defect responsible for the poor replication of this virus. Rather, the double mutant virus is severely defective in its ability to accumulate late viral transcripts in the nucleus through either reduced transcription or decreased stability of the nuclear pool of viral RNA. It seems likely that this defect contributes to the poor replication of this virus.
Late viral gene expression is significantly reduced in HeLa cells infected with the E1B-55K/E4orf3 double mutant virus.
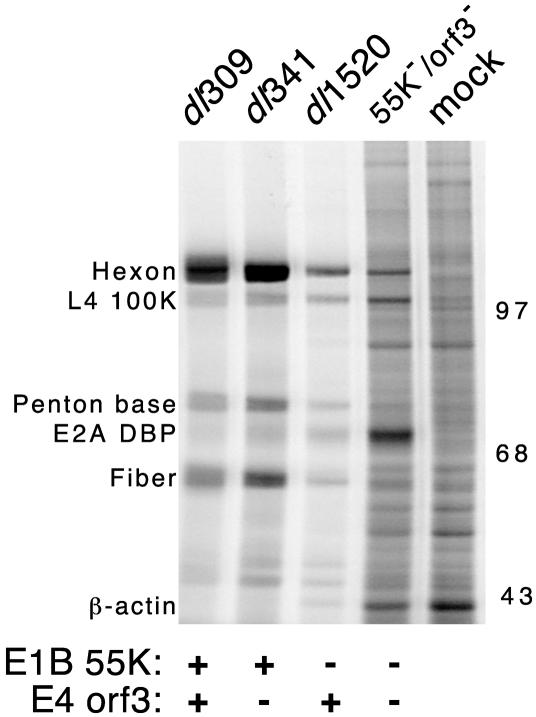
To determine if the apparent defect in mRNA synthesis or nuclear RNA stability observed with the E1B-55K/E4orf3 double mutant virus was reflected in a decrease in viral late protein levels, late viral gene expression was measured. HeLa cells were pulse labeled with 35S-labeled amino acids 36 h after infection with each of the viruses indicated in Fig. 6, and the newly synthesized proteins were visualized after separation by SDS-PAGE. Cells infected with the wild-type virus and the E4orf3 mutant virus synthesized viral late proteins at the greatest rate. As expected, cells infected with the E1B-55K mutant virus synthesized viral late proteins at a lower rate than wild-type virus-infected cells.
FIG. 6.
E1B-55K/E4orf3 double mutant virus-infected cells express reduced levels of late viral proteins and increased levels of cellular proteins. HeLa cells were mock infected or infected with the indicated viruses at a multiplicity of 20 PFU per cell. The status of the relevant viral genes is indicated below the gel. At 36 h postinfection, cells were labeled with 35S-labeled amino acids for 1 h. Proteins from 105 cells (per lane) were separated by SDS-PAGE. Radioactive proteins were visualized by phosphorescence imaging. The approximate migration and mass (in kilodaltons) of molecular size standards are indicated to the right of the gel. The positions of prominent viral and cellular proteins were determined with adenovirus virion standards labeled with [14C]amino acids or by immunoprecipitation and are indicated to the left of the image. The gel shown is representative of three independent experiments.
Two striking observations can be made about protein synthesis in cells infected with the E1B-55K/E4orf3 double mutant virus at late times of infection. First, consistent with the severely reduced levels of late viral mRNA, little viral late protein synthesis was apparent in the double mutant virus-infected cells. When newly synthesized hexon was recovered by immunoprecipitation from infected cells that were pulse labeled at 36 or 48 h postinfection, hexon synthesis was reduced approximately 5- to 10-fold in E1B-55K/E4orf3 double mutant virus-infected cells compared to E1B-55K single mutant virus-infected cells (data not shown). Second, the double mutant virus-infected cells continued to synthesize host proteins at nearly the same rate as mock-infected cells. This is evident in the greater overall density throughout the lane as well as the increased number of well-resolved bands that are also seen in the adjacent lane from mock-infected cells.
These data suggest that the E1B-55K/E4orf3 double mutant virus is profoundly defective in its ability to shut off host gene expression. An additional point of interest is the overexpression of the E2A 72-kDa DBP in the E1B-55K mutant and E1B-55K/E4orf3 double mutant virus-infected cells. Other investigators have noted that this E2A gene is overexpressed in abortive infections or during an infection that fails to proceed through the late phase (17). We previously noted that the rate of DBP synthesis was greater in S-phase cells infected with the cell cycle-restricted E1B-55K mutant or E4orf6 mutant virus than in G1-phase cells infected with same viruses. Because the S-phase-infected cells produce more virus than G1-phase-infected cells, elevated levels of DBP are not strictly correlated with a nonproductive infection (22). Nonetheless, the basis or consequences of dysregulated expression of the E2A gene is not well understood. Furthermore, in the double mutant virus-infected cells, synthesis of the L4-100K protein was not as severely diminished as that of the hexon protein. This is evident by comparing the relative labeling intensity of these two proteins in Fig. 6. The identity of these proteins was established by immunoprecipitation with a monoclonal antibody specific for the L4-100K protein (12) and rabbit antiserum raised against adenovirus type 5 virions (data not shown).
From these collective findings, we conclude that in the context of an E1B-55K mutant background, an important role for the E4orf3 protein is to promote efficient synthesis of late viral transcripts while concomitantly, or possibly consequently, inhibiting host translation. Additionally, these data do not exclude the possibility that the E4orf3 protein contributes to host cell shutoff by inhibiting nuclear export of host cellular messages.
POD morphology and number are unaffected by infection with the E1B-55K/E4orf3 double mutant virus.
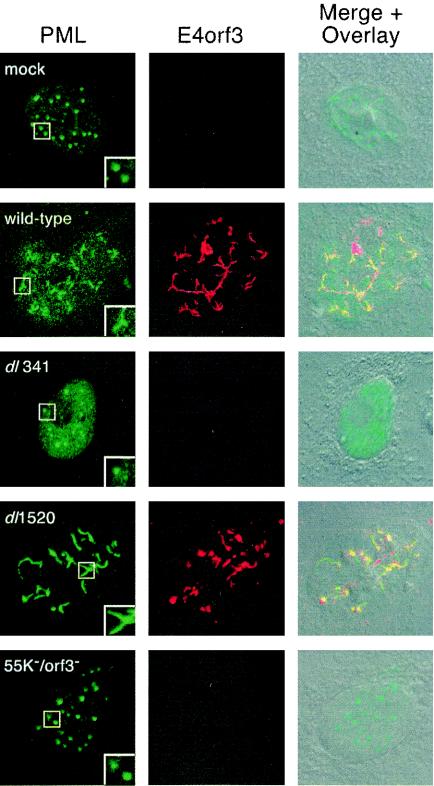
Expression of the E4orf3 protein alters the morphology of subnuclear structures known as promyelocytic leukemia (PML) oncogenic domains (PODs). These nuclear domains are discrete interchromosomal accumulations of several proteins, of which the PML protein is one of the most widely studied. During an adenovirus infection, a characteristic effect of the E4orf3 protein is to change the discrete punctate appearance of the PODs to an elongated or track-like appearance (Fig. 7, insets of wild-type- and dl1520-infected cells stained for PML). It has also been reported that the E1B-55K protein transiently associates with POD-associated proteins PML (35) and Daxx (71).
FIG. 7.
E1B-55K/E4orf3 double mutant virus fails to reorganize PML protein. A549 cells were mock infected, infected with the wild-type virus dl309, the E4orf3 mutant virus dl341, the E1B-55K mutant virus dl1520, or the E1B-55K/E4orf3 double mutant virus 55K−/orf3− at a multiplicity of 10 PFU per cell. At 48 h postinfection, double-label immunofluorescence was used to visualize PML protein (green) with the mouse monoclonal antibody 5E10 and the E4orf3 protein (red) with rat monoclonal antibody 6A11. The merged red and green images are overlaid on a differential interference contrast image of the cell. The micrographs were acquired as a single optical slice with confocal laser scanning microscopy. The inset in each PML image is a twofold enlargement illustrating the characteristic morphology of the indicated PML-containing nuclear bodies.
To determine if there were gross changes in POD morphology in the absence of two viral proteins that target POD constituents, the localization of the PML protein was evaluated in mock-infected A549 cells and A549 cells infected with the relevant viruses. At 48 h after infection, the localization of both the PML protein and the E4orf3 protein was determined by double-label immunofluorescence, as shown in Fig. 7. In the mock-infected cell nucleus, the PML protein was concentrated in punctate, well-organized foci. In the wild-type and E1B-55K mutant virus-infected cells, however, the PML protein had reorganized into more diffuse, track-like structures. The PML and E4orf3 proteins frequently colocalized in these track-like structures, as shown by the yellow structures in the merged images for the wild-type and E1B-55K mutant virus-infected cells. Additionally, although the E4orf3 protein was predominantly nuclear, we noted that a portion of the E4orf3 protein appeared in the cytoplasm of some infected A549 cells in a perinuclear fashion (Fig. 7 and data not shown).
In contrast to the localization observed in wild-type virus-infected and E1B-55K mutant virus-infected cells, the PML protein remained in the characteristic punctate structures observed in mock-infected cells or after infection with the E4orf3 single mutant virus or the E1B-55K/E4orf3 double mutant virus. It is possible that there were fewer punctate PML nuclear bodies in the E4orf3 mutant virus-infected cells than in the double mutant virus-infected cells, although this difference was not precisely determined. Nonetheless, these results demonstrate that the E1B-55K/E4orf3 double mutant virus is indeed defective in its ability to reorganize POD structure.
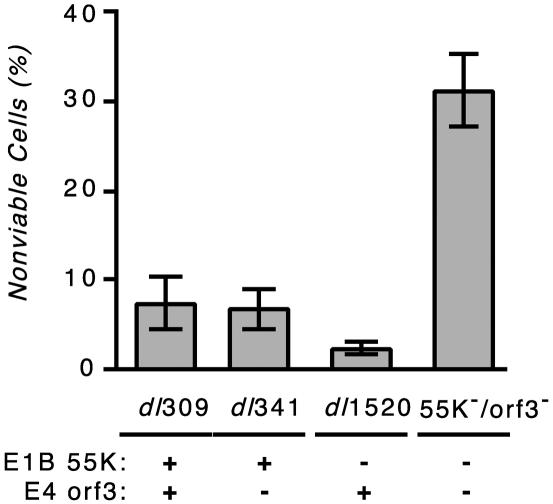
Cells infected with the E1B-55K/E4orf3 double mutant virus die more rapidly than cells infected with the wild-type or single mutant viruses.
We noted that at 3 days after infection, HeLa cells infected with the double mutant virus displayed a more heterogeneous appearance than cells infected with the other viruses. Therefore, HeLa cells were infected with the viruses indicated in Fig. 8 at a multiplicity of 10 PFU per cell, and cell viability was measured at 3 days postinfection by trypan blue dye exclusion. No cell death was observed in mock-infected cells (data not shown), and very little cell death was measured in wild-type virus-infected or single mutant virus-infected cultures (Fig. 8). Strikingly, at the same time after infection, the double mutant virus-infected cell population reproducibly contained a significant number (>30%) of nonviable cells. This outcome was unexpected because the double mutant virus replicated so poorly in HeLa cells (Fig. 1). This result, which demonstrates that virus-mediated cell killing and virus replication can be separated, suggests a role for the E4orf3 protein in preserving the integrity of the infected cell in an E1B-55K mutant virus infection. Studies are under way to determine why the double mutant virus-infected cells were dying.
FIG. 8.
E1B-55K/E4orf3 double mutant virus kills cells more rapidly than the wild-type or parental mutant virus. HeLa cells were infected with the indicated viruses at a multiplicity of 10 PFU per cell. The status of the relevant viral genes is indicated below the histogram. At 72 h postinfection, the fraction of nonviable cells was measured by trypan blue dye uptake. The results shown are the averages of three to five independent infections performed in three independent experiments. Fewer than 1% of the mock-infected cells were trypan blue dye positive at 72 h postinfection. Error bars indicate the standard error of the mean.
DISCUSSION
Mutant adenoviruses that lack only the E4orf3 protein have been shown to replicate to wild-type levels. For this reason, the contribution of the E4orf3 protein to a wild-type adenovirus infection has been difficult to elucidate. This study demonstrates important and diverse roles for the E4orf3 protein that are apparent in the absence of E1B-55K function and thus critical in a cell cycle-restricted adenovirus infection. In accordance with the findings of other investigators, we show that E4orf3 prevents the formation of viral DNA concatemers in the absence of E1B-55K (Fig. 3). Surprisingly, the accumulation of viral DNA concatemers, per se, is not the basis for the severely defective nature of the E1B-55K/E4orf3 double mutant virus (Fig. 4). Perhaps a more significant function for virus replication is the ability of the E4orf3 protein to promote the synthesis or nuclear stability of late viral RNA in an E1B-55K mutant background (Fig. 5). Finally, and again only within the context of an E1B-55K mutant virus, the E4orf3 protein acts to suppress host translation (Fig. 6).
The E1B-55K/E4orf3 double mutant virus replicates to lower levels than the wild-type or either parental mutant virus (Fig. 1). Two characteristics of the infected-cell population contribute to this reduced virus yield. First, the E1B-55K/E4orf3 double mutant virus replicated in approximately 4% of successfully infected HeLa cells (62). Second, the amount of progeny virus per productive cell, or burst size, was 20- to 40-fold less for the E1B-55K/E4orf3 double mutant virus than the wild-type virus. Thus, in an E1B-55K-deficient background, the E4orf3 protein promotes efficient viral replication by increasing both the burst size and the probability that an infected cell will produce virus. This property of the E4orf3 protein is cell cycle dependent, because an S-phase cell is approximately 2.5-fold more likely than a G1-phase cell to permit replication of a cell cycle-restricted virus (62).
Some properties of the E1B-55K/E4orf3 double mutant virus analyzed here are shared by dl1016, a virus with deletions in the E1B-55K and E4orf1, E4orf2, and E4orf3 genes (9). In their study of dl1016 and other viruses defective in the E1B and E4 genes, Bridge and Ketner concluded that the E4orf3 protein and a complex of the E1B-55K and E4orf6 proteins act in parallel to promote late viral mRNA biogenesis (9). Also studying dl1016, Weitzman and associates suggested that the E4orf3 protein and E1B-55K-E4orf6 protein complex act in a compensatory manner to prevent the repair of double-stranded DNA breaks and block the formation of viral DNA concatemers (64). Our findings agree with those of both groups of investigators. The elimination of E4orf3 function in an E1B-55K mutant virus background leads to a significant reduction in the levels of late viral RNA in both the nucleus and cytoplasm (Fig. 5). Additionally, we observed the production of viral DNA concatemers in cells infected with the E1B-55K/E4orf3 double mutant virus (Fig. 3B).
However, there are apparently some differences between the multiple mutant dl1016 and the E1B-55K/E4orf3 double mutant virus analyzed here. The double mutant virus studied in this work appears to be less defective at directing viral DNA synthesis (Fig. 3A) than dl1016, which was shown to accumulate 40-fold less viral DNA than wild-type virus-infected cells (9). Although both dl1016 and the E1B-55K/E4orf3 double mutant virus are severely defective at directing late viral protein synthesis measured 24 h after infection (9), the E1B-55K/E4orf3 double mutant virus is profoundly defective in its ability to shut off host translation (Fig. 6), even compared to the multiple mutant dl1016 (data not shown). This difference suggests that the E4orf1 or E4orf2 protein may affect protein synthesis during an E1B-55K mutant virus infection. The work of Javier and associates demonstrated that the E4orf1 protein interacts with cellular targets that serve to activate the phosphatidylinositol 3-kinase pathway, an important regulator of a variety of cellular activities, including translation (18). However, the significance of E4orf1 in the outcome of a productive lytic infection has yet to be described. Studies are under way to investigate the role of these proteins in the context of other viral mutations.
It seems likely that either inefficient expression of late viral genes or the concatenation of viral DNA would significantly impair virus replication. Because the DNA sequence recognized during encapsidation of the viral genome must be close to a free end and because the viral capsid can only accommodate a small excess of DNA beyond a monomer length (reviewed in reference 47), it is reasonable to expect that concatenation of the nascent viral DNA is sufficient to impair the production of virus. However, a surprising finding reported here is that the E1B-55K/E4orf3 double mutant virus did not replicate any better in MO59J cells than in MO59K cells despite the fact that viral DNA concatemers did not accumulate in MO59J cells but did in MO59K cells (Fig. 4B and C). Although it remains to be determined how the formation of viral DNA concatemers affects the replication of dl1016, Evans and Hearing have recently shown that the ability of E4orf3 to promote virus replication can be genetically uncoupled from its ability to prevent viral DNA concatenation (15). These investigators created several E4orf6 mutant viruses bearing amino acid substitutions in the E4orf3 protein that directed efficient viral DNA replication and virus replication but were unable to block the formation of viral DNA concatemers.
A prominent defect observed during replication of the E1B-55K/E4orf3 double mutant virus was the failure to accumulate late viral RNA in both the cytoplasm and nucleus (Fig. 5). Because the cytoplasmic-to-nuclear late mRNA ratio in the double mutant virus-infected cells was the same as in the E1B-55K single mutant virus-infected cells, the loss of E4orf3 function does not appear to exacerbate the defect in viral mRNA transport associated with the loss of E1B-55K. Rather, in agreement with the conclusions of Bridge and Ketner, the E4orf3 protein appears to be important for the synthesis or nuclear stability of late viral RNA when E1B-55K is absent. E4orf6 mutant viruses that are also defective in the E4orf3 gene display a similar defect (10, 29). For these reasons, the E4orf3 protein has been considered to provide a function that compensates for or overlaps that of the E1B-55K-E4orf6 protein complex (10, 15, 29). These results, together with the poor growth of the E1B-55K/E4orf3 double mutant virus even when DNA concatenation was prevented, lead us to suggest that the critical role of E4orf3 during the replication of a cell cycle-restricted virus is its ability to promote the synthesis or nuclear stability of late viral RNA.
The E4orf3 protein maintains diverse activities within the infected cell that alter viral RNA metabolism. Both the E4orf6 and E4orf3 proteins stimulate the accumulation of late mRNA by stimulating constitutive splicing (42), although E4orf6 promotes exon exclusion, while E4orf3 promotes exon inclusion (44). Perhaps in the absence of E1B-55K-E4orf6 function, deletion of the E4orf3 gene in a cell cycle-restricted virus leads to inefficiently spliced viral late messages, the outcome of which is rapid degradation in the nucleus.
It seems likely that RNA splicing occurs throughout most of the cell cycle. However, the activity of some splicing factors, such as members of the SR protein family, are regulated by cell cycle-dependent changes in phosphorylation (45). The E4orf3 protein may act in the S-phase-infected cell to enhance the activity of the factors needed for efficient viral RNA processing. This would explain the ability of the E4orf3 protein to enhance S-phase replication of both the E4orf6 mutant (22) and E1B-55K mutant (62) viruses. A likely target for the E4orf3 protein is the PODs. The proteins in these subnuclear structures undergo profound biochemical changes, such as sumoylation and phosphorylation, in a cell cycle-dependent manner (16). It is possible that by disrupting POD organization, the E4orf3 protein releases factors necessary for efficient late viral RNA processing. Again, however, this activity of E4orf3 is required only in the absence of E1B-55K-E4orf6 function, as the E4orf3 single mutant virus is phenotypically indistinguishable from the wild-type virus.
Another significant finding revealed by this work is the profound inability of the E1B-55K/E4orf3 double mutant virus to shut off host protein synthesis (Fig. 6). At late times of infection, host translation is effectively blocked by the wild-type virus in most cell types. The E1B-55K single mutant virus blocks host translation to a lesser extent, perhaps to a level consistent with the limited amount of late gene expression. It has been suggested that adenovirus primarily inhibits host translation through the action of the late viral protein L4-100K (13, 28). The L4-100K protein displaces the mitogen-activated protein kinase-activating kinase Mnk1 from the eIF-4F cap-binding complex (13), leading to reduced phosphorylation of eIF-4E and a decrease in cap-dependent translation (reviewed in reference 61). Although it seems reasonable that the E1B-55K/E4orf3 double mutant virus fails to block host translation because of the failure to promote late viral gene expression, the L4-100K protein appeared to be synthesized at rates approaching that measured in cells infected with the E1B-55K single mutant virus (Fig. 6). Nonetheless, it is conceivable that the L4-100K protein synthesized in the double mutant virus-infected cells was unable to repress translation and that differences between the L4-100K proteins synthesized in mutant and wild-type virus-infected cells were not revealed by SDS-PAGE.
We (21) and others (48, 51) have suggested that virus replication and cell killing are closely linked, such that efficient cell killing requires efficient virus replication. As the E1B-55K/E4orf3 double mutant virus replicated to levels nearly 1,000-fold less than that of the wild-type virus, it was quite surprising to see more cell death among cells infected with the double mutant virus than with the wild-type virus (Fig. 8). The failure of the E1B-55K/E4orf3 double mutant virus to block host translation may underlie the increased cell death. It has been suggested that many viruses block host gene expression to suppress the antiviral response (38). By failing to suppress host translation, the double mutant virus-infected cells may have become more susceptible to the host antiviral response, resulting in cell death. It is also possible that dysregulated expression of viral genes in the E1B-55K/E4orf3 double mutant virus infection, such as aberrant overexpression of the E2A DBP (Fig. 6), may be responsible for the increased cell death. Studies are currently under way to further characterize the death observed in the double mutant virus-infected cells as well as to determine the mechanism(s) of this death.
The E4 region expresses multiple activities that are redundant with the activities encoded by many other viral genes. Because the E4 promoter was found to be nearly as active as the E1A promoter in some cell types (14), it is conceivable that products of the E4 region could direct a productive infection in the absence of E1 function. The work of Marton and associates (39) and O'Connor and Hearing (43) suggests that the E4orf6/7 protein can functionally compensate for the activity of E1A. Ketner and associates suggested that the E4orf3 and E4orf6 proteins act primarily to enhance the efficiency of viral replication when other viral proteins are limiting (40). Examples of cryptic E4 function in an E1-deficient background can be derived from the study of E1-deficient adenovirus vectors used for gene therapy. For example, in cells infected with an E1-deficient virus, the E4orf3 protein sustained the expression of a nonviral transgene (2). The E4 region enabled an adenovirus vector to prolong the survival of primary human endothelial cells (55), and the E4 region of an E1-defective adenovirus vector enhanced leukocyte adhesion to and migration through infected endothelial cells (54). It will be of considerable interest to determine the pattern of viral gene expression in naturally infected human cells during limited virus replication. Perhaps a role for the highly conserved E4orf3 protein will be most evident under these circumstances.
Acknowledgments
This work was supported in part by Public Health Service grant CA 77342 and a supplement from the National Cancer Institute. Tissue culture reagents and services were provided by the Tissue Culture Core Laboratory, and confocal laser scanning microscopy was performed through the Micromed facility, both services of the Comprehensive Cancer Center of Wake Forest University, which is supported in part by a grant from the National Cancer Institute, CA 12197.
We gratefully acknowledge Pat Hearing (SUNY Stony Brook) for the E4orf3 mutant virus dl341, Arnie Berk (UCLA) for the E1B-55K mutant virus dl1520D, Thomas Dobner (Universitat Regensburg) for the E4orf3 rat monoclonal antibody 6A11, Luitzen De Jong (University of Amsterdam) for the PML mouse monoclonal antibody 5E10, and Ryan Topping (Wake Forest University) for creation of the E4orf3 expression plasmid pRT1. We thank Ken Grant of the Micromed facility for assistance with confocal laser scanning microscopy, Kuie Pao Tsai for technical assistance, and Doug Lyles and Griff Parks for providing valuable advice on the work in progress and on the manuscript.
REFERENCES
- 1.Allalunis-Turner, M. J., G. M. Barron, R. S. Day 3rd, K. D. Dobler, and R. Mirzayans. 1993. Isolation of two cell lines from a human malignant glioma specimen differing in sensitivity to radiation and chemotherapeutic drugs. Radiat. Res. 134:349-354. [PubMed] [Google Scholar]
- 2.Armentano, D., M. P. Smith, C. C. Sookdeo, J. Zabner, M. A. Perricone, J. A. St George, S. C. Wadsworth, and R. J. Gregory. 1999. E4ORF3 requirement for achieving long-term transgene expression from the cytomegalovirus promoter in adenovirus vectors. J. Virol. 73:7031-7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1993. Current protocols in molecular biology. Greene Publishing Associates, Inc.-John Wiley and Sons, Inc., New York, N.Y.
- 4.Babiss, L. E., and H. S. Ginsberg. 1984. Adenovirus type 5 early region 1b gene product is required for efficient shutoff of host protein synthesis. J. Virol. 50:202-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babiss, L. E., H. S. Ginsberg, and J. E. Darnell, Jr. 1985. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol. Cell. Biol. 5:2552-2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barker, D. D., and A. J. Berk. 1987. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology 156:107-121. [DOI] [PubMed] [Google Scholar]
- 7.Ben-Israel, H., and T. Kleinberger. 2002. Adenovirus and cell cycle control. Front. Biosci. 7:d1369-d1395. [DOI] [PubMed] [Google Scholar]
- 8.Boyer, J., K. Rohleder, and G. Ketner. 1999. Adenovirus E4 34k and E4 11k inhibit double strand break repair and are physically associated with the cellular DNA-dependent protein kinase. Virology 263:307-312. [DOI] [PubMed] [Google Scholar]
- 9.Bridge, E., and G. Ketner. 1990. Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174:345-353. [DOI] [PubMed] [Google Scholar]
- 10.Bridge, E., and G. Ketner. 1989. Redundant control of adenovirus late gene expression by early region 4. J. Virol. 63:631-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carney, J. P., R. S. Maser, H. Olivares, E. M. Davis, M. Le Beau, J. R. Yates, 3rd, L. Hays, W. F. Morgan, and J. H. Petrini. 1998. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93:477-486. [DOI] [PubMed] [Google Scholar]
- 12.Cepko, C. L., and P. A. Sharp. 1983. Aberrant distribution of human adenovirus type 2 late proteins in monkey kidney cells. J. Virol. 46:302-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuesta, R., Q. Xi, and R. J. Schneider. 2000. Adenovirus-specific translation by displacement of kinase Mnk1 from cap-initiation complex eIF4F. EMBO J. 19:3465-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dery, C. V., C. H. Herrmann, and M. B. Mathews. 1987. Response of individual adenovirus promoters to the products of the E1A gene. Oncogene 2:15-23. [PubMed] [Google Scholar]
- 15.Evans, J. D., and P. Hearing. 2003. Distinct roles of the Adenovirus E4 ORF3 protein in viral DNA replication and inhibition of genome concatenation. J. Virol. 77:5295-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett, R. D., P. Lomonte, T. Sternsdorf, R. van Driel, and A. Orr. 1999. Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci. 112:4581-4588. [DOI] [PubMed] [Google Scholar]
- 17.Fessler, S. P., and C. S. Young. 1998. Control of adenovirus early gene expression during the late phase of infection. J. Virol. 72:4049-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frese, K. K., S. S. Lee, D. L. Thomas, I. J. Latorre, R. S. Weiss, B. A. Glaunsinger, and R. T. Javier. 2003. Selective PDZ protein-dependent stimulation of phosphatidylinositol 3-kinase by the adenovirus E4-ORF1 oncoprotein. Oncogene 22:710-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gimona, M., J. Vandekerckhove, M. Goethals, M. Herzog, Z. Lando, and J. V. Small. 1994. Beta-actin specific monoclonal antibody. Cell. Motil. Cytoskeleton 27:108-116. [DOI] [PubMed] [Google Scholar]
- 20.Goodrum, F. D., and D. A. Ornelles. 1997. The early region 1B 55-kilodalton oncoprotein of adenovirus relieves growth restrictions imposed on viral replication by the cell cycle. J. Virol. 71:548-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goodrum, F. D., and D. A. Ornelles. 1998. p53 status does not determine outcome of E1B 55-kilodalton mutant adenovirus lytic infection. J. Virol. 72:9479-9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodrum, F. D., and D. A. Ornelles. 1999. Roles for the E4 orf6, orf3, and E1B 55-kilodalton proteins in cell cycle-independent adenovirus replication. J. Virol. 73:7474-7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gozes, I., and C. J. Barnstable. 1982. Monoclonal antibodies that recognize discrete forms of tubulin. Proc. Natl. Acad. Sci. USA 79:2579-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haber, J. E. 1998. The many interfaces of Mre11. Cell 95:583-586. [DOI] [PubMed] [Google Scholar]
- 25.Halbert, D. N., J. R. Cutt, and T. Shenk. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56:250-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harada, J. N., A. Shevchenko, D. C. Pallas, and A. J. Berk. 2002. Analysis of the adenovirus E1B-55K-anchored proteome reveals its link to ubiquitination machinery. J. Virol. 76:9194-9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harlow, E., B. R. Franza, Jr., and C. Schley. 1985. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J. Virol. 55:533-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayes, B. W., G. C. Telling, M. M. Myat, J. F. Williams, and S. J. Flint. 1990. The adenovirus L4 100-kilodalton protein is necessary for efficient translation of viral late mRNA species. J. Virol. 64:2732-2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang, M. M., and P. Hearing. 1989. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J. Virol. 63:2605-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jones, N., and T. Shenk. 1979. Isolation of adenovirus type 5 host range deletion mutants defective for transformation of rat embryo cells. Cell 17:683-689. [DOI] [PubMed] [Google Scholar]
- 31.Jones, N., and T. Shenk. 1978. Isolation of deletion and substitution mutants of adenovirus type 5. Cell 13:181-188. [DOI] [PubMed] [Google Scholar]
- 32.Kafatos, F. C., C. W. Jones, and A. Efstratiadis. 1979. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 7:1541-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Konig, C., J. Roth, and M. Dobbelstein. 1999. Adenovirus type 5 E4orf3 protein relieves p53 inhibition by E1B 55-kilodalton protein. J. Virol. 73:2253-2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lees-Miller, S. P., R. Godbout, D. W. Chan, M. Weinfeld, R. S. Day, 3rd, G. M. Barron, and J. Allalunis-Turner. 1995. Absence of p350 subunit of DNA-activated protein kinase from a radiosensitive human cell line. Science 267:1183-1185. [DOI] [PubMed] [Google Scholar]
- 35.Leppard, K. N., and R. D. Everett. 1999. The adenovirus type 5 E1b 55K and E4 Orf3 proteins associate in infected cells and affect ND10 components. J. Gen. Virol. 80:997-1008. [DOI] [PubMed] [Google Scholar]
- 36.Leppard, K. N., and T. Shenk. 1989. The adenovirus E1B 55 kd protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 8:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Linke, S. P. 1998. Cancer. Has the smart bomb been defused? Nature (London) 395:13, 15. [DOI] [PubMed] [Google Scholar]
- 38.Lyles, D. S. 2000. Cytopathogenesis and inhibition of host gene expression by RNA viruses. Microbiol. Mol. Biol. Rev. 64:709-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marton, M. J., S. B. Baim, D. A. Ornelles, and T. Shenk. 1990. The adenovirus E4 17-kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA-binding properties and stimulating E1A-independent accumulation of E2 mRNA. J. Virol. 64:2345-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medghalchi, S., R. Padmanabhan, and G. Ketner. 1997. Early region 4 modulates adenovirus DNA replication by two genetically separable mechanisms. Virology 236:8-17. [DOI] [PubMed] [Google Scholar]
- 41.Nevels, M., B. Tauber, E. Kremmer, T. Spruss, H. Wolf, and T. Dobner. 1999. Transforming potential of the adenovirus type 5 E4orf3 protein. J. Virol. 73:1591-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nordqvist, K., K. Ohman, and G. Akusjarvi. 1994. Hum. adenovirus encodes two proteins which have opposite effects on accumulation of alternatively spliced mRNAs. Mol. Cell. Biol. 14:437-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O'Connor, R. J., and P. Hearing. 2000. The E4-6/7 protein functionally compensates for the loss of E1A expression in adenovirus infection. J. Virol. 74:5819-5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohman, K., K. Nordqvist, and G. Akusjarvi. 1993. Two adenovirus proteins with redundant activities in virus growth facilitates tripartite leader mRNA accumulation. Virology 194:50-58. [DOI] [PubMed] [Google Scholar]
- 45.Okamoto, Y., H. Onogi, R. Honda, H. Yasuda, T. Wakabayashi, Y. Nimura, and M. Hagiwara. 1998. cdc2 kinase-mediated phosphorylation of splicing factor SF2/ASF. Biochem. Biophys. Res. Commun. 249:872-878. [DOI] [PubMed] [Google Scholar]
- 46.Ornelles, D. A., and T. Shenk. 1991. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 65:424-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ostapchuk, P., and P. Hearing. 2003. Regulation of adenovirus packaging. Curr. Top. Microbiol. Immunol. 272:165-185. [DOI] [PubMed] [Google Scholar]
- 48.Petit, T., K. K. Davidson, C. Cerna, R. A. Lawrence, D. D. Von Hoff, C. Heise, D. Kirn, and E. Izbicka. 2002. Efficient induction of apoptosis by ONYX-015 adenovirus in human colon cancer cell lines regardless of p53 status. Anticancer Drugs 13:47-50. [PubMed] [Google Scholar]
- 49.Petrini, J. H. 1999. The mammalian Mre11-Rad50-nbs1 protein complex: integration of functions in the cellular DNA-damage response. Am. J. Hum. Genet. 64:1264-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pilder, S., M. Moore, J. Logan, and T. Shenk. 1986. The adenovirus E1B 55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6:470-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Portella, G., S. Scala, D. Vitagliano, G. Vecchio, and A. Fusco. 2002. ONYX-015, an E1B gene-defective adenovirus, induces cell death in human anaplastic thyroid carcinoma cell lines. J. Clin. Endocrinol. Metab. 87:2525-2531. [DOI] [PubMed] [Google Scholar]
- 52.Querido, E., P. Blanchette, Q. Yan, T. Kamura, M. Morrison, D. Boivin, W. G. Kaelin, R. C. Conaway, J. W. Conaway, and P. E. Branton. 2001. Degradation of p53 by adenovirus E4orf6 and E1B55K proteins occurs via a novel mechanism involving a Cullin-containing complex. Genes Dev. 15:3104-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Querido, E., R. C. Marcellus, A. Lai, R. Charbonneau, J. G. Teodoro, G. Ketner, and P. E. Branton. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 71:3788-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rafii, S., S. Dias, S. Meeus, K. Hattori, R. Ramachandran, F. Feuerback, S. Worgall, N. R. Hackett, and R. G. Crystal. 2001. Infection of endothelium with E1(−)E4(+), but not E1(−)E4(−), adenovirus gene transfer vectors enhances leukocyte adhesion and migration by modulation of ICAM-1, VCAM-1, CD34, and chemokine expression. Circ. Res. 88:903-910. [DOI] [PubMed] [Google Scholar]
- 55.Ramalingam, R., S. Rafii, S. Worgall, D. E. Brough, and R. G. Crystal. 1999. E1(−)E4(+) adenoviral gene transfer vectors function as a “pro-life” signal to promote survival of primary human endothelial cells. Blood 93:2936-2944. [PubMed] [Google Scholar]
- 56.Reich, N. C., P. Sarnow, E. Duprey, and A. J. Levine. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128:480-484. [DOI] [PubMed] [Google Scholar]
- 57.Robinson, A. J., H. B. Younghusband, and A. J. Bellett. 1973. A circular DNA-protein complex from adenoviruses. Virology 56:54-69. [DOI] [PubMed] [Google Scholar]
- 58.Ruben, M., S. Bacchetti, and F. Graham. 1983. Covalently closed circles of adenovirus 5 DNA. Nature (London) 301:172-174. [DOI] [PubMed] [Google Scholar]
- 59.Sarnow, P., P. Hearing, C. W. Anderson, N. Reich, and A. J. Levine. 1982. Identification and characterization of an immunologically conserved adenovirus early region 11,000 Mr protein and its association with the nuclear matrix. J. Mol. Biol. 162:565-583. [DOI] [PubMed] [Google Scholar]
- 60.Sarnow, P., C. A. Sullivan, and A. J. Levine. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120:510-517. [DOI] [PubMed] [Google Scholar]
- 61.Schneider, R. J., and I. Mohr. 2003. Translation initiation and viral tricks. Trends Biochem. Sci. 28:130-136. [DOI] [PubMed] [Google Scholar]
- 62.Shepard, R. N., and D. A. Ornelles. 2003. E4orf3 is necessary for enhanced S-phase replication of cell cycle-restricted subgroup C adenoviruses. J. Virol. 77:8593-8595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Steegenga, W. T., N. Riteco, A. G. Jochemsen, F. J. Fallaux, and J. L. Bos. 1998. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene. 16:349-357. [DOI] [PubMed] [Google Scholar]
- 64.Stracker, T. H., C. T. Carson, and M. D. Weitzman. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature (London) 418:348-352. [DOI] [PubMed] [Google Scholar]
- 65.Stuurman, N., A. de Graaf, A. Floore, A. Josso, B. Humbel, L. de Jong, and R. van Driel. 1992. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 101:773-784. [DOI] [PubMed] [Google Scholar]
- 66.Tauber, B., and T. Dobner. 2001. Adenovirus early E4 genes in viral oncogenesis. Oncogene 20:7847-7854. [DOI] [PubMed] [Google Scholar]
- 67.Van der Ploeg, L. H., D. C. Schwartz, C. R. Cantor, and P. Borst. 1984. Antigenic variation in Trypanosoma brucei analyzed by electrophoretic separation of chromosome-sized DNA molecules. Cell 37:77-84. [DOI] [PubMed] [Google Scholar]
- 68.Weiden, M. D., and H. S. Ginsberg. 1994. Deletion of the E4 region of the genome produces adenovirus DNA concatemers. Proc. Natl. Acad. Sci. USA 91:153-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wienzek, S., and M. Dobbelstein. 2001. Viral and cellular factors that target the promyelocytic leukemia oncogenic domains strongly activate a glucocorticoid-responsive promoter. J. Virol. 75:5391-5397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zantema, A., J. A. Fransen, A. Davis-Olivier, F. C. Ramaekers, G. P. Vooijs, B. DeLeys, and A. J. Van der Eb. 1985. Localization of the E1B proteins of adenovirus 5 in transformed cells, as revealed by interaction with monoclonal antibodies. Virology 142:44-58. [DOI] [PubMed] [Google Scholar]
- 71.Zhao, L. Y., A. L. Colosimo, Y. Liu, Y. Wan, and D. Liao. 2003. Adenovirus E1B 55-kilodalton oncoprotein binds to Daxx and eliminates enhancement of p53-dependent transcription by Daxx. J. Virol. 77:11809-11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu, X. D., B. Kuster, M. Mann, J. H. Petrini, and T. Lange. 2000. Cell-cycle-regulated association of RAD50/MRE11/NBS1 with TRF2 and human telomeres. Nat. Genet. 25:347-352. [DOI] [PubMed] [Google Scholar]