A growing body of evidence shows that so-called long noncoding RNAs (lncRNAs) often produce short peptides from small open reading frames (smORFs) (1). Whether and how smORF-encoded peptides fulfill specific functions remain poorly understood. Recent studies in flies (2) and mammals (3) have revealed that transcripts annotated as lncRNAs encode smORF peptides that bind to, and inhibit, the sarco/endoplasmic reticulum calcium adenosine triphosphatase (SERCA), an ion pump that is a key player in handling calcium in striated muscles. On page 271 of this issue, Nelson et al. (4) report that a lncRNA-encoded small peptide competes with SERCA-inhibitory peptides, thereby favoring heart contractility in mammals. These findings open new ways to understand cardiac function and pathologies, and show that smORF peptides act as versatile regulators of protein activity.
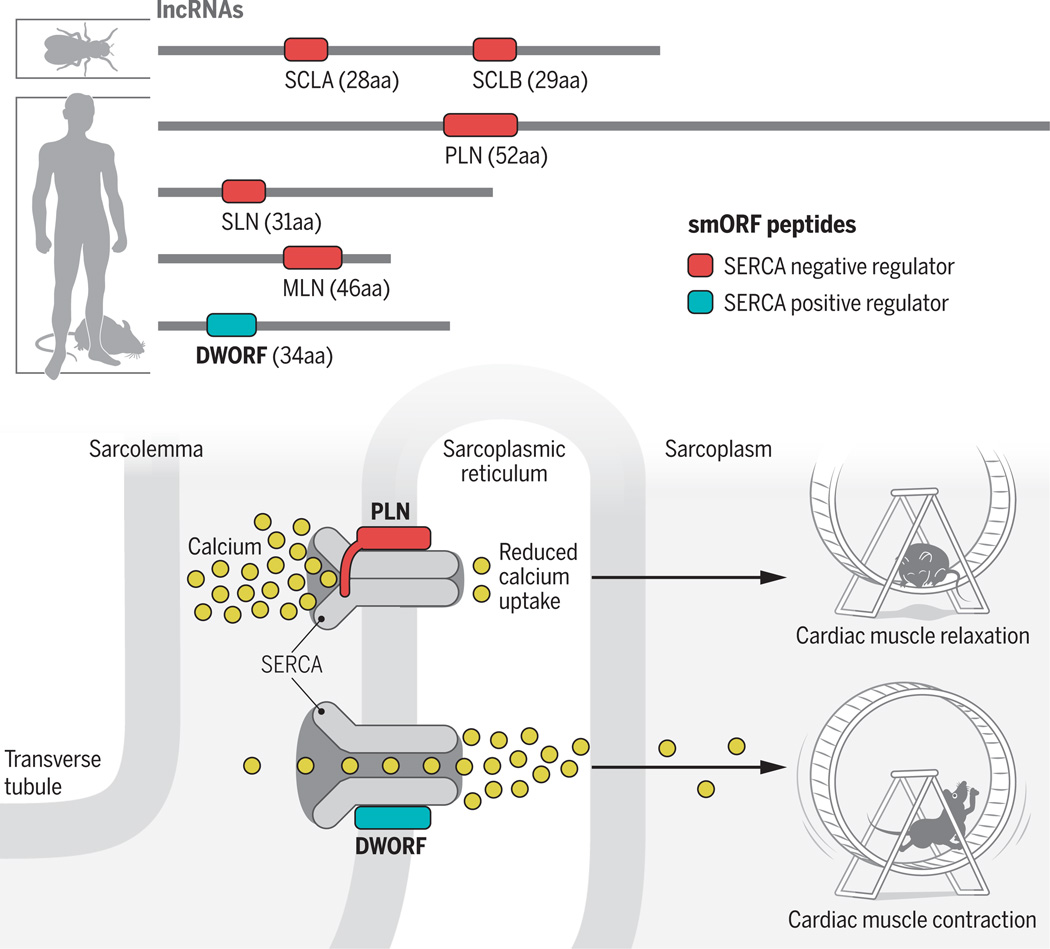
Muscle contraction relies on bursts of calcium released from the sarcoplasmic reticulum into the cytosol. The released calcium activates movement of the molecular motor myosin along actin filaments, enabling a “power stroke” mechanism. The SERCA pump moves calcium from the cytosol back into the sarcoplasmic reticulum, allowing actomyosin relaxation. Cycles of contraction and relaxation thus underlie muscular function. In the early 1970s, biochemical approaches in mammalian muscle cells revealed the inhibitory activity of a 52–amino acid peptide called phospholamban (PLN; also called PLB) (5). PLN is a transmembrane α-helix that fits a groove in the SERCA structure, favoring a conformational state that reduces calcium uptake (6, 7). A second, unrelated peptide of 31 amino acids, sarcolipin (SLN), was later identified in mammals (8) and shown to similarly bind to and inhibit the SERCA (6, 9). Both PLN and SLN are translated from distinct smORFs in transcripts that would be predicted as lncRNAs by current genomic annotation methods.
Whereas the SERCA protein from the fruit fly Drosophila melanogaster bears >70% sequence identity with the human protein, no annotated genes in Drosophila encode SLN- or PLN-like peptides. However, profiling lncRNAs allowed the identification of a muscle-specific transcript (pncr003:2L) in Drosophila that produces two related smORF peptides (2). Computer simulations suggest that these peptides, called sarcolamban A and B (SCLA and SCLB), fit the groove of the SERCA and, consistently, they bind to Drosophila and human SERCA. Furthermore, mutant fruit flies lacking SCL peptides display severe defects in muscle function, including cardiac arrhythmias (2). These results suggested that additional SERCA regulatory peptides could be hidden in the large number of apparently noncoding RNAs in mammals. Indeed, recent work has identified a 46–amino acid smORF peptide, myoregulin (MLN), produced from a transcript specifically expressed in skeletal muscles in mice and humans (3). Again, a similar conformation allows MLN to bind the SERCA and to decrease calcium uptake. Genetic experiments demonstrated the importance of MLN in vivo: Reducing the expression of MLN leads to improved performance in treadmill tests when compared to control mice (3). Therefore, lncRNAs produce a variety of smORF peptides that inhibit SERCA activity, providing tissue-specific control of striated muscle physiology.
Nelson et al. now provide evidence that a smORF peptide positively regulates SERCA activity in mammals. The authors identified an alleged lncRNA, predominantly expressed in the heart muscle of mice and humans, and demonstrated that it produces a 34–amino acid smORF peptide, called DWORF. Genome editing [with clustered regularly interspaced short palindromic repeats (CRISPR)–Cas9] leading to frameshifts in the DWORF peptide sequence resulted in reduced SERCA activity, delayed calcium clearance from the cytoplasm, and slowed relaxation of muscle fibers. Conversely, overexpression of DWORF in the heart increased calcium load in the sarcoplasmic reticulum and accelerated cytoplasmic calcium decay, thereby enhancing contractility. Although DWORF peptides bind to the SERCA protein, they do not intrinsically activate the SERCA. Instead, DWORF displaces the binding of MLN, PLN, or SLN peptides, relieving their inhibitory activity. The DWORF peptide thus appears as an antagonist of negative regulators, competing for binding to the SERCA.
Both the rationale of the antagonistic activities of these peptides and their intimate mechanisms of action remain to be elucidated. Whereas releasing inhibition may favor fast control and fine-tuning of protein activity, the dynamics of exchanges between regulatory peptides and the SERCA—which are both embedded in the sarcoplasmic reticulum membrane—are not known. In addition, PLN phosphorylation is sufficient to prevent its inhibitory activity, providing fast and signal-dependent control (10). Moreover, how DWORF prevents the binding of inhibitory peptides needs to be clarified. Although the DWORF peptide lacks sequence similarity with SLN, PLN, or MLN, it may—like SCL—dock into the same groove in the SERCA. In that case, how could a similar binding event inhibit (SLN, PLN, and MLN) SERCA activity, or not (DWORF)? Perhaps DWORF binds to a different domain of the SERCA, thus masking the regulatory groove. Structural studies, ideally including time-resolved methods, should further our understanding of muscle contraction and might help in the development of smORF-peptide–inspired therapeutics to prevent heart failure.
From a broader perspective, the findings of Nelson et al. highlight the importance of smORF peptides in the control of animal development and physiology, expanding the functional repertoire of protein-coding regions of our genome. Although still in its infancy, the emerging field of smORF peptides suggests that they represent an overlooked reservoir of versatile protein-binding interfaces, well suited to regulate the activity of various protein complexes (1–4, 11). Future work should explore the full range of smORF peptide activities and their role in the function and evolution of living organisms.
Figure. Competing peptides.
Large RNAs annotated as noncoding produce a family of smORF peptides that regulate activity of the SERCA calcium pump, from flies to mice and humans. The DWORF peptide prevents the binding of SLN, PLN, and MLN inhibitory peptides, resulting in increased calcium uptake into the sarcoplasmic reticulum, which improves muscle contractility.
REFERENCES
- 1.Saghatelian A, Couso JP. Nat. Chem. Biol. 2015;11:909. doi: 10.1038/nchembio.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Magny EG, et al. Science. 2013;341:1116. doi: 10.1126/science.1238802. [DOI] [PubMed] [Google Scholar]
- 3.Anderson DM, et al. Cell. 2015;160:595. doi: 10.1016/j.cell.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson BR, et al. Science. 2016;251:271. [Google Scholar]
- 5.Tada M, Kirchberger MA, Katz AM. J. Biol. Chem. 1975;250:2640. [PubMed] [Google Scholar]
- 6.Winther AM, et al. Nature. 2013;495:265. doi: 10.1038/nature11900. [DOI] [PubMed] [Google Scholar]
- 7.Akin BL, Hurley TD, Chen Z, Jones LR. J. Biol. Chem. 2013;288:30181. doi: 10.1074/jbc.M113.501585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Odermatt A, et al. Genomics. 1997;45:541. doi: 10.1006/geno.1997.4967. [DOI] [PubMed] [Google Scholar]
- 9.Toyoshima C, et al. Nature. 2013;495:260. doi: 10.1038/nature11899. [DOI] [PubMed] [Google Scholar]
- 10.MacLennan DH, Kranias EG. Nat. Rev. Mol. Cell Biol. 2003;4:566. doi: 10.1038/nrm1151. [DOI] [PubMed] [Google Scholar]
- 11.Zanet J, et al. Science. 2015;349:1356. doi: 10.1126/science.aac5677. [DOI] [PubMed] [Google Scholar]