Abstract
CEACAM1a glycoproteins are members of the immunoglobulin (Ig) superfamily and the carcinoembryonic antigen family. Isoforms expressing either two or four alternatively spliced Ig-like domains in mice have been found in a number of epithelial, endothelial, or hematopoietic tissues. CEACAM1a functions as an intercellular adhesion molecule, an angiogenic factor, and a tumor cell growth inhibitor. Moreover, the mouse and human CEACAM1a proteins are targets of viral or bacterial pathogens, respectively, including the murine coronavirus mouse hepatitis virus (MHV), Haemophilus influenzae, Neisseria gonorrhoeae, and Neisseria meningitidis, as well as Moraxella catarrhalis in humans. We have shown that targeted disruption of the Ceacam1a (MHVR) gene resulting in a partial ablation of the protein in mice (p/p mice) led to reduced susceptibility to MHV-A59 infection of the modified mice in the BALB/c background. We have now engineered and produced a Ceacam1a−/− mouse that exhibits complete ablation of the CEACAM1a protein in every tissue where it is normally expressed. We report that 3-week-old Ceacam1a−/− mice in the C57BL/6 genetic background are fully resistant to MHV-A59 infection by both intranasal and intracerebral routes. Whereas virus-inoculated wild-type +/+ C57BL/6 mice showed profound liver damage and spinal cord demyelination under these conditions, Ceacam1a−/− mice displayed normal livers and spinal cords. Virus was recovered from liver and spinal cord tissues of +/+ mice but not of −/− mice. These results indicate that CEACAM1a is the sole receptor for MHV-A59 in both liver and brain and that its deletion from the mouse renders the mouse completely resistant to infection by this virus.
The carcinoembryonic antigen (CEA) family comprises a large number of immunoglobulin (Ig)-like genes clustered on human chromosome 19q13.2 (40). The Ceacam1a gene is thought to represent the ancestor of this family, as it is well conserved in its gene structure, alternatively spliced isoforms, its expression patterns, and its functions in rodents and humans (15). The nomenclature of this large family has recently been redefined and standardized (3). The mouse genome contains two Ceacam-like genes, Ceacam1 and Ceacam2 (formerly known as Bgp1 and Bgp2) (28, 29). Although the gene structures are highly homologous, the proteins they encode exhibit a very different pattern of expression, with CEACAM1a protein most abundant in liver, intestine, kidney, and hematopoietic cells (4), whereas CEACAM2 is present in kidney and pancreas (16, 34). The mouse CEACAM1a isoforms contain either two or four extracellular alternatively spliced Ig-like domains that are tethered to the cell membrane via their transmembrane domain and display two different intracytoplasmic domains designated short (S; 10 amino acids [aa]) or long (L; 73 aa). The long cytoplasmic domain is produced by insertion of the 53-bp exon 7 of the Ceacam1a gene that switches the open reading frame of the encoded transcript. This results in an extended cytoplasmic tail by translation of 63 aa, containing inhibitory tyrosine-based immunoreceptor motifs (ITIM) that are well conserved across species (24, 33).
CEACAM1a expression in tissues of mice and humans is widespread, although not ubiquitous. CEACAM1a proteins are abundantly expressed at the surface of epithelial cells of the gastrointestinal and respiratory tracts, particularly in the bile canaliculi, the intestine, the proximal tubules of the kidney, and the lung (12, 25, 30, 31). These glycoproteins are also found on small vascular endothelial cells and in hematopoietic cells (such as activated T cells, B lymphocytes, neutrophils, macrophages, monocytes, platelets, and thymic stromal cells) (6, 10, 12, 14, 26, 27). CEACAM1a isoforms are also displayed on epithelial cells of reproductive tissues such as the uterus, the breast, and the prostate (18, 19, 38). Moreover, the proteins are expressed at low levels in the glial cells of the nervous system (12, 35). In these tissues, CEACAM1a exhibits a number of functions. Briefly, CEACAM1a acts as a cell adhesion molecule, an angiogenic factor, a tumor suppressor, and a signal regulatory protein (3).
Furthermore, human CEACAM1a is a receptor for a number of bacterial pathogens, such as Escherichia coli, Salmonella enterica serovar Typhimurium, Neisseria gonorrhoeae, Neisseria meningitidis, and Haemophilus influenzae, as well as Moraxella catarrhalis (13, 17, 23, 42, 43). Likewise, murine CEACAM1a proteins serve as receptors for a viral pathogen, murine coronavirus mouse hepatitis virus (MHV) (8). Two Ceacam1 alleles in various mouse strains have been identified. The Ceacam1a allele confers susceptibility to MHV infection and is present in most inbred mouse strains, as well as in outbred mice (8). Adult SJL inbred mice carry the Ceacam1b allele and are fully resistant to MHV infection (2, 21, 36). These two alleles differ in 27 of the 108 aa of the first Ig domain of CEACAM1 (7). Mutational analyses showed that the virus binds between aa 34 to 52 in the N-terminal domain of murine CEACAM1a (9, 32, 44). The crystal structure of soluble murine CEACAM1a domain 1 linked to domain 4 showed that the C-C′ loop (aa 35 to 44) has a unique stable convoluted conformation with a projecting Ile41 that is postulated to be important for binding to the viral spike glycoprotein (39). Binding of the spike glycoprotein to CEACAM1a at 37°C induces conformational changes in the spike protein that probably lead to fusion of the viral envelope with the host cell membrane and virus infection (41, 45).
We recently developed a mouse strain with a partial ablation of the CEACAM1a proteins (labeled p/p mice). In this BALB/c mouse model, the isoforms with four Ig domains were reduced in expression by 90 to 95%, whereas expression of those with two Ig domains was slightly increased. Upon intranasal (i.n.) inoculation of these mice with MHV-A59, the p/p mice did not show obvious clinical signs of infection. They had significantly fewer and smaller liver lesions than their wild-type littermate controls. Therefore, these genetically altered p/p mice exhibited decreased susceptibility to MHV-A59 infection (5).
We have now generated two mouse strains (2D2 and 11H11) on a C57BL/6 background that have a complete ablation of the CEACAM1a proteins (the strains are designated −/−). Ceacam1a−/− mice were inoculated with MHV-A59 by i.n. and intracerebral (i.c.) routes, and histopathology, titers of infectious virus, and expression of viral RNA in liver and brain were compared with those of wild-type +/+ mice. In addition, we have assessed the effects of i.c. inoculation of p/p BALB/c mice with MHV-A59. These experiments showed that the virus caused demyelination in the spinal cords of p/p mice without clinical signs but that the demyelinated lesions were smaller than those in wild-type +/+ mice. In contrast, the Ceacam1a−/− mice were fully resistant to MHV infection and had no virus replication or lesions in liver, brain, or spinal cord. These data show that CEACAM1a is the sole receptor for MHV-A59 in C57BL/6 mice.
MATERIALS AND METHODS
Generation of the Ceacam1a−/− mouse lines.
Details of the procedures involved in the generation of the Ceacam1a−/− mouse lines and their primary analyses and phenotypes will be presented in detail in another manuscript (T Dai et al., unpublished data). Briefly, a targeting vector was prepared in which the first two exons of the Ceacam1a gene were replaced by a pTK-neor cassette, thereby eliminating the exon coding for the initiator ATG codon as well as the second exon containing the viral binding domain (5). This vector was electroporated into R1 embryonic stem (ES) cells that were subjected to G418 analog selection. Two ES cell lines (designated 2D2 and 11H11) positive for the integration event were verified for genomic locus integrity and microinjected into C57BL/6 mouse blastocysts. Chimeric males were mated with C57BL/6 female mice, and heterozygous and homozygous Ceacam1a mice were derived (the mouse lines were designated 2D2 and 11H11). Experiments were performed in the second and third backcrosses of both the 2D2 and 11H11 Ceacam1a−/− mice. Care of the mice was according to the standards defined by the Canadian Council on Animal Care.
Genotyping.
Genotyping was performed with <1 cm of tails clipped from 3-week-old pups; genomic DNA was prepared with a QIAamp DNA kit (QIAGEN). Approximately 5 μg of genomic DNA was cleaved with EcoRI restriction endonuclease and separated on 0.75% agarose gels. The DNA was transferred to GeneScreen Plus membranes (NEN-Life Science Products, Boston, Mass.) and hybridized at 42°C for 18 h with 2 × 106 to 4 × 106 dpm of random-primed [α-32P]dATP-labeled restriction fragments (29). A 93-bp BamHI-HindIII fragment cleaved from within the Ceacam1a promoter in a region located outside of the targeting vector was used as a probe. Membranes were washed at a final stringency of 65°C in a solution of 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate (SDS). Alternatively, the mice were genotyped by PCR amplification of their genomic DNA in a final volume of 15 μl containing a 3× dilution of Vent polymerase buffer supplied by the manufacturer, 500 μM of deoxynucleoside triphosphates, 5 ng of the Ceacam1a-specific oligonucleotides (PN8, 5′CTGCCCCTGGCGCTTGGA; PN5, 5′TACATGAAATCGCACAGTCGC)/μl, 5 ng of the neor-specific oligonucleotides (neoforward, 5′CGGTGCCCTGAATGAACTGC; neoreverse, 5′GCCGCCAAGCTCTTCAGCAA)/μl, and 0.4 U of Vent polymerase. Amplifications proceeded through 30 cycles of 20 s of denaturation at 94°C, 30 s of binding at 57°C, and 30 s of elongation at 72°C. The wild-type +/+ mice exhibited a Ceacam1a fragment of 250 bp upon agarose gel electrophoresis, whereas the homozygous −/− mice had a 550-bp neo fragment and the heterozygous +/− mice showed both fragments.
Sampling and preparation of tissues.
The mice were anesthetized with Avertin (Aldrich Chemical Company, Milwaukee, Wis.) and then sacrificed by either cervical dislocation or exsanguination. Tissues were removed, washed in phosphate-buffered saline (PBS), and either snap-frozen on dry ice or liquid nitrogen for subsequent DNA, RNA, protein, or viral titer analyses. Some tissues were washed and fixed in 3.7% paraformaldehyde-PBS or 10% phosphate-buffered formalin and processed for immunohistochemistry.
RNA preparation and Northern analysis.
Mouse tissues were retrieved and snap-frozen on dry ice. The tissues were then powdered with a mortar and pestle kept at −80°C, and the RNA was extracted with materials provided in the RNAqueous kit (Ambion) following the manufacturer's recommendation. Five micrograms of total RNA was subjected to electrophoresis on formaldehyde-agarose gels and transferred to Hybond N+ (Amersham). The membrane was hybridized with a full-length 32P-labeled Ceacam1a cDNA for 18 h at 42°C and washed in a solution of 0.1× SSC plus 0.1% SDS at 65°C. Membranes were exposed to X-ray film for 18 to 96 h.
Antibodies (Abs), immunoblotting, and immunoprecipitations.
Fresh tissues were excised from 2- to 6-month-old Ceacam1a+/+, Ceacam1a+/−, or Ceacam1a−/− mice, snap-frozen on dry ice, and powdered with a mortar and pestle. The powder was resuspended in 500 to 1,000 μl of lysis buffer (20). Proteins were separated on SDS-8% polyacrylamide gel electrophoresis gels and transferred to Immobilon membranes (Millipore, Nepean, Ontario, Canada). Expression of the CEACAM1a isoforms was detected by immunoblotting 75 to 200 μg of total proteins with the anti-CEACAM1a-specific rabbit polyclonal Ab 231 or 2456. Immune complexes were visualized with an ECL detection system (Amersham Pharmacia Biotech, Baie d'Urfé, Quebec, Canada). The control used in the experiments was a cell lysate from CEACAM1a-transfected NIH 3T3 cells (data not shown).
Histological analyses.
Fixed tissues were dehydrated in ethanol and paraffin embedded. Tissue sections were counterstained with hematoxylin and eosin according to standard histological procedures.
MHV preparations and i.n. and i.c. inoculation of mice.
The MHV-A59 virus strain used in these experiments was propagated in the spontaneously transformed 17 Cl 1 line of BALB/c 3T3 cells as previously described (11). The supernatant medium was collected 24 h after inoculation, centrifuged to remove cellular debris, aliquoted, quickly frozen, and stored at −80°C. Titers of infectious virus were determined by plaque assay of 17 Cl 1 cells (11). Three-week-old Ceacam1a−/− and Ceacam1a+/+ mice were inoculated i.n. with 10 μl of virus containing 106 to 108 PFU in Dulbecco's PBS. To obtain concentrated virus for the high-titer inoculum, virions were purified and concentrated by sucrose density ultracentrifugation as previously described (37). Control, uninfected +/+ or −/− mice were sham inoculated i.n. with 10 μl of PBS. The mice were observed daily for clinical signs of illness, such as lethargy, ruffled fur, hunched posture, or paralysis. Mice anesthetized intraperitoneally with Avertin were inoculated i.c. with 103 to 106 PFU of the MHV-A59 virus in 10 μl with a 27-gauge needle and a 1-ml syringe fitted to a Tridak Stepper repetitive delivery device. Samples were evaluated in the same fashion as those from the i.n. inoculated mice.
Analyses of tissues of inoculated mice.
Mice were sacrificed at intervals of 4, 7, 14, 30, 45, and 60 days postinoculation (dpi). Serum samples were collected for evaluation of anti-virus Abs. The livers, brains, and spinal cords were processed to determine the yield of infectious virus and to study histopathology. To quantitate the infectious virus in the liver and the brain, portions of the liver or brain removed at necropsy were rinsed in PBS, weighed, homogenized in Dulbecco's modified Eagle medium with 10% fetal bovine serum, and rapidly frozen and thawed at 37°C three times. Cell debris was removed by centrifugation. The virus titer per gram of liver or brain in the supernatant was determined by plaque assay of 17 Cl 1 cells as described previously (11). Tissues were fixed in neutral buffered formalin or 4% paraformaldehyde, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Sections were examined by light microscopy, and the numbers and sizes of lesions in comparable areas of the liver and brain sections were determined.
Reverse transcription-PCR (RT-PCR) analyses of viral RNA in brain and liver.
To detect viral RNA in infected tissues, RNA was isolated following Invitrogen's TRIzol reagent protocol. Briefly, approximately 100 mg of infected mouse tissue was homogenized and solubilized in 1 ml of TRIzol at 20°C for 5 min. After chloroform extraction, RNA was precipitated from the top aqueous phase. RNA, resuspended in H2O, was used as a template for RT with AMV reverse transcriptase (Promega, Madison, Wis.). PCR to detect viral RNA was performed with Taq DNA polymerase (Promega). Primers to amplify the nucleocapsid and leader region of the coronavirus MHV-A59 mRNA for the E1 glycoprotein genome were designed according to sequences from the National Center for Biotechnology Information (accession number X00509). First rounds of PCR were performed with the forward primer 5′GTACGTACCCTCTCAACTCT and the reverse primer 5′CCCATCAGGTGTTTTAAAAG. The heminested PCRs were performed with the forward primer 5′TACATGAAATCGCACAGTCGC. Some samples were positive in the first rounds of PCR, and other samples required heminested PCR to detect viral RNA. Both positive- and negative-control samples were included in this assay.
Detection of anti-virus Abs.
Mouse sera were tested for anti-virus Abs with an enzyme-linked immunosorbent assay (ELISA) in Immulon-HP 96-well microtiter plates coated with 105 PFU per well of purified MHV-A59 diluted in 50 μl of sodium carbonate buffer (pH 9.5). Plates for the ELISA were blocked overnight in B3 buffer (150 mM NaCl, 50 mM Tris, 0.8 mM EDTA, 0.05% Tween [vol/vol], 0.01% bovine serum albumin [wt/vol] [pH 7.4]) plus 5% bovine serum albumin. Anti-virus Ab titers in mouse serum in B3 buffer were assayed with anti-mouse horseradish peroxidase-labeled anti-mouse Ig Ab and 2,2 N′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS) substrate (Kirkegaard and Perry Laboratories, Gaithersburg, Md.). Absorbance was measured at 405 nm and plotted versus serum dilution.
RESULTS
Generation of the Ceacam1a−/− mice.
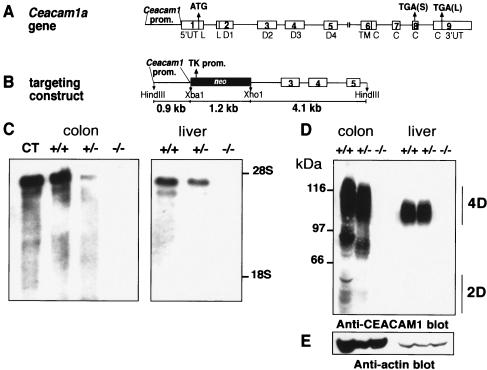
The strategy leading to complete abrogation of CEACAM1a expression in mice was based on the removal of the first two exons of the Ceacam1a gene (Fig. 1A and B). The initiator ATG codon is positioned in the first exon, and many functions associated with the CEACAM1a protein depend on the presence of the first Ig-like domain encompassed within the second exon. For this purpose, an Xba1-Xho1 restriction fragment carrying these two exons was removed from the gene, and a cassette with the TK (thymidine kinase) promoter and the neor gene was inserted (Fig. 1B). The targeting vector was electroporated into mouse R1 ES stem cells, and chimeric mice were generated by microinjection of two ES cell lines (2D2 and 11H11) into C57BL/6 mouse blastocysts. Eight chimeric male mice were obtained, four of which transmitted the Ceacam1a+/−-targeted allele through the germ line. The heterozygous Ceacam1a+/− progeny mice were mated to produce homozygous (−/−) mice. The frequency of germ line transmission was calculated to be 22% on varied backgrounds (C57BL/6, BALB/c, and 129Sv). Mating of heterozygous mice produced expected Mendelian ratios of Ceacam1a−/− offspring (+/+, 1.0; +/−, 1.8; −/−, 0.9). CEACAM1a is expressed in ovary and prostate, yet its ablation did not alter the sex ratio of the progeny (males, 52%; females, 48%).
FIG. 1.
Northern and Western analyses of Ceacam1a−/− mice. (A) The Ceacam1a gene comprises nine exons (numbered boxes) preceded by the Ceacam1a promoter (Prom.). Exon 1 contains the initiator ATG codon at the end of the 5′ untranslated region (5′UT) and the beginning of the half-leader (L) sequence. The second exon encompasses the second half-leader sequence and the first Ig domain (D1). Exons 3, 4, and 5 encode three constant-type Ig domains (D2, D3, and D4). Exon 6 contains the transmembrane domain (TM) and part of the cytoplasmic domain (C), the rest being distributed in exons 7, 8, and 9. Two different stop codons [TGA(S)] and [TGS(L)] terminate the translation of either the CEACAM1a-S or CEACAM1a-L isoforms. (B) A targeting vector was engineered whereby a TK-neor selection cassette was inserted into the XbaI site at the beginning of the 5′UT and an XhoI site located in intron 2. The selection cassette removed the first two exons as well as the initiator ATG codon. It was flanked on the 5′ side by a 0.9-kb Ceacam1a promoter fragment and on the 3′ side by a 4.1-kb fragment containing exons 3, 4, and 5. (C) Northern analyses of colon and liver RNA from mouse progeny. Total RNA was prepared from colon and liver of several mice from each litter, and 5 μg of RNA was separated on formaldehyde-agarose gels. Northern blots were produced and subjected to hybridization with the 32P-labeled full-length Ceacam1a cDNA. Samples from wild-type +/+ and heterozygous +/− mice produced a 4-kb RNA fragment corresponding to the Ceacam1a transcript. The intensity of the RNA fragment in the +/− mice was approximately half that of the control RNA from wild-type +/+ mice. No Ceacam1a transcript was revealed in the homozygous −/− mice even after prolonged exposure of the membranes. CT, hybridization of total RNA prepared from a BALB/c mouse colon. (D) Tissues were excised from 2-month-old Ceacam1a+/+, +/−, or −/− mice, snap-frozen on dry ice, and powdered with a mortar and pestle. The powder was resuspended in 500 μl of lysis buffer. Samples of 200 μg of proteins were separated on SDS-8% PAGE gels and transferred to Immobilon membranes. Expression of the CEACAM1a isoforms was detected by immunoblotting of total proteins with the anti-CEACAM1a-specific rabbit polyclonal Ab 2456. Immune complexes were visualized with an ECL detection system. 4D, CEACAM1a isoforms containing four Ig domains; 2D, CEACAM1a isoforms with two Ig domains. The band appearing at approximately 97 kDa is a degradation product. (E) The membrane shown in panel D was cleaned and reblotted with an anti-actin Ab.
Abrogation of CEACAM1a expression in Ceacam1a-targeted mice.
The complete abrogation of CEACAM1a expression was verified by Northern and Western blotting. Total RNA was prepared from colon and liver of several mice from each litter and analyzed with formaldehyde-agarose gels. Northern blots were produced and subjected to hybridization with the 32P-labeled full-length Ceacam1a cDNA (24). The wild-type +/+ and heterozygous +/− mice produced a 4-kb RNA fragment corresponding to the Ceacam1a transcript. The intensity of the RNA fragment in the +/− mice was approximately half of that in the control RNA produced from wild-type mice. No Ceacam1a transcript was revealed in the homozygous −/− mice even after prolonged exposure of the membranes, indicating that the gene inactivation strategy completely abrogated Ceacam1a transcription (Fig. 1C). We then examined the expression of the CEACAM1a protein isoforms in mouse colon and liver tissues by immunoblotting total proteins from several different mice with anti-CEACAM1a polyclonal Abs (Fig. 1D). In these and other tissues (data not shown), expression of all CEACAM1a isoforms in the homozygous −/− mice was completely eliminated relative to expression in the wild-type +/+ or heterozygous +/− mice (Fig. 1D). Actin protein levels in these tissues were constant (Fig. 1E).
Absence of CEACAM1a expression in tissues of Ceacam1a-deficient mice.
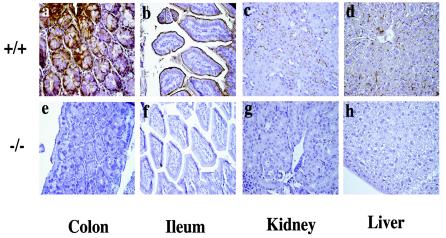
By routine histology, no histological differences were noted in the colon, small intestine, liver, kidney, prostate, ovaries, uterus, brain, lungs, heart, and spleen of −/− mice compared to those of control +/+ mice (Fig. 2 and data not shown). Paraffin-embedded tissue sections were immunostained with anti-CEACAM1a polyclonal (Ab 2456) and monoclonal (MAb-CC1) (data not shown) Abs. When immunostained with an anti-CEACAM1a polyclonal Ab, tissues from wild-type +/+ animals exhibited strong expression on the luminal membrane of surface and crypt cells of the colon (Fig. 2a and b). Bile canaliculi of the liver and proximal tubules of the kidney were also strongly positive for CEACAM1a in +/+ mice (Fig. 2c and d). In contrast, in −/− mice, colonic (Fig. 2e) or intestinal (Fig. 2f) epithelial cells revealed no staining of the crypts, even after a longer development time. This finding was repeated for hepatocytes (Fig. 2h) and the collecting tubules in the kidney (Fig. 2g) of −/− mice. These tissues were also not immunostained with anti-CEACAM1a polyclonal Abs. Other tissues that normally express CEACAM1a (small intestine, endometrium, ovary, prostate, stomach, spleen, thymus, and lung) also displayed no immunostaining in the −/− mice (data not shown).
FIG. 2.
CEACAM1a immunostaining of tissue sections from wild-type +/+ and Ceacam1a−/− mice. Paraformaldehyde-fixed colon, ileum, kidney, and liver tissues were dehydrated in ethanol and paraffin embedded. Sections of 6-μm thickness were stained with the anti-CEACAM1a polyclonal Ab 2456 and counterstained with hematoxylin. (a to d) Tissues from wild-type mice exhibited strong expression at the luminal membrane surface and crypt colonic and intestinal epithelia (a and b). Proximal tubules of the kidney and bile canaliculi of the liver were also strongly positive for CEACAM1a in +/+ mice (c and d). (e to h) In contrast, in Ceacam1a−/− mice, colonic (e) or intestinal (f) epithelial cells revealed no staining of the crypts, in spite of a longer development time. This was also seen for the collecting tubules in the kidney (g) and hepatocytes (h) of Ceacam1a−/− mice that were not labeled with anti-CEACAM1a Abs.
General health status of the offspring.
The Ceacam1a-targeted mice were viable and healthy under a pathogen-free environment. We have maintained a sizeable colony (approximately 350 individuals) of targeted mice for 1 year on the BALB/c, 129Sv, and C57BL/6 backgrounds and have not noticed any reduction in fertility, bone or cartilage abnormalities, tumors, or abnormal behavior. However, these mice do exhibit distinct phenotypes relative to liver insulin resistance and clearance (Dai et al., unpublished) and in in vivo T-cell physiology that will be reported elsewhere (R. Atallah et al., unpublished data).
Results of i.n. inoculation of Ceacam1a−/− mice with MHV-A59.
To define whether the Ceacam1a-targeted mice were susceptible or resistant to MHV infection, we subjected wild-type +/+ and −/− mice from both the 2D2 and 11H11 lines to infection with the MHV-A59 virus. Mouse litters were generated by crossing Ceacam1a+/− siblings, crossing +/+ mice with −/− mice, or mating −/− mice. Experiments were performed with the second and third backcrosses of both lines of Ceacam1a−/− mice. Genotyping was performed as described above on DNA prepared from mouse tails. Control mice were sham -inoculated, and both wild-type +/+ and −/− mice were inoculated i.n. with 10 μl of virus containing 106 PFU of MHV-A59 (Table 1). The mice were examined every day for clinical signs of disease and, if warranted, were sacrificed immediately. Groups of +/+ and −/− mice were sacrificed at 4, 14, 30, 45, and 60 dpi. Blood was collected to test for Abs to viral antigens (serocoversion), and liver and spinal cord tissues were processed for pathology. Liver and brain RNA was tested for the presence of viral RNA that is indicative of viral infection.
TABLE 1.
Intranasal inoculation of Ceacamla−/− mice on C57BL/6 background with 106 PFU of MHV-A59
| dpi and genotype | No. of micea | Anti-virus antibody titerb | Liver pathologyc | Spinal cord demyelination | Viral RNA in brain by RT-PCR | Virus titer per g of e:
|
|
|---|---|---|---|---|---|---|---|
| Liver | Brain | ||||||
| 4 | |||||||
| +/+ | 4 | <100 | 2+ to 4+ | 0/3 | 3/3 | 4.7 ± 0.6 | 5.0 ± 1.3 |
| −/− | 5 | <100 | 0 | 0/5 | 0/5 | <2 | <2 |
| 14 | |||||||
| +/+ | 3 | 175-600 | 1+ | 2/3 | 3/3 | <2 | <2 |
| −/− | 4 | <100-300 | 0 | 0/4 | 0/3 | <2 | <2 |
| 30 | |||||||
| +/+ | 4 | 350-600 | 0 | 4/4 | 1/1 | <2 | <2 |
| −/− | 5 | <100 | 0 | 0/5 | 0/5 | <2 | <2 |
| 45 | |||||||
| +/+ | 1 | 2,300 | 0 | 0/1 | NDd | <2 | <2 |
| −/− | 5 | <100 | 0 | 0/5 | 0/1 | <2 | <2 |
| 60 | |||||||
| −/− | 5 | <100 to 175 | 0 | 0/5 | NDd | <2 | <2 |
The number of mice represents the total from both 2D2 and 11H11 lines.
Values are ranges of serum titers (optical density, 0.8) in ELISA of samples from replicate animals; a value of <100 indicates that the titer was below the range of detection.
Grading system for liver pathology: 0, no lesions: 1, 1 to 10% tissue destruction; 2, 11 to 20% tissue destruction; 3, 21 to 40% tissue destruction; 4, 41 to 100% tissue destruction.
ND, not determined.
Values are geometric mean titers per log10 PFU per gram of tissue; a value of <2 indicates that the titer was below the limit of detection.
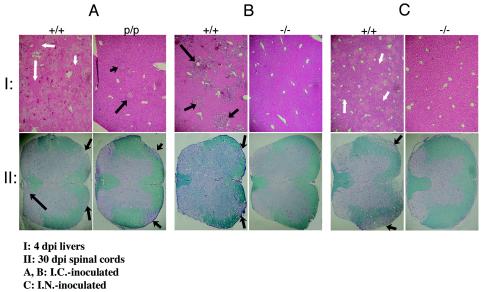
At 4 and 14 dpi, the health of the +/+ mice was compromised, as they showed many of the clinical symptoms described in Materials and Methods, whereas the −/− mice showed no signs of illness. Ultimately, five infected +/+ animals survived and recovered from the infection. Mice sacrificed between 30 and 60 dpi did not demonstrate overt clinical symptoms. As seen in the results shown in Table 1, infected +/+ mice sacrificed between 14 and 45 dpi generally exhibited high anti-virus serum Ab titers. Conversely, only two of the Ceacam1a−/− mice produced an Ab response of relatively low titer to viral antigens. Only one of four −/− mice had seroconverted at 14 dpi, none had mounted an immune response at 30 dpi or 45 dpi, and just one of five −/− mice tested weakly positive at 60 dpi. Liver histology was examined for the presence of viral lesions, and a grading system was established that took into consideration the numbers and sizes of the lesions. At early time points after infection (4 and 14 dpi) (Table 1), all +/+ mice exhibited liver pathology. Indeed, +/+ mice sacrificed at 4 dpi showed lesions involving from 10% (liver pathology grade, 2+) up to more than 90% (4+) of cells (Fig. 3B and C, row I). Some +/+ mice sacrificed at 14 dpi had resolving lesions affecting up to 10% (1+) of cells. After the +/+ mice recovered from signs of infection by the 30- to 60-dpi period, the liver lesions were no longer detectable. None of the −/− mice demonstrated lesions in their liver at any of the time points examined (Table 1 and Fig. 3B and C, row I).
FIG. 3.
(A) Liver and spinal cord lesions in BALB/c +/+ and Ceacam1a partial knockout (p/p) mice on the BALB/c background following i.c. inoculation with 103 PFU MHV-A59. Arrows indicate areas of extensive liver destruction in wild-type +/+ mice (harvested 4 dpi) and demyelination in spinal cords (harvested 30 dpi) as evidenced by hematoxylin and eosin staining and Luxol fast blue staining. Liver damage was present in p/p mice but was much less severe than that in the wild-type +/+ mice. Demyelination was also present in both mice but was less severe in the p/p mice compared to the wild-type +/+ mice. (B) Extensive liver and spinal cord lesions in Ceacam1a+/+ mice following i.c. inoculation with 106 PFU MHV-A59. Livers from Ceacam1a−/− and wild-type +/+ C57BL/6 mice were collected 4 dpi, and spinal cords were collected at 30 dpi. Arrows indicate demyelination that was observed in the spinal cords of wild-type +/+ mice. The Ceacam1a−/− mice did not show any signs of liver damage or spinal cord demyelination. (C) Three-week-old Ceacam1a−/− and wild-type +/+ C57BL/6 mice were i.n. inoculated with 106 PFU of MHV-A59 and sacrificed at 4 dpi. Liver sections were processed for histology, and liver lesions were quantified. Liver lesions were extensive in wild-type (+/+) mice and absent in the CEACAM1a-deficient mice (−/−). Demyelination was observed in the spinal cords of the wild-type +/+ C57BL/6 mice but absent in the Ceacam1a−/− mice.
Spinal cords from these mice were also prepared for histology and analyzed for evidence of demyelination, as indicated by loss or destruction of myelin detected by Luxol fast blue staining and the presence of asymetrical lesions (Fig. 3). At 14 and 30 dpi, two of three and four of four wild-type +/+ mice, respectively, no longer had intact spinal cords and their myelin sheets were very thin, whereas none of the Ceacam1a−/− mice showed any abnormalities in these structures at any time point. Changes in spinal cord histology in the CEACAM1a-positive +/+ mice were generally correlated with the expression of viral RNA detected in the brains of affected animals. No infectious virus was detected in the brain or liver of the inoculated −/− mice, and no viral RNA from either tissue was detected by RT-PCR, even when nested PCR primer sets were used.
When Ceacam1a−/− mice were inoculated i.n. with 108 PFU of the same virus (Table 2), none of the −/− mice showed clinical signs of disease or liver pathology (Fig. 3C, row I). In contrast, livers of +/+ mice were completely devastated (liver pathology grade, 4+) upon i.n. inoculation with just 106 viral PFU. Titers of virus recovered from livers and brains of affected +/+ animals ranged from 8.4 × 102 to 4.6 × 106 PFU/g of tissue, but no infectious virus was detected in the tissues of −/− mice. These results indicate that the Ceacam1a−/− mice were completely resistant to i.n. MHV-A59 infection with at least 100-fold more than the lethal dose sustained by wild-type +/+ mice.
TABLE 2.
Intranasal inoculation of Ceacamla−/− mice on C57BL/6 background with MHV-A59a
| dpi and genotype | No. of mice | Liver pathologyb | Virus titer per g of c
|
|
|---|---|---|---|---|
| Liver | Brain | |||
| 4 | ||||
| +/+ | 5 | 4+ | 5.5 ± 1.0 | 5.7 ± 0.9 |
| −/− | 3 | 0 | <2 | <2 |
| 7 | ||||
| +/+ | 4 | 3+ | 3.6 ± 0.5 | 6.4 |
| −/− | 3 | 0 | <2 | <2 |
Mice (+/+) were inoculated with 106 PFU of virus, and −/− mice were infected with 108 PFU of virus. Mice from the 2D2 line were tested.
Grading system for liver pathology: 0, no lesions: 1, 1 to 10% tissue destruction; 2, 11 to 20% tissue destruction; 3, 21 to 40% tissue destruction; 4, 41 to 100% tissue destruction.
Values are geometric mean titers per log10 PFU per gram of tissue; a value of <2 indicates that the titer was below the limit of detection.
Results of i.c. inoculation of Ceacam1a p/p mice with MHV-A59.
We had previously generated another genetically altered mouse MHVR mutant (designated p/p mice) in the BALB/c background. These animals exhibited greatly reduced expression (>95%) of the CEACAM1a receptor bearing four Ig domains while the other isoforms containing two Ig domains were overexpressed (5). When inoculated i.n. with 106 PFU of the MHV-A59 virus, the p/p mice developed small liver lesions at 3 dpi, compared to their wild-type +/+ littermates that formed abundant, large lesions and soon succumbed to the disease. The lesions in the p/p mice completely disappeared at 7 dpi, and these animals completely recovered from the infection (5).
To gauge the susceptibility of the brains and spinal cords of p/p mice to MHV-A59 without requiring prior amplification of virus inoculated at peripheral sites, we directly inoculated 103 PFU of the MHV-A59 virus i.c. and monitored the mice for the appearance of disease, altered liver and spinal cord histology, seroconversion, and production of infectious virus or viral RNA (Table 3). The p/p mice showed only transient and mild signs of disease, unlike wild-type +/+ mice, which showed more-extensive neurological signs. Furthermore, 3 of 10 +/+ mice died between 4 and 14 dpi. Wild-type +/+ mice that had been inoculated i.c. and sacrificed at 4 dpi showed significantly altered liver histology (liver pathology grade, 4+) with a larger number of large lesions than the wild-type mice inoculated i.n. with the same virus. The Ceacam1a p/p mice examined at 4 dpi also exhibited liver lesions, but these were consistently fewer and smaller (2+) than those seen in the wild-type +/+ mice (Fig. 3A row I). No demyelination of the spinal cord was observed in any mouse sacrificed at 4 dpi, but viral RNA was detected in liver and brain of all the +/+ and p/p mice. By 14 dpi, all surviving wild-type +/+ mice had medium-sized liver lesions (2+), whereas no liver lesions were found in the p/p mice. On day 14, spinal cord demyelination was observed in all wild-type +/+ mice and in three of four p/p mice, although the lesions were less extensive in p/p mice. In wild-type +/+ mice at 14 dpi, viral RNA was detected in the brains of five of five mice and the livers of three of five mice, whereas the p/p mice showed no viral RNA in liver (Table 3). At 30 and 45 dpi, none of the wild-type +/+ or p/p mice exhibited liver lesions, but most (four of five +/+ and nine of nine p/p) had spinal cord demyelination (Fig. 3A, row II). Anti-virus Abs were detected in the sera of three of three of the p/p mice sacrificed at 30 dpi and in two of two of the p/p mice sacrificed at 45 dpi. Viral RNA was detected in brains of two of three wild-type mice at day 30 and in one of one mouse sacrificed at day 45, but viral RNA was not detected in brains of p/p mice at these times. No viral RNA was detected in livers of wild-type +/+ or p/p mice at days 30 and 45 (Table 3). These experiments show that MHV-A59 could infect cells in the spinal cords of p/p mice, although infection caused less-severe demyelination than in wild-type mice.
TABLE 3.
Intracerebral inoculation of Ceacamla p/p mice on BALB/c background with 103 PFU of MHV-A59
| dpi and genotype | No. of micea | Liver pathologyb | Anti-virus antibody titerc | Spinal cord demyelinationd | Viral RNA in:
|
Virus titer per g of e:
|
||
|---|---|---|---|---|---|---|---|---|
| Brain | Liver | Brain | Liver | |||||
| 4 | ||||||||
| +/+ | 5 | 2+ to 4+ | ND | 0/5 | 5/5 | 5/5 | 5.3 ± 0.9 | 4.7 ± 0.5 |
| p/p | 5 | 1+ to 3+ | ND | 0/5 | 5/5 | 5/5 | 3.5 ± 0.4 | 4.1 ± 0.8 |
| 14 | ||||||||
| +/+ | 5 | 1+ to 2+ | ND | 5/5 | 5/5 | 3/5 | <2 | <2 |
| p/p | 4 | 0 to 1+ | ND | 3/4 | 3/4 | 0/4 | <2 | <2 |
| 30 | ||||||||
| +/+ | 3 | 0 | ND | 2/3 | 2/3 | 0/3 | <2 | <2 |
| p/p | 6 | 0 | 600-800 | 6/6 | 0/5 | 0/5 | <2 | <2 |
| 45 | ||||||||
| +/+ | 2 | 0 | ND | 2/2 | 1/1 | 0/2 | <2 | <2 |
| p/p | 3 | 0 | 150-700 | 3/3 | 0/1 | 0/3 | <2 | <2 |
Three +/+ mice died between days 4 and 14.
Grading system for liver pathology: 0, no lesions: 1, 1 to 10% tissue destruction; 2, 11 to 20% tissue destruction; 3, 21 to 40% tissue destruction; 4, 41 to 100% tissue destruction.
Values are ranges of serum titers (optical density, 0.8) in ELISA of samples from replicate animals; a value of <100 indicates that the titer was below the range of detection.
The demyelination of the spinal cords of p/p mice was less severe than that of +/+ mice at the same time points.
Values are geometric mean titers per log10 PFU per gram of tissue; a value of <2 indicates that the titer was below the limit of detection.
Results of i.c. inoculation of Ceacam1a−/− mice with MHV-A59.
To determine whether the virus infected the p/p mouse spinal cord via the CEACAM1a receptor, we inoculated Ceacam1a−/− mice and wild-type +/+ C57BL/6 mice with 106 PFU of MHV-A59 i.c. The results of two independent experiments are summarized in Table 4. Seven of 16 CEACAM1a-positive +/+ C57BL/6 mice inoculated with virus were sacrificed or died by 4 dpi and had severe liver lesions (4+). At 7 dpi, eight of the nine surviving wild-type +/+ mice were sacrificed because they showed severe signs of illness, including ataxia or paralysis. Livers showed extensive virus-induced damage (4+), and spinal cords of seven of eight mice showed demyelination. Virus was recovered from the livers and brains of the wild-type mice, with titers ranging from 102 to 105 PFU per gram of liver and 102 to 105 PFU per gram of brain. On 14 dpi, the one surviving wild-type +/+ mouse was sacrificed because it was moribund. Liver damage (2+) was less extensive than in animals at 7 dpi, but the spinal cord showed extensive demyelination.
TABLE 4.
Intracerebral inoculation of Ceacamla−/− mice on C57BL/6 background with 106 PFU of MHV-A59
| dpi and genotype | No. of micea | Liver pathologyd | Spinal cord demyelination | Anti-virus antibody titere | Virus titer per g of f
|
|
|---|---|---|---|---|---|---|
| Liver | Brain | |||||
| 4 | ||||||
| +/+ | 7b | 4+ | ND | ND | 7b | 7b |
| −/− | 7 | 0 | 0/7 | <100 | <2 | <2 |
| 7 | ||||||
| +/+ | 8 | 4+ | 7/8 | 125-600 | 4.5 ± 0.4 | 5.2 ± 0.9 |
| −/− | 6 | 0 | 0/6 | <100 to 125 | <2 | <2 |
| 14 | ||||||
| +/+ | 1 | 2+ | 1/1 | ND | <2 | <2 |
| −/− | 3 | 0 | 0/3 | ND | <2 | <2 |
| 30 | ||||||
| +/+ | 1c | 0 | 0/1 | <100 | <2 | <2 |
| −/− | 8 | 0 | 0/8 | <100 | <2 | <2 |
| 45 | ||||||
| +/+ | 1c | 0 | 0/1 | <100 | ||
| −/− | 12 | 0 | 0/12 | <100 to 250 | <2 | <2 |
Mice tested were from the 2D2 and 11H11 lines; all mice received the same inoculum.
Seven +/+ mice were sacrificed or died on or before 4 dpi.
Mock-inoculated mice. No MHV-inoculated wild-type mice survived past day 14.
Grading system for liver pathology: 0, no lesions: 1, 1 to 10% tissue destruction; 2, 11 to 20% tissue destruction; 3, 21 to 40% tissue destruction; 4, 41 to 100% tissue destruction.
Values are ranges of serum titers (optical density, 0.8) in ELISA of samples from replicate animals; a value of <100 indicates that the titer was below the range of detection.
Geometric mean titer per log10 PFU per gram of tissue; a value of <2 indicates that the titer was below the limit of detection.
In marked contrast to the lethal liver and neurological disease in wild-type +/+ mice, 36 of 36 Ceacam1a−/− mice inoculated i.c. with MHV-A59 survived (Table 4). All seven −/− mice harvested at 4 dpi showed no signs of disease, had no liver or spinal cord lesions, and had neither viral RNA nor infectious virus in liver or brain. Similar results were obtained with −/− mice sacrificed at days 14, 30, and 45. Low titers of anti-virus Abs were detected in a few i.c.-inoculated −/− mice at days 7 and 45 (Table 4).
These data show that 3-week-old Ceacam1a−/− mice in the C57BL/6 background are fully resistant to very high doses of the MHV-A59 virus administered i.c. or i.n. and indicate that expression of murine CEACAM1a is essential for MHV-A59 infection of liver or brain of C57BL/6 mice.
DISCUSSION
The newly generated Ceacam1a−/− mouse lines showed complete abrogation of expression of the CEACAM1a protein in all tissues that normally express it. We examined the response of two genetically altered mouse models to MHV infection via two different routes of inoculation. We had previously characterized the p/p mouse response to i.n. inoculation with MHV-A59 and found that the mice replicated virus in the liver in small amounts; they showed only mild clinical signs of disease, recovered, and completely cleared the virus (5). Viral transmission by the i.n. route first requires viral replication in the nasal epithelial cells and then dissemination to other organs via either the bloodstream or infected leukocytes or neural routes (1, 22). In the present study, we showed that i.c. inoculation also produced hepatic lesions in p/p mice, probably through viremia. Intracerebral inoculation efficiently induced small demyelinating lesions in the p/p spinal cords that were still apparent in mice sacrificed at 45 dpi.
In contrast to the p/p mice that were partially susceptible to MHV-A59 infection but recovered, mice devoid of the CEACAM1a receptors (−/−) did not produce any obvious clinical signs of disease after i.n. inoculation with up to 108 PFU of MHV-A59 virus. The −/− mice showed no pathology in liver or spinal cord tissues, and no infectious virus or viral RNA could be detected in target tissues. Only 2 of the 19 inoculated −/− mice responded to the inoculum virus as a foreign antigen and raised anti-virus Abs. These results therefore clearly demonstrate that murine CEACAM1a is the sole receptor for MHV-A59 in C57/BL6 mice in vivo. The two mouse models discussed herein were produced on different genetic backgrounds (p/p mice in a BALB/c background and −/− mice in a C57BL/6 background). Yet, the data are consistent with results obtained with cell lines by a number of different groups. Furthermore, both the BALB/c and C57BL/6 wild-type mice carry only the Ceacam1a allele and express the same types of isoforms in the different tissues examined.
Nédellec et al. identified the product of a second Ceacam-like gene, Ceacam2, as a weak MHV receptor (28). In fact, hamster cells (BHK) transfected with cDNA encoding murine CEACAM2 can be infected with MHV-A59 and form multinucleated syncytia (28). However, the virus-neutralizing activity of the soluble CEACAM2 protein was 10,000-fold lower than that of the soluble four-Ig-domain-expressing CEACAM1a protein (46). Therefore, the question remained open as to whether CEACAM2 could serve as an alternate receptor in vivo and even completely substitute for the CEACAM1a MHV receptor in Ceacam1a−/− mice. The fact that the Ceacam1a−/− mice are completely resistant to infection from very high doses of MHV-A59 strongly suggests that CEACAM2 does not act as an alternative receptor for MHV in vivo. CEACAM2 is expressed in wild-type C57BL/6 and BALB/c mice as well as in Ceacam1a−/− mice (data not shown). However, its tissue distribution is strikingly different than that of CEACAM1a, being mostly present in kidney, pancreas, and macrophages and essentially absent in liver and brain (28, 34).
CEACAM1a is a multifunctional protein; abrogating the expression of this protein leads to a number of phenotypic abnormalities. Preliminary results obtained from studies of Ceacam1a−/− mice indicate that they exhibit defects in insulin clearance from the liver and are hyperinsulinemic and insulin resistant (Dai et al., unpublished). In addition, the−/− mice accumulate lipid in their liver, suggesting a serious dysfunction in the proper utilization or storage of fatty acids and triglycerides and/or possibly a deregulation in their peroxidation (N. Leung et al., unpublished data). The −/− mice also show signs of immune dysfunction. The development of the T lymphocytes is normal, the numbers of CD4+ versus CD8+ T lymphocytes are very similar in the thymus and spleen of the +/+ and −/− mice, and the numbers of IgM-positive B lymphocytes in the spleen are almost identical in the wild-type +/+ and knockout −/− mice. However, the T lymphocytes are functionally impaired in proliferation, with altered cytokine secretion levels (Atallah et al., unpublished). The hepatic or immune dysfunctions might be expected to potentiate viral infection. In spite of these deficiencies, the C57BL/6 Ceacam1a−/− mice remained entirely resistant to massive doses of MHV-A59 virus. Thus, murine CEACAM1a proteins are required for infection of both brain and liver of 3-week-old C57BL/6 mice with MHV-A59, and the CEACAM1a proteins are the only receptors for MHV-A59 in these animals.
Acknowledgments
We thank Michel L. Tremblay for fruitful discussions and the McGill Transgenic Core Facility personnel for their help. We also wish to convey our appreciation to the personnel of both the Animal Care Center of McGill University and the Center for Laboratory Animal Care at the University of Colorado Health Sciences Center for their excellent care of the animals.
This work was supported by the Canadian Institutes of Health Research (grant MOP 42501 to N.B.) and the National Institutes of Health (grant AI25231 to K.V.H.).
Nicole Beauchemin is a senior scientist from the Fonds de la Recherche en Santé du Québec.
REFERENCES
- 1.Barthold, S. W. 1988. Olfactory neural pathway in mouse hepatitis virus nasoencephalitis. Acta Neuropathol. (Berlin) 76:502-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barthold, S. W., and A. L. Smith. 1987. Response of genetically susceptible and resistant mice to intranasal inoculation with mouse hepatitis virus JHM. Virus Res. 7:225-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beauchemin, N., P. Draber, G. Dveksler, P. Gold, S. Gray-Owen, F. Grunert, S. Hammarstrom, K. V. Holmes, A. Karlsson, M. Kuroki, S. H. Lin, L. Lucka, S. M. Najjar, M. Neumaier, B. Obrink, J. E. Shively, K. M. Skubitz, C. P. Stanners, P. Thomas, J. A. Thompson, M. Virji, S. von Kleist, C. Wagener, S. Watt, and W. Zimmermann. 1999. Redefined nomenclature for members of the carcinoembryonic antigen family. Exp. Cell Res. 252:243-249. [DOI] [PubMed] [Google Scholar]
- 4.Beauchemin, N., and S. H. Lin. 1998. Role of C-CAM as a tumor suppressor, p. 155-175. In C. P. Stanners (ed.), Cell adhesion and communication mediated by the CEA family. Harwood Academic Publishers, Amsterdam, The Netherlands.
- 5.Blau, D. M., C. Turbide, M. Tremblay, M. Olson, S. Letourneau, E. Michaliszyn, S. Jothy, K. V. Holmes, and N. Beauchemin. 2001. Targeted disruption of the Ceacam1 (MHVR) gene leads to reduced susceptibility of mice to mouse hepatitis virus infection. J. Virol. 75:8173-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coutelier, J.-P., C. Godfraind, G. S. Dveksler, M. Wysocka, C. B. Cardellichio, H. Noël, and K. V. Holmes. 1994. B lymphocyte and macrophage expression of carcinoembryonic antigen-related adhesion molecules that serve as receptors for the murine coronavirus. Eur. J. Immunol. 24:1383-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dveksler, G. S., C. W. Dieffenbach, C. B. Cardellichio, K. McCuaig, M. N. Pensiero, G. S. Jiang, N. Beauchemin, and K. V. Holmes. 1993. Several members of the mouse carcinoembryonic antigen-related glycoprotein family are functional receptors for the coronavirus mouse hepatitis virus-A59. J. Virol. 67:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dveksler, G. S., M. N. Pensiero, C. B. Cardellichio, R. K. Williams, G. S. Jiang, K. V. Holmes, and C. W. Dieffenbach. 1991. Cloning of the mouse hepatitis virus (MHV) receptor: expression in human and hamster cell lines confers susceptibility to MHV. J. Virol. 65:6881-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dveksler, G. S., M. N. Pensiero, C. W. Dieffenbach, C. B. Cardellichio, A. A. Basile, P. E. Elia, and K. V. Holmes. 1993. Mouse hepatitis virus strain A59 and blocking antireceptor monoclonal antibody bind to the N-terminal domain of the cellular receptor. Proc. Natl. Acad. Sci. USA 90:1716-1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ergün, S., N. Kilic, G. Ziegeler, A. Hansen, P. Nollau, J. Götze, J. H. Wurmbach, A. Horst, J. Weil, M. Fernando, and C. Wagener. 2000. CEA-related cell adhesion molecule 1 (CEACAM1): a potent angiogenic factor and a major effector of vascular endothelial growth factor (VEGF). Mol. Cell 5:311-320. [DOI] [PubMed] [Google Scholar]
- 11.Frana, M. F., J. N. Behnke, L. S. Sturman, and K. V. Holmes. 1985. Proteolytic cleavage of the E2 glycoprotein of murine coronavirus: host-dependent differences in proteolytic cleavage and cell fusion. J. Virol. 56:912-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godfraind, C., S. G. Langreth, C. B. Cardellichio, R. Knobler, J. P. Coutelier, M. Dubois-Dalcq, and K. V. Holmes. 1995. Tissue and cellular distribution of an adhesion molecule in the carcinoembryonic antigen family that serves as a receptor for mouse hepatitis virus. Lab. Investig. 73:615-627. [PubMed] [Google Scholar]
- 13.Gray-Owen, S. D., C. Dehio, A. Haude, F. Grunert, and T. F. Meyer. 1997. CD66 carcinoembryonic antigens mediate interactions between Opa-expressing Neisseria gonorrhoeae and human polymorphonuclear phagocytes. EMBO J. 16:3435-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greicius, G., E. Severinson, N. Beauchemin, B. Obrink, and B. Singer. 2003. CEACAM1 is a potent regulator of B cell receptor complex-induced activation. J. Leukoc. Biol. 74:126-134. [DOI] [PubMed] [Google Scholar]
- 15.Hammarstrom, S. 1999. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin. Cancer Biol. 9:67-81. [DOI] [PubMed] [Google Scholar]
- 16.Han, E., D. Phan, P. Lo, M. N. Poy, R. Behringer, S. M. Najjar, and S. H. Lin. 2001. Differences in tissue-specific and embryonic expression of mouse Ceacam1 and Ceacam2 genes. Biochem. J. 355:417-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill, D. J., and M. Virji. 2003. A novel cell-binding mechanism of Moraxella catarrhalis ubiquitous surface protein UspA: specific targeting of the N-domain of carcinoembryonic antigen-related cell adhesion molecules by UspA1. Mol. Microbiol. 48:117-129. [DOI] [PubMed] [Google Scholar]
- 18.Hsieh, J.-T., W. Luo, W. Song, Y. Wang, D. I. Kleinerman, N. T. Van, and S.-H. Lin. 1995. Tumor suppressive role of an androgen-regulated epithelial cell adhesion molecule (C-CAM) in prostate carcinoma cell revealed by sense and antisense approaches. Cancer Res. 55:190-197. [PubMed] [Google Scholar]
- 19.Huang, J., J. F. Simpson, C. Glackin, L. Riethorf, C. Wagener, and J. E. Shively. 1998. Expression of biliary glycoprotein (CD66a) in normal and malignant breast epithelial cells. Anticancer Res. 18:3203-3212. [PubMed] [Google Scholar]
- 20.Huber, M., L. Izzi, P. Grondin, C. Houde, T. Kunath, A. Veillette, and N. Beauchemin. 1999. The carboxyl-terminal region of biliary glycoprotein controls its tyrosine phosphorylation and association with protein-tyrosine phosphatases SHP-1 and SHP-2 in epithelial cells. J. Biol. Chem. 274:335-344. [DOI] [PubMed] [Google Scholar]
- 21.Knobler, R. L., M. V. Haspel, and M. B. Oldstone. 1981. Mouse hepatitis virus type 4 (JHM strains) induced fatal central nervous system disease. I. Genetic control and murine neuron as the susceptible site of the disease. J. Exp. Med. 153:832-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavi, E., P. S. Fishman, M. K. Highkin, and S. R. Weiss. 1988. Limbic encephalitis after inhalation of murine coronavirus. Lab. Investig. 58:31-36. [PubMed] [Google Scholar]
- 23.Leusch, H.-G., Z. Drzeniek, Z. Markos-Pusztai, and C. Wagener. 1991. Binding of Escherichia coli and Salmonella strains to members of the carcinoembryonic antigen family: differential binding inhibition by aromatic α-glycosides of mannose. Infect. Immun. 59:2051-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCuaig, K., M. Rosenberg, P. Nédellec, C. Turbide, and N. Beauchemin. 1993. Expression of the Bgp gene and characterization of mouse colon biliary glycoprotein isoforms. Gene 127:173-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCuaig, K., C. Turbide, and N. Beauchemin. 1992. mmmCGM1a: a mouse carcinoembryonic antigen gene family member, generated by alternative splicing, functions as an adhesion molecule. Cell Growth Differ. 3:165-174. [PubMed] [Google Scholar]
- 26.Moller, M. J., R. Kammerer, F. Grunert, and S. von Kleist. 1996. Biliary glycoprotein (BGP) expression on T cells and on a natural-killer-cell sub-population. Int. J. Cancer 65:740-745. [DOI] [PubMed] [Google Scholar]
- 27.Morales, V. M., A. Christ, S. M. Watt, H. S. Kim, K. W. Johnson, N. Utku, A. M. Texieira, A. Mizoguchi, E. Mizoguchi, G. J. Russell, S. E. Russell, A. K. Bhan, G. J. Freeman, and R. S. Blumberg. 1999. Regulation of human intestinal intraepithelial lymphocyte cytolytic function by biliary glycoprotein (CD66a). J. Immunol. 163:1363-1370. [PubMed] [Google Scholar]
- 28.Nédellec, P., G. S. Dveksler, E. Daniels, C. Turbide, B. Chow, A. A. Basile, K. V. Holmes, and N. Beauchemin. 1994. Bgp2, a new member of the carcinoembryonic antigen-related gene family, encodes an alternative receptor for mouse hepatitis viruses. J. Virol. 68:4525-4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nédellec, P., C. Turbide, and N. Beauchemin. 1995. Characterization and transcriptional activity of the mouse biliary glycoprotein 1 gene, a carcinoembryonic antigen-related gene. Eur. J. Biochem. 231:104-114. [DOI] [PubMed] [Google Scholar]
- 30.Ocklind, C., and B. Obrink. 1982. Intercellular adhesion of rat hepatocytes. Identification of a cell surface glycoprotein involved in the initial adhesion process. J. Biol. Chem. 257:6788-6795. [PubMed] [Google Scholar]
- 31.Prall, F., P. Nollau, M. Neumaier, H. D. Haubeck, Z. Drzeniek, U. Helmchen, T. Loning, and C. Wagener. 1996. CD66a (BGP), an adhesion molecule of the carcinoembryonic antigen family, is expressed in epithelium, endothelium, and myeloid cells in a wide range of normal human tissues. J. Histochem. Cytochem. 44:35-41. [DOI] [PubMed] [Google Scholar]
- 32.Rao, P. V., S. Kumari, and T. M. Gallagher. 1997. Identification of a contiguous 6-residue determinant in the MHV receptor that controls the level of virion binding to cells. Virology 229:336-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ravetch, J. V., and L. L. Lanier. 2000. Immune inhibitory receptors. Science 290:84-89. [DOI] [PubMed] [Google Scholar]
- 34.Robitaille, J., L. Izzi, E. Daniels, B. Zelus, K. V. Holmes, and N. Beauchemin. 1999. Comparison of expression patterns and cell adhesion properties of the mouse biliary glycoproteins Bgp1 and Bgp2. Eur. J. Biochem. 264:534-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawa, H., K. Kamada, H. Sato, S. Sendo, A. Kondo, I. Saito, M. Edlund, and B. Obrink. 1994. C-CAM expression in the developing rat central nervous system. Brain Res. Dev. Brain Res. 78:35-43. [DOI] [PubMed] [Google Scholar]
- 36.Stohlman, S. A., and J. A. Frelinger. 1978. Resistance to fatal central nervous system disease by mouse hepatitis virus strain JHM I. Genetic analysis. Immunogenetics 6:277-281. [PubMed] [Google Scholar]
- 37.Sturman, L. S., K. V. Holmes, and J. Behnke. 1980. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J. Virol. 33:449-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svalander, P. C., P. Odin, B. O. Nilsson, and B. Öbrink. 1990. Expression of cellCAM-105 in the apical surface of rat uterine epithelium is controlled by ovarian steriod hormones. J. Reprod. Fertil. 88:213-221. [DOI] [PubMed] [Google Scholar]
- 39.Tan, K., B. D. Zelus, R. Meijers, J. H. Liu, J. M. Bergelson, N. Duke, R. Zhang, A. Joachimiak, K. V. Holmes, and J. H. Wang. 2002. Crystal structure of murine sCEACAM1a[1,4]: a coronavirus receptor in the CEA family. EMBO J. 21:2076-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson, J. A., F. Grunert, and W. Zimmermann. 1991. Carcinoembryonic antigen gene family: molecular biology and clinical perspectives. J. Clin. Lab. Anal. 5:344-366. [DOI] [PubMed] [Google Scholar]
- 41.Tsai, J. C., B. D. Zelus, K. V. Holmes, and S. R. Weiss. 2003. The N-terminal domain of murine coronavirus spike glycoprotein determines the CEACAM1 receptor specificity of the virus strain. J. Virol. 77:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Virji, M., D. Evans, J. Griffith, D. Hill, L. Serino, A. Hadfield, and S. M. Watt. 2000. Carcinoembryonic antigens are targeted by diverse strains of typable and non-typable Haemophilus influenzae. Mol. Microbiol. 36:784-795. [DOI] [PubMed] [Google Scholar]
- 43.Virji, M., K. Makepeace, D. J. Ferguson, and S. M. Watt. 1996. Carcinoembryonic antigens (CD66) on epithelial cells and neutrophils are receptors for Opa proteins of pathogenic neisseriae. Mol. Microbiol. 22:941-950. [DOI] [PubMed] [Google Scholar]
- 44.Wessner, D. R., P. C. Shick, J. H. Lu, C. B. Cardellichio, S. E. Gagneten, N. Beauchemin, K. V. Holmes, and G. S. Dveksler. 1998. Mutational analysis of the virus and monoclonal antibody binding sites in MHVR, the cellular receptor of the murine coronavirus mouse hepatitis virus strain MHV-59. J. Virol. 72:1941-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zelus, B. D., J. H. Schickli, D. M. Blau, S. R. Weiss, and K. V. Holmes. 2003. Conformational changes in the spike glycoprotein of murine coronavirus are induced at 37oC either by soluble murine CEACAM1 receptors or by pH 8. J. Virol. 77:830-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zelus, B. D., D. R. Wessner, R. K. Williams, M. N. Pensiero, F. T. Phibbs, M. deSouza, G. S. Dveksler, and K. V. Holmes. 1998. Purified, soluble recombinant mouse hepatitis virus receptor, Bgp1b, and Bgp2 murine coronavirus receptors differ in their mouse hepatitis virus binding and neutralizing activities. J. Virol. 72:7237-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]