Abstract
Cytotoxic T lymphocytes (CTL) are associated with control of immunodeficiency virus infection but also select for variants that escape immune recognition. Declining frequencies of epitope-specific CTL frequencies have been correlated with viral escape in individual hosts. However, escape mutations may give rise to new epitopes that could be recognized by CTL expressing appropriate T-cell receptors and thus still be immunogenic when escape variants are passed to individuals expressing the appropriate major histocompatibility complex class I molecules. To determine whether peptide ligands that have been altered through escape can be immunogenic in new hosts, we challenged naïve, immunocompetent macaques with a molecularly cloned simian immunodeficiency virus (SIV) bearing common escape mutations in three immunodominant CTL epitopes. Responses to the altered peptides were barely detectable in fresh samples at any time after infection. Surprisingly, CTL specific for two of three escaped epitopes could be expanded by in vitro stimulation with synthetic peptides. Our results suggest that some escape variant epitopes evolving in infected individuals do not efficiently stimulate new populations of CTL, either in that individual or upon passage to new hosts. Nevertheless, escape variation may not completely abolish an epitope's immunogenicity. Moreover, since the mutant epitope sequences did not revert to wild type during the study period, it is possible that low-frequency CTL exerted enough selective pressure to preserve epitope mutations in viruses replicating in vivo.
In recent years, there has been increasing interest in AIDS vaccine approaches that elicit cytotoxic T lymphocytes (CTL), which recognize and eliminate cells infected with human immunodeficiency virus (HIV) (26). Unlike antibodies, effective CTL responses can be directed against epitopes derived from any viral protein, raising the possibility that CTL can be targeted to regions that are more conserved than the viral envelope. Current vaccine modalities can elicit potent CTL responses against multiple viral epitopes (25). Indeed, many lines of evidence indicate that cell-mediated immunity plays a major role in control of virus replication. Several studies have suggested an association between certain major histocompatibility complex (MHC) class I and class II alleles and control of viral replication or susceptibility to disease (6, 7, 11, 12, 15-17, 28, 36, 38, 39). CTL are also implicated in the initial control of immunodeficiency virus infection, since they appear in close temporal association with the reduction in peak viremia in both HIV-infected humans (5, 22) and simian immunodeficiency virus (SIV)-infected macaques (23). Antibody-mediated depletion of CD8+ cells in infected macaques resulted in dramatically increased virus loads in both acute and chronic infection (14, 27, 37).
However, the plasticity of the viral genome also allows the generation of mutants that escape CTL recognition. Certain high-frequency CTL exert intense selective pressure on virus sequences, as revealed by the nearly total extinction of CTL-susceptible sequences from the actively replicating virus population within a few weeks of infection (2, 32). Escape from CTL has been observed in several studies of infected humans (12, 18, 21, 34, 35, 41) and macaques (2, 8, 30, 32, 40). Moreover, one report has shown that an HIV-1 escape mutant can be transmitted vertically (11), while other studies in vaccinated macaques have suggested that the evolution of escape mutants may be associated with a loss of containment of viral replication (4, 31). It therefore seems likely that escape from CTL responses occurs in most infected individuals (32).
The apparent ubiquity of CTL escape may greatly complicate the design of CTL-based vaccines. The evolution of escape variants during infection of a single host may play a key part in viral persistence and therefore in the ultimate failure of immune containment and progression to AIDS. However, some investigators have suggested that T-cell receptor repertoires can recognize multiple epitope variants, so that CTL responses can coevolve along with viral escape variants in infected individuals (13). If T-cell receptor populations can recognize new variant epitopes arising within a single host, it seems plausible that variant epitope sequences could also be recognized efficiently in new hosts. Escape could also create “neo-epitopes,” novel sequences that are immunogenic to naïve T cells in individuals expressing the appropriate MHC class I molecules.
The most rigorous test of the immunogenicity of epitopes altered through escape is to challenge a fully intact immune system with an escape mutant virus. Therefore, we identified common escape mutations that accumulated in immunodominant epitopes of SIVmac239 in infected macaques expressing the high-frequency MHC class I molecules Mamu-A*01 and Mamu-B*17. Together, these molecules bind three immunodominant CTL epitopes in SIVmac239: Gag181-189CM9 (CTPYDINQM, Gag CM9) and Tat28-35SL8 (STPESANL, Tat SL8) are bound by Mamu-A*01, and Nef165-173IW9 (IRYPKTFGW, Nef IW9) is bound by Mamu-B*17. We have previously shown that the acute-phase response in Mamu-A*01 Mamu-B*17 double-positive macaques is dominated by CTL that recognize these three epitopes (33). We introduced common escape mutations into the SIVmac239 molecular clone and challenged macaques expressing both Mamu-A*01 and Mamu-B*17 with the mutant virus. CTL responses directed against the mutant epitopes were extremely low frequency or undetectable in fresh samples from each of the infected animals. In the absence of these responses, a completely new immunodominance hierarchy was established. Our results suggest it is unlikely that “escaped” epitopes will be recognized in newly infected individuals expressing appropriate MHC class I molecules.
MATERIALS AND METHODS
Determination of consensus escape sequences.
Viral RNA was isolated from plasma obtained from macaques at the Wisconsin National Primate Research Center that were chronically infected with SIVmac239 that expressed either Mamu-A*01 (n = 11) or Mamu-B*17 (n = 4) with the QIAamp viral RNA minikit (Qiagen, Valencia, Calif.) by a modification of the manufacturer's protocol: 1 ml of plasma was centrifuged for 60 min at >17,000 × g in order to pellet virus, after which 860 μl of plasma was discarded, and virus was resuspended in the remaining volume. The manufacturer's protocol was then followed. The suggested volume of 140 μl of plasma. viral RNA was then amplified with the One-Step reverse transcription -PCR kit (Qiagen), generating amplicons that spanned regions of ≈500 bp around the three epitopes of interest. Cycling conditions were as follows: 50°C for 1 h, 95°C for 15 min, followed by 35 cycles of 95°C for 30 s, 55°C or 59°C for 30 s, and 68°C for 2 min, and an extension step of 68°C for 20 min. Amplicons produced in this manner were either directly sequenced or cloned into pCR2.1TOPO (Invitrogen, Carlsbad, Calif.) for sequencing on an ABI 377 automated sequencer. Consensus escape sequences were defined as the most common epitope sequence found in the population of clones from all Mamu-A*01- or Mamu-B*17-positive animals analyzed.
Virus stocks.
The construction and characterization of triple epitope mutant (3×) SIV has been described previously (9).
Animals and infections.
The animals in this study were Indian rhesus macaques (Macaca mulatta) from the Wisconsin National Primate Research Center colony. They were typed for MHC class I alleles Mamu-A*01, Mamu-A*02, Mamu-A*08, Mamu-A*11, Mamu-B*01, Mamu-B*03, Mamu-B*04, Mamu-B*17, and Mamu-B*29 by sequence-specific PCR (20). The animals were cared for according to the regulations and guidelines of the University of Wisconsin Institutional Animal Care and Use Committee. Animals were challenged intravenously with 100 50% tissue culture infectious doses of either wild-type or 3× SIV. Plasma virus load was determined by the branched-chain DNA assay (Bayer Diagnostics, Emeryville, Calif.).
Intracellular cytokine staining assay.
The intracellular cytokine staining assay was performed essentially as described in detail previously (40). Briefly, 500,000 freshly isolated peripheral blood mononuclear cells (PBMC) were incubated for 1.5 h at 37°C in 200 μl of R10 (RPMI 1640 containing 10% fetal calf serum and antibiotics) with anti-CD28 and anti-CD49d and synthetic peptides (pools of 15-mers or single peptides representing minimal optimal CTL epitopes) based on the wild-type or 3× SIV protein sequence. Then 10 μg of brefeldin A per ml was added to prevent protein transport from the Golgi apparatus, and the cells were incubated a further 5 h at 37°C. Cells were then washed and stained for surface expression of CD4 and CD8 markers and fixed overnight in 1% paraformaldehyde at 4°C. The following day, cells were permeabilized in buffer containing 1% saponin and stained for expression of the cytokines gamma interferon (IFN-γ) and interleukin-2 (IL-2) before being fixed in 1% paraformaldehyde for 2 h at 4°C. Events were then collected on a FACSCalibur flow cytometer (Becton Dickinson, San Jose, Calif.) with CellQuest software and analyzed with FlowJo 4.4.3for Macintosh (Treestar, Ashland, Oreg.).
MHC class I tetramer assay.
We stained 5 million cells (fresh PBMC or bulk CTL cultures) with 2 μg of tetramer for 1 h at 37°C in a volume of 200 μl of R10. Then 3μl of anti-human CD3-fluorescein isothiocyanate conjugate (BD Pharmingen, San Diego, Calif.) and 6 μl of anti-CD8-peridinin chlorophyll protein (Becton Dickinson) were added, and the mixture was incubated for a further 45 min at room temperature. Cells were then washed twice in R10 or flow cytometry buffer (phosphate-buffered saline containing 2% fetal bovine serum and 5% bovine serum albumin) and fixed for at least 30 min at 4°C in 1% paraformaldehyde. Flow cytometry data were collected and analyzed as above.
CTL lines.
Peptide-specific CTL were cultured from fresh whole blood samples with autologous and/or congeneic B-lymphoblastoid cell lines as stimulators. CD8-positive cells were enriched from whole blood with anti-CD8 whole-blood microbeads on an AutoMACS cell sorter (Miltenyi Biotec, Auburn, Calif.) according to the manufacturer's protocol. Stimulator B-lymphoblastoid cell lines were pulsed with 1 μg of peptide (corresponding to the Gag CM9, Tat SL8, or Nef IW9 sequence of wild-type or 3× SIV) 1 to 2 h at 37°C, washed, and irradiated (3,000 rads). Stimulators were then mixed with purified CD8-positive PBMC at a ratio of 2:1 B-lymphoblastoid cell lines to CD8 in R15 (RPMI 1640 containing 15% fetal calf serum and antibiotics) augmented with 10 ng of recombinant human interleukin-7 (Sigma, St. Louis, Mo.) per ml and incubated for 48 h. Cells were fed with R15 containing 50 U of IL-2 (proleukin; Chiron Therapeutica, Emeryville, Calif.) per ml every 3 to 5 days thereafter and restimulated with peptide-pulsed, irradiated B-lymphoblastoid cell lines every 7 to 10 days. Bulk CTL lines were first used in tetramer assays after 17 days in culture.
RESULTS
Identification of consensus CTL escape mutations and construction of 3× SIV.
In order to identify common escape mutations, we analyzed sequences of immunodominant CTL epitopes bound by the high-frequency MHC class I molecules Mamu-A*01 and Mamu-B*17 in macaques that expressed either of these molecules and were chronically infected with SIVmac239. We reasoned that sequences present during chronic infection, when virus loads were typically greater than 106 viral RNA copies/ml of plasma, would represent viruses that had both effectively escaped from CTL responses and attained sufficient replicative fitness to allow high-titer growth. The variant epitope sequences found most often in Mamu-A*01 or Mamu-B*17 single-positive macaques were chosen for use in our mutagenesis strategy (Table 1). Each of the mutant peptides showed at least a 90% reduction in binding to MHC class I. Indeed, almost all of the consensus epitope mutations were in residues thought to affect MHC-peptide binding. The only exception was the change at position 6 in the mutant Nef IW9 epitope (see Table 1), which likely affects T-cell receptor recognition. However, the mutations in the Gag CM9 and Nef IW9 epitopes do not occur in primary anchor residues and therefore do not completely eliminate peptide binding to MHC class I; the 50% inhibitory concentration for the mutant epitopes was 354 nM for CAPYDINQM and 339 nM for TRYPKIFGW (9). In contrast, the leucine-to-glutamine mutation in the Tat SL8 epitope sequence alters the C-terminal anchor of the peptide, essentially abolishing binding to Mamu-A*01 (50% inhibitory concentration of 25,535 nM) (9). We produced a cloned virus encoding all of these mutations (3× SIV), which attained titers similar to that of wild-type SIVmac239 in both tissue culture and infected macaques but appeared to have a fitness defect in comparison to SIVmac239 both in vitro and in vivo (9).
TABLE 1.
Epitopes and mutants
| Epitope | Sequence | MHC | Likely effect of mutation(s) |
|---|---|---|---|
| Gag CM9 | CTPYDINQM | A*01 | |
| Gag CM9 mutanta | CAPYDINQM | P2: secondary anchor | |
| Tat SL8 | STPESANL | A*01 | |
| Tat SL8 mutant | PTPESANQ | P1: binding? P8: C-terminal anchor | |
| Nef IW9 | IRYPKTFGW | B*17 | |
| Nef IW9 mutant | TRYPKIFGW | P1: secondary anchor? P6: TCR |
Substitutions within Gag CM9 were associated with isoleucine-to-valine substitutions upstream and downstream of the epitope sequence. Since these substitutions were likely required to restore fitness to the Gag CM9 escape virus (10), they were included in the mutagenesis strategy. We identified no extraepitopic mutations occurring in association with the consensus variants of Tat SL8 or Nef IW9.
3× SIV mutations alter immunodominance relationships in acute infection.
To determine whether the altered epitope sequences in 3× SIV could be recognized by immunocompetent, naïve hosts, we infected four Mamu-A*01 Mamu-B*17 double-positive macaques intravenously with 100 50% tissue culture infectious doses of the mutant virus. One Mamu-A*01 Mamu-B*17 double-positive macaque was infected by the same route with the same dose of wild-type SIVmac239 (Table 2). We measured the total cellular immune response to 3× SIV during acute infection by staining PBMC obtained at 4 weeks postinfection for intracellular cytokine accumulation upon stimulation with pools of overlapping synthetic peptides representing the entire SIVmac239 proteome. Acute-phase CTL in Mamu-A*01 Mamu-B*17 double-positive animals previously infected with wild-type SIVmac239 (n = 5) targeted an average of 9.6 peptide pools (range, 7 to 15) (33). Mamu-A*01 Mamu-B*17 double-positive animals infected with 3× SIV showed at least as broad a CTL repertoire, targeting between 10 and 20 distinct peptide pools (Fig. 1a).
TABLE 2.
Animals used in this study
| Animal | MHC class I genotypea | Virusb | Peak virus loadc (106 copies/ml) | Setpoint virus loadd (106 copies/ml) |
|---|---|---|---|---|
| B | A*01 A*02 B*17 B*29 | Wild type | 4.6 | 0.0515 |
| D | A*02 | 3× | 13.0 | 4.6e |
| E | A*01 A*11 B*01 B*17 B*29 | 3× | 4.7 | 1.3 |
| F | A*01 A*11 B*17 B*29 | 3× | 4.1 | 0.0096 |
| G | A*01 A*02 B*17 B*29 | 3× | 3.4 | 0.0244 |
| H | A*01 B*17 B*29 | 3× | 1.6 | 0.0006 |
Animals were typed for Mamu-A*01, -A*02, -A*08, -A*11, -B*01, -B*03, -B*04, -B*17, and-B*29 by PCR with sequence-specific primers (PCR-SSP) as described in Materials and Methods. The alleles listed gave positive results in replicate PCR-SSP assays.
Animals were inoculated intravenously with 100 50% tissue culture infections doses of wild-type SIVmac239 or epitope mutant SIVmac239 (3×).
Virus loads are shown as viral RNA copies per milliliter of plasma, as determined by the branched-chain DNA assay, and were reported previously (9).
Geometric mean virus load between 12 and 40 weeks postinfection, based on ≥5 separate time points.
Animal D died 30 weeks postinfection.
FIG. 1.
(a) Breadth of CTL responses to 3× SIV during acute infection. Freshly isolated PBMC were stimulated with pools of synthetic peptides representing the entire SIV proteome. The number of pools that stimulated IFN-γ production by CD8-positive lymphocytes is indicated for each viral protein. (b) Expected immunodominant CTL responses are not detected in acute 3× SIV infection. Frequencies of CTL (CD8+ IFN-γ+ lymphocytes) recognizing various SIV peptide pools and proteins were summed to give the total SIV-specific response. The percentage of the total response contributed by each specificity is shown for animals F, G, and H as pie graphs. The frequencies of CTL responding to each protein or pool are given in the table to the right of each graph. Unfortunately, data from 3× SIV-infected animal E are unavailable due to technical problems.
We have previously shown that acute-phase CTL populations in Mamu-A*01 Mamu-B*17 double-positive animals are dominated by cells specific for peptide pools containing the epitopes Gag CM9 (Gag162-211), Tat SL8 (Tat1-51), and Nef IW9 (Nef126-176), with an average of 50% of total SIV-specific CTL targeting these three pools (33). In contrast, we detected virtually no CTL targeting these regions in three Mamu-A*01 Mamu-B*17 double-positive animals infected with 3× SIV (Fig. 1b). The sole exception to this trend was a response to Tat1-51 in animal F, but this response is unlikely to be due to Tat SL8-specific CTL, since the epitope peptides (STPESANL or PTPESANQ) alone were not recognized (data not shown). Indeed, we detected no cytokine production above background levels when stimulating cells with individual peptides representing the wild-type or mutant epitope sequences for any of the three epitopes that were altered in 3× SIV (data not shown), indicating that the mutant epitopes are not effectively recognized even near the peak of the CTL response.
Instead of the expected immunodominant responses, we found a striking expansion of CTL specific for the Vif protein. In each animal for which data were available, between three and four Vif peptide pools were recognized, out of a total of five pools for the whole protein (Fig. 1a). Moreover, these Vif-specific CTL accounted for 19 to 71% of all CTL responding to 3× SIV (Fig. 1b). In animals F and G, Env was also frequently targeted by CTL that recognized four to eight pools and represented 28 to 39% of the total response. The MHC molecules restricting these CTL responses were not determined, but since no two animals responded to the same set of pools in Vif or Env, at least some of these responses are likely to be restricted by molecules other than Mamu-A*01 or Mamu-B*17.
Low-frequency T cells bind Gag CM9 and Nef IW9 tetramers.
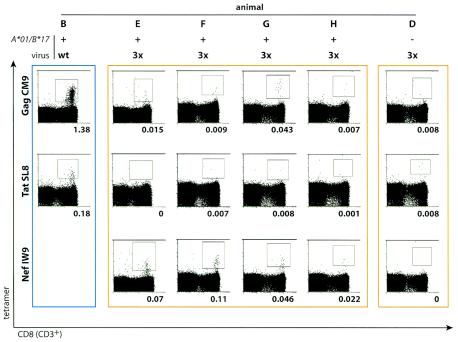
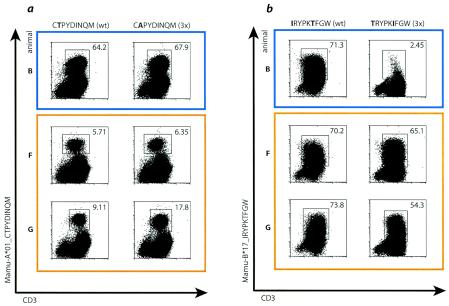
Since we could not detect CD8-positive cells reacting to the expected immunodominant peptides in a functional assay, we used another approach, staining fresh PBMC isolated during the acute phase (3 to 8 weeks postinfection) with MHC class I tetramers loaded with the wild-type peptides Gag CM9, Tat SL8, and Nef IW9. PBMC from animal D, which expressed neither Mamu-A*01 nor Mamu-B*17, were included in these experiments as a negative control. We were not able to produce tetramers loaded with the mutant peptides because in each case, the escape mutations resulted in a greater than 90% reduction in peptide binding to MHC class I. As expected, animal B, which was infected with wild-type SIVmac239, had a detectable population of CD3- and CD8-positive cells that bound the Gag CM9 tetramer (1.38%) (Fig. 2). We also detected small tetramer-positive populations in some macaques infected with 3× SIV. Animal G had small but distinct populations of cells that bound the tetramers loaded with Gag CM9 (0.043% CD3/CD8) and Nef IW9 (0.046% CD3/CD8). Animal F had a larger population of Nef IW9 tetramer-positive cells (0.11% CD3/CD8). Animals E and H also had small populations of CD3- and CD8-positive cells that bound the Nef IW9 tetramer. However, we could not detect staining above background with the Tat SL8 tetramer in any 3× SIV-infected animal. The lack of staining with the Tat SL8 tetramer in animal F also confirmed that IFN-γ secretion stimulated by the peptide pool Tat1-51 in this animal was not due to recognition of the Tat SL8 peptide.
FIG. 2.
Small populations of PBMC from 3× SIV-infected animals bind MHC class I tetramers folded with Gag CM9 or Nef IW9 peptides. Freshly isolated PBMC were obtained during acute infection and stained with MHC class I tetramers loaded with the wild-type peptide Gag CM9, Tat SL8, or Nef IW9. Numbers below each histogram show the percentage of CD3+ CD8+ lymphocytes that bound tetramer. The blue border indicates the animal infected with wild-type SIV; orange borders indicate animals infected with 3× SIV. Animals B, E, F, G, and H express Mamu-A*01 and Mamu-B*17. Animal D, infected with 3× SIV, expresses neither molecule and was included as a negative control.
Together, the tetramer data suggest that Mamu-A*01 Mamu-B*17 double-positive macaques had only small populations of CTL that recognized the altered sequences of the previously immunodominant peptides in the acute phase of 3× SIV infection, supporting our observations in the cytokine staining assay. These responses could have been primed by the mutant epitope sequences, a plausible conclusion because the infecting virus was clonal. On the other hand, it is also possible that low-frequency reversion of 3× SIV epitope sequences to wild-type epitope sequences primed responses to the wild-type epitope sequences. The tetramer and intracellular cytokine staining data alone cannot distinguish between these possibilities.
Lack of CTL responses to mutant epitopes in fresh PBMC during early chronic infection.
We measured the responses to mutant and wild-type epitopes after resolution of primary viremia by staining for cytokine production at 10 weeks postinfection. As expected, animal B, infected with wild-type SIVmac239, had high frequencies of CD8-positive cells that secreted IFN-γ in response to the wild-type peptides Gag CM9 (CTPYDINQM), Tat SL8 (STPESANL), and Nef IW9 (IRYPKTFGW) (Fig. 3, blue bars), in accord with our previous observations of Mamu-A*01 Mamu-B*17 double-positive macaques (33). Animals B and G, which both expressed Mamu-A*02, also recognized a Nef-derived epitope that we previously identified as immunodominant in early infection of Mamu-A*02-positive animals (40). Interestingly, cells from animal B also recognized the mutant peptides CAPYDINQM and, to a lesser extent, TRYPKIFGW. In contrast, there were almost no significant responses (frequency greater than twice background) to either the mutant or wild-type epitope sequences in the majority of animals infected with 3× SIV (Fig. 3). The sole exception was animal E, which had CD8-positive cells that recognized the inoculum (mutant) Nef epitope (TRYPKIFGW) and the wild-type epitope (IRYPKTFGW) almost equally well. Together, these data suggest that, while the mutant peptides appear to be poorly immunogenic, wild-type or 3× SIV-infected animals can harbor CTL populations that recognize the wild-type and mutant epitopes.
FIG. 3.
CTL that recognize mutant or wild-type sequences of altered epitopes are undetectable in 3× SIV-infected animals' PBMC. Synthetic peptides representing minimal optimal CTL epitopes were used to stimulate freshly isolated PBMC obtained 10 weeks postinfection. Animal B (blue bars), infected with wild-type SIV, made high-frequency CTL responses restricted by Mamu-A*01, Mamu-A*02, and Mamu-B*17. In contrast, none of the expected Mamu-A*01- or Mamu-B*17-restricted immunodominant responses were detected in animals infected with 3× SIV except for a strong response in animal E (yellow bars), which recognized the mutant and wild-type Nef IW9 peptides. Mamu-A*02-positive animal G (red bars) also recognized the wild-type Nef YY9 peptide.
Cellular immune responses to 3× SIV and viral evolution during chronic infection.
To determine whether extremely low frequency CTL specific for the altered peptides expanded over time, we stained PBMC isolated at ≥40 weeks postinfection for cytokine production upon stimulation with the set of peptides spanning the entire SIVmac239 proteome. Throughout the chronic phase, each animal except E maintained virus loads below 100,000 viral RNA copies/ml of plasma, in accord with our previous observations of animals that coexpress Mamu-A*01 and Mamu-B*17 (33). Animal E was sacrificed at 40 weeks postinfection with a virus load of >106 copies/ml. Responses in each infected animal were broadly specific, similar to our acute-phase observations. Animal B, infected with wild-type SIVmac239, recognized 17 pools in the CD8 compartment (Fig. 4a, blue bars). Among the animals infected with 3× SIV, E, G, and H recognized similar numbers of pools with CD8-positive cells (11 to 16 pools, Fig. 4a). In contrast, CD8-positive cells from animal F recognized only five pools. There appeared to be no relationship between the breadth (number of pools recognized) of CD8 responses and the level of setpoint viremia, similar to results obtained in a recent study of CTL responses in 57 HIV-infected subjects (1).
FIG. 4.
(a) Wild-type and 3× SIV-infected Mamu-A*01 Mamu-B*17 double-positive animals make broad CD8 T-cell responses during chronic infection. PBMC were isolated during late chronic infection (≥40 weeks postinfection) and stimulated in intracellular cytokine staining with peptide pools spanning the entire SIV proteome. The number of peptide pools in each protein that stimulated an IFN-γ response are totaled for each animal as in Fig. 1. Animal E PBMC were tested at necropsy, 40 weeks postinfection. (b) Contribution of chronic-phase CTL specificities to total SIV-specific response in 3× SIV-infected animals. IFN-γ responses were evaluated in intracellular cytokine staining as in Fig. 1b above.
In animal B, infected with wild-type SIVmac239, responses to pools containing the Tat SL8 and Nef IW9 epitopes had diminished below the level of detection, while Gag162-211-specific CTL frequencies remained above 1% of all CD8-positive lymphocytes, in accord with previous observations of Mamu-A*01- and Mamu-B*17-positive animals (Fig. 4b). The Tat SL8 and Nef IW9 epitope sequences accrued mutations in animal B during chronic infection; this likely accounts for the diminution in these epitope-specific CTL after 40 weeks (data not shown). In animals infected with 3× SIV, the Vif-specific CTL that dominated the acute-phase responses had diminished in chronic infection, perhaps as a result of escape mutations within Vif-derived CTL epitopes (data not shown). The overall frequencies of virus-specific CTL were also lower in 3× SIV-infected animals than in wild-type SIV-infected animal B.
Responses to the regions containing Gag CM9, Tat SL8, and Nef IW9 remained mostly undetectable during the late chronic phase in animals infected with 3× SIV (Fig. 4b). Escape mutations in these epitopes also persisted throughout the study period (9). Surprisingly, we observed an expansion of CTL reacting to peptide pools Gag162-211 and Nef126-176 in three out of four of these animals. However, all these reactivities cannot be accounted for solely by responses to the expected immunodominant epitopes. Responses to the wild-type and mutant Nef IW9 peptides were just above the threshold of detection in animals F and H (data not shown). In animal G, 0.01% of circulating CD8+ cells recognized Gag162-211, while the mutant epitope CAPYDINQM was recognized by 0.009% (data not shown). In contrast, there was no response to the wild-type peptide CTPYDINQM. Thus, responses to the altered peptides remained near the threshold of detection in fresh PBMC throughout 3× SIV infection.
In vitro stimulation reveals CTL populations that recognize mutant epitopes.
Since responses to the mutant epitopes were difficult to detect in freshly isolated PBMC, we further investigated the ability of 3× SIV-infected animals to recognize the mutant peptides by expanding CTL in vitro, stimulating purified CD8+ cells with B-lymphoblastoid cell lines expressing Mamu-A*01 and Mamu-B*17 loaded with the wild-type or mutant peptides. We cultured bulk CTL lines for at least 17 days and stained each line with tetramers as described above for fresh PBMC.
After 17 days in culture, we had expanded CTL from each 3× SIV-infected animal that bound Gag CM9 and Nef IW9 but not Tat SL8 tetramers. CTL lines grown from 3× SIV-infected animals bound tetramer whether they were stimulated with wild-type or mutant peptides, suggesting that the low-frequency CTL populations detected in fresh PBMC recognized both the autologous (mutant) and wild-type sequences (Fig. 5a and b). Additionally, both wild-type and mutant Gag CM9 peptides efficiently expanded CTL from wild-type SIV-infected animal B (Fig. 5a). In contrast, however, Nef IW9-specific CTL lines generated from animal B only bound tetramer if they had been raised with the wild-type peptide (Fig. 5b). These results suggest that, if animal B has populations of CTL that recognize the mutant Nef IW9 epitopes, these populations are distinct from those that recognize the autologous, wild-type epitopes. Moreover, the in vitro expansion of CTL from 3× SIV-infected animals that bind tetramers loaded with Gag CM9 and Nef IW9 confirms that CTL with these specificities are indeed present at a low frequency in these animals.
FIG. 5.
In vitro expansion of CTL from 3× SIV- and wild-type SIV-infected animals. CTL lines were raised from chronically infected animals as described in Materials and Methods. After 17 days in culture, cells were stained with anti-CD3antibody and tetramers folded with wild-type Gag CM9 or Nef IW9 peptide. The sequences of the peptides used to raise the CTL lines are shown above the panels. Blue borders show animal infected with wild-type SIV. Orange borders show animals infected with 3× SIV. (a) Wild-type (CTPYDINQM) and mutant (CAPYDINQM) peptides can effectively expand CTL from animals infected with either virus. (b) 3× SIV-infected animals harbor CTL populations that can be stimulated with peptides representing wild-type (IRYPKTFGW) or mutant (TRYPKIFGW) Nef IW9 sequences. However, only the wild-type peptide can effectively stimulate CTL from wild-type SIV-infected animal B.
DISCUSSION
Efforts to design antibody-based vaccines for HIV have been frustrated by the mutability of the viral envelope gene. Consequently, there has been increasing interest in developing effective CTL responses against AIDS viruses. Unfortunately, the same high rate of mutation and plasticity of the viral genome that result in escape from antibody responses allow the same extremely rapid escape from CTL. A key question, therefore, is whether escape gives rise to neo-epitopes that can still be recognized by CTL bearing the appropriate T-cell receptors. Here, we rigorously tested this possibility and have shown that viruses bearing common escape mutations in immunodominant epitopes interfere with the development of specific CTL responses in naïve hosts. Macaques infected with 3× SIV, which encoded escape mutations in epitopes predicted to be immunodominant, did not efficiently recognize the altered peptides. CTL specific for the mutant peptides were barely detectable in fresh peripheral blood. Since 3× SIV replicated to high titer during acute infection and 3× SIV-infected animals made other strong CTL responses, we reason that it is unlikely that these animals' poor reactivity to the altered epitopes can be explained by the fitness defect of 3× SIV, which we have examined in detail elsewhere (9). Interestingly, CTL that recognized both wild-type and mutant peptide sequences for two of three variant epitopes could be expanded in vitro from 3× SIV-infected macaques. Similarly, wild-type and mutant Gag CM9 peptides could stimulate CTL from animal B, infected with the wild-type virus. In contrast, only the wild-type peptide could stimulate the expansion of Nef IW9-specific CTL from this animal.
It was previously hypothesized that CTL responses and virus sequences could coevolve in individuals infected with HIV (13), but the degree to which epitope mutations could be recognized in new hosts remained unclear. Indeed, it is difficult to comprehensively address this question in HIV-infected humans. Since the vast majority of infected individuals are only identified after symptoms of retroviral infection appear, and because the viral inoculum itself likely comprises several viral quasispecies, it is difficult to unambiguously characterize the infecting sequence(s). Synthetic peptide sets used to assay T-cell responses may therefore differ from autologous viruses at critical residues, especially in variable regions of the viral proteome (3). Transmitted strains of HIV and SIV have also undergone selection at other sites, particularly those in env related to coreceptor usage. These changes may affect viral pathogenicity, and therefore the development of immune responses, upon transmission to new hosts (19, 24). We attempted to control for these variables as much as possible by infecting macaques with a molecularly cloned virus, 3× SIV, derived from the well-characterized SIVmac239. The use of clonal inoculum viruses also enabled us to measure cellular immune responses with synthetic peptides whose sequences corresponded exactly to the infecting viruses.
These results have several important implications for the understanding of T-cell responses to highly variable immunodeficiency viruses. First, they suggest that CTL epitope sequences evolving under CTL pressure in vivo are unlikely to be recognized when passed to individuals expressing the same or related MHC class I molecules. For example, HIV-1 escape variants that had been selected for by HLA-B27-restricted CTL in pregnant women were not effectively recognized by their infected HLA-B27-positive offspring (11). Furthermore, in animals infected with 3× SIV, we found little or no T-cell reactivity to the variant epitopes. These results suggest that, as the AIDS epidemic progresses, CTL epitopes bound by high-frequency MHC class I molecules, such as HLA-A2, might accrue enough escape mutations to be effectively lost from circulating virus strains. Indeed, recent studies have found evidence of such adaptive evolution of HIV on the population scale (29). It remains unclear whether, and to what extent, HIV-1 sequences are accumulating CTL escape mutations as the epidemic progresses, but the frequent evolution of escape variants may jeopardize vaccines that rely on induction of immunodominant CTL restricted by common HLA molecules unless escape mutations often revert upon transmission of viral variants to new hosts.
Interestingly, however, the failure to develop “canonical” immunodominant CTL responses in 3× SIV infection may not necessarily have grave consequences for control of virus replication. We have previously reported that animals expressing Mamu-A*01 and Mamu-B*17 are predisposed to unusually effective control of SIVmac239 infection (33). One might assume that the nearly total elimination of immunodominant responses in Mamu-A*01 Mamu-B*17 double-positive animals by infection with 3× SIV would prevent this control, but surprisingly, three of four 3× SIV-infected animals maintained virus loads below 100,000 copies/ml throughout the study period.
We have recently shown that escape from Gag CM9-specific CTL is associated with a catastrophic cost to viral fitness, which is partially overcome by mutations in the gag open reading frame outside the epitope sequence (10). Since 3× SIV encodes the same mutations in gag, it likely has a similar fitness disadvantage with respect to wild-type SIVmac239. It is therefore striking that epitope mutations were maintained in the Mamu-A*01 Mamu-B*17 double-positive animals. Selection in the 3× SIV-infected animals exerted by the low-frequency CTL specific for the previously immunodominant epitopes may preserve the engineered escape mutations. Maintenance of these mutations may keep the virus population in a state of suboptimal replicative fitness, rendering the infection easier to control via previously subdominant responses. In this model, reversion to CTL-susceptible epitope sequences may occur at a low frequency throughout infection, but revertant viruses would rapidly be eliminated by CTL that recognize the revertant epitopes.
We have shown that escape mutations arising in one host are unlikely to stimulate vigorous CTL responses when the variants are transmitted to new hosts with similar MHC alleles. CTL escape may thus represent a strategy of viral immune evasion that transcends the natural history of infection of a single host. Moreover, these results imply that one cannot determine a priori the immunodominant immune responses in an individual without knowing the sequence of the infecting virus. Vaccine strategies that rely on a few immunodominant CTL epitopes may therefore be doomed to failure, as escape mutations within these epitopes accumulate in circulating virus strains. However, this pessimistic view is potentially ameliorated by the surprising result that even very low frequency CTL can exert detectable selective pressure on immunodeficiency viruses. In fact, the ability of CTL to select for variants with reduced fitness may provide a criterion by which to evaluate the effectiveness of immune responses. If escape variation does exact a cost to overall viral fitness (9, 42), this cost could be exploited in the design of CTL-based vaccines. Vaccine approaches might be designed to include CTL responses that drive the virus population toward low replicative fitness by selecting for mutations whose effects are particularly deleterious. Additionally, potent CTL responses could be targeted to structurally or functionally constrained regions in which mutations would not be tolerated.
Acknowledgments
This work was supported by National Institutes of Health grants RO1-AI-46366, RO1-AI-49120, and RO1-AI-52056 to D.I.W. and P51 RR0001676-43to the WNPRC. D.I.W. is an Elizabeth Glaser Scientist.
REFERENCES
- 1.Addo, M. M., X. G. Yu, A. Rathod, D. Cohen, R. L. Eldridge, D. Strick, M. N. Johnston, C. Corcoran, A. G. Wurcel, C. A. Fitzpatrick, M. E. Feeney, W. R. Rodriguez, N. Basgoz, R. Draenert, D. R. Stone, C. Brander, P. J. Goulder, E. S. Rosenberg, M. Altfeld, and B. D. Walker. 2003. Comprehensive epitope analysis of human immunodeficiency virus type 1 (HIV-1)-specific T-cell responses directed against the entire expressed HIV-1 genome demonstrate broadly directed responses, but no correlation to viral load. J. Virol. 77:2081-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 3.Altfeld, M., M. M. Addo, R. Shankarappa, P. K. Lee, T. M. Allen, X. G. Yu, A. Rathod, J. Harlow, K. O'Sullivan, M. N. Johnston, P. J. Goulder, J. I. Mullins, E. S. Rosenberg, C. Brander, B. Korber, and B. D. Walker. 2003. Enhanced detection of human immunodeficiency virus type 1-specific T-cell responses to highly variable regions by using peptides based on autologous virus sequences. J. Virol. 77:7330-7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 5.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrington, M., and R. E. Bontrop. 2002. Effects of MHC class I on HIV/SIV disease in primates. AIDS 16(Suppl. 4):S105-S114. [DOI] [PubMed] [Google Scholar]
- 7.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748-1752. [DOI] [PubMed] [Google Scholar]
- 8.Evans, D. T., D. H. O'Connor, P. Jing, J. L. Dzuris, J. Sidney, J. da Silva, T. M. Allen, H. Horton, J. E. Venham, R. A. Rudersdorf, T. Vogel, C. D. Pauza, R. E. Bontrop, R. DeMars, A. Sette, A. L. Hughes, and D. I. Watkins. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino-acid variation in simian immunodeficiency virus env and Nef. Nat. Med. 5:1270-1276. [DOI] [PubMed] [Google Scholar]
- 9.Friedrich, T. C., E. J. Dodds, L. J. Yant, L. Vojnov, R. Rudersdorf, C. Cullen, D. T. Evans, R. C. Desrosiers, B. R. Mothe, J. Sidney, A. Sette, K. Kunstman, S. Wolinsky, M. Piatak, J. Lifson, A. L. Hughes, N. Wilson, D. H. O'Connor, and D. I. Watkins. 2004. Reversion of CTL escape-variant immunodeficiency viruses in vivo. Nat. Med. 10:275-281. [DOI] [PubMed] [Google Scholar]
- 10.Friedrich, T. C., C. A. Frye, L. J. Yant, D. H. O'Connor, N. A. Kriewaldt, M. Benson, L. Vojnov, E. J. Dodds, C. Cullen, R. Rudersdorf, A. L. Hughes, N. Wilson, and D. I. Watkins. 2004. Extraepitopic compensatory substitutions partially restore fitness to simian immunodeficiency virus variants that escape from an immunodominant cytotoxic-T-lymphocyte response. J. Virol. 78:2581-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 12.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland-Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 13.Haas, G., U. Plikat, P. Debre, M. Lucchiari, C. Katlama, Y. Dudoit, O. Bonduelle, M. Bauer, H. G. Ihlenfeldt, G. Jung, B. Maier, A. Meyerhans, and B. Autran. 1996. Dynamics of viral variants in HIV-1 Nef and specific cytotoxic T lymphocytes in vivo. J. Immunol. 157:4212-4221. [PubMed] [Google Scholar]
- 14.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8(+) T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, J. J. Goedert, C. Winkler, S. J. O'Brien, C. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405-411. [DOI] [PubMed] [Google Scholar]
- 16.Kaslow, R. A., C. Rivers, J. Tang, T. J. Bender, P. A. Goepfert, R. El Habib, K. Weinhold, and M. J. Mulligan. 2001. Polymorphisms in HLA class I genes associated with both favorable prognosis of human immunodeficiency virus (HIV) type 1 infection and positive cytotoxic T-lymphocyte responses to ALVAC-HIV recombinant canarypox vaccines. J. Virol. 75:8681-8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keet, I. P., J. Tang, M. R. Klein, S. LeBlanc, C. Enger, C. Rivers, R. J. Apple, D. Mann, J. J. Goedert, F. Miedema, and R. A. Kaslow. 1999. Consistent associations of HLA class I and II and transporter gene products with progression of human immunodeficiency virus type 1 infection in homosexual men. J. Infect. Dis. 180:299-309. [DOI] [PubMed] [Google Scholar]
- 18.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimata, J. T., L. Kuller, D. B. Anderson, P. Dailey, and J. Overbaugh. 1999. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat. Med. 5:535-541. [DOI] [PubMed] [Google Scholar]
- 20.Knapp, L. A., E. Lehmann, L. Hennes, M. E. Eberle, and D. I. Watkins. 1997. High-resolution HLA-DRB typing using denaturing gradient gel electrophoresis and direct sequencing. Tissue Antigens 50:170-177. [DOI] [PubMed] [Google Scholar]
- 21.Koenig, S., A. J. Conley, Y. A. Brewah, G. M. Jones, S. Leath, L. J. Boots, V. Davey, G. Pantaleo, J. F. Demarest, C. Carter, et al. 1995. Transfer of HIV-1-specific cytotoxic T lymphocytes to an AIDS patient leads to selection for mutant HIV variants and subsequent disease progression. Nat. Med. 1:330-336. [DOI] [PubMed] [Google Scholar]
- 22.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuroda, M. J., J. E. Schmitz, W. A. Charini, C. E. Nickerson, M. A. Lifton, C. I. Lord, M. A. Forman, and N. L. Letvin. 1999. Emergence of CTL coincides with clearance of virus during primary simian immunodeficiency virus infection in rhesus monkeys. J. Immunol. 162:5127-5133. [PubMed] [Google Scholar]
- 24.Liu, S. L., J. E. Mittler, D. C. Nickle, T. M. Mulvania, D. Shriner, A. G. Rodrigo, B. Kosloff, X. He, L. Corey, and J. I. Mullins. 2002. Selection for human immunodeficiency virus type 1 recombinants in a patient with rapid progression to AIDS. J. Virol. 76:10674-10684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McMichael, A. J., and T. Hanke. 2003. HIV vaccines 1983-2003. Nat. Med. 9:874-880. [DOI] [PubMed] [Google Scholar]
- 26.McMichael, A. J., and S. L. Rowland-Jones. 2001. Cellular immune responses to HIV. Nature 410:980-987. [DOI] [PubMed] [Google Scholar]
- 27.Metzner, K. J., X. Jin, F. V. Lee, A. Gettie, D. E. Bauer, M. Di Mascio, A. S. Perelson, P. A. Marx, D. D. Ho, L. G. Kostrikis, and R. I. Connor. 2000. Effects of In vivo CD8(+) T cell depletion on virus replication in rhesus macaques immunized with a live, attenuated simian immunodeficiency virus vaccine. J. Exp. Med. 191:1921-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 97:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 30.Mortara, L., F. Letourneur, H. Gras-Masse, A. Venet, J. G. Guillet, and I. Bourgault-Villada. 1998. Selection of virus variants and emergence of virus escape mutants after immunization with an epitope vaccine. J. Virol. 72:1403-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mortara, L., F. Letourneur, P. Villefroy, C. Beyer, H. Gras-Masse, J. G. Guillet, and I. Bourgault-Villada. 2000. Temporal loss of Nef-epitope CTL recognition following macaque lipopeptide immunization and SIV challenge. Virology 278:551-561. [DOI] [PubMed] [Google Scholar]
- 32.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493-499. [DOI] [PubMed] [Google Scholar]
- 33.O'Connor, D. H., B. R. Mothe, J. T. Weinfurter, S. Fuenger, W. M. Rehrauer, P. Jing, R. R. Rudersdorf, M. E. Liebl, K. Krebs, J. Vasquez, E. Dodds, J. Loffredo, S. Martin, A. B. McDermott, T. M. Allen, C. Wang, G. G. Doxiadis, D. C. Montefiori, A. Hughes, D. R. Burton, D. B. Allison, S. M. Wolinsky, R. Bontrop, L. J. Picker, and D. I. Watkins. 2003. Major histocompatibility complex class I alleles associated with slow simian immunodeficiency virus disease progression bind epitopes recognized by dominant acute-phase cytotoxic-T-lymphocyte responses. J. Virol. 77:9029-9040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, et al. 1991. Human immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453-459. [DOI] [PubMed] [Google Scholar]
- 35.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saah, A. J., D. R. Hoover, S. Weng, M. Carrington, J. Mellors, C. R. Rinaldo, Jr., D. Mann, R. Apple, J. P. Phair, R. Detels, S. O'Brien, C. Enger, P. Johnson, and R. A. Kaslow. 1998. Association of HLA profiles with early plasma viral load, CD4+ cell count and rate of progression to AIDS following acute HIV-1 infection. Multicenter AIDS Cohort Study. AIDS 12:2107-2113. [DOI] [PubMed] [Google Scholar]
- 37.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 38.Tang, J., C. Costello, I. P. Keet, C. Rivers, S. Leblanc, E. Karita, S. Allen, and R. A. Kaslow. 1999. HLA class I homozygosity accelerates disease progression in human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retroviruses 15:317-324. [DOI] [PubMed] [Google Scholar]
- 39.Tang, J., S. Tang, E. Lobashevsky, A. D. Myracle, U. Fideli, G. Aldrovandi, S. Allen, R. Musonda, and R. A. Kaslow. 2002. Favorable and unfavorable HLA class I alleles and haplotypes in Zambians predominantly infected with clade C human immunodeficiency virus type 1. J. Virol. 76:8276-8284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogel, T. U., T. C. Friedrich, D. H. O'Connor, W. Rehrauer, E. J. Dodds, H. Hickman, W. Hildebrand, J. Sidney, A. Sette, A. Hughes, H. Horton, K. Vielhuber, R. Rudersdorf, I. P. De Souza, M. R. Reynolds, T. M. Allen, N. Wilson, and D. I. Watkins. 2002. Escape in one of two cytotoxic T-lymphocyte epitopes bound by a high-frequency major histocompatibility complex class I molecule, Mamu-A*02: a paradigm for virus evolution and persistence? J. Virol. 76:11623-11636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolinsky, S. M., B. T. Korber, A. U. Neumann, M. Daniels, K. J. Kunstman, A. J. Whetsell, M. R. Furtado, Y. Cao, D. D. Ho, and J. T. Safrit. 1996. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science 272:537-542. [DOI] [PubMed] [Google Scholar]
- 42.Yang, O. O., P. T. Sarkis, A. Ali, J. D. Harlow, C. Brander, S. A. Kalams, and B. D. Walker. 2003. Determinants of HIV-1 mutational escape from cytotoxic T lymphocytes. J. Exp. Med. 197:1365-1375. [DOI] [PMC free article] [PubMed] [Google Scholar]