Abstract
Regulation of chromatin through histone acetylation is an important step in gene expression. The Gcn5 histone acetyltransferase is part of protein complexes, e.g., the SAGA complex, that interact with transcriptional activators, targeting the enzyme to specific promoters and assisting in recruitment of the basal RNA polymerase transcription machinery. The Ada2 protein directly binds to Gcn5 and stimulates its catalytic activity. Drosophila contains two Ada2 proteins, Drosophila Ada2a (dAda2a) and dAda2b. We have generated flies that lack dAda2b, which is part of a Drosophila SAGA-like complex. dAda2b is required for viability in Drosophila, and its deletion causes a reduction in histone H3 acetylation. A global hypoacetylation of chromatin was detected on polytene chromosomes in dAda2b mutants. This indicates that the dGcn5-dAda2b complex could have functions in addition to assisting in transcriptional activation through gene-specific acetylation. Although the Drosophila p53 protein was previously shown to interact with the SAGA-like complex in vitro, we find that p53 induction of reaper gene expression occurs normally in dAda2b mutants. Moreover, dAda2b mutant animals show excessive p53-dependent apoptosis in response to gamma radiation. Based on this result, we speculate that dAda2b may be necessary for efficient DNA repair or generation of a DNA damage signal. This could be an evolutionarily conserved function, since a yeast ada2 mutant is also sensitive to a genotoxic agent.
The Ada2 protein was first described for yeast, in which its inactivation could relieve the toxicity caused by overexpression of the strong transcriptional activation domain in the viral VP16 protein (6). This provided one of the earliest pieces of evidence for the existence of transcriptional adapters or coactivators. Ada2 proteins were subsequently shown to exist in protein complexes containing the Gcn5 histone acetyltransferase (23). Histone acetylation has been linked to transcriptional activation for a long time, given the observation that actively transcribed regions tend to be hyperacetylated, whereas transcriptionally silent regions generally are hypoacetylated (3). With the identification of the first histone acetyltransferase (HAT) as the Tetrahymena homolog of Gcn5 (13) and the subsequent demonstration that other transcriptional regulators, such as the coactivators p300 and CBP, steroid receptor coactivators, and the TFIID subunit TAF1, also possess intrinsic HAT activity, the link between acetylation and transcription was firmly established (12). In addition, an important role for histone acetylation in DNA replication and repair is emerging (27, 38).
Gcn5 is evolutionarily conserved and is present in several multiprotein complexes, such as the SAGA and ADA complexes in yeast and the PCAF, STAGA, and TFTC complexes in mammals (17). In yeast, Gcn5 is contained in a trimeric subcomplex with the Ada2 and Ada3 proteins within SAGA and ADA complexes (21, 24). Ada2 binds to Gcn5, and deletion of Ada2 results in a loss of Gcn5 from both SAGA and ADA complexes (21). Ada2 contains a zinc finger ZZ domain (40) and a SANT domain (1). The Ada2 SANT domain stimulates the HAT activity of Gcn5 and is involved in substrate recognition (9, 47). Transcriptional activators are believed to recruit SAGA to target genes by interacting with the Tra1 subunit (11). The SAGA complex also facilitates TATA binding protein interaction with the promoter through the Spt3 subunit (20, 29, 46).
The Drosophila melanogaster genome contains two Ada2 genes, Drosophila Ada2a (dAda2a) and dAda2b, and the corresponding proteins are present in different Gcn5-containing complexes (28, 36). A Drosophila SAGA-like complex contains dAda2b but not the dAda2a protein (28, 36). We have taken a genetic approach to analyze the function of the dAda2b protein in the development of a multicellular organism. No other SAGA-specific mutant has yet been described for Drosophila. We find that dAda2b is required for viability and normal histone H3 acetylation levels. In the absence of dAda2b, we observe increased apoptosis in response to radiation-induced DNA damage. We speculate that Ada2/Gcn5 may affect DNA damage recognition or repair, since yeast strains lacking Ada2/Gcn5 are also sensitive to a genotoxic agent.
MATERIALS AND METHODS
Plasmid DNA.
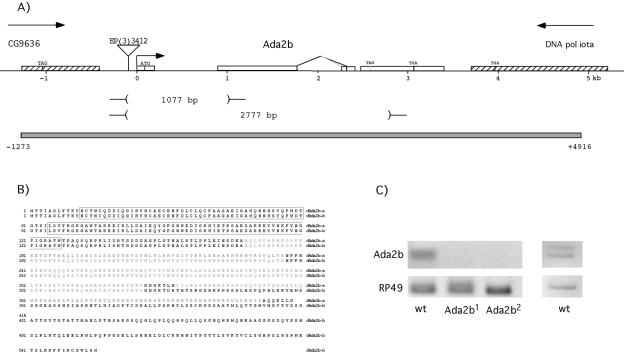
The entire expressed sequence tag (EST) clone LD24527 has been sequenced by the Berkeley Drosophila Genome Project and contains a full-length dAda2b isoform a cDNA (dAda2b-a). T. Kusch and J. Workman kindly provided us with a dAda2b isoform b cDNA. We generated a dAda2b genomic rescue construct as follows. The bacterial artificial chromosome clone BACR45A07 was digested with BamHI, and the fragments were subcloned into the pBluescript vector. Clones containing the dAda2b gene were selected by colony hybridization with a dAda2b-a cDNA probe. A fragment extending from 1,273 nucleotides upstream of the dAda2b start site to 1,521 nucleotides downstream of the last dAda2b exon was subcloned into the P-element vector pCaSper 4 by using the restriction enzymes XhoI and NotI. This sequence encompasses the entire last exon of the upstream gene CG9636, the complete dAda2b locus, and 1,221 nucleotides from the last exon in the downstream gene DNA pol iota (Fig. 1).
FIG. 1.
The Drosophila Ada2b locus. (A) The dAda2b locus is drawn to scale. It spans 3.4 kb and contains four exons (open boxes). The alternative splicing that occurs in exon 3 is indicated, together with the stop codons in exon 4 that terminate the alternative reading frames. Stippled boxes indicate the last exons of the flanking genes CG9636 and DNA pol iota. The location of the P-element EP(3)3412 is indicated by a triangle. Two of the deletions generated by imprecise excision of this P element are shown. The gray box indicates the genomic sequence present in the transgene that rescues the dAda2b alleles. (B) Comparison of the two protein isoforms generated by alternative splicing. Isoform a is present in the EST clone LD 24527 and is the same protein sequence as described in reference 36. The b isoform was described in reference 28. The conserved ZZ and SANT domains are boxed, and Ada boxes 1 to 3 are shaded gray. (C) RT-PCR results with RNA prepared from wild-type (wt) or dAda2b1 or dAda2b2 homozygous mutant pupae are shown at the left. The upper panel shows 930- and 1,008-bp products generated with an Ada2b primer pair, and the lower panel shows the RP49 product (control). With a different primer pair, the alternative splice site usage in dAda2b exon 3 is evident (upper right panel). Isoform a generates a 262-bp product, whereas isoform b gives rise to a 184-bp product.
A dGcn5 full-length cDNA was assembled by fusing a 5′ fragment from EST clone LD05304 with a 3′ fragment from EST clone LD31889 at a unique and common XhoI site. The dGcn5 open reading frame was amplified by PCR and cloned into the BamHI-EcoRI sites of the pGEX 5X-1 vector to generate a glutathione S-transferase (GST)-dGCN5 construct.
Generation of dAda2b mutants and P-element transformation.
The P element EP(3)3412 is inserted 87 bp upstream of the dAda2b gene, as determined by the Berkeley Drosophila Genome Project. We mobilized the P element by using a transposase source and scored for loss of the white marker gene. One hundred fifty independently derived white-eyed strains were tested for complementation with deficiency 3(R)dsx5, which has a deletion that includes dAda2b. Fourteen lethal lines were established, including the dAda2b1 and dAda2b2 alleles used in this study. The dAda2b alleles were balanced over TM3,Sb, and over TM3,Ubx-lacZ, TM3,Ubi-GFP, or TM6,Tb to enable selection of homozygous mutant embryos and larvae. The dAda2b alleles were also recombined onto an FRT-containing chromosome, and germ line clones were generated as previously described (18). No eggs were obtained. However, the lethality and fertility of the FRT dAda2b stock could be rescued by the genomic rescue transgene, demonstrating that dAda2b is required for oogenesis. The genomic rescue construct was injected into w1118 flies according to standard procedures (43), and an insertion on the X chromosome was used in rescue experiments.
Southern blotting and PCR.
The 14 lethal strains obtained by the imprecise-excision scheme were characterized by Southern blotting and PCR. Genomic DNAs were prepared from the wild type and the 14 lethal strains; digested with either NdeI, HindIII, or BamHI; transferred to nylon filters; and probed with a 2.0-kb EcoRI-BglII fragment containing the full-length dAda2b-a cDNA by standard procedures. To identify the deletion breakpoints, PCR on genomic DNA was performed with the following primer pairs. Primer Ada2-2 (5′-AGTTTTTAATCCTGACCACCGC, bp −588 to −610 relative to the dAda2b transcription start site) was used in conjunction with primer Ada2-6 (5′-GTTCCGGTCACCTAACACCTC, bp +1938 to +1958 relative to the transcription start site) to amplify the dAda2b1 allele. The dAda2b2 allele was amplified with primers Ada2-2 and Ada2-7 (5′-ACATATCCTAGCGGCATTCAGTC, bp +3469 to + 3492 relative to + 1). The PCR products were sequenced with primer Ada2-2 to determine the deletion endpoints.
RT-PCR.
Total RNA was isolated from w1118 (wild type) and Ada2b1 and Ada2b2 mutant pupae by use of TRIzol reagent (Invitrogen). Five micrograms of total RNA was reverse transcribed with SuperScript II RNase H-free reverse transcriptase (RT) (Invitrogen). A dAda2b 3′-end-specific primer (Ada2 RT-F1, 5′-GTGCAAGGAAGAGCTTGGCG) and the ribosomal protein 49 (RP49) 3′-end-specific primer (RP49-R, 5′-TCTCGCCGCAGTAAAC) were used for first-strand synthesis. PCR was performed with Ada2b primers Ada2 RT-F1 (located in exon 3) and the forward primer Ada2 RT-R1 (5′-GAACTGCGCATTCGACAGTCAG, located at the end of exon 2) or Ada2 RT-R3 (5′-ATTCGAGTGCACTGCGCGGAG, located in exon 1) and with the RP49 primer pair RP49-R and RP49-F (5′-TGACCATCCGCCCAGCATACA) for 20 cycles at an annealing temperature of 56°C.
In situ hybridization and immunohistochemistry.
Embryos were collected from dAda2b alleles balanced over TM3,Ubx-lacZ and from Oregon R or w1118 flies (wild-type controls) on apple juice plates and aged appropriately. Ovaries (10 per probe) were dissected in Ringer's solution, fixed in 4% paraformaldehyde, and devitellinized in methanol. RNA in situ hybridization with digoxigenin-labeled antisense RNA probes was performed as previously described (25, 48).
Immunohistochemistry was performed essentially as described previously (44). In brief, embryos were dechorionated in bleach, fixed in 4% formaldehyde, and divitellinized in methanol. Embryos were blocked in block solution (0.5% bovine serum albumin and 0.1% Triton X-100 in phosphate-buffered saline [PBS]) and incubated with primary antibodies at 4°C overnight. The primary antibodies used were rat dAda2b (1:200; kindly provided by T. Kusch and J. Workman), rabbit Ac-H3 K9 (1:200, Upstate), rabbit Ac-H3 K14 (1:100, Upstate), rabbit tetra-Ac-H4 (1:200; Upstate), and rabbit Ac-H4 K8 (1:100; Upstate). To distinguish dAda2b homozygous embryos, a mouse β-galactosidase antibody (1:2,000; Promega), recognizing LacZ expressed on the balancer chromosome, was mixed with the primary antibodies. Embryos were washed in block solution and incubated with Cy3-conjugated anti-rat (1:200) or anti-rabbit (1:500) and Cy2-conjugated anti-mouse (1:200) secondary antibodies (Jackson Laboratories) at room temperature for 2 h. After being washed in PBS-0.1% Triton X-100 solution, embryos were rinsed in PBS-50% glycerol and then mounted in 80% glycerol.
Polytene stainings.
Third-instar larvae from wild-type (Oregon R) or Ada2/Ada2 homozygous larvae were dissected in PBS-0.1% Triton X-100. Polytene chromosomes were prepared and stained essentially as described previously (51). Salivary glands were fixed in 2% formaldehyde in PBS-0.1% Triton X-100-0.2% NP-40 for 30 s, followed by 2 min in 50% acetic acid-1% formaldehyde. Polytene chromosomes were chromosomes as described previously (51). The slides were washed for 30 min in PBS-0.1% Triton X-100, transferred to blocking solution (0.1 M maleic acid, 0.15 M NaCl, 1% Boehringer blocking reagent), and incubated for 30 min at room temperature. The slides were then incubated overnight at 4°C with a rabbit polyclonal primary antibody (rabbit Ac-H3 K9 [1:200; Upstate], rabbit Ac-H3 K14 [1:200; Upstate], rabbit tetra-Ac-H4 [1:20; Upstate], or rabbit Ac-H4 K8 [1:75; Upstate]). The slides were washed twice for 10 min each in 0.1 M maleic acid-0.15 M NaCl-0.3% Tween 20 and blocked for 30 min. As a secondary antibody, a donkey anti-rabbit antibody conjugated with Cy3 was used (Jackson Laboratories); it was diluted 1:400 and incubated in room temperature for 2 h. The squashes were counterstained with DAPI (4′,6′-diamidino-2-phenylindole) (1 μg/ml) and washed twice for 10 min each before mounting with Vectashield (Vector). Several nuclei from at least three slides of each type were analyzed by using a Zeiss Axiophot microscope equipped with a KAPPA DX30C charge-coupled device camera.
GST pull-down assays.
Expression of the GST-dGcn5 fusion protein in Escherichia coli BL21(pLysS) cells was induced with IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 37°C, followed by purification on glutathione-Sepharose beads (Amersham Pharmacia). The dAda2b-a cDNA was in vitro translated with the TNT T7 coupled reticulocyte lysate system (Promega) in the presence of in vitro translation grade redivue l-[35S]methionine (Amersham Pharmacia). In vitro-translated dAda2b protein (20 μl) was mixed with equal amounts of purified GST or GST-dGcn5 protein on glutathione-Sepharose beads and incubated at room temperature for 1 h. The beads were washed three times in NTEN buffer (20 mM Tris [pH 8.0], 100 mM NaCl, 1 mM EDTA, 0.5% NP-40). Proteins bound to the beads were eluted by addition of sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer and analyzed by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis followed by exposure to an FLA 3000 phosphorimager (Fuji).
Radiation-induced apoptosis assay and reaper enhancer detection.
Wild-type, dAda2b, p53, or dAda2b p53 homozygous mutant third-instar larvae were mock treated or X irradiated with 800 or 4,000 rads. After a 4-h recovery at room temperature, wing imaginal disks were dissected, directly incubated in a solution of PBS and 1.6 μg of acridine orange per ml for 5 min, and then washed in PBS twice. Samples were mounted in PBS. Images were obtained on a Zeiss Axioplan 2 conventional fluorescence microscope. At the lower radiation dose, fewer cells entered apoptosis, but the difference between wild-type and mutant tissue was more readily detected. The number of apoptotic cells per disk after 800 rads of irradiation was counted in three disks of each genotype.
We generated the dAda2b p53 recombinant chromosome by crossing the dAda2b2 allele with the p53 null p535A-1-4. Progeny were crossed to dAda2b/TM6 Tb to check for the presence of dAda2b. Stocks were established, and the presence of the p53 mutation was assayed by PCR with primers p53-C1 and p53-C2, described in a personal communication to Flybase from Kent Golic (http://flybase.org/). Two dAda2b p53/TM6 Tb stocks were examined for apoptosis following irradiation.
A heatshock-reaper (hs-reaper) transgene (kindly provided by Hermann Steller) was used to induce apoptosis independently of DNA damage. An insertion on the second chromosome was crossed into dAda2b mutants. Flies of the genotype hs-reaper/+; dAda2b/TM6 Tb were used to generate dAda2b/dAda2b homozygous mutant larvae containing the hs-reaper transgene and compared with larvae from the homozygous hs-reaper stock. To induce reaper expression, wandering third-instar larvae were heat shocked at 37°C for 15 min and left to recover for 1 h at 25°C, and then the wing imaginal disks were dissected. The disks were incubated in acridine orange as described above, and the number of apoptotic cells in three disks of each genotype was counted. To ensure that transgene copy number did not influence our results, we repeated the experiment with a stock homozygous for hs-reaper (hs-reaper; dAda2b/TM6 Tb), and we found no difference from the wild-type hs-reaper stock.
The reaper enhancer transgene contains a 150-bp radiation-responsive enhancer from the reaper gene, including a p53 response element, which drives lacZ reporter gene expression (10). An insertion on the second chromosome was crossed into dAda2b mutant flies. Embryos were collected, either mock treated or X irradiated with 4,000 rads, left to recover for 3 h at room temperature, and fixed as previously described (32). The embryos were stained with a rabbit β-galactosidase antibody (1:300; Cappel) that recognizes LacZ expressed from the reaper transgene as well as Ubx-lacZ present on the balancer chromosome, using the Vectastain ABC Elite kit.
Yeast MMS assay.
A Saccharomyces cerevisiae ada2 mutant strain (Δada2 his3Δ200 leu2Δ1 ura3-52 trp1Δ) and its isogenic wild-type parent were kindly provided by S. Berger. The gcn5 and ada3 mutant strains were obtained from the Research Genetics collection of deletion mutants (52) and compared to the wild-type strain BY4743 (MATa/MATα his3Δ 1his3Δ1 leu2Δ/leu2Δ ura3Δ/ura3Δ met15Δ/MET15 lys2Δ/LYS2). Tenfold serial dilutions of log-phase cultures were spotted onto yeast extract-peptone-dextrose medium lacking methyl methanesulfonate (MMS) or containing 0.02% MMS. Pictures of the plates were taken after 3 days at 30°C.
RESULTS
Generation of dAda2b mutants.
The dAda2b gene is located at cytological position 84F4 and consists of four exons spanning 3.4 kb (Fig. 1A). A P-element transposon insertion 87 nucleotides upstream of the dAda2b putative transcription start site has been mapped by the Berkeley Drosophila Genome Project. This insertion [EP(3)3412] is homozygous viable and does not cause any visible phenotype. We generated imprecise excisions of the P element by introduction of a transposase source, which resulted in 14 lethal lines. Southern blotting and PCR analysis of genomic DNA showed that eight of these have disruptions in the upstream gene CG9636, whereas six of the lethal lines contain deletions in the dAda2b gene (Fig. 1 and data not shown). The dAda2b1 allele contains a 1,077-bp deletion encompassing the transcription start site and the entire first exon, including the translation start codon, whereas the dAda2b2 allele contains a 2.77-kb deletion spanning almost the entire gene.
Comparison of EST sequence data with the published cDNA sequence (28) indicated that two alternative splice acceptor sites might be used in the third exon. We performed RT-PCR experiments with RNAs prepared from embryos, larvae, pupae, and S2 tissue culture cells. We found that both splice sites are utilized, but we observed no difference in splice site usage during development (Fig. 1C and data not shown). The alternative splice sites result in different reading frames and generate proteins of 418 and 555 amino acids with divergent carboxy termini (Fig. 1B).
We determined the lethal phase of dAda2b mutants by use of a green fluorescent protein-expressing balancer chromosome that allowed us to select homozygous mutant animals. dAda2b1 and dAda2b2 homozygotes die during early pupal stages, most often before head eversion occurs, without any other gross morphological abnormalities (data not shown). Ada2b mRNA was undetectable in homozygous mutant pupae (Fig. 1C), suggesting that dAda2b1 and dAda2b2 are null alleles.
A transgenic line containing a genomic piece of DNA encompassing the dAda2b locus fully rescues the lethality of the dAda2b alleles, demonstrating that the lethality is caused by loss of dAda2b gene product (Fig. 1A).
Embryonic expression of dAda2b and dGcn5.
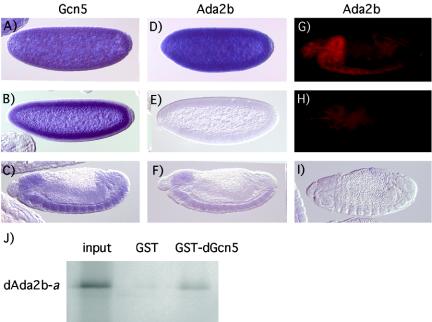
Ada2 proteins are present in many Gcn5-containing complexes. To confirm that Drosophila Ada2b and Gcn5 are able to interact, a GST pull-down assay was performed. As shown in Fig. 2J, bacterially expressed GST-dGcn5 efficiently binds to in vitro-translated dAda2b protein. We next compared the embryonic expression pattern of dAda2b to that of dGcn5 by in situ hybridization. Both dAda2b and dGcn5 are maternally contributed, as their transcripts can be detected at the precellular stage (Fig. 2A and D). We confirmed this finding by staining ovaries with the dAda2b and dGcn5 probes. Both transcripts are present in nurse cells of stage 10 egg chambers and are dumped in the oocyte when the nurse cells degenerate at late oogenesis (data not shown).
FIG. 2.
Drosophila Ada2b and Gcn5 interact and are expressed in similar patterns during embryogenesis. (A to I) Lateral views of embryos hybridized with digoxigenin-labeled RNA probes or with a dAda2b antibody. Anterior is to the left and dorsal is up. (A to C) Hybridization of a dGcn5 probe to wild-type embryos. Precellular (A), cellularized (B), and advanced-stage embryos (C) are shown. In early embryos the message is ubiquitously distributed due to its maternal contribution. In late-stage embryos, expression is strongest in the CNS, but is also present in the gut. (D to F) Wild-type embryos incubated with a dAda2b probe. Ubiquitous staining is observed in precellular embryos (D). Maternal dAda2b mRNA is absent from cellularized embryos (E). In advanced-stage embryos, expression is strongest in the CNS, but low levels of message are also found in the gut (F). (G and H) Stage 16 embryos stained with a dAda2b antibody. In wild-type embryos (G), strong staining is observed in the CNS. Most of the dAda2b protein has disappeared by this stage of development in dAda2b mutant embryos (H). (I) In dAda2b mutant embryos, no zygotic dAda2b mRNA can be detected. (J) Interaction between dAda2b and dGcn5 proteins in vitro. The dAda2b isoform a cDNA was in vitro translated and incubated with GST (control) or a GST-dGcn5 fusion protein. Ten percent of the input was run on a gel together with eluates of GST and GST-dGcn5 resins. The dAda2b-a protein is efficiently retained by GST-dGcn5.
In wild-type embryos undergoing cellularization, no maternal dAda2b mRNA remained, whereas that of dGcn5 persisted (Fig. 2B and E). In advanced-stage embryos, zygotic expression of both dAda2b and dGcn5 was strongest in the central nervous system (CNS) and was present at lower levels in most other tissues (Fig. 2C and F). These results suggest that dAda2b and dGcn5 are found in many of the same cells during embryo development. As expected, in dAda2b homozygous embryos, no dAda2b mRNA could be detected after stage 5, when the maternal message is no longer present (Fig. 2I and data not shown). On the other hand, we found that the maternal protein persists until late in stage 15, at which point reduced dAda2b protein levels are found in homozygous mutant embryos (Fig. 2H). In order to investigate the contribution of dAda2b to early embryo development, we attempted to remove maternal dAda2b in germ line clones. However, dAda2b homozygous germ cells arrest at an early stage of oogenesis, and for this reason we were unable to assess dAda2b function in early embryo development.
Reduced histone H3 acetylation levels in dAda2b mutant animals.
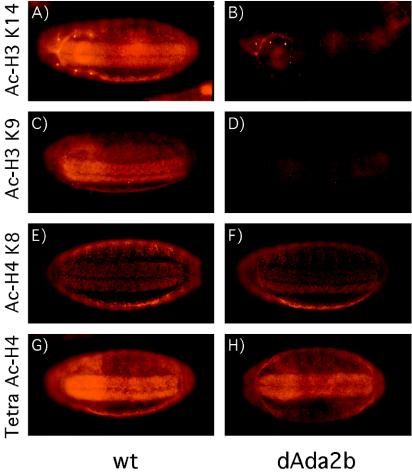
Since yeast Ada2 is required for maintaining Gcn5 in the SAGA complex and stimulates the catalytic activity of Gcn5, we investigated histone acetylation levels in dAda2b mutants. We performed whole-mount immunostainings of Drosophila embryos by using antibodies directed against different acetylated forms of histone H3 and histone H4. No difference between wild-type and dAda2b mutant embryos was observed until late stage 15, when dAda2b protein levels become reduced (Fig. 2). At this stage, anti-acetylated H3 and H4 antibodies labeled mainly the CNS and epidermis (Fig. 3). We found a dramatic decrease in staining intensity in dAda2b mutant embryos compared to the wild type when using antibodies directed against acetylated lysine 14 or lysine 9 in histone H3 (Fig. 3A to D). By contrast, anti-acetylated H4 lysine 8 and a tetra-acetylated histone H4 antibody gave comparable signals in wild-type and dAda2b mutant embryos (Fig. 3E to H).
FIG. 3.
Reduced histone H3 acetylation in dAda2b mutant embryos. Antibodies that recognize different acetylated forms of histone H3 and histone H4 were used to stain Drosophila embryos. All four antibodies show strongest signals in the CNS and epidermis. Ventral views of stage 16 embryos are shown, with anterior to the left. Homozygous dAda2b mutant embryos were identified by the absence of Ubx-lacZ expression, which is present in embryos containing a balancer chromosome. (A and B) Wild-type (wt) (A) and dAda2b mutant (B) stage 16.1 embryos were stained with an antibody recognizing histone H3 acetylated at lysine 14. A substantial reduction in staining intensity was found in dAda2b mutant embryos. (C and D) wt (C) and dAda2b mutant (D) stage 16.1 embryos incubated with an antibody directed against acetylated histone H3 lysine 9. The staining intensity is diminished in dAda2b mutant embryos compared to wt embryos. (E and F) wt (E) and mutant (F) stage 16.2 embryos stained with an acetylated histone H4 lysine 8 antibody. The staining pattern and intensity are indistinguishable between wt and dAda2b mutant embryos. (G and H) Stage 16.2 wt (G) and dAda2b mutant (H) embryos incubated with a tetra-acetylated histone H4 antibody. Comparable staining intensities were observed in wt and mutant embryos.
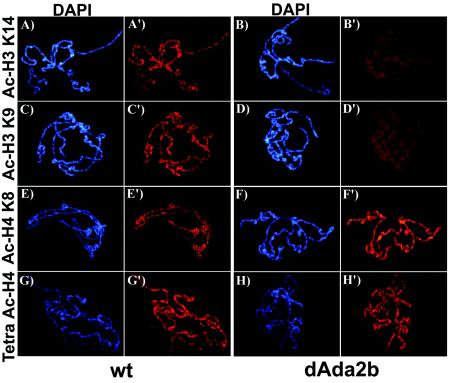
We wanted to extend these findings to the acetylation state of the chromosomes. For this purpose, we immunostained polytene chromosomes prepared from third-instar larval salivary glands with the same antibodies. As shown in Fig. 4, use of anti-acetylated H3 and H4 antibodies resulted in banded patterns on the polytene chromosomes, demonstrating regions enriched in these histone modifications. As previously observed (39, 49), acetylated H3 and H4 predominantly localized to euchromatic bands that stain strongly with the DNA dye DAPI (Fig. 4). Interbands and puffs, which are thought to correspond to regulatory regions and highly transcribed regions, respectively, stained weakly or not at all with DAPI and were mostly devoid of acetylated histones (Fig. 4). In accordance with what we observed in embryos, acetylated H3 K14 and K9 levels were severely reduced on dAda2b mutant chromosomes (Fig. 4A to D′). Interestingly, diminished staining was detected on entire chromosomes, not at merely a few loci, demonstrating that global histone H3 acetylation is dependent on dAda2b. Comparison of acetylated H4 K8 and tetra-acetylated H4 stainings between wild-type and dAda2b mutant chromosomes revealed no obvious differences (Fig. 4E to H′). These results suggest that dAda2b is required for normal histone H3 acetylation levels but does not contribute significantly to histone H4 acetylation. This conclusion is in agreement with data from yeast showing that Gcn5 preferentially acetylates histone H3 (22). We propose that the dAda2b protein is required for full catalytic activity of dGcn5 or is necessary for proper targeting of dGcn5 to chromatin.
FIG. 4.
Histone H3 acetylation on salivary gland polytene chromosomes is reduced in dAda2b mutants. Polytene chromosomes were prepared from wild-type (wt) and dAda2b homozygous mutant third-instar larval salivary glands, stained with antibodies directed against different acetylated forms of histone H3 or H4, and counterstained with DAPI. For wt (A and A′) and dAda2b mutant (B and B′) chromosomes incubated with an anti-acetylated histone H3 lysine 14 antibody, the staining intensity was greatly diminished on dAda2b mutant chromosomes compared to wt. A reduction in staining intensity was also found on dAda2b mutant chromosomes incubated with anti-acetylated H3 K9 (D and D′) compared to the wt (C and C′). By contrast, we observed similar staining intensities for wt and dAda2b mutant chromosomes incubated with anti-acetylated H4 K8 (E to F′) or a tetra-acetylated H4 antibody (G to H′).
dAda2b is not required for p53 function.
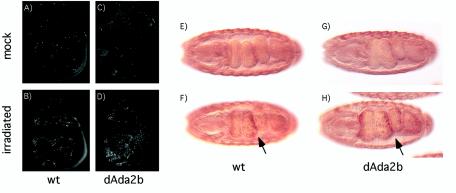
It is believed that the SAGA complex is recruited to target genes through interactions with transcriptional activators. For example, it has been shown that the acidic activation domains in VP16 and p53 can interact with both mammalian and Drosophila SAGA complexes (16, 28, 50). To determine whether dAda2b contributes to p53 function in vivo, we analyzed the p53-dependent induction of apoptosis in response to X rays. It has previously been shown that in response to irradiation, p53 binds to a radiation-responsive enhancer in the reaper gene, and the resulting induction of reaper expression leads to apoptosis (10, 37). This induction of apoptosis strictly depends on p53, as in p53 mutants, no apoptosis is observed following irradiation (30, 45). We treated wild-type and dAda2b mutant larvae with ionizing radiation and visualized apoptotic cells in wing disks with the vital dye acridine orange. In untreated wing disks, very few stained cells were observed in either the wild type or dAda2b mutants (Fig. 5A and B). However, following irradiation, there was a substantial increase in the amount of apoptosis in wild-type wing disks (439 ± 44 stained cells) (Fig. 5C). Unexpectedly, in dAda2b mutant disks, an even greater amount of apoptosis was observed (873 ± 32 stained cells) (Fig. 5D). To extend these observations, we introduced a transgene containing a 150-bp radiation-responsive enhancer from the reaper gene into dAda2b homozygous mutant embryos (10). This enhancer contains a p53-response element and is dependent on p53 for activity (10, 45). Enhancer-driven reporter gene expression was monitored at stage 16 of embryogenesis, when maternal dAda2b protein levels are diminished. At this stage, the enhancer was active even in the absence of irradiation, but its activity was further increased by gamma radiation (compare Fig. 5E and F). A similar increase in reporter gene expression was observed in dAda2b mutant embryos following irradiation (Fig. 5G and H). In summary, we conclude that p53 function following radiation-induced DNA damage does not require the dAda2b gene.
FIG. 5.
Ada2b mutants are hypersensitive to radiation-induced apoptosis and can induce expression of the p53 target gene reaper. (A to D) Wild-type (wt) and dAda2b mutant third-instar larvae were mock treated or irradiated with 800 rads. Wing imaginal disks were dissected after 4 h and incubated in the vital dye acridine orange, which preferentially stains apoptotic cells. Very few apoptotic cells are observed in mock-treated wt (A) and dAda2b mutant (C) wing disks. Following irradiation, extensive apoptosis is observed in the wt wing disk (B). More apoptotic cells are found in dAda2b mutant disks than in the wt after irradiation (D). (E to H) Expression of a p53-dependent radiation-responsive enhancer from the reaper gene does not require dAda2b. Embryos were incubated with a β-galactosidase antibody that recognizes reaper-driven lacZ expression and identifies dAda2b homozygous mutant embryos by distinguishing Ubx-lacZ expression from the balancer chromosome. Stage 16.2 embryos are shown from a ventral view, with anterior to the left. In the absence of irradiation, lacZ expression is strong in the epidermis but largely absent from the gut in both wt (E) and dAda2b mutant (G) embryos. Expression in the gut is strongly induced in both wt (F) and dAda2b mutant (H) embryos following irradiation (arrows).
Increased radiation-induced apoptosis in dAda2b mutants is p53 dependent.
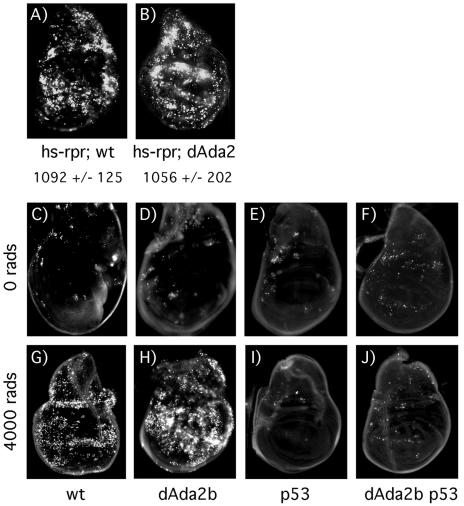
Increased apoptosis in dAda2b mutant wing disks in response to gamma radiation could result from less efficient DNA repair in the mutant, forcing more cells into apoptosis. Inefficient DNA repair might result from gene regulatory defects in dAda2b mutant cells or from a more direct involvement of dAda2b in DNA repair. Alternatively, dAda2b normally has an inhibitory role in generation of a DNA damage signal or in controlling p53 function, resulting in increased apoptosis in the mutant. To begin an investigation of which step in the path to apoptosis dAda2b might affect, we induced reaper expression independently of DNA damage by using a reaper transgene under the control of a heat shock promoter. As shown in Fig. 6A and B, no significant difference in the amount of apoptosis between wild-type and dAda2b mutant larvae expressing hs-reaper was observed. This result indicates that dAda2b is acting upstream of reaper induction in response to irradiation. We then examined whether the increased apoptosis resulting from irradiation in dAda2b mutants is dependent on p53. For this purpose, we generated a recombinant chromosome containing the dAda2b2 and p535A-1-4 alleles. As expected, in the p53 single mutant, no apoptosis was observed in response to irradiation (Fig. 6E and I). In dAda2b p53 double mutant wing disks, a small amount of apoptosis was observed before irradiation (Fig. 6F). Importantly, no further increase in apoptosis was observed following irradiation (Fig. 6J). The additional apoptosis observed in dAda2b single mutants (compare Fig. 5B with D and Fig. 6G with H) is therefore p53 dependent.
FIG. 6.
dAda2b acts upstream of reaper in p53-dependent apoptosis. (A and B) Wild-type (wt) and dAda2b mutant third-instar larvae containing an hs-reaper transgene (hs-rpr) were heat shocked, and wing imaginal disks were incubated in acridine orange after a 1-h recovery period. Similar numbers of apoptotic cells are observed in wt (A) (1,092 ± 125 stained cells) and dAda2b mutant (B) (1,056 ± 202 stained cells) wing disks containing the hs-reaper transgene. (C to J) Radiation-induced apoptosis in dAda2b mutants is p53 dependent. wt (C and G), dAda2b mutant (D and H), p53 mutant (E and I), and dAda2b p53 double mutant (F and J) third-instar larvae were mock treated or irradiated with 4,000 rads. After 4 h, wing imaginal disks were dissected and incubated in acridine orange. Very few apoptotic cells are observed in mock-treated wt (C), dAda2b mutant (D), and p53 mutant (E) wing disks. Slightly more apoptotic cells are found in mock-treated dAda2b p53 double mutant wing disks (F). In response to 4,000 rads of irradiation, massive apoptosis is observed in wt wing disks (G), and even more is observed in dAda2b mutant wing disks (H). As expected, no apoptosis is found in p53 mutant wing disks (I). In dAda2b p53 double mutant wing disks (J), there is no increase in the amount of apoptosis compared to mock-treated disks (F).
Yeast Ada2 is sensitive to DNA damage.
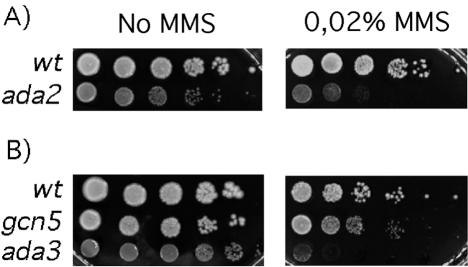
If dAda2b regulates DNA repair or is involved in the generation of a DNA damage signal, this function may be evolutionarily conserved. To investigating this possibility, we asked whether a yeast strain lacking Ada2p is also sensitive to DNA-damaging agents. We plated serial dilutions of ada2 mutant and isogenic wild-type yeast cells on medium containing MMS and compared their growth to that on rich medium lacking the drug. As shown in Fig. 7 and as previously observed, ada2 mutant cells were slow growing even on rich medium. A further impairment of growth was noted on MMS-containing medium (Fig. 7A). This result suggests that the Saccharomyces ada2 mutant is sensitive to DNA damage, as is the Drosophila Ada2b mutant.
FIG. 7.
A yeast ada2 mutant strain is sensitive to DNA damage. (A) Tenfold serial dilutions of isogenic wild-type (wt) and ada2 mutant yeast strains were plated on rich medium without MMS and on medium containing 0.02% MMS. (B) Growth of wt yeast on 0.02% MMS was compared to that of gcn5 mutant and ada3 mutant yeast. The strain lacking Ada3p is highly sensitive to MMS, whereas the gcn5 mutant grows a little more slowly than the wt.
We then tested whether strains lacking the Ada2p-associated proteins Gcn5p and Ada3p exhibit a similar sensitivity to MMS. Figure 7B shows that although gcn5 mutant cells had a plating efficiency similar to that of the wild type, the colonies were smaller and therefore were growing more slowly than their wild-type counterparts. The ada3 mutant strain was the most sensitive to MMS, as its plating efficiency was severely affected.
Taken together, our results show that Ada2 and its associated proteins are sensitive to genotoxic agents in both yeast and flies, indicating that an Ada2-Gcn5 complex may facilitate efficient DNA repair or the generation of a DNA damage signal.
DISCUSSION
The Drosophila Ada2b protein is present in a SAGA-like HAT complex (28, 36). In accordance with this finding, we show that dAda2b and dGcn5 directly interact and are expressed in similar patterns during embryogenesis. We have demonstrated that Ada2b is required for viability in Drosophila. Although dAda2b protein levels become greatly reduced in mutant embryos at stage 15, the animals survive until the pupal stage. Concomitant with reduced dAda2b protein amounts, we observed a significant reduction in acetylation of lysines 14 and 9 in histone H3 (Fig. 3). Lysine 14 acetylation is most severely affected, since a reduced staining intensity was observed even in dAda2b heterozygous embryos compared to the wild type (data not shown). The preference for H3 K14, and the selective reduction in histone H3 over histone H4 acetylation, fits the substrate specificity of Gcn5 (22). Furthermore, Gcn5's presence in the SAGA complex is dependent on Ada2 in yeast (21). We therefore assume that the loss of histone H3 acetylation in dAda2b mutants results from diminished dGcn5 activity, either as a consequence of reduced catalytic activity of the enzyme or due to a failure to target dGcn5 to chromatin.
The reduced histone H3 acetylation levels appear to persist during development, as third-instar larval salivary gland chromosomes retained a similar reduction in H3 acetylation (Fig. 4). Interestingly, chromatin acetylation is globally affected in dAda2b mutants, rather than affecting a subset of genes. Therefore, dAda2b is not likely to function solely as an adaptor that brings dGcn5 to only some promoter regions through associations with transcriptional activators. Instead, it is probable that dAda2b has a function similar to that of yeast Ada2, which is to retain Gcn5 within the SAGA complex and to stimulate its catalytic activity. This result also suggests that the SAGA complex, or at least a dAda2b-dGcn5 complex, has functions in addition to transcription-coupled gene-specific acetylation. Some heterochromatic regions with low gene density that stain with DAPI contain acetylated histone H3. These regions exhibit a decrease in H3 acetylation similar to that of other parts of the chromosomes (Fig. 4).
Drosophila Gcn5 is present in complexes in addition to the dAda2b-containing SAGA-like complex (28, 36). Our results indicate that the fraction of dGcn5 that associates with dAda2b makes a significant contribution to the overall levels of histone H3 acetylation and that association with dAda2a cannot substitute for dGcn5's function within the SAGA complex. Interestingly, normal histone H3 acetylation appears to be dispensable for development from at least mid-embryogenesis until pupation. Furthermore, we do not know if the lethality observed at pupation is caused by reduced histone acetylation or by another function of dAda2b. In addition to its role in pupal development, dAda2b is also required for oogenesis. Females with dAda2b homozygous germ cells lack developed ovaries.
All Ada2 proteins share a ZZ zinc finger and a SANT domain in their N termini (28, 36). The minimal Gcn5 interaction domain includes both the ZZ domain and the SANT domain (5, 14, 15), and the SANT domain additionally stimulates Gcn5's catalytic activity (9, 47). We observed that the dAda2b gene is alternatively spliced, generating proteins with different C termini. Both protein isoforms contain the ZZ and SANT domains, but the longer isoform lacks another conserved element, Ada box 3 (36). However, we found no evidence for differential usage of the two proteins during development (Fig. 1). The Ada2-Gcn5 interaction, although conserved in evolution, is species specific. For example, human Ada2a cannot interact with yeast Gcn5 (15), and both Drosophila Ada2a and Ada2b fail to interact with yeast Gcn5 (36). As might be expected from these results, we found that dAda2b fails to complement a yeast Ada2 mutant (not shown).
It is believed that the SAGA complex is recruited to target genes through interactions with sequence-specific transcription factors (12). At the promoter, the Gcn5 subunit acetylates histone H3 and perhaps other proteins, whereas the Spt 3 subunit facilitates preinitiation complex assembly by interacting with TATA binding protein (4, 7, 8, 20, 29, 46). Several yeast activator proteins, including Gcn4, Pho4, and Gal4, require the SAGA complex for activity and recruit it to target genes in vivo (4, 6, 8, 19, 26, 29). The activator proteins VP16 and Drosophila p53 can precipitate the SAGA-like complex from a Drosophila S2 cell nuclear extract (28). We tested whether this interaction is of significance for the in vivo function of the Drosophila p53 protein. We found that p53-induced apoptosis and reaper gene expression in response to DNA damage are not impaired in dAda2b mutants (Fig. 5). This result indicates that the Ada2/Gcn5 subcomplex of SAGA is not necessary for p53 function, but it does not exclude the possibility that p53 activity requires other SAGA components. In fact, many yeast genes show a differential requirement for the Ada2/Gcn5 and Spt3 subcomplexes of SAGA (19, 31).
Unexpectedly, we found increased apoptosis in response to gamma radiation in dAda2b mutants. Irradiation causes DNA damage, which leads to activation of ATM/ATR kinases that phosphorylate a variety of substrates, including the Chk2 kinase, ultimately resulting in p53 activation (2, 35, 42). In Drosophila, p53 activation brings about apoptosis but does not lead to a G1 cell cycle arrest, which is another possible outcome in mammalian cells (30, 45). It is possible that the reason for increased apoptosis in dAda2b mutants is that dAda2b normally has an inhibitory role in the generation of a DNA damage signal or in controlling p53 function. Another possibility is that DNA repair is not as efficient in dAda2b mutants as in the wild type, leading more cells to enter the apoptosis program. The role of dAda2b in DNA repair could be indirect. For example, dAda2b could be necessary for proper expression of a DNA repair factor. Alternatively, dAda2b might have a more direct function in DNA repair, presumably through chromatin modification. We found that in dAda2b mutants, excessive apoptosis occurs after irradiation but not before irradiation or when apoptosis is induced by an alternative means (Fig. 5 and 6). Furthermore, the extra apoptosis is p53 dependent (Fig. 6). For this reason, we find it unlikely that increased apoptosis in dAda2b mutants is the consequence of a higher sensitivity to stress in general. Instead, we believe that dAda2b has a role in the p53-dependent pathway that is activated by DNA damage.
All eukaryotes respond to DNA lesions with activation of ATM/ATR kinases, although the downstream component p53 is absent from unicellular organisms such as the yeast S. cerevisiae (33). If the involvement of dAda2b in the DNA damage response is upstream of p53 activation, one would predict this function to be conserved in evolution. In fact, we demonstrate that yeast strains lacking Ada2p, or Gcn5p or Ada3p, are sensitive to the genotoxic agent MMS (Fig. 7). It is possible, therefore, that histone acetylation facilitates DNA repair or generates a DNA damage signal. Intriguingly, an increase in radiation-induced apoptosis of a magnitude similar to what we observe in dAda2b mutants was observed in larvae in which the C terminus of a histone variant, H2Av, is missing (34). The H2Av C terminus contains a conserved serine that is phosphorylated in response to DNA double-strand breaks (41). Apparently, the absence of this phosphorylation event reduces DNA repair efficiency, which increases the amount of apoptosis twofold (34). Perhaps histone acetylation likewise contributes to the generation of a DNA damage signal, and maybe this is why chromatin is globally acetylated by Ada2/Gcn5. Future studies will be aimed at investigating this possibility.
Acknowledgments
We thank Tom Kusch and Jerry Workman for reagents and for sharing unpublished results, John Abrams and Hermann Steller for providing us with reaper transgenes, Shelley Berger for yeast strains, Stefan Åström for advice on yeast work, Monika Björk for help with injections, and Achim Haecker for comments on the manuscript.
This work was supported by grants from the Swedish Research Council (to M.M. and J.L.) and the Swedish Cancer Society (to M.M.).
REFERENCES
- 1.Aasland, R., A. F. Stewart, and T. Gibson. 1996. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci. 21:87-88. [PubMed] [Google Scholar]
- 2.Abraham, R. T. 2001. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 15:2177-2196. [DOI] [PubMed] [Google Scholar]
- 3.Allfrey, V. G., R. Faulkner, and A. E. Mirsky. 1964. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA 51:786-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barbaric, S., H. Reinke, and W. Horz. 2003. Multiple mechanistically distinct functions of SAGA at the PHO5 promoter. Mol. Cell. Biol. 23:3468-3476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barlev, N. A., R. Candau, L. Wang, P. Darpino, N. Silverman, and S. L. Berger. 1995. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J. Biol. Chem. 270:19337-19344. [DOI] [PubMed] [Google Scholar]
- 6.Berger, S. L., B. Pina, N. Silverman, G. A. Marcus, J. Agapite, J. L. Regier, S. J. Triezenberg, and L. Guarente. 1992. Genetic isolation of ADA2: a potential transcriptional adaptor required for function of certain acidic activation domains. Cell 70:251-265. [DOI] [PubMed] [Google Scholar]
- 7.Bhaumik, S. R., and M. R. Green. 2002. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 22:7365-7371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhaumik, S. R., and M. R. Green. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev. 15:1935-1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer, L. A., M. R. Langer, K. A. Crowley, S. Tan, J. M. Denu, and C. L. Peterson. 2002. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell 10:935-942. [DOI] [PubMed] [Google Scholar]
- 10.Brodsky, M. H., W. Nordstrom, G. Tsang, E. Kwan, G. M. Rubin, and J. M. Abrams. 2000. Drosophila p53 binds a damage response element at the reaper locus. Cell 101:103-113. [DOI] [PubMed] [Google Scholar]
- 11.Brown, C. E., L. Howe, K. Sousa, S. C. Alley, M. J. Carrozza, S. Tan, and J. L. Workman. 2001. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 292:2333-2337. [DOI] [PubMed] [Google Scholar]
- 12.Brown, C. E., T. Lechner, L. Howe, and J. L. Workman. 2000. The many HATs of transcription coactivators. Trends Biochem. Sci. 25:15-19. [DOI] [PubMed] [Google Scholar]
- 13.Brownell, J. E., J. Zhou, T. Ranalli, R. Kobayashi, D. G. Edmondson, S. Y. Roth, and C. D. Allis. 1996. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 84:843-851. [DOI] [PubMed] [Google Scholar]
- 14.Candau, R., and S. L. Berger. 1996. Structural and functional analysis of yeast putative adaptors. Evidence for an adaptor complex in vivo. J. Biol. Chem. 271:5237-5245. [DOI] [PubMed] [Google Scholar]
- 15.Candau, R., P. A. Moore, L. Wang, N. Barlev, C. Y. Ying, C. A. Rosen, and S. L. Berger. 1996. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol. Cell. Biol. 16:593-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Candau, R., D. M. Scolnick, P. Darpino, C. Y. Ying, T. D. Halazonetis, and S. L. Berger. 1997. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene 15:807-816. [DOI] [PubMed] [Google Scholar]
- 17.Carrozza, M. J., R. T. Utley, J. L. Workman, and J. Cote. 2003. The diverse functions of histone acetyltransferase complexes. Trends Genet. 19:321-329. [DOI] [PubMed] [Google Scholar]
- 18.Chou, T. B., and N. Perrimon. 1996. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics 144:1673-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dudley, A. M., C. Rougeulle, and F. Winston. 1999. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes Dev. 13:2940-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eisenmann, D. M., K. M. Arndt, S. L. Ricupero, J. W. Rooney, and F. Winston. 1992. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes Dev. 6:1319-1331. [DOI] [PubMed] [Google Scholar]
- 21.Grant, P. A., L. Duggan, J. Cote, S. M. Roberts, J. E. Brownell, R. Candau, R. Ohba, T. Owen-Hughes, C. D. Allis, F. Winston, S. L. Berger, and J. L. Workman. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 11:1640-1650. [DOI] [PubMed] [Google Scholar]
- 22.Grant, P. A., A. Eberharter, S. John, R. G. Cook, B. M. Turner, and J. L. Workman. 1999. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 274:5895-5900. [DOI] [PubMed] [Google Scholar]
- 23.Hampsey, M. 1997. A SAGA of histone acetylation and gene expression. Trends Genet. 13:427-429. [DOI] [PubMed] [Google Scholar]
- 24.Horiuchi, J., N. Silverman, G. A. Marcus, and L. Guarente. 1995. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol. Cell. Biol. 15:1203-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang, J., T. Hoey, and M. Levine. 1991. Autoregulation of a segmentation gene in Drosophila: combinatorial interaction of the even-skipped homeo box protein with a distal enhancer element. Genes Dev. 5:265-277. [DOI] [PubMed] [Google Scholar]
- 26.Kuo, M. H., E. von Baur, K. Struhl, and C. D. Allis. 2000. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 6:1309-1320. [DOI] [PubMed] [Google Scholar]
- 27.Kurdistani, S. K., and M. Grunstein. 2003. Histone acetylation and deacetylation in yeast. Nat. Rev. Mol. Cell Biol. 4:276-284. [DOI] [PubMed] [Google Scholar]
- 28.Kusch, T., S. Guelman, S. M. Abmayr, and J. L. Workman. 2003. Two Drosophila Ada2 homologues function in different multiprotein complexes. Mol. Cell. Biol. 23:3305-3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larschan, E., and F. Winston. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev. 15:1946-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, J. H., E. Lee, J. Park, E. Kim, J. Kim, and J. Chung. 2003. In vivo p53 function is indispensable for DNA damage-induced apoptotic signaling in Drosophila. FEBS Lett. 550:5-10. [DOI] [PubMed] [Google Scholar]
- 31.Lee, T. I., H. C. Causton, F. C. Holstege, W. C. Shen, N. Hannett, E. G. Jennings, F. Winston, M. R. Green, and R. A. Young. 2000. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 405:701-704. [DOI] [PubMed] [Google Scholar]
- 32.Lilja, T., D. Qi, M. Stabell, and M. Mannervik. 2003. The CBP coactivator functions both upstream and downstream of Dpp/Screw signaling in the early Drosophila embryo. Dev. Biol. 262:294-302. [DOI] [PubMed] [Google Scholar]
- 33.Lowndes, N. F., and J. R. Murguia. 2000. Sensing and responding to DNA damage. Curr. Opin. Genet. Dev. 10:17-25. [DOI] [PubMed] [Google Scholar]
- 34.Madigan, J. P., H. L. Chotkowski, and R. L. Glaser. 2002. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 30:3698-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Motoyama, N., and K. Naka. 2004. DNA damage tumor suppressor genes and genomic instability. Curr. Opin. Genet. Dev. 14:11-16. [DOI] [PubMed] [Google Scholar]
- 36.Muratoglu, S., S. Georgieva, G. Papai, E. Scheer, I. Enunlu, O. Komonyi, I. Cserpan, L. Lebedeva, E. Nabirochkina, A. Udvardy, L. Tora, and I. Boros. 2003. Two different Drosophila ADA2 homologues are present in distinct GCN5 histone acetyltransferase-containing complexes. Mol. Cell. Biol. 23:306-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ollmann, M., L. M. Young, C. J. Di Como, F. Karim, M. Belvin, S. Robertson, K. Whittaker, M. Demsky, W. W. Fisher, A. Buchman, G. Duyk, L. Friedman, C. Prives, and C. Kopczynski. 2000. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell 101:91-101. [DOI] [PubMed] [Google Scholar]
- 38.Peterson, C. L., and J. Cote. 2004. Cellular machineries for chromosomal DNA repair. Genes Dev. 18:602-616. [DOI] [PubMed] [Google Scholar]
- 39.Pile, L. A., and D. A. Wassarman. 2000. Chromosomal localization links the SIN3-RPD3 complex to the regulation of chromatin condensation, histone acetylation and gene expression. EMBO J. 19:6131-6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ponting, C. P., D. J. Blake, K. E. Davies, J. Kendrick-Jones, and S. J. Winder. 1996. ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem. Sci. 21:11-13. [PubMed] [Google Scholar]
- 41.Redon, C., D. Pilch, E. Rogakou, O. Sedelnikova, K. Newrock, and W. Bonner. 2002. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 12:162-169. [DOI] [PubMed] [Google Scholar]
- 42.Rouse, J., and S. P. Jackson. 2002. Interfaces between the detection, signaling, and repair of DNA damage. Science 297:547-551. [DOI] [PubMed] [Google Scholar]
- 43.Rubin, G. M., and A. C. Spradling. 1982. Genetic transformation of Drosophila with transposable element vectors. Science 218:348-353. [DOI] [PubMed] [Google Scholar]
- 44.Samakovlis, C., G. Manning, P. Steneberg, N. Hacohen, R. Cantera, and M. A. Krasnow. 1996. Genetic control of epithelial tube fusion during Drosophila tracheal development. Development 122:3531-3536. [DOI] [PubMed] [Google Scholar]
- 45.Sogame, N., M. Kim, and J. M. Abrams. 2003. Drosophila p53 preserves genomic stability by regulating cell death. Proc. Natl. Acad. Sci. USA 100:4696-4701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sterner, D. E., P. A. Grant, S. M. Roberts, L. J. Duggan, R. Belotserkovskaya, L. A. Pacella, F. Winston, J. L. Workman, and S. L. Berger. 1999. Functional organization of the yeast SAGA complex: distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 19:86-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sterner, D. E., X. Wang, M. H. Bloom, G. M. Simon, and S. L. Berger. 2002. The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J. Biol. Chem. 277:8178-8186. [DOI] [PubMed] [Google Scholar]
- 48.Tautz, D., and C. Pfeifle. 1989. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98:81-85. [DOI] [PubMed] [Google Scholar]
- 49.Turner, B. M., A. J. Birley, and J. Lavender. 1992. Histone H4 isoforms acetylated at specific lysine residues define individual chromosomes and chromatin domains in Drosophila polytene nuclei. Cell 69:375-384. [DOI] [PubMed] [Google Scholar]
- 50.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394:498-502. [DOI] [PubMed] [Google Scholar]
- 51.White, R. A. H. 1998. Immunolabelling of Drosophila, p. 215-240. In D. B. Roberts (ed.), Drosophila, a practical approach, 2nd ed. IRL Press, Oxford, United Kingdom.
- 52.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, R. W. Davis, et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285:901-906. [DOI] [PubMed] [Google Scholar]