Abstract
The sodium/iodide symporter (NIS) is a plasma membrane protein that mediates active iodide transport in thyroid and mammary cells. It is a prerequisite for radioiodide treatment of thyroid cancer and a promising diagnostic and therapeutic tool for breast cancer. We investigated the molecular mechanisms governing NIS expression in mammary cells. Here we report that Nkx-2.5, a cardiac homeobox transcription factor that is also expressed in the thyroid primordium, is a potent inducer of the NIS promoter. By binding to two specific promoter sites (N2 and W), Nkx-2.5 induced the rNIS promoter (about 50-fold over the basal level). Interestingly, coincident with NIS expression, Nkx-2.5 mRNA and protein were present in lactating, but not virgin, mammary glands in two human breast cancer samples and in all-trans retinoic acid (tRA)-stimulated MCF-7 breast cancer cells. A cotransfected dominant-negative Nkx-2.5 mutant abolished tRA-induced endogenous NIS induction, which shows that Nkx-2.5 activity is critical for this process. Remarkably, in MCF-7 cells, Nkx-2.5 overexpression alone was sufficient to induce NIS and iodide uptake. In conclusion, Nkx-2.5 is a novel relevant transcriptional regulator of mammary NIS and could thus be exploited to manipulate NIS expression in breast cancer treatment strategies.
Active transport of iodide into the thyroid gland is an essential, rate-limiting step in thyroid hormone biosynthesis. It is mediated by a specific Na+/I− symporter (NIS) located at the basolateral membrane of thyroid follicular cells (10).
Radioiodide therapy is the mainstay of thyroid cancer treatment. In fact, radioactive iodide is transported into cancer cells, where it exerts a local destructive effect. However, a loss of differentiation, including the loss of iodide transport, is frequent in thyroid carcinoma and results in resistance to radioiodide treatment and a poor prognosis (11). Thyroid-stimulating hormone, which stimulates NIS gene expression in the thyroid, is used to increase the NIS level and to improve the efficacy of radioiodide treatment (31). NIS levels in thyroid tissues can also be modulated by iodide (44) and transforming growth factor β1 (16).
Iodide also accumulates in the salivary glands, gastric mucosa, and lactating mammary glands (10). Iodide uptake in normal breast tissue was reported over 40 years ago; iodide in breast milk is concentrated 6- to 15-fold compared with that in plasma, and about 20% is organified as a result of the effect of peroxidase expressed in breast alveolar cells (38, 42). NIS-mediated iodide uptake in the mammary gland during lactation (40) is used by the nursing newborn for the biosynthesis of thyroid hormones, which are essential for the correct development of the nervous system, skeletal muscle, and lungs (26). Recently, NIS was found to be expressed in about 50 to 80% of invasive breast cancers (10), raising the possibility that radioiodide may be used for the diagnosis and treatment of breast cancer.
Gene transfer of NIS is a way to promote iodide uptake by several human tumors, e.g., gliomas (4), prostate cancer (37), melanoma, and ovarian, liver, and colon cancer (23). Tissue-specific promoters can be used to target NIS to malignant cells, thereby maximizing tissue-specific cytotoxicity and sparing nonmalignant cells. However, the NIS protein must undergo a complex maturation program to be functional, and thus the physiological milieu is critical for effective radioiodine treatment.
Retinoic acid (RA) increases iodide uptake in some differentiated thyroid cancers (33, 34). Moreover, in MCF-7 breast cancer cells, all-trans RA (tRA) stimulates iodide uptake after RA receptor (RAR)-induced up-regulation of the NIS gene (17). In neither case is the molecular mechanism governing NIS expression known.
Several regulatory regions in the rat (12, 27, 43) and human (1, 30, 39, 45) NIS promoters have been characterized. The paired-domain-containing factor Pax-8 binds to two sites in the rat NIS (rNIS) upstream enhancer, which contains a cyclic AMP response element-like sequence that mediates thyroid-specific transcription (27). Thyroid-specific transcription factor 1 (TTF-1) binds to the proximal rNIS promoter region, between positions −245 and −230, and stimulates its activity (12).
Nkx-2.5 is a homeobox-containing transcription factor originally identified as a potential vertebrate homologue of the Drosophila gene tinman (18). It belongs to the NK2 class of homeobox proteins, which have a tyrosine residue at amino acid 54 of the homeodomain and a conserved 23-amino-acid NK2-specific domain (3). Nkx-2.5 is expressed in the heart (15, 18), in heart progenitor cells, and in the thyroid primordium during development (22). We recently demonstrated that Nkx-2.5 induces the expression of the human type 2 deiodinase promoter, a selenoenzyme that is critical for thyroid hormone metabolism (9).
Whereas the control of NIS expression in the thyroid gland has been widely studied, little is known about the factors that regulate NIS transcription in the mammary gland. We have examined the regulation of NIS transcription in a breast carcinoma cell line and found that Nkx-2.5, by binding to two specific sites, is a potent inducer of the NIS promoter. We also show that Nkx-2.5 expression is critical in RA-induced NIS up-regulation in MCF-7 cells as well as in mammary glands during lactation. The overexpression of Nkx-2.5, which alone induced iodide uptake in breast cancer cells, may eventually be exploited as a treatment strategy for cancer.
MATERIALS AND METHODS
Plasmids and expression constructs.
The expression plasmids for mouse Nkx-2.5 and Nkx-N188K are described elsewhere (9). The luciferase reporter plasmids NisLuc2, NisLuc4, NisLuc5 (27), and TPO-Luc (13), as well as the chloramphenicol acetyltransferase (CAT) constructs Tg-CAT (36), E1B-CAT (7), and C5-CAT (24), were kindly provided by R. Di Lauro.
The construct NisLuc5Delta (140 bp) was generated by digesting the NisLuc5 plasmid with SacI-PstI. The NisLuc5-Nm construct, containing a TGA to CGC mutation in the N2 site in the context of the NisLuc5 construct, was generated by a PCR with oligonucleotides N4s and N5r (Table 1) and with NisLuc5 as a template. The resulting fragment, which contained a mutation in the N2 site (TGAG to CGCG) within 470 nucleotides (nt) of the rNIS promoter, was subcloned into the pGL-3 basic vector (Promega, Madison, Wis.). The NisLuc5-Wm construct, containing a mutation in the W site (TTCC to TGAC) within the 470-nt promoter, was generated by a PCR with oligonucleotides rN6Ms and rN6Mr and was subcloned into the pGL-3 basic vector. The construct NisLuc5-2m, harboring the N2 and W mutated sites, was generated by subcloning the PCR product obtained with the N4s and N6r oligonucleotides (Table 1) into pGL-3 basic. All generated plasmids were sequenced in both orientations for sequencing control. TESS (http://www.cbil.upenn.edu/tess) was used to predict transcription factor binding sites on the NIS promoter.
TABLE 1.
Oligonucleotides used for EMSA and PCR assays
| Oligonucleotide | Sequence (5′-3′) | Orientation |
|---|---|---|
| N1 | CCTCATAACTCCGCTTCCTAAATTCCTTAG | Sense |
| N2 | CAAGAGAACCTGAGTGCCTCCCAC | Sense |
| N3 | AGTTCCTTCTCCCAAGCTGCGGAG | Sense |
| W | AGACACGAGTGTTCCCCCACCCCG | Sense |
| NKE-2 | CCTTTGAAGTGGGGGCCTCTTGAGGCAA | Sense |
| D | GGCTGTCAAGGGTATTAGTTT | Sense |
| N4s | CTAAAGCAGAAGAAAGAAATTCTCCAAGAGAACCCGCGTGCCTCCCAC | Sense |
| N5r | CGGGCCTGCAGGGGTGCGGGCAGT | Antisense |
| N6r | CGGGCCTGCAGGGGTGCGGGCAGTCGGGGTGGGGTCACACTCGTGTC | Antisense |
| rN6Ms | GAGACACGAGTGTGACCCCACCCCGAC | Sense |
| hmGTX-6.2-1s | GGCCAGCAGATCTTCGCGCTGGAGAA | Sense |
| hmGTX-6.2-2r | AGCGCCAAGTTCGAGGGTTTGTGCTT | Antisense |
| NK1s | CGGAATTCCTTCCCCAGCCCTGCGCTCACACCCAC | Sense |
| Nkx-p1 | CTTCAAGCCAGAGGCCTACG | Sense |
| Nkx-p2 | CCGCCTCTGTCTTCTCCAGCTC | Antisense |
| Nis2s | ACCCAGGAAACTCGTGATTATCT | Sense |
| Nis3r | AGGGCACCGTAATAGAGATAGGA | Antisense |
| hNkx-303s | CCCAGCCAAGGACCCTAGA | Sense |
| hNkx-389r | GCGTTGTCCGCCTCTGTCT | Antisense |
| hNIS-2089s | CCATCCTGGATGACAACTTGG | Sense |
| hNIS-2187r | AAAAACAGACGATCCTCATTG | Antisense |
| hNIS-728s | GAAACTAAGGCCCAGGGAGGAG | Sense |
| hNIS-250r | AGGACAGGCTATCTGGGTGGC | Sense |
| BetaACT-2 | GTCAGGCAGCTCGTAGCTCT | Antisense |
| BetaACT-1 | TCGTGCGTGACATTAAGGAG | Sense |
| BetaACT-2 | GTCAGGCAGCTCGTAGCTCT | Antisense |
| mNkx-303s | CCCAGCCAAAGACCCTCGG | Sense |
| mNkx-389r | GCGCCATCCGTCTCGGCTT | Antisense |
| mNISs | TTTGCCGACACCTTTTATGCT | Sense |
| mNISr | GCGAGGTCCCACCACAGTAA | Antisense |
DNA transfection and CAT and luciferase expression assays.
HeLa cells (60% confluent) were transiently cotransfected with reporter CAT and luciferase plasmids by the calcium phosphate precipitation method. For each 60-mm-diameter dish, 3 μg of luciferase reporter vector was cotransfected with 0.2 μg of Nkx-2.5-expressing vector and 0.3 μg of CMV-CAT vector as an internal control. For an uninduced control, the pFLAG-CMV-2 empty vector was used in the same quantity as the Nkx-2.5 vector. Luciferase activities were measured 48 h after transfection, and differences in transfection efficiencies were corrected relative to the CAT activity level. Each construct was analyzed in duplicate in at least three separate transfections. Data (luciferase/CAT or CAT/luciferase ratios) are reported as means ± standard deviations (SD).
Bosc-23 cells (clone number CRL-11270; American Type Culture Collection), grown in 100-mm-diameter dishes to 70% confluence, were transiently transfected with Lipofectamine (GIBCO BRL, Gaithersburg, Md.). Ten micrograms of Nkx-2.5 vector was mixed with 1.6 ml of Optimem and 40 μl of Lipofectamine for 45 min and then added to the cells in 6 ml of total Optimem. After 48 h, the cells were washed and harvested by being scraped into 2 ml of 1× phosphate-buffered saline (PBS), pH 7.4. After centrifugation at 500 × g, the pellets were frozen at −80°C until they were required for the preparation of nuclear extracts.
MCF-7 cells were cotransfected in a 60-mm-diameter plate with 3 μg of the NisLuc reporter vector, 0.4 μg of CMV-CAT as an internal control, and 0.2 μg of CMV-FLAG or an Nkx-N188K-expressing vector by the use of Lipofectamine as described above. Thirty-six hours after transfection, the plates were stimulated for 12 h with 1 μM tRA or with an identical volume of absolute ethanol as a control. The cells were washed, harvested by being scraped into 2 ml of 1× PBS, pH 7.4, and centrifuged at 500 × g, after which the pellets were frozen at −80°C. For time course experiments, cells were stimulated at different times with 1 μM tRA and were harvested with Trizol reagent (Life Technologies, Ltd., Paisley, Scotland) for RNA preparation.
Semiquantitative EMSA and antibody interference assay.
The putative Nkx-2.5 binding sequences within the rNIS promoter were evaluated by electrophoretic mobility shift assays (EMSAs) with double-stranded end-labeled oligonucleotides harboring the sequences (one strand) from nt −282 to −251 (N1), −446 to −423 (N2), −399 to −372 (N3), and −250 to −230 (W) (Table 1). As a positive control for Nkx-2.5 binding, we used NKE-2, the high-affinity Nkx-2.5-binding oligonucleotide described for the rat ANF promoter (20, 35), or the Nkx-2.5 binder oligonucleotide D, described for the human type 2 deiodinase promoter (9). Antisense oligonucleotides were labeled with a T4 polynucleotide kinase (New England Biolabs, Boston, Mass.) and radioactive [γ-32P]ATP. Double-stranded oligonucleotides were purified by passage through NICK columns containing Sephadex G-50 DNA-grade resin (Amersham-Pharmacia, Piscataway, N.J.).
Nuclear extracts were prepared as previously described (9) from transfected Bosc-23 or MCF-7 cells or rat heart tissue. EMSAs were performed at room temperature with 30-μl reaction mixtures containing final concentrations of 20 mM Tris-HCl (pH 7.5), 75 mM KCl, 1 mM dithiothreitol, 10% glycerol, and 1 mg of poly(dI-dC)/ml. Binding reaction mixtures containing 10 μg of nuclear extracts and 10 fmol of the probe were equilibrated for 30 min at room temperature and separated by electrophoresis through a 6% polyacrylamide gel containing 0.5× Tris-borate-EDTA.
For antibody interference assays, 2 μl of anti-FLAG antibody (Sigma Chemical Co., Saint Louis, Mo.) or a preimmune antiserum (NR) was added to protein extracts and equilibrated at 4°C for 30 min before the addition of the probe. The binding specificity was assessed by competition with a 100-fold molar excess of cold competitors incubated with the proteins 15 min before the addition of the probe.
RT-PCR assays.
mRNAs were extracted from all tissues and MCF-7 cells by the use of Trizol reagent (Life Technologies, Ltd.). cDNAs were prepared by the use of Superscript III (Life Technologies, Ltd.) according to the manufacturer's instructions. The reverse transcriptase (RT) products were used for PCRs with Nkx-2.5-specific, NIS-specific, or NK-6.2-specific oligonucleotides. To exclude genomic DNA contamination, we designed the primers to span one intron. For Nk-6.2 amplification, total RNAs were extracted from MCF-7 cells after 6 and 12 h of treatment with 1 μM tRA or vehicle. The cDNAs were amplified (30 cycles) with hmGtx-6.2-1s and hmGtx-6.2-2r (Table 1), and the PCR products were analyzed by Southern blotting with a human Nk-6.2 cDNA-labeled fragment as a probe.
After electrophoresis, the PCR products were transferred onto nylon membranes and hybridized with an [α-32P]dCTP randomly labeled Nkx-2.5 cDNA probe. All human tissues (thyroid, heart, and breast cancer samples) used for this study were obtained according to protocols approved by the Institutional Review Board of the University of Naples “Federico II.” The RT-PCR bands were quantified by densitometric analysis (PhosphorImager GS525; Bio-Rad, Richmond, Calif.).
Iodide uptake assays.
MCF-7 cells were grown in 60-mm-diameter dishes with Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. For measurements of iodide uptake, the medium was removed and the cells were washed with 0.5 ml of Hanks' balanced salt solution (HBSS). The cells were then incubated for 2 h at 37°C with 2 ml of HBSS containing 10 μM NaI and carrier-free Na125I to give a specific activity of 10 mCi/mmol (assay buffer). Half of the reactions received assay buffer supplemented with 30 μM KClO4, a specific inhibitor of I− uptake, as a control for specificity. After a 1-h incubation, the cells were washed twice with ice-cold HBSS and harvested with HBSS. Radioactivity was quantitated in a γ-counter (LS5000CE; Beckman) and normalized to the total amount of proteins, as measured by a Bio-Rad assay. The results are expressed as amounts of 125I accumulated per microgram of protein.
mRNA preparation and Northern blot analysis.
Total RNAs were extracted from cells or frozen tissues by the use of Trizol (Life Technologies, Ltd.) according to the manufacturer's instructions. After isolation, the total RNAs were spectrophotometrically quantified at 260 nm and subjected to poly(A)+ mRNA purification by use of an Oligotex mRNA kit (Qiagen, Chatsworth, Calif.). Ten micrograms of poly(A)+ RNA was resolved by electrophoresis through a formaldehyde gel and subjected to Northern blotting on a Hybond N membrane (Amersham-Pharmacia) as described by the manufacturer. For Nkx-2.5 detection, blots were hybridized with a cDNA fragment corresponding to human Nkx-2.5 obtained by digesting pFLAG-Nkx2.5 with EcoRI and XbaI and were then labeled with a Prime-Lt II random primer labeling kit (Stratagene). The signal was revealed by autoradiography using X-Omat film (Kodak).
Western blot analysis.
Nuclear extracts from MCF-7-TetON cells, prepared as described above, were resolved by sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis. The gel was transferred onto an Immobilon P membrane (Millipore, Bedford, Mass.) for 12 h at a constant current of 50 mA. Immunodetection of FLAG-tagged Nkx-2.5 was performed with a monoclonal anti-FLAG antibody (M2; Sigma) diluted 1:3,000 in Tween 20-tPBS buffer containing 5% nonfat milk (Bio-Rad), and the filter was treated with a 1:3,000 dilution of goat anti-mouse immunoglobulin G conjugated to horseradish peroxidase (Amersham).
Real-time PCR.
Real-time PCR was used to detect variations in the numbers of Nkx-2.5, NIS, and β-actin gene copies as described elsewhere (14). The cDNAs were amplified by PCR in an ABI Prism 7700 sequence detector (PE Biosystems, Foster City, Calif.) with the fluorescent double-stranded DNA-binding dye SYBR Green (Applied Biosystems). Specific primers for the Nkx-2.5, NIS, and β-actin genes (Table 1) were designed to work under the same cycling conditions (95°C for 10 min followed by 40 cycles at 95°C for 15 s and 60°C for 1 min), generating products of comparable sizes (about 100 bp for each amplification). Primer combinations were positioned to span an exon-exon junction to avoid genomic DNA interference. For each reaction, standard curves for reference genes were constructed based on six fourfold serial dilutions of cDNA. All samples were run in triplicate.
The template concentration was calculated from the cycle number when the amount of PCR product passed a threshold established in the exponential phase of the PCR. The relative amounts of gene expression were calculated with β-actin expression as an internal standard (calibrator). The results, expressed as N-fold differences in target gene expression, were determined as follows: N · target = 2(ΔCt sample − ΔCt calibrator).
Establishment of a doxycycline-regulated, Nkx-2.5-expressing MCF-7 cell line.
An Nkx-2.5 inducible cell line was generated in MCF-7 cells by use of the TetON system (Clontech, Palo Alto, Calif.). A DNA fragment corresponding to an NH2-terminally flagged Nkx-2.5 was subcloned into a tetracycline-inducible vector, pTRE-2 (Clontech), to obtain pTRE-Nkx2.5. The oligonucleotides CMV-FLAG and NK2r (Table 1) were used to amplify a 998-bp fragment that was digested with XbaI and subcloned into PvuII and XbaI sites in pTRE-2. MCF-7-TetON cells were purchased from Clontech and grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 100 μg of penicillin-streptomycin/ml, and 300 μg of G418/ml in a 5% CO2 incubator at 37°C.
To obtain doxycycline-inducible Nkx-2.5-expressing MCF-7 clones, we cotransfected pTRE-Nkx2.5 (3 μg) and a pHyg resistance vector (10 μg) into MCF-7-TetON cells by the use of Lipofectamine (GIBCO BRL). Two days after transfection, the cultures were grown in the presence of 0.3 mg of hygromycin/ml. Seventy clones were selected and screened by Western blot analysis for the ability to express Nkx-2.5 in the presence, but not in the absence, of doxycycline (1 μg/μl). Positive clones were then characterized by Northern blotting for Nkx-2.5 mRNA expression with 30 μg of total mRNA. Of the positive clones, clones 19 and 35 were selected for further studies.
ChIP.
Briefly, approximately 8 × 106 cells were fixed with 1% formaldehyde in growth medium at 37°C for 30 min. Glycine was added to a final concentration of 0.125 M to stop cross-linking. Sonicated, cross-linked chromatin was centrifuged at 13,400 × g for 30 min to remove insoluble material. The soluble chromatin was then diluted 10-fold in dilution buffer and used directly for chromatin immunoprecipitation (ChIP). One percent of the total DNA was taken to be used as a positive control for PCR assays (denoted the input DNA). Six optical density (A260) units of chromatin from MCF-7 and clone 19 cells was mixed with 30 μl of anti-FLAG antibody-conjugated agarose beads (EZview Red Anti-FLAG M2 affinity gel; Sigma) and incubated at 4°C for 2 h. After several rounds of washing, bound DNA-protein complexes were eluted by incubation with 1% SDS-0.1 M NaHCO3 elution buffer. Formaldehyde cross-links were reversed by incubation in 200 mM NaCl at 65°C. The samples were extracted twice with phenol-chloroform and precipitated with ethanol. DNA fragments were recovered by centrifugation, resuspended in 50 μl of H2O, and used for PCR amplifications with oligonucleotides hNIS-728s and hNIS-250r (Table 1).
RESULTS
Nkx-2.5 transactivates a subset of thyroid-specific promoters in HeLa cells.
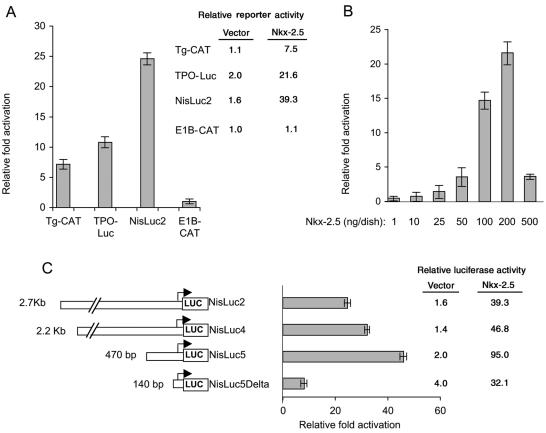
We previously reported that the transcription factor Nkx-2.5, in synergy with GATA-4, activates the promoter of human type 2 deiodinase (9), which is expressed in the heart and the thyroid. Because Nkx-2.5 is expressed in thyroid precursor cells (22), we suggested that it might mediate the transcriptional activation of other thyroidal genes. In addition, Nkx-2.5 belongs to the NK-2 family of transcription factors, which includes TTF-1, and the consensus binding sites of the two transcription factors are virtually identical (28). We evaluated whether Nkx-2.5 activated various thyroid-specific promoters in HeLa cells. As shown in Fig. 1A, Nkx-2.5 activated a subset of thyroid-specific promoters constituted by Tg-CAT (7-fold over the basal level), TPO-Luc (10.8-fold), and NisLuc2 (24.6-fold), but not a control E1B construct (Fig. 1A). The effect of Nkx-2.5 was largest on the NisLuc2 reporter, which contains the 2.7-kb 5′-flanking region of the rNIS gene. This was surprising because the NIS promoter, consistent with reported data (12), responded poorly to TTF-1, i.e., there was a 2.0-fold maximal induction under our conditions (data not shown), which is substantially lower than the robust effects exerted by TTF-1 on the Tg and TPO promoters (5).
FIG. 1.
Nkx-2.5 transactivates NIS and other thyroid-specific promoters in a dose-dependent fashion. (A) The indicated reporter plasmids (3 μg) were transiently cotransfected in HeLa cells with a plasmid encoding the Nkx-2.5 transcription factor (0.2 μg/60-mm-diameter dish) or CMV-FLAG (0.2 μg/60-mm-diameter dish) together with 0.3 μg of CMV-CAT (for luciferase reporter constructs) or RSV-Luc (for CAT reporter constructs) as an internal control. The relative fold induction in activity for each reporter plasmid was determined by calculating the Nkx-2.5/vector ratio when the vector (CMV-FLAG) activity was set at 1. The relative activity of each CAT or luciferase reporter plasmid with cotransfected CMV-FLAG (vector) or Nkx-2.5, expressed in arbitrary units, is also indicated. The results are shown as means ± SD of the CAT/luciferase (Tg-CAT and E1B) and luciferase/CAT (TPO-Luc and NisLuc2) ratios from at least three separate experiments, which were done in duplicate. (B) Nkx-2.5 transactivates the NIS promoter in a dose-dependent fashion. HeLa cells were cotransfected with the luciferase reporter construct NisLuc2 (3 μg/60-mm-diameter dish) containing the 2.7-kb 5′-flanking region of the rNIS gene and with the indicated increasing amounts of the Nkx-2.5 plasmid in the presence of 0.3 μg of CMV-CAT to normalize for transfection efficiency. The luciferase activity for each point transfected with CMV-FLAG was set at 1. (C) Deletion analyses of NIS promoter. Luciferase reporter constructs (3 μg/60-mm-diameter dish) containing various lengths of the NIS promoter were cotransfected into HeLa cells with 0.2 μg of Nkx-2.5 expression plasmid or CMV-FLAG (set at 1 in the experiment)/60-mm-diameter dish and 0.3 μg of CMV-CAT/60-mm-diameter dish. The relative activity of each luciferase reporter plasmid with cotransfected Nkx-2.5 or CMV-FLAG, expressed in arbitrary units, is also indicated. Deletion of the promoter region between −470 and −140 significantly decreased the activation of the promoter activity by Nkx-2.5.
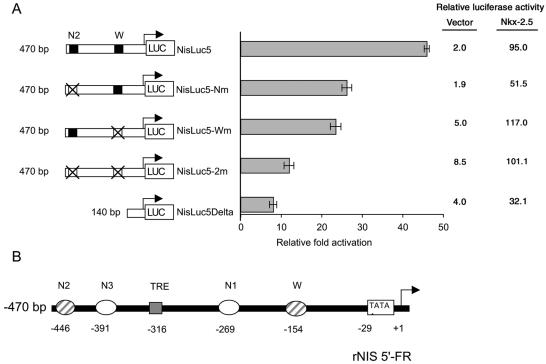
A dose-response curve obtained with increasing amounts of transfected Nkx-2.5 showed that the NISLuc2 promoter was induced in a dose-dependent fashion (Fig. 1B). To define the sequence requirements that mediate the transcriptional response, we analyzed a series of 5′-truncated rNIS promoter constructs. Deletions from the 5′ end revealed that, within the 2.7-kb 5′-flanking DNA examined, the 470-nt DNA fragment reproducibly induced maximal CAT activity (46-fold over the basal level) (Fig. 1C). An additional 5′ deletion, comprising only 140 bp of the 5′-flanking region of the rNIS gene, drastically reduced the response of the promoter (8- versus 46-fold), indicating that most of the Nkx-2.5-responsive elements are situated between nt −470 and −140 of the rNIS 5′-flanking region. A computer-assisted analysis of this region, using transcription element search software, revealed several Nkx-2.5 consensus binding sites, including the previously reported W site (12), which is critical for the TTF-1 response, as well as three new putative binding sites, which we named N1, N2, and N3 (see Fig. 3B and Table 1).
FIG. 3.
Effects of mutations in the N2 and W Nkx-2.5 binding sites of the rNIS promoter on the functional response to transiently expressed Nkx-2.5 in HeLa cells. (A) The N2 and W sites were mutated individually or together, by site-directed mutagenesis, in the context of the 470-bp rNIS promoter construct (see Materials and Methods). Three-microgram samples of the indicated reporter constructs were cotransfected into HeLa cells with 0.2 μg of Nkx-2.5 vector or CMV-FLAG/60-mm-diameter dish and with 0.3 μg of the CMV-CAT plasmid as an internal control. The relative fold induction in activity for each luciferase reporter plasmid was determined by calculating the Nkx-2.5/vector ratio when the vector (CMV-FLAG) activity was set at 1. The relative activity of each luciferase reporter plasmid with cotransfected Nkx-2.5 or CMV-FLAG, expressed in arbitrary units, is also indicated. The data shown are means ± SD of the luciferase/CAT ratios from four separate experiments performed in duplicate. A single mutation of the N2 or W site only partially reduced the transactivation response to Nkx-2.5. A disruption of both the N2 and W sites or their deletion drastically reduced the reporter gene activity. (B) Schematic representation of rNIS proximal promoter region. Critical binding sites (TRE and W), as designated in original papers, together with the putative Nkx-2.5 binding sites, are schematically shown. TRE, TSH response element. The numbers below each site indicate the distances from the transcriptional start site (+1).
Analysis of putative Nkx-2.5 binding sites in the rNIS promoter.
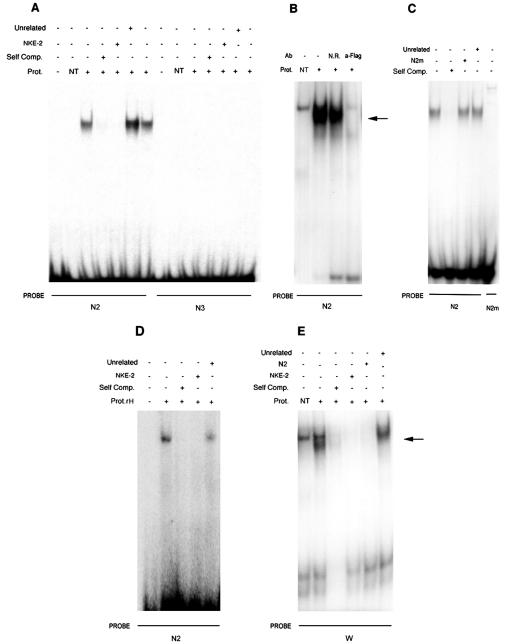
Using EMSAs, we examined the binding of a FLAG-tagged Nkx-2.5 protein with oligonucleotide probes corresponding to the predicted binding sites. A shifted band was visible when nuclear extracts from Nkx-2.5-transfected Bosc-23 cells were added to the radiolabeled N2 probe (Fig. 2A), whereas no complex was observed with oligonucleotides N1 (data not shown) and N3 (Fig. 2A). With the N2 oligonucleotide, a weaker band corresponding to endogenous Nkx-2.5 (9) also appeared in nontransfected cells (Fig. 2B). Nkx-2.5 binding to N2 is specific because it was efficiently displaced by an excess of unlabeled oligonucleotide or NKE-2 (Fig. 2A), which corresponds to the high-affinity Nkx-2.5 binding site derived from the proximal ANF promoter (35). Conversely, the Nkx-2.5/N2 complex was not competed by an unrelated oligonucleotide (Fig. 2A to C) or by a mutated N2 oligonucleotide (N2m, which contains a TGAG to CGCG mutation in the Nkx-2.5 binding core motif) (Fig. 2C). Moreover, radiolabeled N2m did not form a specific complex when incubated with Nkx-2.5-containing nuclear extracts (Fig. 2C). An antibody competition interference assay (see Materials and Methods) revealed a specific anti-FLAG displaceable protein, corresponding to Nkx-2.5, in the radiolabeled N2 complex (Fig. 2B). A band corresponding to Nkx-2.5 was also observed when rat heart nuclear proteins were incubated with the N2 oligonucleotide (Fig. 2D), indicating that endogenous Nkx-2.5 can bind to the N2 site. We also tested by EMSA whether the W oligonucleotide (previously identified as a TTF-1 binder) could bind to the Nkx-2.5 protein. The W oligonucleotide formed a specific complex with Nkx-2.5 that was specifically displaced by the appropriate competitors (Fig. 2E).
FIG. 2.
EMSA of interaction of Nkx-2.5 protein with three putative binding sites within the rNIS 5′ proximal promoter. (A) Ability of two putative Nkx-2.5 binding sites, N2 and N3, to form protein-DNA complexes with nuclear extracts from Nkx-2.5-transfected cells. Nuclear extracts (10 μg) from transfected cells with (+) or without (NT) the Nkx-2.5 expression plasmid were added to radiolabeled oligonucleotides as indicated. The specificity of the complex was verified by competition analysis. A constant amount (100-fold excess) of cold oligonucleotides was added to the mix reaction as indicated in Materials and Methods. (B) Antibody interference assay of Nkx-2.5 binding to the N2 site. Nuclear extracts with transfected Nkx-2.5 were preincubated with an anti-FLAG (α-FLAG) or unrelated (N.R.) antibody for 30 min before EMSA analysis. Preincubation with the anti-FLAG antibody prevented the formation of the Nkx-2.5-DNA complex, although a supershifted band is not visible (lane 4). (C) Mutational analysis of the N2 oligonucleotide. The labeled N2 oligonucleotide was incubated with nuclear extracts (10 μg) from Bosc-23 cells transfected with (+) the Nkx-2.5 expression plasmid, and the interaction was competed with a 100-fold molar excess of unlabeled N2 (self competition; lane 2), the N2m oligonucleotide (lane 3), or an unrelated oligonucleotide (lane 4). The N2m probe, which contains a TGAG to CGCG mutation, did not compete for the binding of N2 to Nkx-2.5 protein or was able to bind nuclear extracts from Nkx-transfected cells when it was radiolabeled (lane 5). (D) Interaction of cardiac protein with the N2 oligonucleotide. Endogenous Nkx-2.5 in rat cardiac nuclear extracts binds N2. Nuclear extracts (10 μg) from rat hearts were assayed for the capacity to form complexes with N2. A 100-fold molar excess of the indicated competitors was used as a control for binding specificity. (E) Binding of the TTF-1-responsive W oligonucleotide to Nkx-2.5 protein. Radiolabeled W was mixed with nuclear extracts (10 μg) from transfected cells with (+) or without (NT) the Nkx-2.5 expression plasmid. A competition analysis was performed to assess the specificity of the complex by adding a constant amount (100-fold excess) of the indicated cold oligonucleotides.
The N2 and W sites are required for a complete functional response of rNIS to Nkx-2.5.
As reported above, Nkx-2.5 binds to N2 and W. We measured the functional contribution of these sites to the transactivation of the rNIS promoter by disrupting one or both of them in the context of the 470-bp construct. A mutation of N2 (NisLuc5-Nm) only partially reduced the transactivation response to Nkx-2.5 (Fig. 3A). A comparable result was obtained with a deletion of the W site (NisLuc5-Wm) (Fig. 3A). Only when both N2 and W were mutated within the −470 promoter (NisLuc5-2m) was the promoter response drastically reduced (Fig. 3A), resulting in a mere 11-fold induction by Nkx-2.5. This value is comparable to that obtained with the minimally responsive NisLuc5Delta construct (eightfold). These experiments indicate that the 470-bp rNIS proximal promoter region contains at least two Nkx-2.5 binding sites, N2 and W, located approximately 440 and 150 bp, respectively, upstream of the transcription start site (Fig. 3B), and that these sites are critical for the NIS promoter response to Nkx-2.5.
Nkx-2.5 mRNA is not expressed in human and rat adult thyroids but is expressed in breast carcinomas.
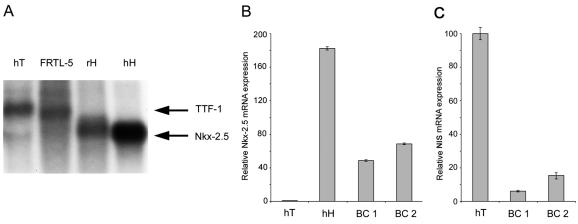
Nkx-2.5 mRNA is abundantly expressed in the mouse thyroid primordium, first in the primitive pharyngeal floor and subsequently in the group of cells that migrate anteroventrally to form the thyroid anlage (29). Given the unexpected role of Nkx-2.5 in activating thyroid-specific promoters, we measured Nkx-2.5 expression in the adult thyroid. Northern blotting with 10 μg of poly(A)+ RNA from human adult thyroid and FRTL-5 cells (derived from the rat thyroid) revealed a band of about 2.2 kb, corresponding to Nkx-2.5 mRNA, only in rat and human hearts (Fig. 4A, lanes 3 and 4), not in thyroids (lanes 1 and 2), which showed only a larger band. By reprobing the same filter with a TTF-1-specific probe, we identified the larger band in the thyroid samples as TTF-1 (data not shown). When we used real-time PCR with Nkx-2.5-specific oligonucleotides, there was no significant amplification corresponding to Nkx-2.5 from the human thyroid sample, as opposed to the human heart sample used as a positive control (Fig. 4B). Interestingly, Nkx-2.5 mRNA was expressed in two breast cancer samples (Fig. 4B) that also expressed significant NIS levels (Fig. 4C). Thus, Nkx-2.5 is virtually absent from the adult thyroid but is expressed in some breast cancer specimens. In this scenario, Nkx-2.5 is unlikely to play a role in adult life in sustaining the expression of thyroid-specific genes, including NIS.
FIG. 4.
Expression of Nkx-2.5 mRNA in adult thyroid and breast carcinoma. (A) Northern blot analysis with 10 μg of poly(A)+ RNAs from human thyroids (hT), human hearts (hH), rat hearts (rH), and rat FRTL-5 cells. The blot was probed with a mouse Nkx-2.5 cDNA fragment and exposed for 2 days at −80°C. (B) Nkx-2.5 expression levels in human thyroids, hearts, and breast carcinomas (BC1 and BC2) were measured by real-time PCR analysis (with oligonucleotides hNkx-303s and hNkx-389r [Table 1]). (C) NIS expression levels in breast carcinomas (BC1 and BC2) and in the human thyroid as a positive control were measured by real-time PCR analysis (with oligonucleotides hNis-2089s and hNis-2187r [Table 1]). β-Actin was used as a reference. For each reaction, standard curves for the reference gene were constructed by using six fourfold serial dilutions of cDNA. All samples were run in triplicate and reported as Nkx-2.5 (B) or NIS (C) expression levels relative to those in the thyroid (set at 1 for panel B and at 100 for panel C).
Nkx-2.5 mRNA and protein are present in lactating mammary glands and in RA-stimulated MCF-7 cells.
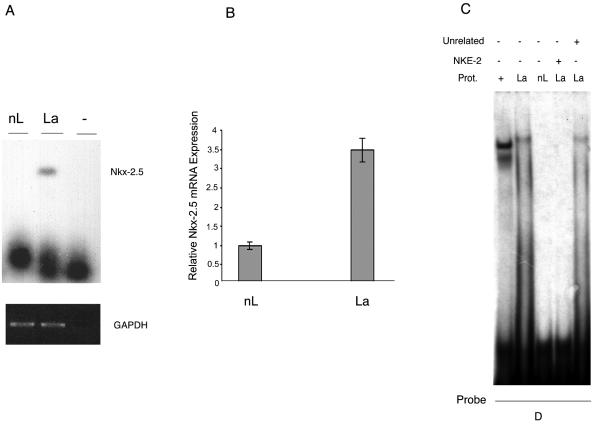
Given the novel finding of Nkx-2.5 expression in breast cancer specimens, we investigated Nkx-2.5 in other NIS-expressing mammary gland models. NIS is virtually absent from nubile rodent mammary glands, but its expression increases toward the end of gestation and is high in lactating mammary glands (40). We evaluated whether lactation-induced NIS was associated with Nkx-2.5 expression. RT-PCR (Fig. 5A) revealed a 405-bp band, corresponding to mNkx-2.5, that was only present in the lactating mammary gland. We verified this result by real-time PCR (Fig. 5B) and found increased Nkx-2.5 mRNA expression (3.5-fold) in lactating versus nonlactating mammary glands. Finally, we used EMSA (an anti-Nkx-2.5 antibody was unavailable) to identify the Nkx-2.5 protein in nuclear extracts from lactating and nonlactating mammary glands. Extracts were incubated with a radiolabeled oligonucleotide derived from the human type 2 deiodinase promoter (probe D), which specifically binds Nkx-2.5 (9). A single shifted complex appeared in the case of lactating tissue (Fig. 5C) but not in the case of the corresponding nonlactating extracts. Competition with NKE-2 and an unrelated oligonucleotide confirmed the specificity of this complex (Fig. 5C).
FIG. 5.
Differential expression of Nkx-2.5 in lactating versus nonlactating mammary glands. (A) RT-PCR analysis of Nkx-2.5 expression in the mouse mammary gland. Total RNAs were extracted from lactating and nonlactating mammary tissues, and cDNAs were prepared as previously described (9). After PCRs (35 cycles) with the mouse Nkx-2.5-specific primers NK1s and NK5r (Table 1), a Southern blot analysis revealed a single Nkx-2.5-corresponding band of 405 bp in the lactating mammary gland (lane 2). GAPDH amplification products (20 cycles) were used as an internal control, viewed by ethidium bromide staining, and photographed under UV light. (B) Real-time PCR assay of Nkx-2.5 expression in cDNAs from lactating and nonlactating mouse mammary glands. Nkx-2.5 mRNA expression was revealed by using the specific oligonucleotides mNkx-303s and mNkx-389r for mNkx-2.5 and the β-actin gene (Table 1). All samples were run in triplicate. The amount of Nkx-2.5 mRNA in the nonlactating mammary gland was set at 1. (C) EMSA of the interaction between the D oligonucleotide (which binds to Nkx-2.5) and nuclear extracts from mammary glands. Competition analysis was performed to verify the specificity of the complex by using a 100-fold molar excess of cold NKE-2 (lane 4) or an unrelated oligonucleotide (lane 5). Lane 1 corresponds to nuclear extracts (1 μg) from Nkx-2.5-transfected Bosc-23 cells used as a positive control for Nkx-2.5-oligo D binding. The autoradiogram exposure time was 16 h.
In conclusion, the results demonstrate that Nkx-2.5 mRNA and protein are present in mammary glands during a physiologically relevant period, i.e., lactation. This finding, together with the effect of Nkx-2.5 on the NIS promoter, suggests that this homeobox protein could affect NIS expression in mammary glands under physiological circumstances that call for an active NIS protein.
Nkx-2.5 participates in RA-induced NIS expression in MCF-7 cells.
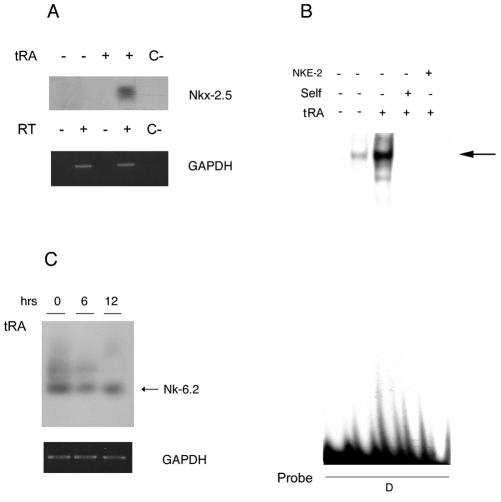
tRA induces NIS mRNA expression and iodide uptake in estrogen receptor-positive MCF-7 cells. NIS mRNA upregulation is time- and dose-dependent, with maximal stimulation at 12 h with 1 μM tRA (17). To determine whether Nkx-2.5 is involved in this induction, we used RT-PCR to analyze MCF-7 cells stimulated with 1 μM RA for 12 h. Semiquantitative RT-PCR and Southern blotting revealed an Nkx-2.5-corresponding 280-bp band in RA-treated MCF-7 cells (Fig. 6A). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) amplification was included as a control for equivalence of the cDNAs. This result indicates that Nkx-2.5 mRNA is induced in MCF-7 cells upon RA stimulation.
FIG. 6.
tRA induces Nkx-2.5 mRNA and protein in MCF-7 cells. (A) Nkx-2.5 mRNA expression in MCF-7 cells stimulated with 1 μM tRA for 12 h. The gels show an RT-PCR analysis with oligonucleotides Nkx-p1 and Nkx-p2 (Table 1) of cDNAs isolated from MCF-7 cells stimulated with 1 μM tRA or vehicle. Nkx-2.5 amplifications (35 cycles) were analyzed by Southern blotting, with a human Nkx-2.5 cDNA labeled fragment as a probe. GAPDH amplification products (20 cycles) were used as an internal control and were viewed by ethidium bromide staining under UV light. The figure shows results for one RT-PCR that was run twice and yielded comparable results. C−, no cDNA. (B) Effect of tRA on Nkx-2.5 protein levels in MCF-7 cells. Nuclear extracts from MCF-7 cells treated with 1 μM tRA (+) or vehicle (−) for 12 h were incubated with labeled oligonucleotide D. The free probe lane (lane 1) contained only labeled oligonucleotide D. The shifted band was competed with a 100-fold molar excess of cold oligonucleotide D (lane 4) or NKE-2 (lane 5). The autoradiogram exposure time was 16 h. (C) Semiquantitative RT-PCR assay to analyze NK-6.2 levels upon tRA treatment. Total RNAs were extracted from MCF-7 cells after treatment with 1 μM tRA for 6 and 12 h. The cDNAs, prepared as described in Materials and Methods, were amplified with the hmGtx-6.2-1s and hmGtx-6.2-2r oligonucleotides (Table 1) (30 cycles), and PCR products were analyzed by Southern blotting, with a human NK-6.2 cDNA labeled fragment as a probe.
We then determined whether Nkx-2.5 protein expression in MCF-7 cells corresponded to the RA-induced mRNA. Nuclear extracts from MCF-7 cells were incubated with radiolabeled oligonucleotide D as an Nkx-2.5 binder. A specific Nkx-2.5/oligonucleotide D complex, which was barely detectable in untreated cells, became clearly evident after tRA treatment (Fig. 6B). This complex was specifically competed by a molar excess of unlabeled self as well as by the NKE-2 oligonucleotide (Fig. 6B) and was absent when a mutated oligonucleotide D was used as a probe (data not shown). Overall, these data indicate that the Nkx-2.5 mRNA and protein are induced by RA treatment in MCF-7 cells.
To assess whether the effect of RA was specific for Nkx-2.5 or whether it corresponded to a general mechanism that activated other NK2 homeobox-containing proteins, we investigated whether tRA induces the expression of NK-6.2 (Gtx), the only other NK2 homeobox gene expressed in MCF-7 cells, as deduced from the MCF-7-specific UniGene database. RT-PCR with NK-6.2-specific oligonucleotides (Table 1) showed an NK-6.2-corresponding band in MCF-7 cells whose level remained unchanged after 6 and 12 h of RA treatment (Fig. 6C). This experiment indicates that, of the two NK2 transcription factors known in this cell type, RA specifically induces Nkx-2.5 mRNA.
RA stimulates the C5-CAT promoter in MCF-7 cells.
Given the effect of Nkx-2.5 on the NIS promoter, we asked whether Nkx-2.5 plays a functional role in RA-dependent NIS induction in MCF-7 cells. To this aim, we evaluated RA induction of the C5-CAT plasmid, which contains five repeated Nkx-2.5 consensus binding sites (25) and responds transcriptionally to Nkx-2.5 (35). Surprisingly, C5-CAT was potently induced by 1 μM tRA (116-fold over the basal level) (Fig. 7), an effect not observed with the E1B control plasmid that lacks the repeated Nkx-2.5-responsive elements. This effect was even higher than that observed with the two NIS promoter constructs, NisLuc2 (2.7 kb) and NisLuc5 (0.5 kb), which were stimulated by RA, consistent with the case for NIS mRNA (Fig. 7). These results show that RA induces a functionally active Nkx-2.5 in MCF-7 cells.
FIG. 7.
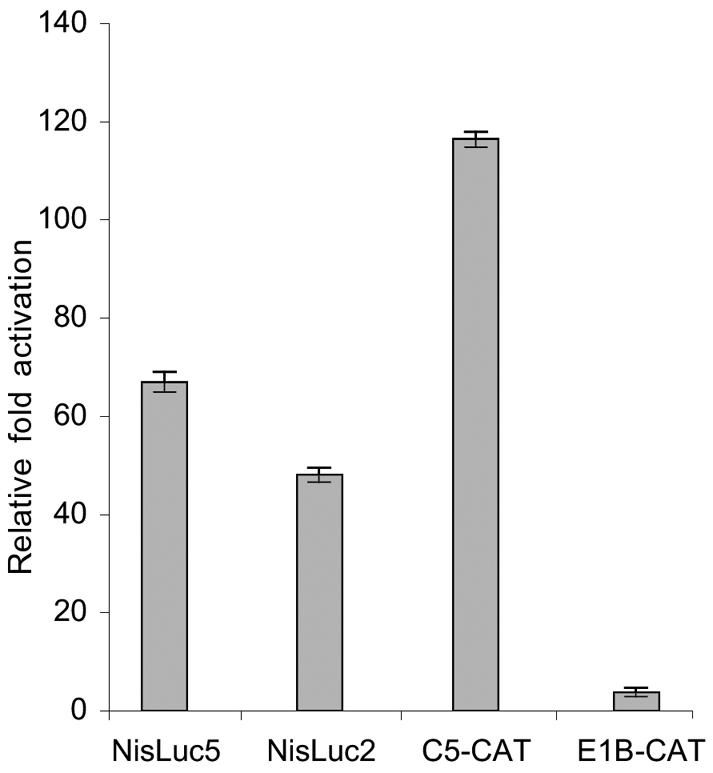
Effect of RA on C5-CAT and NIS promoter constructs. (A) MCF-7 cells were transiently cotransfected with 3 μg of the indicated reporter constructs/60-mm-diameter dish and with 0.3 μg of CMV-CAT or RSV-Luc plasmid by use of the Lipofectamine reagent. Reporter activities were determined 12 h after treatment with 1 μM tRA or equal amounts of vehicle. The results are expressed as the fold induction ± SD by tRA relative to vehicle-treated cells (set at 1 for each construct). Transfections were done in duplicate in three experiments that yielded similar results.
Time course of RA-induced Nkx-2.5 mRNA in MCF-7 cells.
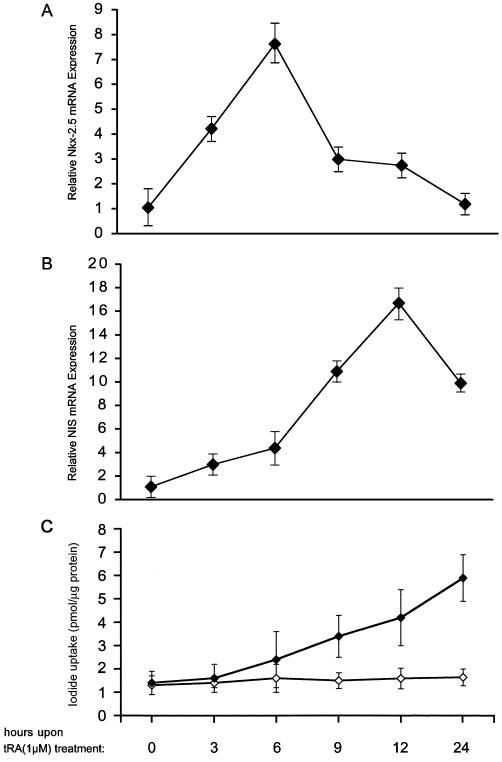
Because RA induces the Nkx-2.5 and NIS proteins in MCF-7 cells, and given the ability of Nkx-2.5 to induce the NIS promoter, we speculated that RA activation of NIS mRNA synthesis might be mediated by Nkx-2.5. In this context, we postulated that RA-induced Nkx-2.5 mRNA synthesis precedes RA-induced NIS mRNA transcription. To test this hypothesis, we used a time course experiment to analyze the RA-induced up-regulation of Nkx-2.5 in parallel with NIS mRNA induction in MCF-7 cells. Real-time PCR was conducted with cDNAs from MCF-7 cells treated with 1 μM RA for various times. In control cells (0 h), Nkx-2.5 basal levels rapidly increased as early as 3 h (4.2-fold) after exposure to 1 μM RA and reached a maximal level at 6 h (7.6-fold) (Fig. 8). Subsequently, Nkx-2.5 mRNA levels started to decrease and reached almost the control level 24 h after RA administration. NIS mRNA levels, analyzed in the same cDNA samples, had a different profile. As reported above, NIS reached its maximal level 12 h after RA stimulation (16.6-fold more than the control), i.e., 6 h later than the Nkx-2.5 mRNA peak (Fig. 8A and B). Iodide uptake showed a linear pattern in the time frame analyzed in this experiment (Fig. 8C), indicating that a mature and active NIS is synthesized in MCF-7 cells upon RA treatment. These findings support our hypothesis that the increased Nkx-2.5 levels induced by RA are related to the subsequent NIS induction.
FIG. 8.
Time course of RA-dependent induction of Nkx-2.5 and NIS mRNA and activity in MCF-7 cells. MCF-7 cells were incubated with 1 μM tRA for the indicated times. Total cDNAs were synthesized as described in the text. Real-time PCR was used to evaluate Nkx-2.5 (A) (oligonucleotides hNkx-303s and hNkx-389r) and NIS (B) (oligonucleotides hNIS-2089s and hNIS-2187r) mRNA levels after RA treatment. Relative template concentrations were calculated by using β-actin expression as an internal standard (calibrator) (see Materials and Methods). For each experiment, normalized copies of the target gene in untreated cells (0 h) were set at 1. The final results are expressed as N-fold differences in target gene expression relative to untreated cells. For each reaction, standard curves for the reference gene were constructed by using six fourfold serial dilutions of cDNA. All samples were run in triplicate, and PCRs were repeated three times with different cDNA preparations. (C) Characterization of iodide uptake by RA-treated MCF-7 cells. Iodide uptake was measured by incubating the cells with 500 μl of HBSS containing 10 μM NaI and carrier-free Na[125I] to give a specific activity of 10 mCi/mmol for 1 h (⧫). Half of the reaction mixture was supplemented with assay buffer plus 30 μM KClO4 (◊) as a control of specific uptake. The counts were measured in a γ-counter and normalized to the total amount of proteins measured by the Bio-Rad assay. The results are expressed as picomoles of incorporated iodide per microgram of protein. Error bars show SD. Each experiment was done in duplicate at least three times, with comparable results.
Nkx-2.5 is required for RA-induced transcription of NIS mRNA.
Having identified a time-consistent variation in Nkx-2.5 and NIS levels, we looked for a causal relationship between Nkx-2.5 induction and increased NIS expression. For this purpose, we examined the effect of an Nkx-2.5 dominant-negative mutant (Nkx-N188K) on RA-induced NIS activation. Nkx-N188K is a naturally occurring missense Nkx-2.5 mutant that has been described for patients with congenital heart disease (2). We recently demonstrated that Nkx-N188K does not transactivate the human type 2 deiodinase promoter, whereas it exerts a dominant-negative effect on the wild-type protein, in part by titrating it away from its target DNA (9).
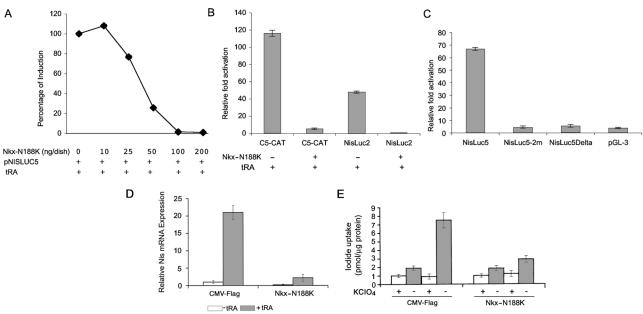
We first evaluated the effect of cotransfected Nkx-N188K on RA stimulation of the 470-bp NIS promoter. MCF-7 cells that were transiently transfected with the NisLuc5 construct were treated with RA in the presence or absence of Nkx-N188K. Nkx-N188K dose-dependently blocked the induction by RA (Fig. 9A), and at 100 ng/dish, it completely blocked RA-induced NIS promoter activation. A complete elimination of the RA effect was also observed when cells were transfected with the longer NisLuc2 promoter construct or with the artificial promoter C5-CAT (Fig. 9B), suggesting that Nkx-2.5 is crucial for the induction of these promoters by RA.
FIG. 9.
Inhibition of RA-induced NIS by dominant-negative Nkx-2.5 mutant (N188K). (A) Dose-response curve showing effect of Nkx-N188K on RA-induced NisLuc5 promoter activity. MCF-7 cells were transiently cotransfected by use of the Lipofectamine reagent with the NisLuc5 construct (3 μg/60-mm-diameter dish) and the indicated amounts of Nkx-N188K or CMV-FLAG as a control. Twelve hours before being harvested, the cells were treated with 1 μM tRA or an equal amount of vehicle. A 100% induction represents the effect induced by RA on the luciferase/CAT ratio when compared to the control without tRA. (B) Inhibition of RA-dependent activation of C5-CAT and NisLuc2 constructs mediated by Nkx-N188K. MCF-7 cells were transiently cotransfected with the indicated reporters (3 μg/60-mm-diameter dish) and Nkx-N188K (100 ng/60-mm-diameter dish) or CMV-FLAG. Twelve hours before being harvested, the cells were treated with 1 μM tRA or vehicle. For the various reporter constructs, the CAT/luciferase or luciferase/CAT ratios obtained without RA were set at 1. The results shown are means ± SD of the CAT/luciferase or luciferase/CAT ratios of at least three separate experiments done in duplicate. (C) The N2 and W sites in the rNIS promoter are required for RA-induced NIS promoter activity. The N2 and W sites were mutated (NisLuc5-2m) by site-directed mutagenesis in the context of the 470-bp rNIS promoter construct, and the reporter constructs obtained were cotransfected into MCF-7 cells. The pGL3 basic vector was transfected as a negative control. Twelve hours before being harvested, the cells were stimulated with 1 μM tRA or vehicle. For each reporter construct, the luciferase/CAT ratio obtained with the vehicle was set at 1. The data shown are means ± SD of the luciferase/CAT ratios from four separate experiments done in duplicate. (D and E) Endogenous NIS response to tRA is obscured by Nkx-N188K. MCF-7 cells were transiently cotransfected with the Nkx-N188K or CMV-FLAG plasmid as a control. (D) Twelve hours before being harvested, the cells were treated with 1 μM tRA or vehicle. NIS expression was measured by real-time PCR analysis. All samples were run in triplicate, and results are reported as NIS expression levels relative to unstimulated CMV-Flag-transfected cells (set at 1). (E) Twelve hours after transfection, the cells were treated with 1 μM tRA or an equal amount of vehicle, and iodide uptake was measured 48 h thereafter. The results are expressed as picomoles of incorporated iodide per microgram of protein. Error bars show the SD. Each experiment was done in duplicate three times, with comparable results.
To evaluate the role played by the Nkx-2.5 binding sites N2 and W in the stimulation induced by RA, we repeated the cotransfection experiments with the NisLuc5-2m and NisLuc5Delta constructs, neither of which contains the N2 or W site, and measured the functional response to RA (Fig. 9C). Interestingly, the Nis-Luc5 response to RA was drastically reduced in the absence of N2 and W (Fig. 9C). NisLuc5-2m resulted in only a fivefold response to RA, which was comparable to that observed with the NisLuc5Delta construct (sixfold). These findings demonstrate that Nkx-2.5 mediates the effect exerted by RA on the NIS promoter and that the integrity of the Nkx-2.5 binding sites is required for this effect.
In order to assess whether RA induces endogenous NIS up-regulation in MCF-7 cells through an Nkx-2.5-dependent mechanism, we blocked the Nkx-2.5 action via its dominant-negative mutant. We measured the effect of RA treatment on endogenous NIS mRNA in MCF-7 cells that were transiently transfected with Nkx-N188K. The expression of Nkx-N188K drastically reduced RA-induced NIS mRNA up-regulation, an effect that was not observed in control transfected cells (Fig. 9D). Iodide uptake was also consistently negatively affected by the dominant-negative Nkx-2.5 isoform (Fig. 9E).
Taken together, these findings show that NIS expression is controlled by endogenous Nkx-2.5 and that RA signaling on the NIS gene occurs via an Nkx-2.5-dependent mechanism.
Forced expression of Nkx-2.5 in MCF-7 cells is sufficient to induce NIS transcription.
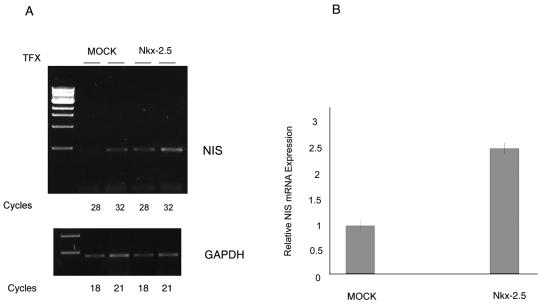
The finding that Nkx-2.5 expressed in MCF-7 cells and in breast tumors is a potent inducer of the NIS gene prompted us to assess the role of a forced overexpression of Nkx-2.5 in mammary tumor cells. We investigated whether Nkx-2.5 induced NIS mRNA transcription in the absence of RA. Total RNAs were extracted from transiently Nkx-transfected MCF-7 cells, and NIS levels were measured by semiquantitative RT-PCR (Fig. 10A). Despite the partial efficiency of transient transfections (about 40% under these conditions [data not shown]), the NIS mRNA levels were increased in Nkx-2.5-transfected cells. We used semiquantitative PCR of GAPDH to verify the equivalence of cDNAs (Fig. 10A). We also measured NIS mRNAs by real-time PCR and found a 2.7-fold higher NIS mRNA level in Nkx-2.5-transfected cells than in control cells (Fig. 10B). This experiment suggested that Nkx-2.5 alone is sufficient to induce NIS mRNA in MCF-7 cells.
FIG. 10.
Stimulation of NIS mRNA in MCF-7 cells transiently transfected with Nkx-2.5. (A) cDNAs from Nkx-2.5- or mock-transfected MCF-7 cells were analyzed by PCRs (28 and 32 cycles) with oligonucleotides Nis2s and Nis3r (Table 1) for NIS expression. A 465-bp DNA fragment corresponding to the NIS cDNA was increased by Nkx-2.5 transfection compared to mock-transfected cells. GAPDH amplification products (18 and 21 cycles) were used as an internal control. The figure shows the results for one RT-PCR, which was run twice and yielded comparable results. (B) Real-time PCR with oligonucleotides hNIS-2089s and hNIS-2187r (Table 1) to evaluate NIS mRNA levels in MCF-7 transiently transfected cells. NIS amplifications were normalized to the expression of β-actin as an internal standard (calibrator). The normalized NIS mRNA level in mock-transfected cells was set at 1. The results are expressed as N-fold differences in target gene expression relative to mock-transfected cells. For each reaction, standard curves for target and reference genes were constructed by using fourfold serial dilutions of cDNAs. All samples were run in triplicate and PCRs were run three times with the same cDNAs.
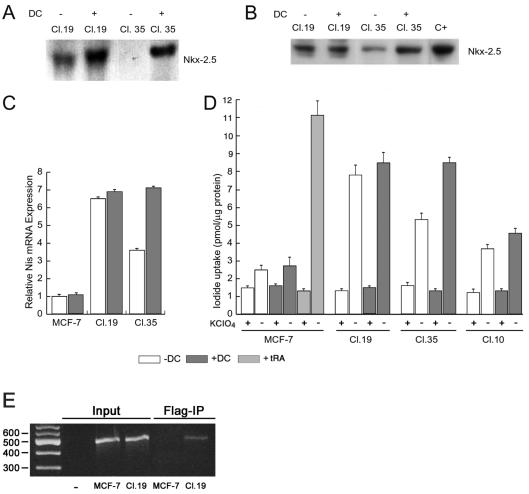
To better characterize the potential of Nkx-2.5 to induce NIS, we generated MCF-7 clones in which Nkx-2.5 gene expression was controlled by a tetracycline-inducible promoter. FLAG-tagged Nkx-2.5 controlled by a tetracycline-responsive promoter was stably cotransfected into the MCF-7-TetON cell line together with a TK-hygro plasmid (see Materials and Methods). After transfection, cells were selected based on their capability of growing in antibiotic-selective medium (see Materials and Methods). Of the 70 clones analyzed, clones 19 and 35 were selected for their ability to express Nkx-2.5 transgene mRNA and protein in the presence of 1 μg of doxycycline/μl (Fig. 11A and B). Clone 19 had elevated levels of Nkx-2.5, regardless of doxycycline treatment, whereas clone 35 had a robust doxycycline-dependent Nkx-2.5 induction at both the mRNA and protein levels (Fig. 11A and B). Having established a system by which to manipulate Nkx-2.5 levels, we used the selected clones to analyze the effect of exogenous Nkx-2.5 on NIS levels.
FIG. 11.
Selection and characterization of MCF-7-TetON clones expressing Nkx-2.5. (A) Northern blot of doxycycline-induced Nkx-2.5 expression in MCF-7-TetON clones. Two clones, 19 and 35, were selected for their ability to express Nkx-2.5 mRNA and protein. After 12 h of doxycycline treatment, total RNAs (30 μg) were resolved in a 0.8% agarose gel, blotted onto a nylon membrane, and probed for Nkx-2.5. (B) Western blot analysis of Nkx-2.5 expression in doxycycline-stimulated MCF-7 clones. After doxycycline treatment, the cells were lysed and 20 μg of nuclear extracts was subjected to SDS gel electrophoresis and immunoblotted with a monoclonal anti-FLAG antibody (M2; Sigma). C+, nuclear extracts from Nkx-2.5-transfected Bosc-23 cells used as a positive control. (C) NIS mRNA levels were measured by real-time PCR in doxycycline-stimulated clones 19 and 35 and in parental MCF-7-TetON cells. Specific primers for NIS (hNIS-2089s and hNIS-2187r) and the β-actin gene were used (Table 1). All samples were run in triplicate. The relative amounts of gene expression were calculated by using the expression of β-actin as an internal standard (calibrator). The normalized NIS mRNA level in unstimulated MCF-7-TetON cells was set at 1. (D) Iodide uptake assay of doxycycline-stimulated clones 19 and 35 and MCF-7-TetON cells treated with 1 μM tRA for 48 h. The cells were treated with 1 μg of doxycycline/μl for 72 h before the assay. The results are expressed as picomoles of incorporated iodide per microgram of protein. Error bars show the SD. Each experiment was done in duplicate at least three times, with comparable results. (E) Binding of Nkx-2.5 protein to human NIS promoter. ChIP was performed with antibodies against FLAG-tagged Nkx-2.5 with clone 19 and with MCF-7 cells used as a negative control. The immunoprecipitates were analyzed by PCRs (Flag-IP) with oligonucleotide primers (hNIS-728s and hNIS-250r) (Table 1) corresponding to the human NIS promoter (nt −728 to −250 from the ATG). Parallel PCRs were performed with total input DNAs obtained from unprecipitated aliquots of similarly treated chromatin samples.
After 12 h of doxycycline treatment, cDNA samples were analyzed by real-time PCR for NIS mRNA levels. The NIS mRNA levels were increased in clone 19 and were not significantly affected by doxycycline. Although the NIS mRNA levels were slightly increased in clone 35, they were similar to those in parental cells under basal conditions. After doxycycline treatment, the NIS levels increased 7.1-fold and reached levels comparable to those measured in clone 19. Interestingly, NIS expression in both clones reflected the Nkx-2.5 levels (Fig. 11A and B). Because RA-induced NIS mRNA increased the iodide uptake in MCF-7 cells, we examined whether Nkx-2.5 mediated a similar effect in the selected clones. It should be noted that the NIS protein must be expressed, targeted, and retained at the plasma membrane surface for active I transport to occur (10). After 72 h of treatment with doxycycline, clones 19 and 35 and the parental MCF-7 cells were assayed for I− uptake as described in Materials and Methods (Fig. 11D). For both selected clones, Nkx-2.5 expression resulted in a specific increase in KClO4-inhibitable I− uptake that was significant compared to that of the parental MCF-7-TetON cells or the RA-treated MCF-7 cells used as a positive control (Fig. 11D). Iodide uptake was also increased in clone 10, in which Nkx-2.5 transgene expression was just barely detectable (data not shown). This indicates that even modest Nkx-2.5 levels can regulate endogenous NIS, as was also observed with unstimulated clone 35.
To gain insight into the mechanism by which Nkx-2.5 promotes NIS expression, we examined the association in vivo between the Nkx-2.5 protein and the NIS promoter by using ChIP. To overcome the lack of a specific anti-Nkx-2.5 antibody, we used MCF-7 cells expressing flagged Nkx-2.5 (clone 19). The transcriptional response of the human promoter to Nkx-2.5 was similar to that of the rNIS promoter (M. Dentice, unpublished results). Interestingly, Nkx-2.5 can physically interact with the NIS promoter, as deduced by the 478-bp product, corresponding to the NIS promoter, obtained from the immunoprecipitate from clone 19 (Fig. 11E) but not from untransfected MCF-7 cells used as a negative control.
Taken together, these data indicate that Nkx-2.5 induces NIS expression in MCF-7 cells by a physical association with the NIS promoter and that a mature and functioning NIS protein is synthesized in these cells as a consequence of Nkx-2.5 expression.
DISCUSSION
In this study, we demonstrated that the transcription factor Nkx-2.5 is a crucial component in the transcriptional regulation of NIS in the mammary gland and in breast cancer cells. Several homeobox genes have been demonstrated to be expressed in mammary glands, and there is compelling evidence that they are important for normal mammary development (21). Although homeobox genes clearly occupy a prominent position in the developmental regulatory hierarchy, the NK-2 subset of homeobox genes has received little attention with respect to mammary gland organogenesis, functional differentiation, and cancer. We provide the first evidence that the Nkx-2.5 homeobox gene plays a role in the mammary gland.
Besides its key role in thyroid physiology, NIS-mediated iodide accumulation is a prerequisite for diagnostic thyroid scintigraphy and for radioiodine treatment of benign and malignant thyroid disease. Because decreased NIS expression and iodide uptake in dedifferentiated thyroid carcinomas lower the efficiency of radioiodide therapy, attempts have been made to stimulate NIS expression in transformed cancer cells. However, the lack of data about the regulation of NIS expression has hampered this approach. Here we report that Nkx-2.5 is a potent inducer of the NIS promoter. Indeed, in HeLa cells it activated the NIS gene about 50-fold.
We also showed that thyroid-specific promoters respond transcriptionally to Nkx-2.5. This was predictable given the partial overlap in the Nkx-2.5 and TTF-1 consensus binding sites. In contrast, we did not expect the high response of the NIS promoter to Nkx-2.5 because among the thyroid promoters tested, NIS was the least responsive to TTF-1 in vitro, i.e., only 2.5-fold over basal values in transfected cells (12) and practically unresponsive to TTF-1 in vivo (32). In this regard, it is noteworthy that the NIS and Nkx-2.5 genes are evolutionarily conserved, as demonstrated by the presence of NIS and Nkx-2.5 orthologs in the Ciona intestinalis genome (8).
The function of Nkx-2.5 in thyroid morphogenesis and differentiation is unknown. Murine Nkx-2.5 begins to be expressed in thyroid precursor cells at embryonic day 8.5 (E8.5), a period coincident with the appearance of TTF-1, Pax-8, and TTF-2 (6). However, unlike that of other thyroid transcription factors, Nkx-2.5 expression disappears from the thyroid around E12.5 (22), before the appearance of thyroid differentiation markers, including NIS (6). It should be noted that expression data are based on mRNA detection by in situ hybridization and that the Nkx-2.5 protein might have a slightly different expression profile. We showed here that Nkx-2.5 mRNA is virtually absent from the adult human thyroid. Overall, these data indicate that Nkx-2.5 is unlikely to be involved in maintaining NIS expression in the adult thyroid, although its expression during development suggests that it could be involved in thyroid commitment.
RA is required for such diverse cellular programs as development, differentiation, and growth. The retinoid signal is transduced by the RARs and their isoforms and by the retinoid X receptors. In MCF-7 cells, RA induces NIS expression (17) and a complex program culminating in the inhibition of cell progression and apoptosis (41). Here we demonstrated that RA efficiently induces Nkx-2.5 expression and activity in MCF-7 cells. Nkx-2.5 expression was very low in MCF-7 cells under basal conditions and rapidly increased upon RA treatment (Fig. 6 and 8). Cotransfection with Nkx-N188K completely eliminated RA-induced NIS activation, indicating that RA signaling on the NIS gene is Nkx-2.5 dependent. In this context, it is not inconceivable that Nkx-2.5 mediates the RA signal transduction pathway by regulating downstream genes.
Retinoids accelerate the morphological differentiation of mammary epithelial cells, thereby increasing the expression of casein, the major milk protein, and lactoferrin (19). In this scenario, Nkx-2.5 induction by retinoids is a novel transcriptional mechanism that increases the expression of downstream differentiation markers, of which NIS is probably not an isolated example. In this context, Nkx-2.5 expression seems to be required for optimal NIS expression in mammary glands during the physiologically relevant period of lactation.
Some breast cancers express NIS, which is responsible for iodide uptake by these malignancies (40). We demonstrated Nkx-2.5 expression in two human breast cancer specimens that were characterized by high NIS expression. It will be interesting to determine whether or not NIS expression is associated with concurrent Nkx-2.5 expression in a large series of human breast cancers.
What is the physiological relevance of Nkx-2.5 expression during lactation? As a general principle, homeobox genes encode transcription factors that play pivotal roles in the determination and maintenance of cell fate and cell identity. In all likelihood, homeobox genes control mammary gland development and function by the sequential activation or inactivation of specific sets of genes at specific developmental transition points. For example, Hoxa9, Hoxb9, and Hoxd9 are important for alveolar development near midpregnancy, whereas Hoxd10 is required at the next major transition point, i.e., the onset of lactation (21). As suggested by our expression data, Nkx-2.5 probably plays a critical role in the mammary gland near or during lactation. Important questions remain about Nkx-2.5 gene function in the diverse phases of mammary gland development and about the precise cellular localization of Nkx-2.5 in the mammary gland. However, our data indicate that Nkx-2.5 induction is involved in inducing NIS expression during lactation.
In mammary glands, sexual hormones that are stimulated during pregnancy, including estrogens, are involved in NIS expression (40). It will be interesting to investigate the direct effect of these hormones on Nkx-2.5 and to determine whether RA directly affects Nkx-2.5 transcription. In this regard, we found several putative RA binding sites in the human Nkx-2.5 promoter region. Preliminary data from our laboratory indicate that RA induces the proximal 1.9 kb of the human Nkx-2.5 promoter. It is feasible that RAR binds to the Nkx-2.5 promoter, thereby activating its transcription and so inducing NIS. Detailed functional promoter studies are necessary to verify this model. However, direct protein-protein interactions occur between RAR and TTF-1 (46), so mechanisms other than transcriptional regulation cannot be excluded.
In conclusion, we report a hitherto unrecognized role of Nkx-2.5 in mammary gland physiology. Our experiments show that Nkx-2.5 is a potent positive regulator of NIS transcription. In breast cancer cells, Nkx-2.5 mediates the effect of RA on NIS regulation, suggesting an additional pathway for retinoid-dependent mammary cell differentiation. Nkx-2.5 plays a primary role in RA-induced NIS transcription and probably in the hormonal regulation of NIS expression in the mammary gland during lactation. The ability of Nkx-2.5 to induce iodide uptake in breast cancer cells may be useful for the diagnosis and treatment of some differentiated breast cancers.
Acknowledgments
We are grateful to Roberto Di Lauro for helpful discussions and guidance and for sharing several reagents. We thank Massimo Santoro and Giuseppe Viglietto for critical reading of the manuscript, Jean Gilder (Scientific Communication) for text editing, and Salvatore Salzano for expert technical assistance.
This work was supported by a CNR grant 2001 to D.S.
REFERENCES
- 1.Behr, M., T. L. Schmitt, C. R. Espinoza, and U. Loos. 1998. Cloning of a functional promoter of the human sodium/iodide-symporter gene. Biochem. J. 331:359-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson, D. W., G. M. Silberbach, A. Kavanaugh-McHugh, C. Cottrill, Y. Zhang, S. Riggs, O. Smalls, M. C. Johnson, M. S. Watson, J. G. Seidman, C. E. Seidman, J. Plowden, and J. D. Kugler. 1999. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J. Clin. Investig. 104:1567-1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, C. Y., and R. J. Schwartz. 1995. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J. Biol. Chem. 270:15628-15633. [DOI] [PubMed] [Google Scholar]
- 4.Cho, J. Y., R. Leveille, R. Kao, B. Rousset, A. F. Parlow, W. E. Burak, Jr., E. L. Mazzaferri, and S. M. Jhiang. 2000. Hormonal regulation of radioiodide uptake activity and Na+/I− symporter expression in mammary glands. J. Clin. Endocrinol. Metab. 85:2936-2943. [DOI] [PubMed] [Google Scholar]
- 5.Civitareale, D., R. Lonigro, A. J. Sinclair, and R. Di Lauro. 1989. A thyroid-specific nuclear protein essential for tissue-specific expression of the thyroglobulin promoter. EMBO J. 8:2537-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Damante, G., G. Tell, and R. Di Lauro. 2001. A unique combination of transcription factors controls differentiation of thyroid cells. Prog. Nucleic Acids Res. Mol. Biol. 66:307-356. [DOI] [PubMed] [Google Scholar]
- 7.De Felice, M., G. Damante, M. Zannini, H. Francis-Lang, and R. Di Lauro. 1995. Redundant domains contribute to the transcriptional activity of the thyroid transcription factor 1. J. Biol. Chem. 270:26649-26656. [DOI] [PubMed] [Google Scholar]
- 8.Dehal, P., Y. Satou, R. K. Campbell, J. Chapman, B. Degnan, A. De Tomaso, B. Davidson, A. Di Gregorio, M. Gelpke, D. M. Goodstein, N. Harafuji, K. E. Hastings, I. Ho, K. Hotta, W. Huang, T. Kawashima, P. Lemaire, D. Martinez, I. A. Meinertzhagen, S. Necula, M. Nonaka, N. Putnam, S. Rash, H. Saiga, M. Satake, A. Terry, L. Yamada, H. G. Wang, S. Awazu, K. Azumi, J. Boore, M. Branno, S. Chin-Bow, R. DeSantis, S. Doyle, P. Francino, D. N. Keys, S. Haga, H. Hayashi, K. Hino, K. S. Imai, K. Inaba, S. Kano, K. Kobayashi, M. Kobayashi, B. I. Lee, K. W. Makabe, C. Manohar, G. Matassi, M. Medina, Y. Mochizuki, S. Mount, T. Morishita, S. Miura, A. Nakayama, S. Nishizaka, H. Nomoto, F. Ohta, K. Oishi, I. Rigoutsos, M. Sano, A. Sasaki, Y. Sasakura, E. Shoguchi, T. Shin-i, A. Spagnuolo, D. Stainier, M. M. Suzuki, O. Tassy, N. Takatori, M. Tokuoka, K. Yagi, F. Yoshizaki, S. Wada, C. Zhang, P. D. Hyatt, F. Larimer, C. Detter, N. Doggett, T. Glavina, T. Hawkins, P. Richardson, S. Lucas, Y. Kohara, M. Levine, N. Satoh, and D. S. Rokhsar. 2002. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298:2157-2167. [DOI] [PubMed] [Google Scholar]
- 9.Dentice, M., C. Morisco, M. Vitale, G. Rossi, G. Fenzi, and D. Salvatore. 2003. The different cardiac expression of the type 2 iodothyronine deiodinase gene between human and rat is related to the differential response of the dio2 genes to Nkx-2.5 and GATA-4 transcription factors. Mol. Endocrinol. 17:1508-1521. [DOI] [PubMed] [Google Scholar]
- 10.Dohan, O., A. De La Vieja, V. Paroder, C. Riedel, M. Artani, M. Reed, C. S. Ginter, and N. Carrasco. 2003. The sodium/iodide symporter (NIS): characterization, regulation, and medical significance. Endocr. Rev. 24:48-77. [DOI] [PubMed] [Google Scholar]
- 11.Dulgeroff, A. J., and J. M. Hershman. 1994. Medical therapy for differentiated thyroid carcinoma. Endocr. Rev. 15:500-515. [DOI] [PubMed] [Google Scholar]
- 12.Endo, T., M. Kaneshige, M. Nakazato, M. Ohmori, N. Harii, and T. Onaya. 1997. Thyroid transcription factor-1 activates the promoter activity of rat thyroid Na+/I− symporter gene. Mol. Endocrinol. 11:1747-1755. [DOI] [PubMed] [Google Scholar]
- 13.Francis-Lang, H., M. Price, M. Polycarpou-Schwarz, and R. Di Lauro. 1992. Cell-type-specific expression of the rat thyroperoxidase promoter indicates common mechanisms for thyroid-specific gene expression. Mol. Cell. Biol. 12:576-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson, U. E., C. A. Heid, and P. M. Williams. 1996. A novel method for real time quantitative RT-PCR. Genome Res. 6:995-1001. [DOI] [PubMed] [Google Scholar]
- 15.Kasahara, H., S. Bartunkova, M. Schinke, M. Tanaka, and S. Izumo. 1998. Cardiac and extracardiac expression of Csx/Nkx2.5 homeodomain protein. Circ. Res. 82:936-946. [DOI] [PubMed] [Google Scholar]
- 16.Kawaguchi, A., M. Ikeda, T. Endo, T. Kogai, A. Miyazaki, and T. Onaya. 1997. Transforming growth factor-beta1 suppresses thyrotropin-induced Na+/I− symporter messenger RNA and protein levels in FRTL-5 rat thyroid cells. Thyroid 7:789-794. [DOI] [PubMed] [Google Scholar]
- 17.Kogai, T., J. J. Schultz, L. S. Johnson, M. Huang, and G. A. Brent. 2000. Retinoic acid induces sodium/iodide symporter gene expression and radioiodide uptake in the MCF-7 breast cancer cell line. Proc. Natl. Acad. Sci. USA 97:8519-8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komuro, I., and S. Izumo. 1993. Csx: a murine homeobox-containing gene specifically expressed in the developing heart. Proc. Natl. Acad. Sci. USA 90:8145-8149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, P. P., M. T. Lee, K. M. Darcy, K. Shudo, and M. M. Ip. 1995. Modulation of normal mammary epithelial cell proliferation, morphogenesis, and functional differentiation by retinoids: a comparison of the retinobenzoic acid derivative RE80 with retinoic acid. Endocrinology 136:1707-1717. [DOI] [PubMed] [Google Scholar]
- 20.Lee, Y., T. Shioi, H. Kasahara, S. M. Jobe, R. J. Wiese, B. E. Markham, and S. Izumo. 1998. The cardiac tissue-restricted homeobox protein Csx/Nkx2.5 physically associates with the zinc finger protein GATA4 and cooperatively activates atrial natriuretic factor gene expression. Mol. Cell. Biol. 18:3120-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lewis, M. T. 2000. Homeobox genes in mammary gland development and neoplasia. Breast Cancer Res. 2:158-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lints, T. J., L. M. Parsons, L. Hartley, I. Lyons, and R. P. Harvey. 1993. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development 119:969. [DOI] [PubMed] [Google Scholar]
- 23.Mandell, R. B., L. Z. Mandell, and C. J. Link, Jr. 1999. Radioisotope concentrator gene therapy using the sodium/iodide symporter gene. Cancer Res. 59:661-668. [PubMed] [Google Scholar]
- 24.Missero, C., G. Cobellis, M. De Felice, and R. Di Lauro. 1998. Molecular events involved in differentiation of thyroid follicular cells. Mol. Cell. Endocrinol. 140:37-43. [DOI] [PubMed] [Google Scholar]
- 25.Missero, C., M. T. Pirro, and R. Di Lauro. 2000. Multiple Ras downstream pathways mediate functional repression of the homeobox gene product TTF-1. Mol. Cell. Biol. 20:2783-2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mountford, P. J., A. J. Coakley, I. R. Fleet, M. Hamon, and R. B. Heap. 1986. Transfer of radioiodide to milk and its inhibition. Nature 322:600. [DOI] [PubMed] [Google Scholar]
- 27.Ohno, M., M. Zannini, O. Levy, N. Carrasco, and R. di Lauro. 1999. The paired-domain transcription factor Pax8 binds to the upstream enhancer of the rat sodium/iodide symporter gene and participates in both thyroid-specific and cyclic-AMP-dependent transcription. Mol. Cell. Biol. 19:2051-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ray, M. K., C. Y. Chen, R. J. Schwartz, and F. J. DeMayo. 1996. Transcriptional regulation of a mouse Clara cell-specific protein (mCC10) gene by the NKx transcription factor family members thyroid transcription factor 1 and cardiac muscle-specific homeobox protein (CSX). Mol. Cell. Biol. 16:2056-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reecy, J. M., X. Li, M. Yamada, F. J. DeMayo, C. S. Newman, R. P. Harvey, and R. J. Schwartz. 1999. Identification of upstream regulatory regions in the heart-expressed homeobox gene Nkx2-5. Development 126:839-849. [DOI] [PubMed] [Google Scholar]
- 30.Ryu, K. Y., Q. Tong, and S. M. Jhiang. 1998. Promoter characterization of the human Na+/I− symporter. J. Clin. Endocrinol. Metab. 83:3247-3251. [DOI] [PubMed] [Google Scholar]
- 31.Saito, T., T. Endo, A. Kawaguchi, M. Ikeda, M. Nakazato, T. Kogai, and T. Onaya. 1997. Increased expression of the Na+/I− symporter in cultured human thyroid cells exposed to thyrotropin and in Graves' thyroid tissue. J. Clin. Endocrinol. Metab. 82:3331-3336. [DOI] [PubMed] [Google Scholar]
- 32.Schmitt, T. L., C. R. Espinoza, and U. Loos. 2001. Transcriptional regulation of the human sodium/iodide symporter gene by Pax8 and TTF-1. Exp. Clin. Endocrinol. Diabetes 109:27-31. [DOI] [PubMed] [Google Scholar]
- 33.Schmutzler, C. 2001. Regulation of the sodium/iodide symporter by retinoids—a review. Exp. Clin. Endocrinol. Diabetes 109:41-44. [DOI] [PubMed] [Google Scholar]
- 34.Schmutzler, C., T. L. Schmitt, F. Glaser, U. Loos, and J. Kohrle. 2002. The promoter of the human sodium/iodide-symporter gene responds to retinoic acid. Mol. Cell. Endocrinol. 189:145-155. [DOI] [PubMed] [Google Scholar]
- 35.Shiojima, I., I. Komuro, T. Oka, Y. Hiroi, T. Mizuno, E. Takimoto, K. Monzen, R. Aikawa, H. Akazawa, T. Yamazaki, S. Kudoh, and Y. Yazaki. 1999. Context-dependent transcriptional cooperation mediated by cardiac transcription factors Csx/Nkx-2.5 and GATA-4. J. Biol. Chem. 274:8231-8239. [DOI] [PubMed] [Google Scholar]
- 36.Sinclair, A. J., R. Lonigro, D. Civitareale, L. Ghibelli, and R. Di Lauro. 1990. The tissue-specific expression of the thyroglobulin gene requires interaction between thyroid-specific and ubiquitous factors. Eur. J. Biochem. 193:311-318. [DOI] [PubMed] [Google Scholar]
- 37.Spitzweg, C., M. K. O'Connor, E. R. Bergert, D. J. Tindall, C. Y. Young, and J. C. Morris. 2000. Treatment of prostate cancer by radioiodine therapy after tissue-specific expression of the sodium iodide symporter. Cancer Res. 60:6526-6530. [PubMed] [Google Scholar]
- 38.Strum, J. M. 1978. Site of iodination in rat mammary gland. Anat. Rec. 192:235-244. [DOI] [PubMed] [Google Scholar]
- 39.Taki, K., T. Kogai, Y. Kanamoto, J. M. Hershman, and G. A. Brent. 2002. A thyroid-specific far-upstream enhancer in the human sodium/iodide symporter gene requires Pax-8 binding and cyclic adenosine 3′,5′-monophosphate response element-like sequence binding proteins for full activity and is differentially regulated in normal and thyroid cancer cells. Mol. Endocrinol. 16:2266-2282. [DOI] [PubMed] [Google Scholar]
- 40.Tazebay, U. H., I. L. Wapnir, O. Levy, O. Dohan, L. S. Zuckier, Q. H. Zhao, H. F. Deng, P. S. Amenta, S. Fineberg, R. G. Pestell, and N. Carrasco. 2000. The mammary gland iodide transporter is expressed during lactation and in breast cancer. Nat. Med. 6:871-878. [DOI] [PubMed] [Google Scholar]
- 41.Teixeira, C., and M. A. Pratt. 1997. CDK2 is a target for retinoic acid-mediated growth inhibition in MCF-7 human breast cancer cells. Mol. Endocrinol. 11:1191-1202. [DOI] [PubMed] [Google Scholar]
- 42.Thorpe, S. M. 1976. Increased uptake of iodide by hormone-responsive compared to hormone-independent mammary tumors in GR mice. Int. J. Cancer 18:345-350. [DOI] [PubMed] [Google Scholar]
- 43.Tong, Q., K. Y. Ryu, and S. M. Jhiang. 1997. Promoter characterization of the rat Na+/I− symporter gene. Biochem. Biophys. Res. Commun. 239:34-41. [DOI] [PubMed] [Google Scholar]
- 44.Uyttersprot, N., N. Pelgrims, N. Carrasco, C. Gervy, C. Maenhaut, J. E. Dumont, and F. Miot. 1997. Moderate doses of iodide in vivo inhibit cell proliferation and the expression of thyroperoxidase and Na+/I− symporter mRNAs in dog thyroid. Mol. Cell. Endocrinol. 131:195-203. [DOI] [PubMed] [Google Scholar]
- 45.Venkataraman, G. M., M. Yatin, and K. B. Ain. 1998. Cloning of the human sodium-iodide symporter promoter and characterization in a differentiated human thyroid cell line, KAT-50. Thyroid 8:63-69. [DOI] [PubMed] [Google Scholar]
- 46.Yan, C., A. Naltner, J. Conkright, and M. Ghaffari. 2001. Protein-protein interaction of retinoic acid receptor alpha and thyroid transcription factor-1 in respiratory epithelial cells. J. Biol. Chem. 276:21686-21691. [DOI] [PubMed] [Google Scholar]