Abstract
The mariner family is probably the most widely distributed family of transposons in nature. Although these transposons are related to the well-studied bacterial insertion elements, there is evidence for major differences in their reaction mechanisms. We report the identification and characterization of complexes that contain the Himar1 transposase bound to a single transposon end. Titrations and mixing experiments with the native transposase and transposase fusions suggested that they contain different numbers of transposase monomers. However, the DNA protection footprints of the two most abundant single-end complexes are identical. This indicates that some transposase monomers may be bound to the transposon end solely by protein-protein interactions. This would mean that the Himar1 transposase can dimerize independently of the second transposon end and that the architecture of the synaptic complex has more in common with V(D)J recombination than with bacterial insertion elements. Like V(D)J recombination and in contrast to the case for bacterial elements, Himar1 catalysis does not appear to depend on synapsis of the transposon ends, and the single-end complexes are active for nicking and probably for cleavage. We discuss the role of this single-end activity in generating the mutations that inactivate the vast majority of mariner elements in eukaryotes.
The Tc1/mariner superfamily of transposons consists of the Tc1 and mariner families of elements in eukaryotes and the more distantly related IS630-like elements in bacteria (25, 39). Members of the superfamily have a single transposase gene expressed in the germ line and/or the soma, transpose via a “cut and paste” DNA intermediate, and duplicate a TA dinucleotide upon insertion. This is probably the most widespread family of transposons in nature: members have been identified in bacteria (IS630), ciliates, fungi, plants, and most animal phyla, from Porifera (sponges) to humans. Although mariner elements are widespread, they are unevenly distributed in closely related species and the vast majority are inactive because of mutations. This suggests that they have an unusual life style that involves a high rate of horizontal transfer to new hosts, followed by a burst of transposition and subsequent vertical inactivation (37, 43).
Homology-dependent gene silencing serves to control the spread of transposons, retroviruses, and other repetitive DNA elements in many eukaryotes. It is mediated by at least three distinct mechanisms, specifically DNA methylation, histone deacetylation, and RNA interference. However, depending on the identity of the organism and the repetitive element in question, these mechanisms are not always completely effective. As noted by McClintock, the rate of transposition in a given germ line can change over time, with cycles of activation and silencing lasting several generations (cited in reference 35). Furthermore, although all three mechanisms of homology-dependent gene silencing operate in plants and are active against many retrotransposons, some DNA transposons, such as the mutator element, appear to escape silencing. In Arabidopsis and maize, a low level of transcription is maintained despite the production of small interfering RNAs (35, 46). Small interfering RNAs derived from the Tc1 element have also been detected in the germ line of Caenorhabditis elegans (52). Even so, new insertions are easily isolated. The system is not completely effective in flies either. Over the last 50 years, the P-element transposon has invaded every wild population of Drosophila melanogaster on the planet (4, 20), and elements such as Himar1 can achieve a huge copy number in individual species such as the horn fly, from which it was isolated (30, 44).
Genetic evidence suggests that the mariner element Mos1 may have a mechanism for self-regulation (38). The Mos1 transposase appears to act specifically at the 5′ end of the element to produce immobilizing mutations, dubbed self-inflicted wounds. It is far from clear whether or not these mutations are an adaptive feature maintained by the unusual life style and evolutionary history of the mariner transposons (see Discussion). Nevertheless, self-inflicted wounds have not been documented for other transposons and it would be interesting to discover the underlying mechanism responsible for them. In the present work, we have characterized the early intermediates of Himar1 transposition and have documented several differences compared to bacterial insertion (IS) elements. The most important difference is that the Himar1 transposase is able to nick and perhaps cleave an isolated transposon end. The mechanistic significance of this difference is introduced below and will be discussed later as an explanation for the generation of self-inflicted wounds.
One of the earliest steps of a transposition reaction is the transposase-mediated synapsis of the transposon ends into paired-end complexes (PECs). These higher-order complexes, which may also incorporate host proteins, are large and have a complicated structure that often reflects the presence of sophisticated regulatory mechanisms (9, 14, 24). All of the catalytic steps of the transposition reaction take place within the PEC, which generally becomes more stable as the reaction progresses and as thermodynamic energy is lost from the system, rendering the reaction essentially irreversible. One consequence of this increasing stability is that early transposition intermediates such as the initial synaptic complex are usually the least stable and the most difficult to study.
Bacterial transposons have been under investigation for many years, but the work is often problematic because the transposase proteins are highly prone to aggregation and because reconstitution in vitro has proven to be difficult. Eukaryotic transposons generally appear to be even more difficult to reconstitute in vitro, and until recently, very little was known about their mechanisms except that which could be assumed by drawing analogies with bacterial elements. Indeed, many of the analogies are perfectly valid because the Tc1/mariner family and the bacterial IS elements descended from a common ancestor and have a characteristic DDE motif in their active sites (39). The DDE motif of the transposases is located in the RNase H-like structural fold that is shared by a diverse group of proteins such as DNA polymerases, RuvC, and human immunodeficiency virus integrase.
One apparently universal feature of the classical DNA transposons from bacteria is that the first nick at the end of the transposon generates the 3′ OH group that is eventually joined to the target site by a direct transesterification mechanism (40). In the nonreplicative, or cut-and-paste, transposons, such as Tn10 and Tn5, cleavage of the second strand is achieved via a hairpin intermediate on the transposon end (7, 28). In contrast, some eukaryotic DNA transposons appear to have a fundamental difference in the polarity of the reaction chemistry. The footprints left behind after the excision of Tam3 from Antirrhinum majus provided indirect evidence that excision of the transposon generates a hairpin end on the flanking DNA (13). Assuming a direct transesterification mechanism, this would require a reversal in the polarity of the reaction chemistry such that the first nick would generate the 5′ P at the end of the transposon. Indeed, first-strand nicking in Himar1 and Mos1 from Drosophila mauritiana generates the 5′ P at the end of the transposon (18; R. Chalmers and N. Buisine, unpublished observation). However, Mos1 does not use a hairpin intermediate, and the mechanism of second-strand cleavage remains unclear.
In this paper, we report the characterization of complexes between the Himar1 transposase and the transposon end. The behavior of the complexes is significantly different from those of the well-characterized DDE transposase family members from bacteria. At least two of the Himar1 complexes detected by electrophoretic mobility shift assays (EMSAs) contained only a single transposon end, suggesting that the synaptic complex is unstable. In contrast to the bacterial transposons, in which catalysis is tightly coupled to synapsis of the transposon ends (3, 17, 49), the Himar1 single-end complexes are active when they are supplemented with Mg2+. Together with differences in the polarity of the reaction chemistry, this suggests that the architecture of the synaptic complex in mariner is radically different from that in bacterial DDE transposons. Indeed, mariner appears to be more similar to V(D)J recombination, which uses a tetrameric core, has the same polarity of the reaction chemistry, and is active in single-end complexes.
MATERIALS AND METHODS
Plasmids, DNA, and bacteria.
The wild type or the C9 hyperactive mutant of Himar1 transposase was expressed as a maltose binding protein (MBP) fusion (1). The C9 mutations Q131R and E137K provide a 50-fold increase in a bacterial mating-out assay (29), but they had very little difference in the in vitro EMSA and activity assays performed here. All of the experiments were performed with both types of protein, except for those shown in Fig. 5, which were performed only with the wild-type protein. The kanamycin-marked mini-mariner transposon (Fig. 1A) was called pMarKan (30). DNA fragments for transposition and EMSAs were prepared by digesting pKL97 and pKL99. For the construction of pKL97, the 5′ end of mariner was copied from p27fh-5′ (30) by a PCR with primers incorporating XhoI sites. The fragment was cloned into XhoI-digested pBluescript II SK(+). pKL99 is identical to pKL97 except that it contains two tandem copies of the XhoI PCR fragment. Transposon end fragments were purified by polyacrylamide gel electrophoresis and recovered by a crush and soak method (48). For the EMSA shown in Fig. 2B, the transposon end was made by annealing the following two synthetic oligonucleotides: KL143 (5′-GTGTGATGGATTATAACAGGTTGGCTGATAAGTCCCCGGTCTGACACATA) and KL144 (5′-AGGTATGTGTCAGACCGGGGACTTATCAGCCAACCTGTTATAATCCATCACAC). DNA preparation and transposase expression were done with the endonuclease I-deficient Escherichia coli strain DH5α. Hydroxyl radical and copper phenanthroline footprinting was performed as described previously (15, 50).
FIG. 5.
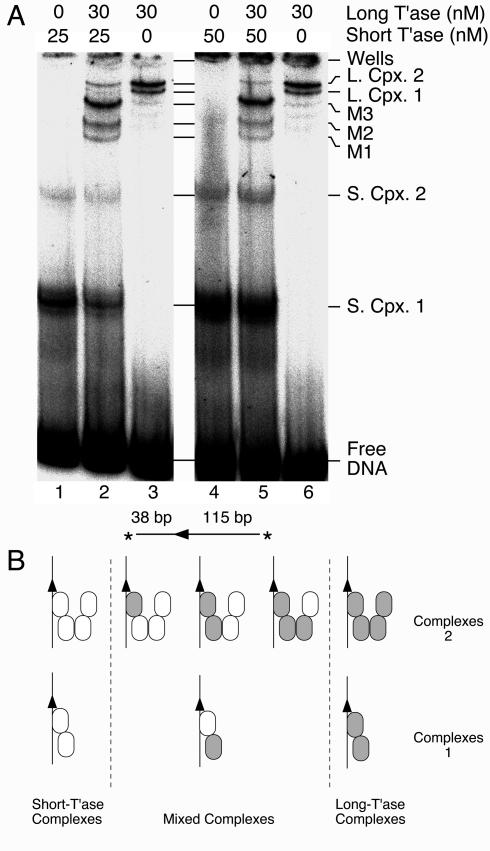
Transposase stoichiometry in Himar1 complexes. (A) Long and short versions of Himar1 transposase were used to form complexes as indicated. The reactions (20 μl) contained 12 fmol of the transposon end, which was generated by SpeI digestion of pKL97. The long transposase was provided by the standard MBP fusion used in all of the other experiments. The short version was the native transposase generated by digestion of the MBP fusion with the protease Factor Xa (see Materials and Methods). Assembly of the complexes was initiated by the addition of transposase, and the reaction was treated with 20 ng of heparin to reduce the background smear on the gel. The gel was 16% acrylamide with a 1:130 acrylamide/bisacrylamide ratio and TAE buffer. T'ase, transposase; L. Cpx., long complex; S. Cpx., short complex; M1-3, mixed complexes. Note that the minor bands running below long complex 1 in lanes 3 and 6 do not have the same mobilities as bands M1, M2, and M3. (B) Tentative model showing the number of transposase monomers in the Himar1 PEC. The transposon end is illustrated by arrowheads, and the long and short versions of transposase are indicated with filled and open ovals, respectively. If complexes 1 and 2 contain a dimer and a tetramer of transposase, respectively, then four mixed complexes are expected. This assumes that the method of detection is sensitive to the absolute numbers of long and short subunits in the multimer but is not sensitive to the relative positions of the individual long and short subunits within a mixed multimer. For example, if the gel was sensitive to all of the possible positions of a given subunit within the multimer, we would expect 2 isomers of the mixed dimer and 14 isomers of the mixed tetramer.
FIG. 1.
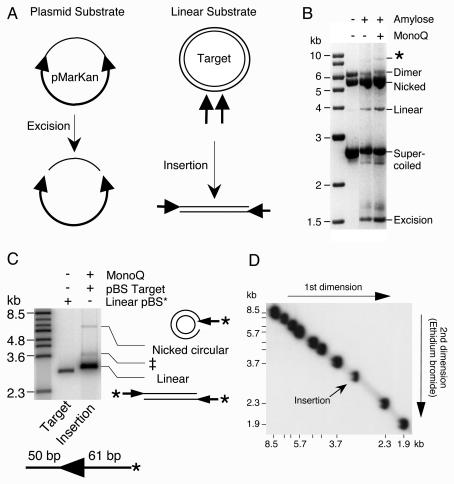
Transposition reactions and further purification of Himar1 transposase. (A) Transposition reactions were performed with either a plasmid substrate or linear transposon ends. pMarKan encodes a mini-transposon in which the 5′ end of Himar1 is cloned as an inverted repeat on either side of a kanamycin resistance marker. This construct was used to assay the transposase activity, as measured by the total amount of excision (left panel). Transposon insertions were measured by use of a linear DNA substrate that also encodes the 5′ end of Himar1. The concerted insertion of a pair of transposon ends into a target plasmid generates a linear product (right panel). Arrowheads, transposon ends; thick lines, transposon; thin lines, plasmid backbone or target DNA. Ampr, ampicillin resistance; Kanr, kanamycin resistance; ori, origin of replication. (B) The total activities of the amylose- and MonoQ-purified transposases were determined by measuring the amounts of transposon excision from the supercoiled plasmid substrate, pMarKan. Each reaction (200 μl) contained 396 fmol (1 μg) of pMarKan and 2 pmol (10 nM) of the respective transposase. Excision of the 2.5-kb transposon leaves behind the plasmid backbone, which contains the origin of replication and the ampicillin resistance gene. A reverse contrast photograph of an ethidium bromide-stained 1.1% agarose-TBE gel is shown. Intramolecular transposon insertions dominate the reaction because of the low concentration of target DNA available in vitro and appear as a smear below the supercoiled substrate. *, intermolecular insertion of the transposon into unreacted substrate plasmid. (C) The MonoQ-purified transposase was tested for its ability to perform concerted insertion of a pair of transposon ends encoded on a linear DNA fragment into a supercoiled target plasmid. The reaction (15 μl) contained 53 fmol of transposon end, 30 fmol (2 nM) of transposase, and 63 fmol (125 ng) of supercoiled pBluescript II as a target. The transposon end fragment was prepared by digesting pKL99 with SfiI and XhoI and was radioactively labeled by end filling. Linear pBluescript II and a phage λ BstEII digestion were end labeled to act as size markers. The products of the reaction are illustrated beside the gel. Thin lines, pBluescript target; thick lines and arrowhead, transposon end. The concerted insertion of transposon ends linearizes the target plasmid. The insertion of a single transposon end produces a lariat structure that comigrates with the nicked circular form of the target plasmid. An autoradiogram of a 0.7% agarose-TBE gel is shown. The size of the transposon arm is shown below the gel. *, position of radioactive label on the transposon arm; ‡, this band is the result of incomplete deproteination of the linear insertion product (34). The flanking DNA is unlabeled. (D) To confirm the linear topology of the insertion product, we mixed a duplicate transposition reaction from the work for panel C with radioactively labeled linear size markers (λ BstEII digest) and analyzed it by two-dimensional gel electrophoresis (11). The TAE-buffered gel had 0.9% agarose in the first dimension and 1.3% agarose plus ethidium bromide in the second dimension. The insertion product migrated on the arc of linear DNA molecules provided by the size markers.
FIG. 2.
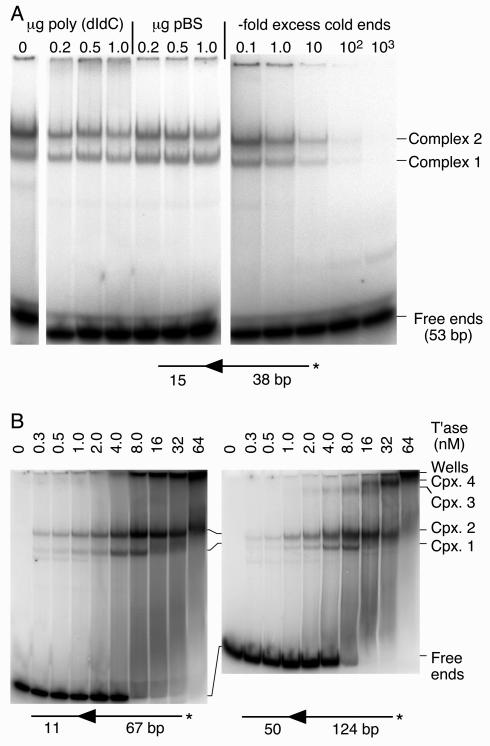
Specific complexes between transposase and 5′ end of Himar1. (A) A linear DNA fragment encoding the 5′ end of Himar1 was used to form complexes with purified transposase (15-μl reactions with 10 fmol of transposon end and 15 fmol [1 nM] of transposase). Assembly of the complexes was challenged with the nonspecific competitor poly(dI-dC) or supercoiled pBluescript II (1 μg of competitor corresponds to a 300-fold excess in nucleotides over the labeled transposon end substrates). The unlabeled (cold) transposon end was also used as a specific competitor (right panel). The competitor DNA was present in the reaction mixture before the addition of transposase. The transposon end fragment was created by annealing oligonucleotides KL143 and KL144. An autoradiogram of an EMSA is shown. *, position of radioactive label on the transposon arm. (B) The complex assembly reaction was titrated with transposase. The DNA fragments were prepared by digesting pKL97 with PstI-SalI or SfiI-XbaI to provide the 67- and 124-bp transposon arms, respectively. The reactions (15 μl) contained 29 and 50 fmol of the short and long fragments, respectively. The maximum amount of transposase (64 nM) corresponds to 960 fmol. An autoradiogram of an EMSA is shown. *, position of radioactive label on the transposon arm. T'ase, transposase; Cpx., complex.
Protein purification.
The fusion between Himar1 transposase and MBP was purified essentially as described in the New England Biolabs pMAL protein fusion purification kit instruction manual and as described previously (1). The pool of peak fractions from the amylose column was diluted with 3 volumes of buffer A (10% glycerol, 25 mM Tris [pH 8.0], 1 mM EDTA, 2 mM dithiothreitol, 10 mM n-decyl-β-d-maltopyranoside) and applied to a 1-ml MonoQ column. The column was equilibrated with buffer A plus 0.05 M NaCl and eluted with a 30-ml gradient of 0.05 to 1 M NaCl. Peak fractions that eluted at 0.13 M NaCl were mixed with a 90% glycerol solution to give a final concentration of 50% and were stored at −20°C.
For some experiments, purified Himar1 transposase was treated with New England Biolabs Factor Xa protease to remove the MBP moiety from the N terminus. The transposase (0.3 mg/ml) was treated with the protease (final concentration, 4 μg/ml) for 6 h at 30°C and then rechromatographed on amylose resin to remove the MBP. The oligomeric state of the native transposase was then assessed by size exclusion chromatography on a Pharmacia Superdex S200 column (20 ml). The size standards were cytochrome c (12.5 kDa), carbonic anhydrase (29 kDa), bovine serum albumin (66 kDa), and apoferritin (440 kDa). The transposase eluted at an apparent molecular mass of 50 kDa, indicating that it exists as a monomer after purification. Since native MBP is a monomer, we therefore assumed that the MBP-transposase fusion used in most experiments also exists as a monomer in solution.
In vitro transposition and EMSA.
The reaction buffer for transposition and EMSA reactions contained 25 mM HEPES (pH 7.9), 10% glycerol, 100 mM NaCl, 20 mM NH4Cl, 250 μg of bovine serum albumin/ml, 2 mM dithiothreitol, and 2 mM CaCl2. When indicated, the reactions were supplemented with 10 mM MgCl2. The plasmid substrate was used at 1 μg per 200-μl reaction, and the reactions were analyzed in 1.1% agarose-Tris-borate-EDTA (TBE) gels. Transposition reactions with linear DNA fragments had 500 cps of radioactively labeled transposon end (absolute amounts are given in the figure legends) and 125 ng (63 fmol) of supercoiled plasmid as a target when it was present. The linear topology of the product of insertions into the supercoiled target was confirmed by two-dimensional agarose gel electrophoresis, with ethidium bromide in the second dimension (10). Transposition reactions were incubated to completion at 30°C (∼5 h), stopped by the addition of a 1/10 volume of 1.5% sodium dodecyl sulfate plus 150 mM EDTA, and heated to 75°C for 10 min. Most EMSAs had 500 cps of radioactively labeled transposon end and were incubated at 30°C for 1.5 h after the addition of transposase, unless noted otherwise. The reaction mixtures were analyzed by being loaded directly onto native 5% polyacrylamide gels in 1× Tris-acetate-EDTA (TAE) at 4°C. The gels were 0.8 mm thick and 25 cm long, and electrophoresis was carried out at 300 V.
RESULTS
Further purification of Himar1 transposase.
Himar1 transposase was first purified as a fusion with MBP by use of an amylose column according to a published protocol (1). The protein was further purified on a MonoQ column as described in Materials and Methods. The transposase was recovered in two peaks, one near the start of the gradient (∼0.13 M NaCl) and the other very late in the gradient (0.6 to 0.7 M NaCl). The late fraction was the most highly purified with respect to other proteins (as judged by Coomassie blue staining) but had little activity. It also had a much higher absorbance at 280 nm than would be expected from the amount of protein visible by Coomassie blue staining. This fraction was found to be fluorescent in the presence of ethidium bromide and SYBR Green, suggesting that the absorbing material was DNA. This was confirmed by a DNase I treatment, which abolished the fluorescence in ethidium bromide and produced an increase in absorption at 260 nm, as expected if free nucleotides were produced. We can therefore conclude that the late transposase fraction remains tightly associated with DNA during purification.
The activity of the early transposase fraction was assayed by using both supercoiled plasmid and linear DNA substrates (Fig. 1). The supercoiled plasmid substrate encodes a mini-mariner construct in which the DNA between the terminal inverted repeats has been replaced with a kanamycin marker. Excision of the transposon from the plasmid leaves behind the 1.5-kb plasmid backbone, which is an end product of the reaction and is an excellent measure of the total transposase activity. When a 10 nM concentration of each transposase was assayed, the MonoQ-purified protein had a threefold higher specific activity than the amylose-purified material (Fig. 1B).
The activity of the MonoQ-purified transposase was further tested in an intermolecular transposition assay (Fig. 1C). The substrate was a linear DNA fragment encoding the terminal 61 bp from the 5′ end of Himar1, together with 50 bp of flanking DNA. The DNA fragment was labeled at the transposon end, and supercoiled pBluescript II was provided as a target. The concerted insertion of a pair of transposon ends was expected to yield a radioactively labeled linear product that was 122 bp larger than the target provided. A product of the correct size was indeed detected (Fig. 1C, rightmost lane) (29). To confirm the expected linear topology of this product, we mixed an identical transposition reaction with a set of DNA molecular weight markers and analyzed it by two-dimensional gel electrophoresis (11). As expected, the Himar1 insertion product migrated on the arc of linear DNA molecules (Fig. 1D).
Two additional minor products were also detected in the insertion assay (Fig. 1C). The uppermost band comigrated with the nicked circular pBluescript II which was present in the reaction as a target. It was produced by the insertion of a single transposon end, which produced a lariat structure. The band just above the linear insertion product was due to incomplete deproteination. Like those from other transposition systems such as Mu and Tn10, the Himar1 insertion product is extremely stable, and deproteination requires very harsh conditions: reactions have to be treated with EDTA and sodium dodecyl sulfate and heated to 75°C for at least 10 min (34; also see Materials and Methods). Together with previous experiments, these results show that the MonoQ-purified MBP-transposase behaves like the native transposase at all steps of the transposition reaction (1, 29, 30).
Specific transposase-transposon end complexes.
An EMSA was performed to analyze complexes formed between the transposase and the transposon end. When a DNA fragment encoding the 5′ end of Himar1 was incubated with the transposase, two major complexes were detected (Fig. 2A). To test whether these complexes were specific, we challenged the assembly of the complex by mixing unlabeled competitor DNA with the labeled transposon end before the addition of transposase. Both complexes were resistant to a very large excess of poly(dI-dC) or pBluescript II DNA. In contrast, both complexes were very sensitive to a challenge with an unlabeled DNA fragment encoding the transposon end. However, if the complexes were allowed to assemble for 15 min, they were resistant to a challenge with unlabeled transposon ends (not shown). This indicates that once they are formed, the complexes are stable and are not in equilibrium, i.e., the transposase does not go through cycles of association and dissociation as simple DNA binding proteins, such as transcriptional repressors, do.
Transposase titration reveals additional complexes.
To investigate the relationship between complexes 1 and 2, we titrated the complex formation reaction with an increasing amount of transposase (Fig. 2B). This was performed with two different DNA fragments, with a 67- or 124-bp transposon arm and different lengths of flanking DNA. Complexes 1 and 2 were both present at the lowest transposase concentration tested. As the transposase concentration was increased, complex 2 began to dominate the reaction. With 16 nM transposase, only complex 2 was present, the free DNA had disappeared, and a significant amount of material had aggregated in the wells. The concentration profile of the complexes exactly coincided with the reported Himar1 insertion activity profile, with a peak at 8 to 10 nM transposase (31).
Transposase titration of the DNA fragment with the 124-bp transposon arm revealed two new bands, complexes 3 and 4 (Fig. 2B, right panel). Presumably, these complexes require the longer transposon arm, either for assembly or to impart stability during electrophoresis. At higher transposase concentrations, complex 4 increased at the expense of complex 3. There appears to be a relationship between the two sets of complexes, as the transition from complex 3 to complex 4 occurred at the same transposase concentration as the transition from complex 1 to complex 2.
Complexes 1 and 2 contain a single transposon end.
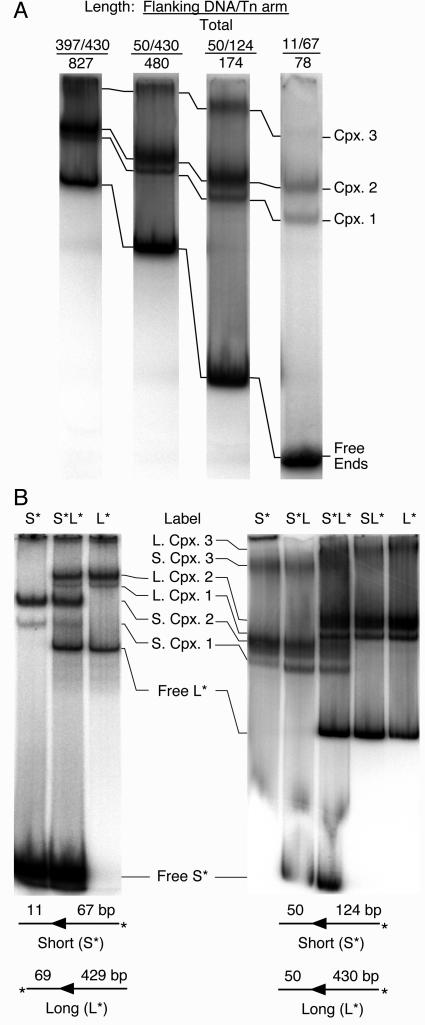
The analysis of complexes formed from mixtures of long and short DNA fragments is a well-established method to determine whether a complex contains one or a pair of transposon arms (8, 47). If a reaction containing a labeled transposon end fragment is supplemented with a longer unlabeled fragment, an additional complex with a decreased mobility will be detected if the complex contains two transposon ends. One prerequisite for this experiment is that complexes containing long and short DNA fragments are sufficiently well resolved during electrophoresis to visualize mixed complexes containing one long and one short fragment. We therefore tested the relative mobilities of complexes assembled with DNA fragments ranging in size from 78 to 827 bp (Fig. 3A). The mobilities of the complexes were clearly dominated by the protein component rather than the length of the DNA fragment used.
FIG. 3.
Stoichiometry of transposon ends in Himar1 complexes. (A) Complexes were assembled in 15-μl reactions with 60 fmol (4 nM) of transposase and transposon end fragments with a range of different lengths. The DNA fragments (with amounts in parentheses) were prepared by digesting pKL97 as follows: 827 bp (50 fmol), NaeI-SapI; 480 bp (33 fmol), SfiI-SapI; 174 bp (22 fmol), SfiI-XbaI; 78 bp (50 fmol), PstI-SalI. Note that all lanes contained 500 cps of label but that the actual amount of DNA (given above) is variable because of differences in the efficiency of labeling and the crush and soak purification. All of the lanes shown came from the same gel, but intervening lanes have been removed. An autoradiogram of an EMSA is shown. (B) Complexes were assembled with either short, long, or a mixture of short and long radioactively labeled transposon end fragments. The sizes of the fragments used are indicated below the panels. The presence of short or long DNA fragments is shown above each lane, with the radioactive label indicated with an asterisk. (Left) The short and long fragments were prepared by digesting pKL97 with SalI-PstI and ApaI-SapI, respectively. Each reaction (20 μl) had approximately 50fmol of transposon end and 80 fmol (4 nM) of transposase. At this ratio of protein to DNA, only complexes 1 and 2 are detected and the background smear is minimal. (Right) The short fragment (31 fmol) was prepared by digesting pKL97 with SfiI-XbaI. The long fragment (47 fmol) was prepared by digesting pKL97 with SfiI-SapI. Each reaction (15 μl) had 120 fmol (8 nM) of transposase so that complexes 1, 2, and 3 were detected; complex 4 was not present because it only appears at the very highest transposase levels (compare with Fig. 2B).
For a determination of the stoichiometry of the transposon ends, the complexes were assembled with various mixtures of short and long DNA fragments that were either labeled or unlabeled. The experiment was performed first under conditions in which complexes 1 and 2 were the major species present (Fig. 3B, left panel). Complexes formed with either the short fragment or the long fragment alone produced a set of short complexes and a set of long complexes, respectively, which were well resolved from each other in the gel (compare the extreme left and right lanes in Fig. 3B, left panel). When short and long fragments were both included in the reaction mixture (central lane), both sets of long and short complexes were produced, but no new bands were detected.
The experiment was next performed under conditions in which complexes 1, 2, and 3 were all detected (Fig. 3B, right panel). As before, the short and long DNA fragments produced sets of short and long complexes, respectively. When the labeled long fragment was supplemented with an unlabeled short fragment, no additional bands were detected. Likewise, when the labeled short fragment was supplemented with an unlabeled long fragment, no additional bands were detected. Finally, when the complexes were formed with a mixture of labeled short and labeled long DNA fragments, both sets of short and long complexes were detected, but no additional bands were present. These results clearly show that complexes 1 and 2 contain only a single transposon end. The result for complex 3 is more difficult to interpret because of the high background in this region of the gel. However, there is no indication of an extra band in the mixed lanes and we can conclude that complex 3 probably also contains a single transposon end.
Complexes 1 and 2 probably arise by decay of the PEC during electrophoresis.
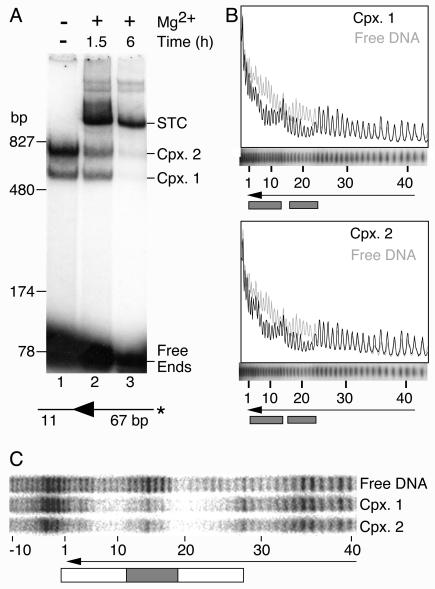
The concerted integration of two Himar1 transposon ends into the TA dinucleotide target implies that the active species in solution is a PEC (30). If the single-end complexes arose by decay of the PEC during the EMSA, then they should disappear when the PEC performs insertions in the presence of Mg2+. A transposition reaction mixture was assembled with transposon ends encoded by a 78-bp DNA fragment and with sufficient transposase to form complexes 1 and 2 (Fig. 4A). When transposition was initiated by the addition of Mg2+, the transposon ends in the PEC inserted into the free transposon ends, which were present in excess and were the only potential target sites available in this reaction. This produced a strand transfer complex (STC), which is extremely stable and requires very harsh conditions to disrupt. The STC appeared to form at the expense of the single-end complexes and the free transposon ends, which all diminished during the course of the reaction. This is a strong indication that the single-end complexes 1 and 2 are precursors of the PEC or that they arise by decay of the PEC during electrophoresis.
FIG. 4.
Complexes 1 and 2 are probably derived from the Himar1 PEC. (A) Transposition reactions were supplemented with 10 mM MgCl2 to initiate the reaction and were incubated at 30°C for the indicated times. An autoradiogram of an EMSA is shown. The smearing of the STC band at the 1.5-h time point makes it appear to have more counts than in the 6-h time point. However, quantification with a phosphorimager showed that there were 1.2-fold more counts in the STC band at the 6-h time point. The DNA fragment was prepared by digesting pKL97 with PstI-SalI. The complexes were assembled in 15-μl reactions with 60 fmol of DNA and 60 fmol of transposase (4 nM). *, position of radioactive label on the transposon arm. Cpx., complex. (B) Hydroxyl radical footprints. Complexes were assembled with the same DNA fragment as that used for panel A, with the radioactive label on the nontransferred strand. The reactions (20 μl) contained 80 fmol of transposase (4 nM) and 37 fmol of transposon end. The reaction mixture was treated with hydroxyl radicals to generate a footprint as described previously (15, 51). Complex 1, complex 2, and the free DNA from nine such reactions were purified in an EMSA and pooled, and the footprint was displayed on a DNA sequencing gel. Traces were generated with Fuji Image Gauge software. The region of the sequencing gel containing the footprint of the complexes is shown aligned below the traces. The arrowhead shows the position of the transposon end. The numbers of base pairs inside the transposon end are indicated. Shaded boxes indicate regions of hydroxyl radical protection. (C) Copper phenanthroline footprints. The complex assembly reaction (100 μl) contained 100 fmol of transposon end and 600 fmol of transposase (30 nM). The DNA was prepared by digesting pKL97 with XbaI-Acc65I and were labeled on the nontransferred stand by end filling. The reaction was treated with 20 ng of heparin (to reduce the background smear) and loaded into five lanes of an EMSA gel. The complexes were treated with copper phenanthroline in the gel as described previously (42), and the footprints were displayed on a DNA sequencing gel. The open box indicates a region of protection extending from the transposon end to bp +28. The shaded box indicates a strong region of protection centered on bp +14.
To further investigate the relationship between the single-end complexes and the PEC, we treated a transposition reaction mixture with hydroxyl radicals to produce a DNA protection pattern. Briefly, a transposition reaction was assembled under standard conditions, treated with hydroxyl radicals, and visualized by an EMSA. The free (uncomplexed) DNA, complex 1, and complex 2 were recovered from the gel, and the footprints were displayed on a denaturing DNA sequencing gel (Fig. 4B). There were two strong patches of hydroxyl radical protection centered on bp +10 and +20 of the transposon end. These patches were in phase with the helical repeat of DNA, indicating that the transposase interacts with one face of the helix. The 10-bp phasing of transposase contacts was similar to that obtained with hydroxyl radicals from bacterial transposon systems such as Tn10, Tn5, and phage Mu (8, 15, 61).
The complexes were also footprinted after separation by EMSA by a treatment of the gel with copper phenanthroline (Fig. 4C). Despite the fact that this technique produced a strong sequence-dependent cleavage pattern on the free DNA, superimposed on this was a region of protection extending from the transposon end to bp +28. Within this region there was an area of strong protection centered on bp +14. Notably, complexes 1 and 2 had almost identical protection patterns.
Since complexes 1 and 2 each contains a single transposon end, the difference in mobility must be due to differences in conformation or in the numbers of transposase monomers that are bound. In either case, the complexes might therefore be expected to have different footprints. The identical hydroxyl radical footprints could be explained if complexes 1 and 2 were both components of the PEC before electrophoresis, when the treatment was performed. However, the copper phenanthroline treatment was performed in the gel after the separation of the complexes. The most likely explanation for the identical footprints is that complex 2 is retarded by additional transposase monomers bound to the transposon end solely by protein-protein interactions.
Transposase multimers.
To determine whether complexes 1 and 2 contain different numbers of transposase monomers, we performed a second mixing experiment with long and short versions of the transposase (Fig. 5A). The long transposase was provided by the standard MBP fusion used in all of the other experiments presented. The short version was provided by the native transposase, which was generated by cleaving off the MBP moiety by use of the Factor Xa site encoded by the expression vector (see Materials and Methods). As before, the rationale for this experiment was that the long and short versions of the transposase would produce a set of long and short complexes when used alone but would produce a set of extra bands when mixed. One prerequisite for this experiment was that the purified transposase was a monomer in solution before the assembly of the complex. This was confirmed by size exclusion chromatography as described in Materials and Methods.
In the experiment with long and short transposon ends described above, a single extra band would have indicated the presence of a PEC. However, the number of extra bands produced in this experiment depended on the numbers of transposase monomers in complexes 1 and 2. For resolution of the various complexes produced in this experiment, it was necessary to change the properties of the standard EMSA gel: the acrylamide concentration was increased from 5 to 16%, and the ratio of acrylamide to bisacrylamide was increased from 19:1 to 130:1. Under these conditions, the wild-type transposase produced a pair of well-separated complexes in the middle of the gel, whereas the MBP fusion produced a pair of complexes close together near the top of the gel (Fig. 5A, compare lanes 4 and 6). When the long and short transposases were mixed, a set of three extra bands was produced (M1, M2, and M3) (lanes 2 and 5). This pattern of bands was extremely reproducible and was observed over a range of different transposase concentrations (for example, compare lanes 1 to 3 with lanes 4 to 6).
The three new bands demonstrated that transposase multimers are present on a single transposon end. Unfortunately, the appearance of three new bands did not immediately suggest the stoichiometry. If complexes 1 and 2 represent transposase monomers and dimers, we would expect one new band in the mixed reaction. In contrast, if complexes 1 and 2 represent dimers and tetramers, we would expect four new bands. Thus, even though the appearance of three new bands clearly indicates that multimers are present, the result is difficult to interpret. The most parsimonious explanation is that there are in reality four new bands but that two of them comigrated. We would therefore like to propose a tentative model for the oligomeric structure of the Himar1 complexes (Fig. 5B). The relationship between these complexes and the active PEC remains an open question. If the active conformation is a tetramer, then complex 1 is essentially half of a PEC, while complex 2 can be viewed as a PEC lacking one of the two transposon ends. Alternatively, if the active PEC contains a transposase dimer, then the tetramer might represent the inhibitory complex that mediates overproduction inhibition (31). Note that the model illustrates only the number of subunits present and is not intended to show the three-dimensional pairwise interactions within the multimer.
Nicking and cleavage in single-end complexes.
For all of the well-characterized transposons from bacteria, the assembly of a synaptic complex containing two transposon ends is an absolute requirement for the catalytic steps of the reaction. In contrast, very little is known about the molecular mechanism of eukaryotic transposons and whether the catalytic steps are coupled to synapsis of the ends.
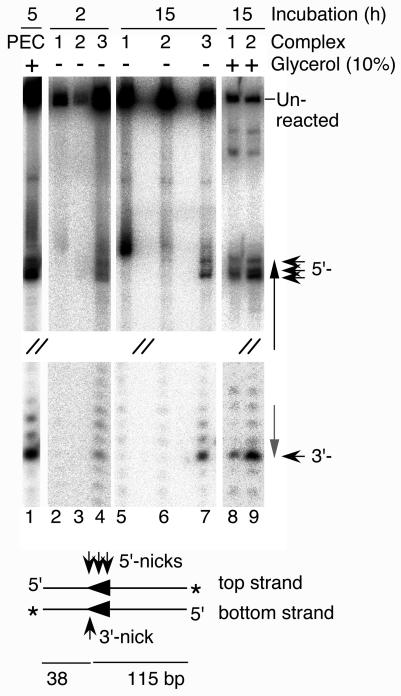
Standard transposition reaction mixtures were assembled in the absence of catalytic metal ions and with sufficient transposase to form complexes 1, 2, and 3. The three complexes were then separated by a standard TAE-buffered EMSA (see Fig. 2B as an example). After electrophoresis, the entire gel was removed from between the glass plates and incubated at 30°C in standard reaction buffer, either with or without 10% glycerol. The gel was next transferred to a solution of EDTA to chelate the Mg2+ and to stop any reaction that may have taken place. After an overnight exposure to photographic film to locate the positions of the single-end complexes, the gel slices were excised from the wet gel and the DNA was recovered. Finally, to reveal the extent of any reaction that had taken place, the DNA was displayed on a denaturing DNA sequencing gel (Fig. 6).
FIG. 6.
Catalysis in single-end complexes. Complexes were assembled in 20-μl reactions in the absence of divalent metal ions with 100 fmol of transposase (5 nM) and 40 fmol of transposon end. The transposon end was generated by SpeI digestion of pKL97. After incubation for 10 min at 30°C, the complexes were separated by EMSA. The gel was incubated in standard reaction buffer (10 mM MgCl2) in the presence or absence of glycerol for 2 or 15 h. Finally, the gel was incubated in EDTA to stop any reaction that may have taken place. The wet gel was exposed to photographic film to locate the complexes, which were then excised from the gel, and the DNAs were recovered by the crush and soak method (48). The DNAs were displayed on a denaturing sequencing gel along with a control transposition reaction performed in solution. The lane containing the control reaction is labeled “PEC” because this is presumably the active species in solution. The middle section of the gel is not shown, as indicated by sets of double forward slashes. In the illustration of the transposon end below the gel, both strands of the DNA are shown. Thick arrowheads, locations of the transposon ends; thin arrowheads, positions of nicks on either strand; asterisk, position of the 3′-end label.
As a control for the activity of the single-end complexes, a standard transposition reaction was included in the gel (Fig. 6, lane 1). In solution, when the active species was presumably a PEC, the nontransferred strand of Himar1 was nicked at several positions around the 5′ end of the transposon (30). In contrast, the transferred strand, which contained the 3′ end of the transposon, was cleaved almost exclusively at the end of the element.
When they were incubated in the absence of glycerol, the three single-end complexes produced different sets of reaction products (Fig. 6). Complex 1 nicked the DNA quite efficiently at an aberrant position on the top strand just outside the transposon end (lane 5). A low level of nicking at this position was also detected in the control reaction, in which the active species is presumably the PEC (lane 1). Complex 2 was almost completely inactive (lane 6). Finally, complex 3 was similar to the control reaction and cleaved both strands of DNA at the transposon end (lane 7).
One potential caveat regarding the activities of the single-end complexes is that the long incubation time may have allowed the components of the complexes to rearrange and to assemble a new PEC. However, two aspects of the results suggest that this was not the case. The first aspect is that the reaction with complex 1 was aberrant: it nicked the top strand just outside the transposon end and failed to nick the bottom strand. The second aspect is that the efficient reassembly of the PEC in lanes 2 to 7 should have been prevented by the absence of glycerol, which was included in the standard transposition reaction because it stimulates the activity 10- to 20-fold.
When the in-gel cleavage experiments were repeated with 10% glycerol in the reaction buffer, complexes 1 and 2 were fully active and able to cleave both strands at the transposon end (Fig. 6, lanes 8 and 9). The presence of glycerol may have promoted reassembly of the PEC. However, even if this were true, it would support the notion that the products detected in the absence of glycerol were from genuine single-end reactions and were not derived from a PEC reassembled in the gel during the incubation period.
DISCUSSION
Himar1 PEC is unstable during electrophoresis.
All of the classical DNA transposition reactions characterized to date take place within a PEC that contains both transposon ends (for examples, see references 8, 18, 47, 53, and 58). PEC assembly is important because it defines the transposon fragment to be moved and ensures a concerted insertion of the transposon ends at the target site. Two aspects of Himar1 transposition provide indirect evidence for the existence of a PEC. The first is that the transposon ends insert into a TA dinucleotide target and that single-end insertions are insignificant compared to double-end insertions (Fig. 1C) (2, 30, 45). The second is that the single-end complexes 1 and 2 disappeared as the STC accumulated (Fig. 4). Taken together, these results suggest that a PEC was present in our transposition reactions but that it dissociated into complex 1, complex 2, or both during electrophoresis.
Stoichiometry of Himar1 transposase complexes.
Up to four complexes were detected when the 5′ end of Himar1 was titrated with transposase (Fig. 2). An analysis of the stoichiometry by the use of long and short transposon ends demonstrated that the major complexes 1 and 2 contained only one transposon end (Fig. 3). The minor complexes 3 and 4 were more difficult to interpret because of the high background level, but there was no indication that they contained more than one transposon end. An analysis of the protein contents of the single-end complexes demonstrated that multimers were present and that complexes 1 and 2 probably correspond to dimers and tetramers (Fig. 5). Multiple single-end complexes appear to be a general feature of this family of transposons. The only other members of the mariner family that have been reconstituted in vitro are Tc1/3, Mos1, and Sleeping Beauty (SB). Mos1 appears to produce a pair of complexes under some circumstances (5) but not others (60). The multiple complexes produced by SB have been studied in more detail (27, 57, 59). Each end of SB encodes a bipartite transposase recognition site that appears to be related to the recombination signal sequences (RSSs) in V(D)J recombination. The presence of more than one transposase recognition site at each end provides a rationale for the existence of multiple complexes. Indeed, titration of the SB transposase produced an increase in complex 2 at the expense of complex 1, as also shown here for Himar1. Together with supporting data, this suggested a model in which SB transposition involves a tetramer of transposase, with a single transposase monomer bound to each of the two recognition sites at each end of the transposon.
The bacterial cut-and-paste transposons, such as Tn10 and Tn5, use a monomer of transposase to cleave both strands of DNA at the transposon end via a hairpin intermediate (7, 28). However, Mos1 does not appear to utilize a hairpin intermediate (18) and the mariner family of transposons may therefore require a dimer of transposase at each end to achieve cleavages of both strands. The recruitment of additional transposase monomers can certainly be used as a model to explain the multiple Himar1 complexes. However, in contrast to SB, which has a bipartite transposase recognition site, Mos1 and Himar1 have simple inverted repeats. All of our data could be accommodated by a model in which the active PEC contains a dimer of transposase while the tetrameric structure proposed for complex 2 mediates overproduction inhibition. This is supported by the activity profile of the in vitro reaction, which has a sharp peak at 10 nM transposase (31), exactly the concentration above which complex 1 disappears from the EMSA gel (Fig. 2B).
Architecture of the Himar1 PEC.
In all of the bacterial transposons examined so far, the catalytic steps of the reaction are tightly coupled to synapsis of the transposon ends. It was therefore slightly surprising to find that the Himar1 single-end complexes have both nicking and cleavage activities (Fig. 6). However, the phenotype of the L124S mutation in the Mos1 element already provided evidence that mariner family transposases are active on a single, unsynapsed transposon end (60). Although this mutant can catalyze a low level of normal transposition with plasmid substrates, the vast majority of the reaction products are cleaved at only one of the two transposon ends. However, this result should be interpreted cautiously, as this phenotype could be explained by several mechanisms other than single-end activity. For example, the apparent single-end cleavage during Tn10 transposition is not due to catalysis without synapsis but results from synapses of the transposon ends on different molecules (11). Another alternative is that if a mutation were to destabilize the synaptic complex, it might dissociate during a conformational change at some intermediate stage of the reaction (16).
The requirement for synapsis in bacterial systems is imposed by the architecture of the PEC: transposase catalyzes the reaction in trans to the transposon end at which it is bound, i.e., a transposase subunit engaged in sequence-specific interactions with one transposon end performs the catalytic steps at the opposite transposon end (3, 17, 49). Single-end activity is precluded because the transposases are unable to dimerize in the absence of the second transposon end. This is most clearly illustrated in the cocrystal structure of the Tn5 transpososome, in which protein-DNA interactions provide two-thirds of the dimer interface (17).
Further insight into the problem of single-end activity can be gained by considering V(D)J recombination, which has been thoroughly characterized (22, 23). V(D)J recombination takes place within a synaptic complex that contains a pair of RSSs, the equivalent of transposon ends. The complex has a tetrameric protein core, within which the Rag1 recombinase acts in trans to catalyze the reaction (54-56). However, even though catalysis is performed in trans, the nicking step in V(D)J recombination is uncoupled from synapsis of the RSS (22, 23, 58). This is possible because Rag1 can dimerize in solution or on a single RSS and does not require the partner DNA segment. The present results suggest that the Himar1 transposase can multimerize on a single transposon end and that the architecture of the mariner synaptic complex therefore has more in common with V(D)J recombination than with bacterial transposons.
It has been suggested that single-end nicking in V(D)J may be an adaptive characteristic that allows a wider array of coding segments to contribute to the immune repertoire (58). Single-end activity may also be adaptive in mariner transposition, as it provides a mechanism for generating the self-inflicted wounds that are proposed to curb the burst of transposition after the invasion of a genome.
Regulation of mariner transposition and generation of self-inflicted wounds.
The distribution of mariner elements in insects has two peculiar features (see references 26, 37, 38, and 44 and references therein). The first is that the presence or absence of mariner is not correlated with the established phylogeny of the host species or strains, suggesting frequent horizontal transfer. The second peculiar feature is that there is a large variation in copy number and that most of the elements are inactive due to large numbers of point mutations and deletions. Indeed, the only active examples of mariner identified in insects to date are the Mos1 elements from D. mauritiana and Drosophila simulans and a newly discovered mariner element from an earwig (6). These observations suggest that after horizontal transfer to a new host, there is a burst of transposition, followed by inactivation of the element and its subsequent loss over time by mutation and genetic drift. This raises an important biological question. If the rate of mariner transposition is sufficient to achieve high copy numbers and to ensure frequent horizontal transfer, why do most genomes contain only inactive elements?
Three mechanisms, operating at the level of the transposase, have been proposed to limit the rate of mariner transposition (26). The first is overproduction inhibition, which has been documented for both mariner and the bacterial IS elements and probably reflects a concentration-dependent aggregation of transposases (12, 26, 31, 36). The second and third mechanisms depend on the presence of defective copies of the element (26, 37, 38). Defective transposase monomers produced by these elements have the potential to assemble into multimers with wild-type transposase and to “poison” the activity of the complex by dominant-negative complementation (19, 26, 36). Finally, if the defective elements retain their transposase binding sites, they will titrate the wild-type elements by acting as a sink to absorb the active transposase, thereby curbing the rate of transposition.
These last two mechanisms for limiting the rate of transposition depend on the generation of mutations within the transposase gene. However, there is a fourth mechanism that depends on the generation of mutations in the terminal inverted repeats that flank the element on either side (38). These mutations were recovered at a very high frequency by an elegant genetic screen involving the excision of peach, a nonautonomous Mos1 element, from the promoter region of the Drosophila white gene. The most common class of mutation was a characteristic short deletion at the 5′ end. These mutations were not the result of bona fide transposition events because they were not recovered at the 3′ end of peach unless the 5′ end had been inactivated beforehand. Instead, it was suggested that the 5′ end of peach is preferred over the 3′ end, and controversially, that transposase acted on a single transposon end to generate these self-inflicted wounds. It was subsequently shown that the 5′ end of the element is indeed a better substrate than the 3′ end in vitro (60). Furthermore, the nicking activity of the single-end complexes described here (Fig. 6) provides a feasible mechanism for the initiation of lesions at a single, isolated transposon end.
There are an estimated 17,000 copies of Himar1 in the horn fly, Haematobia irritans. All of the copies examined contain inactivating mutations in the transposase gene or in the terminal inverted repeats (44). It was recently shown that single-end nicking during V(D)J recombination stimulates homologous recombination in mammalian fibroblasts, although it was not clear whether this is a direct effect or is due to the generation of double-strand breaks during subsequent DNA replication (33). If this is also true in other organisms, the single-end nicking and/or cleavage activity of Himar1 suggests an explanation for why eukaryotes in general have such a high proportion of inactive mariner elements.
DNA repair by homologous recombination can spread mutations among repetitive DNA elements by gene conversion (for example, see reference 21). The failure to complete the process can also generate internal deletions, as is frequently observed in the Drosophila P elements (for examples, see references 32 and 41). It is perhaps not unreasonable to assume that at some point during the invasion of a genome by mariner, bona fide transposition events will be outnumbered by single-end events. The point at which this occurs will depend on a range of different factors, such as the host defenses against transposase expression, the titrating effect of the increasing number of transposon ends, and dominant-negative complementation. In this situation, a wild-type transposon end that interacts well with transposase will experience a higher rate of gene conversion than a mutant end. Since each such event is associated with a risk of conversion to the mutant state, this process has the potential to help purge the genome of active mariner elements.
Acknowledgments
This work was funded by a grant from The Wellcome Trust to R.C. R.C. is a Royal Society University Research Fellow. K.L. was supported a Marie Curie Mobility Research Training Fellowship.
We thank the anonymous referees for many insightful comments.
REFERENCES
- 1.Akerley, B. J., and D. J. Lampe. 2002. Analysis of gene function in bacterial pathogens by GAMBIT. Methods Enzymol. 358:100-108. [DOI] [PubMed] [Google Scholar]
- 2.Akerley, B. J., E. J. Rubin, A. Camilli, D. J. Lampe, H. M. Robertson, and J. J. Mekalanos. 1998. Systematic identification of essential genes by in vitro mariner mutagenesis. Proc. Natl. Acad. Sci. USA 95:8927-8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldaz, H., E. Schuster, and T. A. Baker. 1996. The interwoven architecture of the Mu transposase couples DNA synapsis to catalysis. Cell 85:257-269. [DOI] [PubMed] [Google Scholar]
- 4.Anxolabehere, D., M. G. Kidwell, and G. Periquet. 1988. Molecular characteristics of diverse populations are consistent with the hypothesis of a recent invasion of Drosophila melanogaster by mobile P elements. Mol. Biol. Evol. 5:252-269. [DOI] [PubMed] [Google Scholar]
- 5.Augé-Gouillou, C., M. H. Hamelin, M. V. Demattei, G. Periquet, and Y. Bigot. 2001. The ITR binding domain of the mariner Mos-1 transposase. Mol. Genet. Genomics 265:58-65. [DOI] [PubMed] [Google Scholar]
- 6.Barry, E. G., D. J. Witherspoon, and D. J. Lampe. 2004. A bacterial genetic screen identifies functional coding sequences of the insect mariner transposable element Famar1 amplified from the genome of the earwig, Forficula auricularia. Genetics 166:823-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhasin, A., I. Y. Goryshin, and W. S. Reznikoff. 1999. Hairpin formation in Tn5 transposition. J. Biol. Chem. 274:37021-37029. [DOI] [PubMed] [Google Scholar]
- 8.Bhasin, A., I. Y. Goryshin, M. Steiniger-White, D. York, and W. S. Reznikoff. 2000. Characterization of a Tn5 pre-cleavage synaptic complex. J. Mol. Biol. 302:49-63. [DOI] [PubMed] [Google Scholar]
- 9.Chaconas, G., and R. M. Harshey. 2002. Transposition of phage Mu DNA, p. 384-402. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 10.Chalmers, R., A. Guhathakurta, H. Benjamin, and N. Kleckner. 1998. IHF modulation of Tn10 transposition: sensory transduction of supercoiling status via a proposed protein/DNA molecular spring. Cell 93:897-908. [DOI] [PubMed] [Google Scholar]
- 11.Chalmers, R. M., and N. Kleckner. 1996. IS10/Tn10 transposition efficiently accommodates diverse transposon end configurations. EMBO J. 15:5112-5122. [PMC free article] [PubMed] [Google Scholar]
- 12.Chalmers, R. M., and N. Kleckner. 1994. Tn10/IS10 transposase purification, activation, and in vitro reaction. J. Biol. Chem. 269:8029-8035. [PubMed] [Google Scholar]
- 13.Coen, E. S., T. P. Robbins, J. Almeida, A. Hudson, and R. Carpenter. 1989. Consequences and mechanisms of transposition in Antirrhinum majus, p. 413-436. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 14.Craig, N. L. 2002. Tn7, p. 423-456. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 15.Crellin, P., and R. Chalmers. 2001. Protein-DNA contacts and conformational changes in the Tn10 transpososome during assembly and activation for cleavage. EMBO J. 20:3882-3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crellin, P., S. Sewitz, and R. Chalmers. 2004. DNA looping and catalysis; the IHF-folded arm of Tn10 promotes conformational changes and hairpin resolution. Mol. Cell 13:537-547. [DOI] [PubMed] [Google Scholar]
- 17.Davies, D. R., I. Y. Goryshin, W. S. Reznikoff, and I. Rayment. 2000. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289:77-85. [DOI] [PubMed] [Google Scholar]
- 18.Dawson, A., and D. J. Finnegan. 2003. Excision of the Drosophila mariner transposon Mos1. Comparison with bacterial transposition and V(D)J recombination. Mol. Cell 11:225-235. [DOI] [PubMed] [Google Scholar]
- 19.De Aguiar, D., and D. L. Hartl. 1999. Regulatory potential of nonautonomous mariner elements and subfamily crosstalk. Genetica 107:79-85. [PubMed] [Google Scholar]
- 20.Engels, W. R. 1997. Invasions of P elements. Genetics 145:11-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer, S. E., E. Wienholds, and R. H. Plasterk. 2003. Continuous exchange of sequence information between dispersed Tc1 transposons in the Caenorhabditis elegans genome. Genetics 164:127-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fugmann, S. D., A. I. Lee, P. E. Shockett, I. J. Villey, and D. G. Schatz. 2000. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 18:495-527. [DOI] [PubMed] [Google Scholar]
- 23.Gellert, M. 2002. V(D)J recombination: Rag proteins, repair factors, and regulation. Annu. Rev. Biochem. 71:101-132. [DOI] [PubMed] [Google Scholar]
- 24.Haniford, D. B. 2002. Transposon Tn10, p. 457-483. In N. L. Craig, R. Craigie, M. Gellert, and A. M. Lambowitz (ed.), Mobile DNA II. American Society for Microbiology, Washington, D.C.
- 25.Hartl, D. L., A. R. Lohe, and E. R. Lozovskaya. 1997. Modern thoughts on an ancyent marinere: function, evolution, regulation. Annu. Rev. Genet. 31:337-358. [DOI] [PubMed] [Google Scholar]
- 26.Hartl, D. L., E. R. Lozovskaya, D. I. Nurminsky, and A. R. Lohe. 1997. What restricts the activity of mariner-like transposable elements. Trends Genet. 13:197-201. [DOI] [PubMed] [Google Scholar]
- 27.Ivics, Z., P. B. Hackett, R. H. Plasterk, and Z. Izsvak. 1997. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell 91:501-510. [DOI] [PubMed] [Google Scholar]
- 28.Kennedy, A. K., A. Guhathakurta, N. Kleckner, and D. B. Haniford. 1998. Tn10 transposition via a DNA hairpin intermediate. Cell 95:125-134. [DOI] [PubMed] [Google Scholar]
- 29.Lampe, D. J., B. J. Akerley, E. J. Rubin, J. J. Mekalanos, and H. M. Robertson. 1999. Hyperactive transposase mutants of the Himar1 mariner transposon. Proc. Natl. Acad. Sci. USA 96:11428-11433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lampe, D. J., M. E. A. Churchill, and H. M. Robertson. 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15:5470-5479. [PMC free article] [PubMed] [Google Scholar]
- 31.Lampe, D. J., T. E. Grant, and H. M. Robertson. 1998. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics 149:179-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, C. C., Y. M. Mul, and D. C. Rio. 1996. The Drosophila P-element KP repressor protein dimerizes and interacts with multiple sites on P-element DNA. Mol. Cell. Biol. 16:5616-5622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, G. S., M. B. Neiditch, S. S. Salus, and D. B. Roth. 2004. RAG proteins shepherd double-strand breaks to a specific pathway, suppressing error-prone repair, but RAG nicking initiates homologous recombination. Cell 117:171-184. [DOI] [PubMed] [Google Scholar]
- 34.Lipkow, K. 2003. Molecular analysis of hobo and Himar1 transposition. Ph.D. thesis. Oxford University, Oxford, United Kingdom.
- 35.Lippman, Z., B. May, C. Yordan, T. Singer, and R. Martienssen. 2003. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone Modification. PLoS Biol. 1:E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lohe, A. R., and D. L. Hartl. 1996. Autoregulation of mariner transposase activity by overproduction and dominant-negative complementation. Mol. Biol. Evol. 13:549-555. [DOI] [PubMed] [Google Scholar]
- 37.Lohe, A. R., E. N. Moriyama, D. A. Lidholm, and D. L. Hartl. 1995. Horizontal transmission, vertical inactivation, and stochastic loss of mariner-like transposable elements. Mol. Biol. Evol. 12:62-72. [DOI] [PubMed] [Google Scholar]
- 38.Lohe, A. R., C. Timmons, I. Beerman, E. R. Lozovskaya, and D. L. Hartl. 2000. Self-inflicted wounds, template-directed gap repair and a recombination hotspot. Effects of the mariner transposase. Genetics 154:647-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mizuuchi, K. 1992. Polynucleotidyl transfer reactions in transpositional DNA recombination. J. Biol. Chem. 267:21273-21276. [PubMed] [Google Scholar]
- 41.O'Hare, K., and G. M. Rubin. 1983. Structures of P transposable elements and their sites of insertion and excision in the Drosophila melanogaster genome. Cell 34:25-35. [DOI] [PubMed] [Google Scholar]
- 42.Papavassiliou, A. G. 1994. 1,10-Phenanthroline-copper ion nuclease footprinting of DNA-protein complexes in situ following mobility-shift electrophoresis assays, p. 43-78. In G. G. Kneale (ed.), DNA-protein interactions: principles and protocols. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 43.Robertson, H. M., and D. J. Lampe. 1995. Distribution of transposable elements in arthropods. Annu. Rev. Entomol. 40:333-357. [DOI] [PubMed] [Google Scholar]
- 44.Robertson, H. M., and D. J. Lampe. 1995. Recent horizontal transfer of a mariner transposable element among and between Diptera and Neuroptera. Mol. Biol. Evol. 12:850-862. [DOI] [PubMed] [Google Scholar]
- 45.Rubin, E. J., B. J. Akerley, V. N. Novik, D. J. Lampe, R. N. Husson, and J. J. Mekalanos. 1999. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc. Natl. Acad. Sci. USA 96:1645-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rudenko, G. N., A. Ono, and V. Walbot. 2003. Initiation of silencing of maize MuDR/Mu transposable elements. Plant J. 33:1013-1025. [DOI] [PubMed] [Google Scholar]
- 47.Sakai, J., R. M. Chalmers, and N. Kleckner. 1995. Identification and characterization of a pre-cleavage synaptic complex that is an early intermediate in Tn10 transposition. EMBO J. 14:4374-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sambrook, J., E. F. Fritsch, and T Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 49.Savilahti, H., and K. Mizuuchi. 1996. Mu transpositional recombination: donor DNA cleavage and strand transfer in trans by the Mu transposase. Cell 85:271-280. [DOI] [PubMed] [Google Scholar]
- 50.Schickor, P., and H. Heumann. 1994. Hydroxyl radical footprinting, p. 21-32. In G. G. Kneale (ed.), DNA-protein interactions: principles and protocols. Humana Press, Totowa, N.J.
- 51.Sewitz, S., P. Crellin, and R. Chalmers. 2003. The positive and negative regulation of Tn10 transposition by IHF is mediated by structurally asymmetric transposon arms. Nucleic Acids Res. 31:5868-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sijen, T., and R. H. Plasterk. 2003. Transposon silencing in the Caenorhabditis elegans germ line by natural RNAi. Nature 426:310-314. [DOI] [PubMed] [Google Scholar]
- 53.Surette, M. G., S. J. Buch, and G. Chaconas. 1987. Transpososomes: stable protein-DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell 49:253-262. [DOI] [PubMed] [Google Scholar]
- 54.Swanson, P. C. 2001. The DDE motif in RAG-1 is contributed in trans to a single active site that catalyzes the nicking and transesterification steps of V(D)J recombination. Mol. Cell. Biol. 21:449-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swanson, P. C. 2002. Fine structure and activity of discrete RAG-HMG complexes on V(D)J recombination signals. Mol. Cell. Biol. 22:1340-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Swanson, P. C. 2002. A RAG-1/RAG-2 tetramer supports 12/23-regulated synapsis, cleavage, and transposition of V(D)J recombination signals. Mol. Cell. Biol. 22:7790-7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vigdal, T. J., C. D. Kaufman, Z. Izsvak, D. F. Voytas, and Z. Ivics. 2002. Common physical properties of DNA affecting target site selection of Sleeping Beauty and other Tc1/mariner transposable elements. J. Mol. Biol. 323:441-452. [DOI] [PubMed] [Google Scholar]
- 58.Yu, K., and M. R. Lieber. 2000. The nicking step in V(D)J recombination is independent of synapsis: implications for the immune repertoire. Mol. Cell. Biol. 20:7914-7921.11027262 [Google Scholar]
- 59.Zayed, H., Z. Izsvak, D. Khare, U. Heinemann, and Z. Ivics. 2003. The DNA-bending protein HMGB1 is a cellular cofactor of Sleeping Beauty transposition. Nucleic Acids Res. 31:2313-2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, L., A. Dawson, and D. J. Finnegan. 2001. DNA-binding activity and subunit interaction of the mariner transposase. Nucleic Acids Res. 29:3566-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zou, A. H., P. C. Leung, and R. M. Harshey. 1991. Transposase contacts with Mu DNA ends. J. Biol. Chem. 266:20476-20482. [PubMed] [Google Scholar]