Abstract
Myocyte enhancer factor 2 (MEF2) transcription factors play pivotal roles in cardiac, muscle, and neuron gene expression. All products of MEF2 genes have a common amino-terminal DNA binding and dimerization domain, but the four vertebrate MEF2 gene transcripts are alternatively spliced among coding exons to produce splicing isoforms. In MEF2C alone, alternative splice acceptors in the last exon give forms that include or exclude a short domain that we designate γ. We show that MEF2C is expressed exclusively as γ− isoforms in heart tissue and predominantly as γ− in other adult tissues and in differentiating myocytes. MEF2C γ− isoforms are much more robust than γ+ forms in activating MEF2-responsive reporters in transfected fibroblasts despite indistinguishable expression levels, and they better synergize with MyoD in promoting myogenic conversion. One-hybrid transcription assays using Gal4-MEF2C fusions give similar distinctions between γ− and γ+ isoforms in all cell types tested, including myocytes. Cis effects of γ on MEF2C DNA binding, dimerization, protein stability, or response to CaM or p38 mitogen-activated protein kinase signaling are not apparent, and the isolated γ domain represses transcription when fused to Gal4. One phosphoserine residue is present within the γ domain according to tandem mass spectrometry, and mutation of this residue abolishes γ-mediated transrepression. A similar activity is present in the constitutive γ domain and serine phosphoacceptor of MEF2A. Our findings indicate that γ functions autonomously as a phosphoserine-dependent transrepressor to downregulate transactivation function of MEF2 factors and that alternative splicing and serine phosphorylation converge to provide complex combinatorial control of MEF2C activity.
Myocyte enhancer factor 2 (MEF2) proteins are members of the MADS (MCMI, agamous, deficiens, serum response factor)-box family of transcriptional regulators (5). MEF2 was originally recognized as a sequence-specific DNA binding activity at conserved elements in the promoters of various genes encoding muscle structural proteins and as products of cDNAs encoding proteins related to serum response factor (42, 52). Four distinct vertebrate genes encoding MEF2 forms were subsequently recognized, MEF2A, MEF2B, MEF2C, and MEF2D (7, 27-29). Initial studies of MEF2 largely considered a role in myogenesis, but a wider province is now appreciated. Thus, MEF2 target genes include those encoding structural proteins, transporters and metabolic enzymes of muscle, and effectors of stress signaling (5, 16, 36). In addition, although specific MEF2 gene targets relevant to neuronal function remain to be determined, MEF2 proteins interact directly with neuron-specific transcription factors and have been shown to play a critical role in programmed cell death in this cell type (13, 26, 39).
Regulation of MEF2 function is complex and occurs at many levels. Abundance of MEF2 proteins is controlled at transcriptional (47), translational (4), and degradation (40) steps. The transactivation function of MEF2 proteins is regulated by various means, including through protein-protein interactions with other transcription factors (21, 25, 34, 48) and transcriptional coregulators (19, 31, 33, 45, 50); via phosphorylation by mitogen-activated protein kinases (MAPK) (18, 53) and cyclin-dependent kinase 5 (cdk5) (13); and by determinants of MEF2 and coregulator subcellular localization, including calcium-dependent effects on MEF2 interactors and interactions (9, 19, 32, 38).
The four MEF2 genes are differentially expressed spacially and temporally during development and in mature tissues (12). MEF2 isotype functions partially overlap, but distinct roles for the different genes remain to be fully elucidated and understood. Murine gene disruption studies provide genetic evidence in support of discrete MEF2 isotype-specific functions (24, 37). Thus, mef2-c null mice die at embryonic day 9 due to failure of cardiac development, and the animals also exhibit vascular defects (23, 24). In the mef2-c−/− mouse embryo, there is selective underexpression of only a subset of cardiac contractile proteins whose genes are regulated by MEF2. Since the other mef2 genes are expressed at normal or supraphysiologic levels in the mef2-c null animals, the lack of compensation by these forms indicates a unique role for mef2-c in cardiac development. mef2-a null mice survive to the neonatal period, or in some cases to adulthood, but severe myocardial mitochondrial defects are present that predispose to sudden death (37). Expression of the other mef2 genes is upregulated in these animals, again indicating lack of compensation in vivo for selective MEF2 gene loss. At present it is not clear whether MEF2 isotype-selective functions relate solely to distinctions in temporal or spacial expression or to unique features of the MEF2 protein forms encoded by the different genes.
Each vertebrate MEF2 gene gives rise to multiple isoforms through alternative splicing patterns that are conserved among vertebrates (5). These splicing patterns include use of bona fide alternative exons, a splice versus no-splice option, and use of alternative splice acceptors within one exon. In contrast to extensive work on MEF2 gene and MEF2 protein regulation at the various levels noted above, the differential expression and unique roles and responses of MEF2 splicing variants have remained virtually unexplored (17). No MEF2 protein-protein interactions or MEF2 protein modifications have been identified that involve domains unique to the splicing variants. One genetic study of splicing variants of the sole Drosophila MEF2 gene, Dmef2, has been reported (14). No significant differences were noted in the abilities of the various DMEF2 splicing forms to rescue muscle differentiation defects in a Dmef2 mutant. However, distinctions in splicing variant function may have been obscured by the particular conditions in this study. Furthermore, Dmef2 gene structure and alternative splicing patterns are not analogous to those of the vertebrate MEF2 genes, such that these findings do not specifically inform function of the vertebrate MEF2 splicing forms.
We have initiated a systematic study of the expression and function of MEF2 pre-mRNA splicing variants. Here we describe one critical aspect of the control of MEF2C activity that is exerted by serine phosphorylation within a domain that is included or excluded through alternative splice acceptor site usage in the terminal coding exon of MEF2C. Our findings demonstrate that there are significant differences among MEF2C splicing isoforms in both expression pattern and transactivation function. They also reveal a previously unrecognized phosphorylation-sensitive MEF2 repressor domain and indicate one important mode in which MEF2C is unique among MEF2 genes.
MATERIALS AND METHODS
Plasmid construction.
Plasmid sequences are available from the corresponding author. All construct sequences were verified by dideoxy sequencing (1). [MEF2CPT-IB]3-tk-Luc and [MEF2MEF2A]3-tk-Luc, [−252]-MEF2A-1-Luc, [−150-+75]-jun-Luc, and pCDNA-HA-MKK6EE have been described (G. S. Yu, B. Zhu, and T. Gulick, submitted for publication). pG5Luc (Promega) was modified to include a simian virus 40 (SV40) enhancer fragment from pGL3-Enhancer (Promega) to give pG5Luc[SV40]. The coding region for constitutively active CamK IV lacking the regulatory domain (25) was generated using reverse transcription-PCR with primers 5′-gcgcccaagcttgccaccATGgTCAAAGTCACGGTGCCC-3′ and 5′-gaggtggcggccgctttaGAGCTTCTTTTGAGCGGTATC-3′ (uppercase letters indicate wild-type residues, lowercase letters indicate either mutations or sequence that is extraneous to the sense of interest and used in vector construction, and underlining indicates restriction sites used in subcloning or as signatures), and the HindIII- plus NotI-restricted amplicon was subcloned to give pCDNA3-CamK IV1-317.
MEF2 coding regions were obtained using reverse transcription-PCR with human or murine heart, brain, or skeletal muscle RNA as templates. pET-MEF2C α2.β containing the human MEF2C α2.β isoform coding region has been described, as have pET-MEF2A α1 and pET-MEF2D α1 (Yu et al., submitted). Additional MEF2C isoform coding regions were obtained by fragment swapping from EST IMAGE clones 3076773 (α1), 2009507 (β−) and 3071011 (γ+). MEF2 isoform coding regions from pET28 were introduced into pCDNA3.hygro (Invitrogen) (pCDNA-MEF2C), pM (Clontech) (pM-MEF2C), and pPac (pPac-MEF2C) (T. Gulick, unpublished data). pPac contains the CMV immediate-early promoter driving expression of the cloned insert, as well as the puromycin acyltransferase and EBNA-1 genes expressed from minimal promoters and the Epstein-Barr virus origin of replication, and is stably maintained as an episome at low copy number in mammalian cells under puromycin selection.
Combination PCR on a pET-MEF2C α1.β.γ template was used with vector and mutation-specific complementary primers for mutagenesis of the γ-domain Ser396 and for introduction of a silent mutation to install an XbaI site immediately downstream of the γ domain. The forward primer 5′-ACCCCTTCtAGATACCCACAAC-3′ was used in combination with various reverse primers, including 5′-GTGGGTATCTaGAAGGGGTGGTGGTACGGTCccGggGAGGtgcAACAGGTTCTG-3′(S396A), 5′-GTGGGTATCTaGAAGGGGTGGTGGTACGGTCTCTAGGAGGAcAAACAGG-3′ (S396C), and 5′-GTGGGTATCTaGAAGGGGTGGTGGTACGGTCccGcgGAGGttcAACAGGTTCTG-3′ (S396E). NotI- plus KpnI-restricted amplicons were introduced into pM-MEF2C α1.β.γ. pM-MEF2A α1Δ372-401 was constructed using PCR on a pET-MEF2A α1 template with a reverse vector primer and forward primer 5′-cccgagCCTAGGACAAGCAGCCCTCAGCTCTCTTGTcGAcCGTATGACCCCATCG-3′, followed by introduction of the AvrII- plus NotI-restricted amplicon into pM-MEF2A α1. pM-MEF2A α1S398A was created by using combination PCR with complementary primers for mutagenesis of the γ-domain Ser398 and introduction of a silent mutation to install an XhoI site. The forward primer 5′-CGATTgCACCTC CTCGaGATCGTATG-3′ was used in combination with the reverse primer 5′-ACGATCtCGAGGAGGTGcAATCGGTTC-3′ and vector primers on the pET-MEF2A α1 template, and the AvrII- plus NotI-restricted amplicon was introduced into pM-MEF2A α1. Each MEF2C and MEF2A coding region mutant was also subcloned into pCDNA3.hygro.
Human MEF2C sequences 3′ of the MADS box/MEF2 signature domain were amplified by PCR using forward primers 5′-cccgcgccATGGTGACGTTGAGAAAGAAGGGCC-3′ (α1) or 5′-cccgcgccATGGCATTGAACAAGAAAGAAAACAAAGG-3′ (α2) and reverse vector primer on pM-MEF2C templates. NcoI- and PstI-digested amplicons were reintroduced into pM-MEF2C to give pM-ΔN87-MEF2C constructs. MEF2C open reading frames with the stop codon removed were made by using PCR with a forward vector primer and reverse primer 5′-gcgtgtgcctgcaggTGTTGCCCATCCTTCAGAAAG-3′, and the NcoI- and SbfI-cut amplicons were introduced into a derivative of pCDNA3 containing green fluorescent protein coding sequences to give pCDNA3-MEF2C-EGFP constructs. The MEF2C γ domain with or without the S396A mutation was amplified using PCR with primers 5′-cccgaattccccATGGCTTGCACTAGCACTC-3′ and 5′-cccgcggcggccgctttaTCTAGGAGGAGAAACAGGTTC-3′ (γ) or 5′-cccgcggcggccgctttacCTAGGAGG AGcAACAGGTTC-3′ (γS396A), and the NcoI and NotI-restricted amplicons were subcloned into pM to give pM-[γ]MEF2C and pM-[γS396A]MEF2C, respectively. NheI-NotI inserts from these and the Gal4 DNA binding domain (Gal4DBD)-MEF2C fusions were subcloned into pCDNA3 to give pCDNA-Gal4DBD-[γ]MEF2C and -[γS396A]MEF2C and pCDNA-Gal4DBD-MEF2C isoform expression constructs.
Cultured-cell transfection.
C2C12 cells were maintained and differentiated as described previously (51). HeLa, COS, and 293 cells were maintained in Dulbecco's modified Eagle's medium (DME) with 10% fetal calf serum. Cells were split into 12-well plates 1 day prior to transfection with Superfect (Invitrogen). Triplicate wells received 1.0 μg of reporter plasmid, 0.1 or 0.3 μg of control reporter, and 1.0 μg of expression vector(s) except where otherwise indicated, and cells were harvested for reporter activity determinations 24 to 36 h after transfection. Luciferase readings were corrected for transfection efficiency using β-galactosidase activity from pSV40βGal or Renilla luciferase activity from pRL-tk (Promega). 10T1/2 MyoD-ER cells (gift of Stephen Tapscott, Hutchinson Cancer Research Center) were maintained in DME with 10% charcoal-stripped fetal bovine serum and 500 μg of G418/ml. Cells were stably transfected with pPac-EGFP or the pPac-MEF2C variants and incubated in 5 μg of puromycin/ml to create stable cell lines. Total cell RNA and protein were harvested at serial time points after a change to differentiation medium containing G418 and puromycin and 10 nM estradiol (E2) (Sigma) (3).
Protein analyses.
Western blotting of polyacrylamide gel electrophoresis (PAGE)-separated proteins was performed with horseradish peroxidase chemiluminescence assays (ECL; Amersham). Primary antibodies recognized skeletal muscle myosin heavy chain (2) (MF20; Development Studies Hybridoma Bank, University of Iowa), α-actin (MAB1501R; Chemicon), the Gal4DBD (SC510 and RK5C1; Santa Cruz Biotechnology) or MEF2C (Yu et al., submitted). Immunoprecipitations of cell protein extracts were performed with protein A Sepharose after protein solubilization in buffer containing protein phosphatase inhibitors (1). Precipitated proteins were resolved by sodium dodecyl sulfate (SDS)-PAGE and visualized using silver staining. Bands of interest were excised and submitted for extraction, tryptic digestion, and tandem mass spectrometry (MS/MS) at the Taplin Biological Mass Spectrometry Facility at Harvard Medical School.
RNA analyses.
Murine tissue and C2C12 cell RNA was isolated by conventional procedures (1). RNA was harvested from C2C12 myoblasts daily during growth to confluence in DME with 20% fetal calf serum and daily during differentiation after cell confluence in DME with 2% horse serum. RNase protection assays (RPA) and radiolabeled cRNA probe syntheses were carried out as described previously (51). The template for the mef2-c cRNA probe was a 186-bp BglII-to-HincII fragment from a mouse mef2-c γ− cDNA subcloned into pBS-SK (Stratagene).
RESULTS
One MEF2C alternative splicing pattern is unique among vertebrate MEF2 genes.
The structures of the four vertebrate MEF2 genes, MEF2A, MEF2B, MEF2C, and MEF2D, either have been described (46, 47), were established by us (Yu et al., submitted), or can be inferred from genomic and cDNA sequences in GenBank. All but MEF2B have highly similar gene structures and alternative splicing patterns among coding exons (Fig. 1A and B). In each case, coding exon 1 encodes part of the MADS box, while exon 2 encodes the carboxy terminus of this domain and the adjacent MEF2 signature (5). Alternative third exons 3α1 and 3α2 are present in each gene, as is a short exon β between exons 6 and 7 that is variably included in mRNAs. MEF2D is unique in having one extra coding exon (exon 10) at the 3′ end of the gene. Domains encoded by the alternative α and the β exons of these MEF2 genes have similar primary structures.
FIG. 1.
Vertebrate MEF2 transcripts are alternatively spliced. (A) Schematic of the highly similar structures of three vertebrate MEF2 genes among coding exons. Splicing patterns of MEF2A and MEF2D differ from MEF2C (top) only at the extreme 3′ exons. (B) Schematic of the MEF2C alternative splicing isoforms. Forms are named according which α domain is present (α1 or α2) and whether or not the β or γ domains are present (e.g., α1.γ or α2.β.γ). (C) Sequences flanking and within (bold) the MEF2C γ domain are shown, with two splice acceptors underlined. Human, cow, and rat MEF2C nucleotide sequences are 100% conserved in the region shown. Nucleotide substitutions in opposum (Dv), chicken (Gg), and mouse (Mm) sequences are indicated. (D) Alignment of the carboxy terminus of the MEF2C γ domain with similar regions of human MEF2 and Drosophila Dmef2 gene forms. The cryptic splice acceptor of MEF2C is not conserved in the other MEF2 genes. Residues that do not conform to a splice acceptor polypyrimidine tract (gray boxed characters) or that preclude splicing at this locus (white characters) are indicated.
MEF2C also has an alternative cryptic splice acceptor within exon 9, and we designated the region excluded by use of this alternative γ. Nucleotide sequences within and directly flanking MEF2C γ are >98% conserved among vertebrates (Fig. 1C). In the other vertebrate MEF2 genes and Drosophila Dmef2, nucleotide sequences within the last coding exon do not conform to a splice acceptor (Fig. 1D), such that alternative splicing of this kind is unique to MEF2C. This said, other MEF2 forms and particularly MEF2A have a region that is similar to MEF2C γ. In summary, MEF2A and MEF2D mRNAs each have four potential distinct coding regions. We refer to these and their cognate encoded protein isoforms as α1, α1.β, α2, or α2.β based on their composition with respect to alternative domains. By contrast, there are eight potential MEF2C variants, half of which exclude (γ−) and half of which include (γ+) a γ domain (Fig. 1B).
MEF2C splicing isoforms have distinct transactivation capacities.
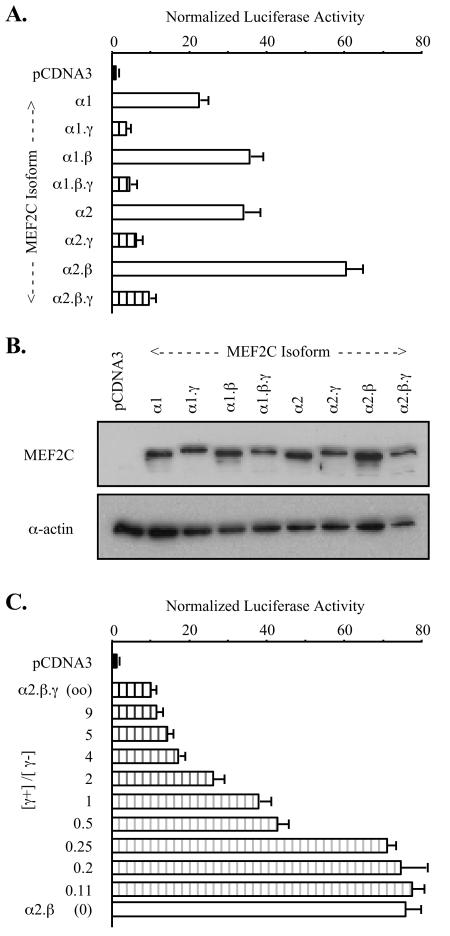
MEF2 mRNAs and MEF2 proteins are widely expressed, yet MEF2 target gene expression is highly restricted (5, 11, 41). This discordance led us to consider selective temporo-spacial expression and differential functions of the splicing variants. To begin to explore this, we tested the activities of the eight MEF2C splicing isoforms on MEF2-responsive reporters in cotransfected cells. COS cells were used in these studies because endogenous MEF2 factor expression and activity are low in this cell type. Results of a representative experiment using a reporter containing three copies of the CPT-IB gene MEF2 element upstream of a minimal HSV tk promoter ([MEF2CPT-IB]3-tk-Luc) are shown in Fig. 2A. Each of the four MEF2C isoforms lacking the γ domain (γ−) was substantially more transcriptionally active than the corresponding γ+ form. The influence of γ was independent of the alternative α or β domains. The activity ratio of γ− to γ+ MEF2C forms in common α and β contexts (e.g., α1.β/α1.β.γ or α2/α2.γ) averaged ∼8 in a series of COS cell transfections. Immunoblots of transfected-cell extracts using a MEF2C-specific antibody (Yu et al., submitted) confirmed that the different isoforms were expressed to equivalent levels (Fig. 2B). Similar distinctions in activities of MEF2C isoforms were apparent when using reporters containing promoters or isolated MEF2 elements from the CPT-IB, MEF2A, or c-jun (16) genes (data not shown). Thus, differences in transactivation given by MEF2C isoforms were MEF2 element and promoter context independent and was not attributable to differential expression or stability.
FIG. 2.
MEF2C splicing isoforms have distinct transactivation capacities. (A) COS-7 cells were cotransfected with [MEF2CPT-IB]3-tk-Luc, control pRL-tk reporter, and pCDNA3 expression vector for the indicated MEF2C splicing isoforms. Firefly luciferase activities were normalized for transfection efficiency by using extract Renilla luciferase and normalizing to activity in pCDNA3-transfected cells (= 1.0). (B) Extracts from cells transfected in panel A were resolved by SDS-PAGE, and MEF2C expression was visualized by immunoblotting with anti-MEF2C antibody that recognizes all MEF2C splicing isoforms (upper panel). Sample loading was normalized using α-actin immunoreactivity (lower panel). (C) COS-7 cells were cotransfected with [MEF2MEF2A]3-tk-Luc, pRL-tk, and either pCDNA3 or the indicated ratios of pCDNA-MEF2C α1.β.γ (γ+) to pCDNA-MEF2Cα1.β (γ−). Each well received a total of 0.3 μg of expression vector. Samples were analyzed as for panel A.
Transcriptional activity and responses to signaling mediated by MEF2 elements can depend on the composition of MEF2 proteins bound to a target element (41, 53). We found that MEF2C splicing variants, including the γ+ and γ− isoforms, can pair promiscuously in MEF2 element binding in vitro (data not shown). This was expected based on prior mapping of MEF2 dimerization and DNA binding functions to the MADS box and MEF2 domains, which are shared among all splicing isoforms (35). Functional consequences were explored by measuring the activity of another MEF2-responsive reporter ([MEF2MEF2A]3-tk-Luc; Yu et al., submitted) in COS cells cotransfected with different ratios of MEF2C α1.β.γ (γ+) and MEF2C α1.β (γ−). Expression of the γ− form alone produced more than 75-fold induction of [MEF2MEF2A]3-tk-Luc, whereas the γ+ form alone induced reporter activity that was less than 10-fold above basal activity (Fig. 2C). Intermediate responses were seen with forced expression of the two forms together, indicating that neither form is dominant and that no unexpected novel transcriptional activity stems from MEF2C splicing isoform coexpression.
MEF2C γ domain function is cell type independent.
MEF2C isoforms were expressed as Gal4p DNA binding domain (Gal4DBD) fusions in tests of transactivation of a Gal4-responsive reporter (pG5Luc). In this one-hybrid system, differences in transactivation produced by the various Gal4DBD-MEF2C fusions recapitulated those of the native MEF2C isoforms. Specifically, Gal4DBD-MEF2C γ− isoforms gave ∼20-fold-higher activity than the corresponding γ+ form in transfected COS cells (Fig. 3A). In a series of transfections, each fusion protein containing the γ domain activated the reporter only slightly above the basal activity in cells expressing the Gal4DBD alone (2- to 10-fold, depending on transfection conditions). Taken together with the potent transactivation given by each γ− isoform, a capacity for a wide dynamic range of function among the MEF2C splicing isoforms was evident. As for the native MEF2C forms, an influence of the γ domain was seen in each alternative α and β context in these fusions, and distinctions in activity were not attributable to differences in levels of expression (Fig. 3B). These observations suggest that the presence or absence of γ does not control MEF2 transcriptional activity via cis effects on formation of a ternary DNA-bound complex. Specifically, γ does not appear to act to blunt MEF2C function by interfering with DNA binding or MEF2 dimerization, nor does deletion of the γ domain give amplified transactivation through enhanced dimerization or DNA binding affinity.
FIG. 3.
MEF2C γ-domain function is cell type independent. (A) COS-7 cells were cotransfected with pG5Luc, control pRL-tk reporter, and pM vectors expressing indicated Gal4DBD-MEF2C splicing isoform fusions. Experiments were analyzed as for Fig. 2, except that normalization was to activity in pM-transfected cells (= 1.0). (B) Extracts from cells transfected in panel A were resolved by SDS-PAGE, and Gal4DBD-MEF2C expression was visualized by immunoblotting with monoclonal antibody that recognizes the Gal4DBD (upper panel). Sample loading was normalized using α-actin immunoreactivity (data not shown). (C) C2C12 myoblasts and myotubes were transfected and analyzed as for panel A.
Having validated the one-hybrid system as an approach for evaluation of MEF2C splicing isoform function, we examined whether distinctions in transactivation generalized to cell types in which MEF2 proteins are endogenously expressed. In HeLa (41) and 293 HEK (53) cells, the pattern of Gal4DBD-MEF2C isoform fusion activities was identical to that seen in COS (data not shown). Thus, all isoforms containing the γ domain were substantially less active than the corresponding γ− forms in activating pG5Luc. Some subtle differences in Gal4DBD-MEF2C fusion activities were apparent among the cell types. For example, γ+ forms gave higher activity relative to the Gal4DBD control in both 293 cells and C2C12 myoblasts than in COS cells. In addition, in C2C12 cells transfected during serum withdrawal-mediated differentiation to myotubes, distinctions in transactivation produced by the γ− versus γ+ isoform fusions were less pronounced than in C2C12 myoblasts or nonmuscle cell types (Fig. 3C). Nonetheless, transactivation by the γ+ isoforms was much lower in all cell types tested, including myocytes.
mef2-c is expressed predominantly as γ− splicing isoforms.
The relative abundance of mef2-c splicing isoforms among murine tissues was examined using RPA. The cRNA probe used to detect γ domain isoforms spanned the mef2-c exon 8-9 junction of a γ− murine mef2-c cDNA (Fig. 4A). In all tissues examined, including skeletal muscle, heart, and brain, where relatively high mef2-c mRNA expression levels were seen, γ− mRNAs were more abundant than γ+ messages (Fig. 4B). The ratio of γ− to γ+ mRNA was similar among tissues studied, with the exception of heart tissue, where abundant γ− message was apparent in the absence of detectable γ+ mRNA. All combinatorial alternative splicing events related to the β and γ domains were evident in tissue mRNAs based on RPA with additional probes (data not shown). We also measured the expression of mef2-c splicing isoform mRNAs in cultured C2C12 cells after serum withdrawal to determine if the ratio of γ− to γ+ mRNA was dynamic during myocyte differentiation. Here, total mef2-c mRNA gradually increased from undetectable to high levels and, as in tissues, γ− mRNAs predominated. The ratio of γ− and γ+ message was constant among these RNA samples (Fig. 4C). However, our results cannot exclude the possibility of predominant expression of γ+ MEF2C message, or a dynamic ratio of γ− to γ+ forms, in tissues or developmental stages not examined here.
FIG. 4.
MEF2C is expressed predominantly as γ− isoforms. (A) Schematics of the MEF2C cDNA region and the γ− template used for cRNA probe synthesis. (B) RPA results for indicated mouse tissue total RNA samples. Protected probe fragment sizes that are specific for γ− (186 bases) and γ+ (117 bases) mRNAs are indicated at the right of the autoradiogram. Reactions each included 5 μg of RNA. (C) RPA using total RNA harvested from C2C12 cells undergoing differentiation. Numbers reference days prior to or after cell confluence, at which time medium serum was changed from 20% fetal calf serum to 2% horse serum. Reactions each included 5 μg of RNA.
MEF2C γ domain Ser phosphorylation controls its function.
Although mRNAs for the various MEF2 alternative splicing isoforms are clearly expressed, the existence of cognate proteins has not been established. We used MS/MS of proteolytic digests of immunoprecipitated MEF2C isoforms to confirm the existence of the variants and for analysis of their modification. These studies and those with overexpressed MEF2 fusions revealed several previously unreported MEF2 phosphoserine and phospho-threonine residues. Among these was a modified Ser residue within the MEF2C γ domain corresponding to position 396 of the α1.β.γ isoform (Fig. 5A). The identical tryptic fragment lacking Ser modification was also identified in samples (Fig. 5B), despite preparation of samples in the presence of a cocktail of protein phosphatase inhibitors.
FIG. 5.
A MEF2C γ-domain Ser is phosphorylated. Extracts were harvested from COS-7 cells transfected with pCDNA-Gal4DBD-MEF2C isoforms in the presence of phosphatase inhibitors. Overexpressed fusions were immunoprecipitated and resolved on SDS-PAGE. Proteins extracted from gel slices were trypsinized, and resulting fragments were resolved using high-performance liquid chromatography and identified by MS/MS. The γ-domain tryptic fragment SEPVSPPR with (A) and without (B) Ser phosphorylation was detected. Insets show tabular data of detected peptide fragment masses. The MEF2C Ser phosphoacceptor corresponds to residue 388 of the α1.γ form, 396 of α1.β.γ, 386 of α2.γ, and 394 of α2.β.γ.
To examine the influence of γ-domain Ser phosphorylation at this residue, the transactivation function of point mutants of MEF2C α1.β.γ (γ+) were tested in transfected COS cells. Initially, S396 was mutated to either Ala or Cys to destroy the phosphorylation potential. In the one-hybrid system, the Gal4DBD-MEF2C α1.β.γS396A and -.γS396C fusions activated pG5Luc more than 50-fold compared to only 4-fold for the analogous wild-type γ+ fusion (Fig. 6A) despite comparable expression levels (Fig. 6B). Similar results were obtained in tests of mutations in the native MEF2C α1.β.γ context. Thus, the [MEF2CPT-IB]3-tk-Luc reporter was activated 20- to 30-fold by MEF2C α1.β.γS396A compared to only 3-fold by the wild-type γ+ form (Fig. 6C). Augmentated transactivation by these mutants was apparent in all cell types examined, including C2C12 cells. γ+ S396E mutants were not less transcriptionally active than the analogous wild-type forms, suggesting that substitution of an acidic Glu residue did not effectively mimic the phosphoserine. Nonetheless, these findings indicate a critical role for phosphorylation of Ser 396 in γ-domain-mediated transcriptional dampening.
FIG. 6.
MEF2C γ-domain Ser phosphorylation controls its function. (A) COS-7 cells were cotransfected as described in the legend to Fig. 3 with pM-MEF2C α1.β or wild-type or indicated point mutants of pM-MEF2C α1.β.γ. Results were analyzed as with Fig. 3. (B) Extracts from cells transfected in panel A were immunoblotted as in Fig. 3. (C) COS-7 cells cotransfected with pCDNA3 constructs expressing indicated MEF2C forms or mutants were analyzed as with Fig. 2.
MEF2C γ-domain function is independent of calmodulin and MAPK signaling and of the MADS and MEF2 domains.
MEF2 factor transactivation functions are known to be regulated by signaling through MAPK and calcium-calmodulin kinase (CaMK) pathways (Fig. 7A). MEF2A and MEF2C are direct substrates for p38 α and β (49, 53) and Erk5 (18) MAPK, and the respective phosphorylations enhance MEF2 transactivation function. Recognized substrate sites for these enzymes are within regions of MEF2A and MEF2C transactivation domains that are common to all splicing isoforms. CaMK IV activity influences MEF2 protein function indirectly through phosphorylation of class II histone deacetylase (HDAC) proteins to dissociate sequestered MEF2 from a complex with HDAC (30).
FIG. 7.
γ-Domain function is independent of p38 MAP and calmodulin kinase signaling and of the MADS box and MEF2 domains. (A) Schematic of known sites of MEF2C phosphorylation (black dots) and of protein kinase, class II HDAC, transcription factor, and coactivator interaction domains (solid bars). The MADS and MEF2 domain interaction with HDAC proteins is abolished with CamK IV-mediated phosphorylation of HDAC. (B) 293 HEK cells were cotransfected with pRL-tk, pG5Luc, the indicated pM-MEF2C isoform fusions, and with or without pCDNA-MKK6EE, followed by incubation in the presence or absence of SB203580 prior to harvesting for analysis as described for Fig. 3. (C) Schematic of Gal4DBD-MEF2C fusions. (D) COS-7 cells were cotransfected and analyzed as for Fig. 3, using constructs expressing the indicated fusions.
We next examined whether γ-domain Ser phosphorylation involved these previously recognized pathways of MEF2C functional regulation. One-hybrid studies of Gal4DBD-MEF2C isoform fusions were conducted with 293 cells cotransfected with pCDNA-MKK6EE or pCDNA-CaMK IV1-317, expressing a constitutively active kinase that activates p38 MAPK (43) or a constitutively active CaMK IV (25), respectively. After transfection, cells were incubated in the presence or absence of the p38 and CaMK IV inhibitors SB203580 and KN62 (8). As shown in Fig. 7B, p38 signaling activity regulated the transactivation functions of γ+ and γ− Gal4-MEF2C fusions, as well as that of the γ+ Ser mutant, to a similar degree on the pG5Luc reporter. This was consistent both with the fact that the p38 α/β docking domain (49) is present in all MEF2C splicing isoforms (Fig. 7A) and with the isolation of both γ− and γ+ partial cDNAs in the original two-hybrid screen that identified MEF2C as a p38 substrate (15). CamK IV signaling activity also controlled the transactivation function of each isoform and the γS396A mutant comparably (data not shown). Furthermore, distinct activities of the isoforms and this mutant were maintained in cells incubated in medium supplemented with both SB203580 and KN62. Thus, modification of MEF2C by p38 MAPK or CaMK IV is neither responsible for nor prerequisite to γ Ser phosphorylation.
One-hybrid studies using Gal4DBD-MEF2C isoform fusions devoid of the N-terminal 87 amino acids of the MADS box and MEF2 signature (ΔN87 MEF2C) were used to determine if γ function involved these domains (Fig. 7C). In transfected COS cells, the ΔN87 MEF2C fusions produced more robust transactivation than the corresponding full-length MEF2C fusions, probably due to relief from HDAC interactions. However, activation of pG5Luc by the ΔN87 MEF2C γ- and γ+ Ser mutant fusions was again much higher than that by the corresponding γ+ fusion (Fig. 7D). In transfected C2C12 cells, the basal activities of ΔN87 isoform fusions were lower than the full-length constructs, perhaps resulting from loss of interactions with myogenic bHLH factors or an important subset of transcriptional coactivators (data not shown). However, independence of γ-domain function from the MADS box and MEF2 domains was also apparent. These findings confirmed that γ does not regulate or require dimerization or DNA binding by intrinsic MADS and MEF2 domains. Further, mechanistic roles for numerous previously described protein interactors with the MADS box and MEF2 signature domain are excluded (19, 21, 22, 31, 33, 34, 45, 50).
The constitutive MEF2A γ domain functions as MEF2C γ.
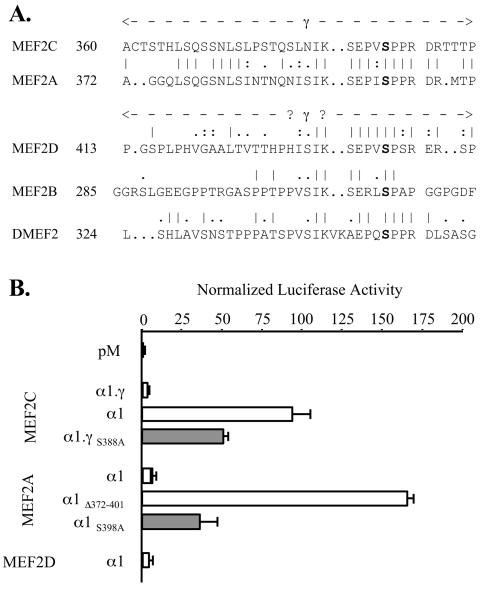
We defined the amino-terminal limit of the γ domain of MEF2A as the residue encoded at the 5′ end of coding exon 9, since the amino acid sequence alignment between the gene forms in this region was somewhat ambiguous. By contrast, a high degree of sequence similarity is apparent between the respective γ carboxy termini (Fig. 8A). To determine if the γ domain of MEF2A also acts to limit transactivation, we compared the transactivation function of Gal4DBD fusions of MEF2A α1 to that of a construct in which residues 372 to 401 (the putative MEF2A γ domain) were deleted (pM-MEF2A α1Δ372-401). As shown in Fig. 8B, this artificial γ− MEF2A fusion produced a ∼20-fold-higher level of stimulation of pG5Luc than the wild-type MEF2A form. Further, mutation of MEF2A α1 Ser 398, analogous to MEF2C α1.β.γ Ser 396, also produced marked enhancement of MEF2A activity. Thus, γ domain function and the effects of γ Ser phosphorylation are evident in both MEF2C and MEF2A. Less conspicuous sequence similarity in the related regions of MEF2D, MEF2B, and DMEF2 (Fig. 8A) leave it uncertain as to whether a functional γ domain exists in these proteins. However, the activities of analogous γ+ isoforms of MEF2C (α1.γ) and MEF2A and MEF2D (each α1) were low and similar, confirming that the MEF2C γ− isoforms have uniquely strong activity among the various MEF2 gene and splicing forms (Fig. 8B).
FIG. 8.
The constitutive MEF2A γ domain functions like MEF2C γ. (A) Alignment of the γ domain of human MEF2C with related regions of human MEF2A, MEF2D, and MEF2B and Drosophila DMEF2. Numbers reference residues in the α1.γ (MEF2C), α1 (MEF2A and MEF2D), or A (Drosophila DMEF2) splicing isoform. (B) COS-7 cells were cotransfected and analyzed as for Fig. 3, using pM constructs expressing the indicated isoform or point or deletion mutant Gal4DBD fusions.
MEF2C γ functions autonomously as a repressor domain.
Transrepression of MEF2-responsive reporters by γ+ isoforms of MEF2C was not seen in our studies in any of a wide variety of cells. This suggested either that γ is not a repressor domain per se but provides for negative modulation of MEF2C transactivation or, alternatively, that an authentic γ repressor function is masked by more-potent activation domain activities of MEF2 under the conditions studied. The latter conclusion was supported by investigations with a fusion of the isolated 32-amino-acid γ domain to the Gal4DBD (Gal4DBD-[γ]MEF2C), in parallel with the analogous fusion containing the S396A mutation (Gal4DBD-[γS396A]MEF2C). Transcription of a Gal4-responsive luciferase reporter with a high basal activity level (pG5Luc[SV40]) was significantly depressed in COS cells expressing Gal4DBD-[γ]MEF2C to 18% of control but not in those expressing Gal4DBD-[γS396A]MEF2C (Fig. 9A). Consistent with this observation, MS/MS detected γ Ser phosphorylation in the Gal4DBD-[γ]MEF2C protein (data not shown). Immunoblots of transfected cells showed that the fusions were stable and were expressed to similar levels (Fig. 9B). Thus, our findings are most consistent with autonomous repression by γ, mediated in trans and controlled by a modification of a Ser residue within γ that requires only γ-domain residues.
FIG. 9.
The MEF2C γ domain functions autonomously as a repressor domain. (A) COS-7 cells were cotransfected and analyzed as for Fig. 3, using constructs expressing the indicated Gal4DBD fusions. (B) Extracts from cells transfected in panel A were immunoblotted as for Fig. 3.
Myogenic conversion of progenitor cells is differentially controlled by MEF2C splicing isoforms.
Functional synergism between MEF2 proteins and myogenic basic helix-loop-helix (bHLH) factors, such as MyoD and myogenin, is well established. Interaction between members of these two families occurs at several levels, including transcriptional cross talk and direct protein-protein interaction (34, 47). Although the latter occur via the MADS and MEF2 domains, which are common to all MEF2 splicing variants, we speculated that the dramatically different transactivation functions of the MEF2C γ− and γ+ isoforms could ramify on myogenic potential. We used 10T1/2 cells transformed with a retrovirus directing expression of a MyoD-estrogen receptor fusion protein (MyoD-ER) (3) to address this question. Estrogen (E2)-dependent MyoD nuclear localization and function are exerted in these cells, providing a system for testing of MEF2C splicing isoforms in the synergistic induction of myogenesis.
MyoD-ER 10T1/2 and control 10T1/2 cells were stably transfected with episomally maintained vectors expressing either the MEF2C α2.β isoform, the α2.β.γ variant, the α2.β.γS394A mutant, or enhanced green fluorescent protein (EGFP) as a control. Endogenous MEF2C protein was undetectable in immunoblots of extracts from the MyoD-ER control cell line stably expressing EGFP. The MEF2C forms and the γ-domain mutant were stably overexpressed to identical high levels with this system (Fig. 10A). The various cell lines were then incubated in medium with or without E2 supplementation to test for MEF2C isoform effects on myogenesis. None of the stably transfected control 10T1/2 cell lines differentiated under any condition studied, as expected (data not shown). By contrast, E2 treatment of the MyoD-ER cells produced clear changes in cell morphology. The rate of morphological change in appearance and of mRNA and protein markers of muscle phenotype differed significantly among cells overexpressing the MEF2C variants. Thus, obvious synergy between MyoD-ER and MEF2C in inducing skeletal muscle myosin heavy chain protein was seen in cells expressing the MEF2C α2.β (γ−) form, but this was barely apparent in those expressing the γ+ variant (Fig. 10B). Cells overexpressing MEF2C α2.β.γS394A showed E2-dependent myogenesis with a time course similar to that of the γ−-expressing cells (Fig. 10C). Taken together, this confirmed attribution of the distinct kinetics of myogenesis to both the γ domain and its Ser phosphorylation. Since MyoD and other myogenic bHLH factors activate mef2-a and mef2-c transcription (44, 47), this system undoubtedly understates functional differences in the MEF2C splicing isoforms. Nonetheless, the biological impact of distinctions in MEF2C splicing isoform transactivation functions is shown categorically.
FIG. 10.
MEF2C isoforms are differentially active in promoting myogenesis. (A) 10T1/2 MyoD-ER cells were stably transfected with pPac expressing green fluorescent protein (GFP), MEF2C α2.β, MEF2C α2.β.γ, or MEF2C α2.β.γS394A. MEF2C isoform and mutant overexpression was confirmed by immunoblotting with antibody to MEF2C (upper panel). Sample loading was normalized using α-actin immunoreactivity (lower panel). (B) Culture medium bathing the cell lines was changed to differentiation medium containing 10 nM E2, followed by harvesting of total cell RNA and protein at the indicated times. Protein extracts were resolved by SDS-PAGE and immunoblotted using monoclonal antibody MF20, which recognizes skeletal muscle myosin heavy chain, a marker of muscle differentiation. (C) 10T1/2 MyoD-ER cells stably transfected with pPac expressing the MEF2Cα2.β.γS394A mutant and control cells expressing the wild-type α2.β or α2.β.γ isoform were incubated in differentiation medium for 3 days before protein extract harvesting and immunoblotting as for panel B.
DISCUSSION
Splicing variants of the various MEF2 gene transcripts are long established, in some cases having been identified during the original cDNA cloning effort (5, 7, 27, 29, 42, 52). Despite this fact, with the exception of PCR-based semiquantitation of tissue expression of the alternative mRNAs, there has been little information reported as to distinct expression or functions of the various MEF2 splicing isoforms (17). Here we have presented one such distinction that involves the convergence of alternative MEF2C pre-mRNA splicing with MEF2C protein modification. The encoded alternative splicing variants include or exclude a trans-acting repression domain whose function depends on serine phosphorylation.
Our work clearly demonstrates that γ-domain transrepression function is autonomous, i.e., does not involve cis effects on other MEF2 protein functions outside of the domain. Further, γ activity does not involve previously characterized MEF2 interactions or modifications. The precise mechanism for repression remains to be elucidated. Among several possibilities, we favor γ phosphoserine-dependent recruitment of a transcriptional corepressor, perhaps by phosphorylation-induced changes in conformation or electrostatic interactions. This would not involve the previously described MEF2 interaction with class II HDAC corepressors, since this has been mapped to the MADS and MEF2 signature domains (31). As one alternative, γ may directly interfere with RNA polymerase II complex assembly or initiation. Although γ phosphoserine-dependent displacement of a coactivator interaction may play some role, this as an exclusive mechanism cannot be reconciled with the transcriptional repression shown by the isolated γ domain in the Gal4DBD-[γ]MEF2C fusion. In any case, previously described MEF2 coactivators are not relevant, since p300 (45) and PGC1 (22) interact with the MADS box and MEF2 signature domain, both of which are functionally independent of γ, and the SRC-MEF2 interaction is retained in carboxy-terminal deletion mutants of MEF2 lacking γ (19).
The demonstrated autonomy of γ-domain transrepression function does not exclude the possibility that γ can control or modulate additional MEF2 functions. However, our studies did not detect any such activities. In particular, we saw no evidence for distinctions between γ+ and γ− forms in terms of DNA binding or dimerization, in protein accumulation or degradation when overexpressed in any of a variety of cell types, or in subcellular or subnuclear localization of MEF2C isoform-EGFP fusions (unpublished data). Further, the magnitude of control of MEF2 transactivation function exerted by p38 MAPK or CaMK IV signaling was not substantially different among the MEF2C splicing variants. This said, the transrepression function of γ was more pronounced in the homologous MEF2 protein context than in isolation, and this may relate to additional roles for this domain.
MS/MS analyses detected MEF2C tryptic fragments containing both phosphorylated and unmodified γ-domain Ser (Ser396 of the α1.β.γ isoform). While fragments with an unmodified residue could have resulted from dephosphorylation of a constitutively modified Ser residue during sample isolation and processing, we favor a scenario in which there is regulated phosphorylation and/or regulated dephosphorylation in response to signaling. Consistent with this hypothesis, differences in the degree of repressor function conferred by γ, as well as the extent of mitigation by γ Ser mutation, were seen in various cells and stages of myocyte differentiation, and we suspect that this is due to a dynamic ratio of modified to unmodified γ Ser. Direct confirmation of an inconstant phosphorylation status at this residue awaits results of our studies with antiphosphopeptide and antipeptide antibodies developed for the MEF2C γ domain. However, regardless of whether control of MEF2C activity occurs at this level, regulation of a functionally critical MEF2C Ser modification is clearly exerted by alternative pre-mRNA splicing to include or exclude this phosphoacceptor site.
Primary sequence near the MEF2C γ phosphoacceptor site suggests that this is a substrate for proline-directed Ser/Thr protein kinase(s) of the mitogen-activated or cyclin-dependent kinase families. Indeed, there is a recent report of cdk5-mediated phosphorylation of MEF2D in neurons at a residue that corresponds to the MEF2C γ Ser phosphoacceptor discussed in our work (13). While we cannot dispute a role for cdk5 in controlling the activity of MEF2D and perhaps other MEF2 isoforms in neurons, activity of this enzyme is considered by most to depend on the p35/p39 coregulators that are expressed exclusively in cells of this tissue (10). Our findings suggest a role for signaling through one or more protein kinases other than cdk5, including enzyme(s) expressed and active in both proliferating and quiescent cells derived from various tissues, including muscle. We saw no significant increase in transactivation by MEF2C γ+ isoforms in cells incubated with various inhibitors of MAP- or cyclin-dependent kinases, such that modification of the γ-domain Ser by multiple different protein kinases appears likely.
Specificity of at least some protein kinases is conferred by a kinase docking site on the target in combination with appropriate phosphoacceptor site(s) (49). Thus, for example, all products of the four vertebrate MEF2 genes have potential Thr phosphoacceptors in a conserved region carboxy-terminal to the β domain, but only those in MEF2A and MEF2C are targets of p38 α/β MAPK (53). A p38 docking domain immediately amino-terminal to β, present in MEF2A and MEF2C, is required for kinase-mediated Thr modification (49). The existence of Ser phosphorylation in the Gal4DBD-[γ]MEF2C fusion expressed in transfected cells suggests that γ alone may be sufficient to dock with the relevant kinase(s) that modify γ Ser. However, the primary structure of γ shows no signficant similarity to other proteins, such that no hint is given as to the identity of the modifying enzyme(s) that might interact here. Alternatively, protein kinase(s) that modify the γ Ser residue may actually interact with the MEF2 holoprotein at a site remote from γ. In this case, our detection of Ser phosphorylation of the overexpressed isolated γ (in the Gal4DBD-[γ]MEF2C fusion) may simply reflect the promiscuous activities of various Pro-directed Ser/Thr kinases.
Given our data, the previously described phenomenon of stimulation of myogenesis by cdk5 activity (20) appears unlikely to involve MEF2 factors directly. However, there are several ways in which this reported observation could be reconciled with cdk5 induction of repressor activity within the γ domain of MEF2 factors. Our finding that MEF2C is expressed predominantly as γ− isoforms provides one intriguing possibility, particularly if there is compensatory induction of MEF2C gene expression in a setting of cdk5-mediated downregulation of MEF2A and MEF2D activities.
A region with primary structure similar to that of MEF2C γ is constitutively present in all splicing isoforms of MEF2A, and we have shown that this domain functions like that of MEF2C. Alignment of MEF2 protein sequences suggests that MEF2D may also have a γ domain as well as the phosphoacceptor Ser (Fig. 8A). Consistent with this, we show that transactivation by MEF2D and MEF2A forms is similar to MEF2C γ+ factors, i.e., that MEF2C γ− forms are uniquely potent. As a consequence, the MEF2C gene is likely to have novel functions compared to MEF2A and MEF2D. Specifically, MEF2C alone encodes variants that are able to escape γ Ser phosphorylation-mediated repression that restrains net transactivation capacity. We speculate that this may explain several enigmatic findings in genetic models and systems previously used to evaluate MEF2 functions. For example, disruption of mef2-c is lethal early in murine embryogenesis (24), whereas mef2-a disruption allows survival beyond birth and a selective muscle mitochondrial defect (37). In each case, expression of the mef2 genes that are not disrupted is upregulated. The selective failure of adequate compensation for loss of mef2-c by either upregulation or forced expression of products of other mef2 genes (24) may be a consequence of absence of γ− forms of mef2 genes other than mef2-c. We also propose that discordance between MEF2 mRNA and MEF2 protein expression levels or MEF2 target gene expression observed among cell types and tissues (11, 41) is partially accounted for by differential expression of splicing isoforms, including MEF2C γ− forms.
The 32-amino-acid γ domain of vertebrate MEF2C proteins is present or absent depending on alternative splice acceptor site usage in the last exon. Carboxy-terminal residues of the γ domain, conserved in MEF2C, MEF2A, and MEF2D, are Ser.Pro.(Pro or Ser).Arg, and it this region that is encoded by the cryptic splice acceptor of MEF2C (Fig. 1D). No unusual codon usage is present in any of the MEF2 gene forms here, and the critical difference in MEF2C involves only the Arg codon (AGA in MEF2C, CGN in MEF2A and MEF2D). This simple substitution creates a splice acceptor in MEF2C with an intron/exon boundary between the second and third residues of the codon. The absence of this cryptic acceptor in MEF2A and MEF2D or in lower eukaryotic genes suggests that acquisition of alternative splicing to give the γ− variant is a relatively recent evolutionary event. The other possibility, that γ+ versus γ− alternative splicing was lost during evolution, appears much less likely, since products of the single MEF2 gene in lower eukaryotes show greatest similarity to those of vertebrate MEF2A, the likely primordial vertebrate MEF2 gene. Although Drosophila mef2 transcripts are alternatively spliced within coding exons (14), Dmef2 gene structure does not resemble that of vertebrate MEF2 genes. We have not yet determined if DMEF2 is a substrate for phosphorylation at the Ser residue corresponding to vertebrate MEF2 γ Ser (e.g., MEF2C α1.β.γ S396) (Fig. 8A). Even if so, however, Dmef2 alternative splicing does not impact inclusion of this site.
The cryptic acceptor in MEF2C coding exon 9 giving γ− forms is used preferentially in the tissue and cultured cell samples examined. We favor the possibility that the relative abundance of MEF2C γ+ to γ− forms may vary at developmental stages, tissues, or conditions not examined here, as is implied by the complete absence of γ+ forms in adult heart. The conservation of nucleotide sequence within and immediately upstream of γ among known vertebrate MEF2C genes is remarkable and suggests that critical determinants of splicing are contained within this short region. Both exon 9 splice acceptors conform well to consensus and lack obvious distinguishing features. If there is largely unregulated preferred use of the cryptic acceptor, a bias of U2AF binding to the downstream polypyrimidine tract or SF1 interaction with the respective branchpoint is likely. Regulated use of the two acceptors would be reminiscent of alternative splicing control by Sx1 or PTB, which can function to selectively displace U2AF from the polypyrimidine tract of one alternative 3′ splice site (6). In either case, the regulation of MEF2C splicing is likely to be very complex, since there is covariation of splicing events involving MEF2C exon β (between exons 6 and 7) and the exon 9 alternative splice acceptors (unpublished data). Given the profound distinctions in MEF2C γ− and γ+ isoform activities and the selective potential for control of the latter by phosphorylation, factors and RNA elements critical to this process are of direct relevance to MEF2 target gene expression.
This report is the first in a series that will address MEF2 alternative splicing and the functions and regulation of the encoded isoforms. Findings presented here make clear that consideration must be given to splicing isoforms when examining functional distinctions among products of the different vertebrate MEF2 genes. We have demonstrated that alternative splicing is critical to the regulation of MEF2C activity and illustrated one way that this converges with protein modification to provide complex combinatorial control of this transcription factor. This work does not challenge previously reported observations regarding posttranscriptional control of MEF2 proteins but introduces one additional crucial level of complexity in the regulation of this important class of transcription factors.
Acknowledgments
This work was supported by grants from the American Heart Association (0150622N), the Juvenile Diabetes Foundation (1998-224), the Clinical Nutrition Research Center at Harvard, and the National Institutes of Health (DK55875, HL72713, and DK02461).
We thank Donald Bloch for manuscript review and help with fluorescence microscopy; Joseph Avruch, John Kyriakis, Andrew Lassar, and Simon Shelley for helpful discussion; Stephen Tapscott for 10T1/2 MyoD-ER cells; Geng-Sheng Yu for plasmid constructs; and Dongmei Cheng, Ross Tomaino, and Steven Gygi for MS/MS analyses.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Bader, D., T. Masaki, and D. A. Fischman. 1982. Immunochemical analysis of myosin heavy chain during avian myogenesis in vivo and in vitro. J. Cell Biol. 95:763-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergstrom, D. A., B. H. Penn, A. Strand, R. L. Perry, M. A. Rudnicki, and S. J. Tapscott. 2002. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell 9:587-600. [DOI] [PubMed] [Google Scholar]
- 4.Black, B. L., J. Lu, and E. N. Olson. 1997. The MEF2A 3′ untranslated region functions as a cis-acting translational repressor. Mol. Cell. Biol. 17:2756-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, B. L., and E. N. Olson. 1998. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 14:167-196. [DOI] [PubMed] [Google Scholar]
- 6.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72:291-336. [DOI] [PubMed] [Google Scholar]
- 7.Breitbart, R. E., C. S. Liang, L. B. Smoot, D. A. Laheru, V. Mahdavi, and B. Nadal-Ginard. 1993. A fourth human MEF2 transcription factor, hMEF2D, is an early marker of the myogenic lineage. Development 118:1095-1106. [DOI] [PubMed] [Google Scholar]
- 8.Davies, S. P., H. Reddy, M. Caivano, and P. Cohen. 2000. Specificity and mechanism of action of some commonly used protein kinase inhibitors. Biochem. J. 351:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Angelis, L., S. Borghi, R. Melchionna, L. Berghella, M. Baccarani-Contri, F. Parise, S. Ferrari, and G. Cossu. 1998. Inhibition of myogenesis by transforming growth factor beta is density-dependent and related to the translocation of transcription factor MEF2 to the cytoplasm. Proc. Natl. Acad. Sci. USA 95:12358-12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhavan, R., and L.-H. Tsai. 2001. A decade of cdk5. Nat. Rev. Mol. Cell Biol. 2:7879-7891. [DOI] [PubMed] [Google Scholar]
- 11.Dodou, E., D. B. Sparrow, T. Mohun, and R. Treisman. 1995. MEF2 proteins, including MEF2A, are expressed in both muscle and non-muscle cells. Nucleic Acids Res. 23:4267-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edmondson, D. G., G. E. Lyons, J. F. Martin, and E. N. Olson. 1994. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development 120:1251-1263. [DOI] [PubMed] [Google Scholar]
- 13.Gong, X., X. Tang, M. Wiedmann, X. Wang, J. Peng, D. Zheng, L. A. C. Blair, J. Marshall, and Z. Mao. 2003. Cdk5-mediated inhibition of the protective effects of transcription factor MEF2 in neurotoxicity-induced apoptosis. Neuron 38:33-46. [DOI] [PubMed] [Google Scholar]
- 14.Gunthorpe, D., K. E. Beatty, and M. V. Taylor. 1999. Different levels, but not different isoforms, of the Drosophila transcription factor DMEF2 affect distinct aspects of muscle differentiation. Dev. Biol. 215:130-145. [DOI] [PubMed] [Google Scholar]
- 15.Han, J., Y. Jiang, Z. Li, V. V. Kravchenko, and R. J. Ulevitch. 1997. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature 386:296-299. [DOI] [PubMed] [Google Scholar]
- 16.Han, T. H., and R. Prywes. 1995. Regulatory role of MEF2D in serum induction of the c-jun promoter. Mol. Cell. Biol. 15:2907-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Janson, C. G., Y. Chen, Y. Li, and D. Leifer. 2001. Functional regulatory regions of human transcription factor MEF2C. Mol. Brain Res. 97:70-82. [DOI] [PubMed] [Google Scholar]
- 18.Kato, Y., M. Zhao, A. Morikawa, T. Sugiyama, D. Chakravortty, N. Koide, T. Yoshida, R. I. Tapping, Y. Yang, T. Yokochi, and J. D. Lee. 2000. Big mitogen-activated kinase regulates multiple members of the MEF2 protein family. J. Biol. Chem. 275:18534-18540. [DOI] [PubMed] [Google Scholar]
- 19.Lazaro, J. B., P. J. Bailey, and A. B. Lassar. 2002. Cyclin D-cdk4 activity modulates the subnuclear localization and interaction of MEF2 with SRC-family coactivators during skeletal muscle differentiation. Genes Dev. 16:1792-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lazaro, J. B., M. Kitzmann, M. A. Poul, M. Vandromme, N. J. C. Lamb, and A. Fernandez. 1997. Cyclin dependent kinase 5, cdk5, a positive regulator of myogenesis in mouse C2 cells. J. Cell Sci. 110:1251-1260. [DOI] [PubMed] [Google Scholar]
- 21.Lee, Y., B. Nadal-Ginard, V. Mahdavi, and S. Izumo. 1997. Myocyte-specific enhancer factor 2 and thyroid hormone receptor associate and synergistically activate the alpha-cardiac myosin heavy-chain gene. Mol. Cell. Biol. 17:2745-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, J., H. Wu, P. T. Tarr, C. Y. Zhang, Z. Wu, O. Boss, L. F. Michael, P. Puigserver, E. Isotani, E. N. Olson, B. B. Lowell, R. Bassel-Duby, and B. M. Spiegelman. 2002. Transcriptional co-activator PGC-1alpha drives the formation of slow-twitch muscle fibres. Nature 418:797-801. [DOI] [PubMed] [Google Scholar]
- 23.Lin, Q., J. Lu, H. Yanagisawa, R. Webb, G. E. Lyons, J. A. Richardson, and E. N. Olson. 1998. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development 125:4565-4574. [DOI] [PubMed] [Google Scholar]
- 24.Lin, Q., J. Schwarz, C. Bucana, and E. N. Olson. 1997. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science 276:1404-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, S., P. Liu, A. Borras, T. Chatila, and S. H. Speck. 1997. Cyclosporin A-sensitive induction of the Epstein-Barr virus lytic switch is mediated via a novel pathway involving a MEF2 family member. EMBO J. 16:143-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mao, Z., A. Bonni, F. Xia, M. Nadal-Vicens, and M. E. Greenberg. 1999. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science 286:785-790. [DOI] [PubMed] [Google Scholar]
- 27.Martin, J. F., J. M. Miano, C. M. Hustad, N. G. Copeland, N. A. Jenkins, and E. N. Olson. 1994. A Mef2 gene that generates a muscle-specific isoform via alternative mRNA splicing. Mol. Cell. Biol. 14:1647-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin, J. F., J. J. Schwarz, and E. N. Olson. 1993. Myocyte enhancer factor (MEF) 2C: a tissue-restricted member of the MEF-2 family of transcription factors. Proc. Natl. Acad. Sci. USA 90:5282-5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McDermott, J. C., M. C. Cardoso, Y. T. Yu, V. Andres, D. Leifer, D. Krainc, S. A. Lipton, and B. Nadal-Ginard. 1993. hMEF2C gene encodes skeletal muscle- and brain-specific transcription factors. Mol. Cell. Biol. 13:2564-2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKinsey, T. A., C. L. Zhang, J. Lu, and E. N. Olson. 2000. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 408:106-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2001. Control of muscle development by dueling HATs and HDACs. Curr. Opin. Genet. Dev. 11:497-504. [DOI] [PubMed] [Google Scholar]
- 32.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2002. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27:40-47. [DOI] [PubMed] [Google Scholar]
- 33.Miska, E. A., C. Karlsson, E. Langley, S. J. Nielsen, J. Pines, and T. Kouzarides. 1999. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 18:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell 83:1125-1136. [DOI] [PubMed] [Google Scholar]
- 35.Molkentin, J. D., B. L. Black, J. F. Martin, and E. N. Olson. 1996. Mutational analysis of the DNA binding, dimerization, and transcriptional activation domains of MEF2C. Mol. Cell. Biol. 16:2627-2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mora, S., and J. E. Pessin. 2000. The MEF2A isoform is required for striated muscle-specific expression of the insulin-responsive GLUT4 glucose transporter. J. Biol. Chem. 275:16323-16328. [DOI] [PubMed] [Google Scholar]
- 37.Naya, F. J., B. L. Black, H. Wu, R. Bassel-Duby, J. A. Richardson, J. A. Hill, and E. N. Olson. 2002. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat. Med. 8:1303-1309. [DOI] [PubMed] [Google Scholar]
- 38.Naya, F. S., and E. Olson. 1999. MEF2: a transcriptional target for signaling pathways controlling skeletal muscle growth and differentiation. Curr. Opin. Cell Biol. 11:683-688. [DOI] [PubMed] [Google Scholar]
- 39.Okamoto, S., D. Krainc, K. Sherman, and S. A. Lipton. 2000. Antiapoptotic role of the p38 mitogen-activated protein kinase-myocyte enhancer factor 2 transcription factor pathway during neuronal differentiation. Proc. Natl. Acad. Sci. USA 97:7561-7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okamoto, S., Z. Li, C. Ju, M. N. Scholzke, E. Mathews, J. Cui, G. S. Salvesen, E. Bossy-Wetzel, and S. A. Lipton. 2002. Dominant-interfering forms of MEF2 generated by caspase cleavage contribute to NMDA-induced neuronal apoptosis. Proc. Natl. Acad. Sci. USA 99:3974-3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ornatsky, O. I., and J. C. McDermott. 1996. MEF2 protein expression, DNA binding specificity and complex composition, and transcriptional activity in muscle and non-muscle cells. J. Biol. Chem. 271:24927-24933. [DOI] [PubMed] [Google Scholar]
- 42.Pollock, R., and R. Treisman. 1991. Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev. 5:2327-2341. [DOI] [PubMed] [Google Scholar]
- 43.Raingeaud, J., A. J. Whitmarsh, T. Barrett, B. Derijard, and R. J. Davis. 1996. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell. Biol. 16:1247-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ridgeway, A. G., S. Wilton, and I. S. Skerjanc. 2000. Myocyte enhancer factor 2C and myogenin up-regulate each other's expression and induce the development of skeletal muscle in P19 cells. J. Biol. Chem. 275:41-46. [DOI] [PubMed] [Google Scholar]
- 45.Sartorelli, V., J. Huang, Y. Hamamori, and L. Kedes. 1997. Molecular mechanisms of myogenic coactivation by p300: direct interaction with the activation domain of MyoD and with the MADS box of MEF2C. Mol. Cell. Biol. 17:1010-1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki, E., J. Lowry, G. Sonoda, J. R. Testa, and K. Walsh. 1996. Structures and chromosome locations of the human MEF2A gene and a pseudogene MEF2AP. Cytogenet. Cell Genet. 73:244-249. [DOI] [PubMed] [Google Scholar]
- 47.Wang, D. Z., M. R. Valdez, J. McAnally, J. Richardson, and E. N. Olson. 2001. The Mef2c gene is a direct transcriptional target of myogenic bHLH and MEF2 proteins during skeletal muscle development. Development 128:4623-4633. [DOI] [PubMed] [Google Scholar]
- 48.Wu, H., F. J. Naya, T. A. McKinsey, B. Mercer, J. M. Shelton, E. R. Chin, A. R. Simard, R. N. Michel, R. Bassel-Duby, E. N. Olson, and R. S. Williams. 2000. MEF2 responds to multiple calcium-regulated signals in the control of skeletal muscle fiber type. EMBO J. 19:1963-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang, S. H., A. Galanis, and A. D. Sharrocks. 1999. Targeting of p38 mitogen-activated protein kinases to MEF2 transcription factors. Mol. Cell. Biol. 19:4028-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Youn, H. D., T. A. Chatila, and J. O. Liu. 2000. Integration of calcineurin and MEF2 signals by the coactivator p300 during T-cell apoptosis. EMBO J. 19:4323-4331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu, G. S., Y. C. Lu, and T. Gulick. 1998. Co-regulation of tissue-specific alternative human carnitine palmitoyltransferase IB gene promoters by fatty acid enzyme substrate. J. Biol. Chem. 273:32901-32909. [DOI] [PubMed] [Google Scholar]
- 52.Yu, Y. T., R. E. Breitbart, L. B. Smoot, Y. Lee, V. Mahdavi, and B. Nadal-Ginard. 1992. Human myocyte-specific enhancer factor 2 comprises a group of tissue-restricted MADS box transcription factors. Genes Dev. 6:1783-1798. [DOI] [PubMed] [Google Scholar]
- 53.Zhao, M., L. New, V. V. Kravchenko, Y. Kato, H. Gram, F. di Padova, E. N. Olson, R. J. Ulevitch, and J. Han. 1999. Regulation of the MEF2 family of transcription factors by p38. Mol. Cell. Biol. 19:21-30. [DOI] [PMC free article] [PubMed] [Google Scholar]