Maize from Tehuacan bears markers of isolated population

A 4-cm-long ancient maize cob dating 5,300 years ago. Image courtesy of Jaime Padilla (Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, Mexico).
The history of maize begins around 9,000 years ago, when humans in central Mexico first began to cultivate a weedy grass known as Balsas teosinte. Archaeological expeditions have recovered the earliest known maize specimens from San Marcos cave in Tehuacan, the oldest of which date to between 5,600 and 5,000 years ago and appear morphometrically to be fully domesticated from their landraces. Miguel Vallebueno-Estrada et al. (pp. 14151–14156) revisited San Marcos cave in 2012 and excavated several additional maize specimens dating to between 5,300 and 4,970 years ago. Using whole-genome shotgun sequencing, the authors characterized the ancient maize genome and compared it with Balsas teosinte and extant maize. The analysis revealed that the oldest known maize from this region was only partially domesticated and belonged to a reduced population of individuals. Furthermore, the high degree of genetic similarity suggests that this population was isolated and strongly influenced by inbreeding. The findings indicate that maize cultivation in the Tehuacan Valley may have progressed from an early phase of small isolated populations to a period in which regional cultivars were crossed, according to the authors. — T.J.
Dispersal patterns of rockfish larvae

Newly recruited splitnose rockfish.
Offspring from bottom-dwelling marine species can disperse across the open ocean to distant destinations, driving population distributions and species persistence. Yet the mechanisms that determine the pathways traveled by the larvae are largely unknown. Because of challenges tied to the continuous tracking of larval movements in the open ocean, Daniel Ottmann et al. (pp. 14067–14072) measured the relatedness of more than 500 juvenile splitnose rockfish (Sebastes diploproa) to study whether individuals that had settled simultaneously to a nearshore habitat along the central Oregon coast were siblings that remained together during dispersal. Splitnose rockfish are live-bearing fish that spend up to a year in the open ocean as larvae and juveniles before settling along the US Pacific Northwest coast. The authors found that more than 11% of new recruits to the area were siblings, suggesting that the young fish remained in a cohesive group throughout their larval and juvenile periods in the open ocean. According to the authors, understanding the dispersal of fish could improve population models and aid conservation of marine organisms across genetic and geopolitical boundaries. — L.C.
Rapid population decline in vertebrates
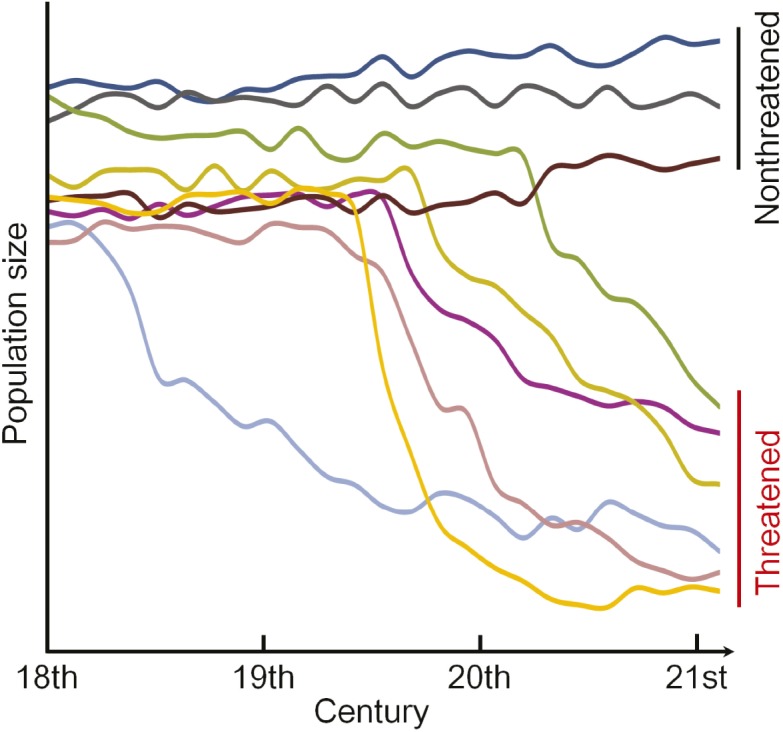
Dynamics of population sizes of many vertebrates.
The forces that drive rapid population decline (RPD) in living species and estimates of the timing of extinction remain unknown for the majority of threatened species. Haipeng Li et al. (pp. 14079–14084) used population genetics modeling to analyze nuclear and mitochondrial data from more than 2,700 vertebrate species. The authors found that on average, RPD in many threatened species began around 123 years ago, in the 19th century, and that the current population sizes in threatened species reflect only a fraction of that of ancestral populations, making up around 5% of ancestral population numbers. Additional analysis revealed that, on average, population sizes in threatened species declined by about 25% every 10 years, and that the ancestral population sizes in threatened species was around 22% smaller than that of nonthreatened species. According to the authors, the relatively smaller population sizes in threatened species might underlie the reduced genetic diversity observed in threatened species, and the RPD of many threatened species might be tied to the spread of industrialization and associated changes in ecosystems worldwide. — C.S.
Magnetic field detection in single neurons
The magnetic fields of neuronal action potentials readily pass through biological tissue and allow for extracellular and noninvasive measurements of action potential dynamics in organisms. However, magnetic techniques used to detect neuronal activity operate at the macroscale or at a level not scalable to functional networks or intact organisms. John Barry et al. (pp. 14133–14138) used a magnetic field sensor consisting of a crystal diamond chip and a surface layer of atomic-scale nitrogen-vacancy (NV) quantum defects to study the magnetic detection of single neuron action potentials in intact organisms. In marine worms and squid, the authors demonstrated that the approach delivered high temporal resolution by providing precise time-resolved measurements of action potentials from individual neurons, correlating magnetic field signals with intracellular voltage measurements, and determining the direction of action potential propagation. The approach worked under ambient conditions and with the NV diamond sensor positioned within 10 micrometers of the biological sample. In the marine worms, extended periods of exposure to the sensor did not induce photodamage or other adverse effects. According to the authors, the technique could have applications in the development of micrometer-scale magnetic imaging for the measurement of functional dynamics in various neuronal processes. — C.S.


