Significance
The cyanobactin prenyltransferases serve as a tool kit for regioselective and chemoselective peptide and protein modifications, in which each enzyme can catalyze the same chemistry on an enormous number of different substrates. Installation of a minimal motif is sufficient to direct modifications on any peptide substrate, which can alter their properties to be more drug-like.
Keywords: RiPP, biosynthesis, prenylation, crystallography
Abstract
The cyanobactin prenyltransferases catalyze a series of known or unprecedented reactions on millions of different substrates, with no easily observable recognition motif and exquisite regioselectivity. Here we define the basis of broad substrate tolerance for the otherwise uncharacterized TruF family. We determined the structures of the Tyr-prenylating enzyme PagF, in complex with an isoprenoid donor analog and a panel of linear and macrocyclic peptide substrates. Unexpectedly, the structures reveal a truncated barrel fold, wherein binding of large peptide substrates is necessary to complete a solvent-exposed hydrophobic pocket to form the catalytically competent active site. Kinetic, mutational, chemical, and computational analyses revealed the structural basis of selectivity, showing a small motif within peptide substrates that is sufficient for recognition by the enzyme. Attaching this 2-residue motif to two random peptides results in their isoprenylation by PagF, demonstrating utility as a general biocatalytic platform for modifications on any peptide substrate.
The rational biosynthesis of drug-like small molecules requires a robust set of intrinsically broadly substrate-tolerant enzymes that can catalyze diverse chemical reactions. Enzymes that modify peptides and proteins are of exceptional interest in drug discovery efforts, and cyanobactin biosynthetic enzymes have been applied to the creation of chemically diverse analogs, including libraries encoding potentially millions of prenylated compounds (1). Cyanobactins (2–5) compose a family of ribosomally synthesized and posttranslationally modified peptides (RiPPs) (6), most of which are macrocyclized by transamidation of their N and C termini. A significant subset of cyanobactins is modified by either forward or reverse isoprenylation on Ser, Thr, or Tyr residues of the macrocycles, or in some cases on the terminal nitrogen of linear peptides (7). The basis of relaxed substrate specificity has been examined for a series of RiPP modification enzymes, revealing that conserved recognition sequences within the leader region of the precursor peptide enable modification of hypervariable core peptides (8–11).
In addition to playing fundamental roles in numerous cellular processes, including signal transduction and trafficking (12, 13), prenylation on small-molecule substrates is critical to bioactivity, and also contributes to the structural diversity of medically relevant secondary metabolites (14–16). Prenylation directs protein substrates to cell membranes and enhances the ATP-independent cell permeability of peptide substrates (17). Consequently, isoprene attachment is of great interest as a modification that can increase cellular uptake (18, 19). Bare peptides are generally considered poor drug candidates, and lipid attachment improves necessary properties, such as the ability to penetrate the blood–brain barrier and absorption, distribution, and metabolism (20). Enzymatic strategies for prenylation are well described in higher eukaryotes and include the modification of protein substrates with either a C15 or C20 isoprenoid catalyzed by protein farnesyl transferases and geranylgeranyl transferases, respectively, or the membrane-integral CaaX proteases (12). There are only limited examples of bacterial peptide prenyltransferases, including the indole geranyl/farnesyl transferase ComQ, which is involved in quorum-sensing pheromone production (12, 21, 22), and the cyanobactin enzymes described here, which catalyze the posttranslational side chain prenylation of peptide natural products (23).
Bioinformatics analysis of metagenome-derived cyanobactin biosynthetic clusters led to the proposal of the TruF family of proteins as candidate prenyltransferases (Fig. 1A and SI Appendix, Fig. S1) (5). Biochemical characterization of LynF (from Lyngbya aestuarii) established the TruF family as broadly substrate-tolerant O-prenyltransferases that catalyze electrophilic alkylation on Tyr residues on cyanobactins, using the five-carbon dimethylallyl pyrophosphate (DMAPP) as the donor (23). Homologs of lynF and truF are prevalent in nearly all cyanobactin biosynthetic clusters and can afford both forward- and reverse-prenylated products (Fig. 1B). Surprisingly, F enzymes are found even in clusters that do not produce a prenylated product (24); for example, the patellamides are not prenylated, but a patF gene is clearly distinguishable in the biosynthetic cluster (25). The crystal structure of unliganded PatF reveals a prenyltransferase barrel fold (26), but this enzyme does not catalyze any biochemical reactions.
Fig. 1.
(A) Chemical structures of representative cyanobactins highlighting relevant chemical features including azoles/azolines (yellow/cyan) installed by heterocyclases and isoprenylation (pink) catalyzed by F enzymes. (B) Forward and reverse prenylation on Tyr residues in substrates.
Analysis of prenylation by LynF reveals that the enzyme uses the mature cyclic peptide as the preferred substrate (23). In other cyanobactin pathway enzymes, substrate permissiveness has been explained in part by the presence of conserved recognition sequences in the leader peptide, which subsequently are proteolytically cleaved during the course of macrocyclization. However, because prenylation occurs after removal of the leader peptide, there is no obvious recognition element in the sequences of the hundreds of prenylated variants identified in cyanobactin pathways. Finally, LynF was able to prenylate several other nonnatural substrates, including N-acetyl– and N-boc–modified d- or l-Tyr, as well as several other linear and cyclic polypeptides (23).
The broad substrate tolerance of F-type prenyltransferases suggests that they can be adapted as a general platform for the enzymatic attachment of isoprenoids to any peptide substrate, but such utility requires knowledge of any dictates for specificity. Toward this goal, we determined several cocrystal structures of PagF from the prenylagaramide biosynthetic pathway of Oscillatoria agardhii (27), in complex with both linear and macrocyclic peptide substrates. The structure reveals a truncation of the α/β prenyltransferase barrel fold observed in bacterial ABBA enzymes and fungal indole PTases (28, 29). However, substrate binding is necessary for the formation of a solvent-occluded active site that is competent for productive isoprenoid transfer. Structural, biochemical, and molecular dynamics simulation data are used to explain the substrate tolerance of PagF and homologs, despite the absence of obvious recognition sequences. These data suggest that an amino terminal Tyr-ψ (where ψ is any aliphatic residue) constitutes the minimal substrate recognition element, and isoprenoid attachment on nonphysiological peptides that contain this N-Tyr-ψ motif by PagF demonstrate the validity of the specificity code. We demonstrate proof of concept for the applicability of this enzyme as a general biotechnological tool for the prenylation of linear and macrocyclic peptides. Finally, these data provide a basis for understanding the distinct substrate preference for the homologous TruF1 for modification on Ser/Thr, rather than on Tyr.
Results
Biochemical Characterization of PagF Activity.
We carried out a complete Michaelis–Menten analysis on PagF using a range of synthetic substrates and the allylic donor DMAPP. Relevant kinetic parameters are reported in Table 1. These data demonstrate that PagF does not efficiently prenylate l-Tyr or small peptides such as N-boc-Tyr and N-acetyl-Tyr (Km = ∼15 mM and Vmax/Km = 1.0 min-1M−1), in contrast to bacterial and fungal ABBA prenyltransferases that can effectively modify the free amino acid l-Trp (14, 16). Instead, like LynF, PagF is a good catalyst only for the prenylation on larger peptides. The catalytic efficiency of PagF relative to N-boc-Tyr increases by 40-fold with a linear Tyr-Tyr-Tyr tripeptide substrate (Km = 0.14 mM). Kinetic properties were further measured with a synthetically prepared native precursor to produce the prenylated natural product, prenylagaramide B, revealing a 4,500-fold increase in catalytic efficiency (Km = 2.2 × 10−4 mM) over N-boc-Tyr. For all competent substrates, the measured Vmax was nearly identical, so that Km differences dominate changes in the catalytic efficiency.
Table 1.
Representative kinetic parameters for PagF with various substrates run in triplicate and volumes occupied by those substrates in the PagF crystal structures
| Substrate | Km, mM | V/K, min−1 mM−1* | Volume, Å3 |
| l-Tyr | 16 | 1.1 | N/A |
| N-acetyl-l-Tyr | 12 | 1.0 | N/A |
| N-boc-l-Tyr | 3.7 | 1.8 | 259 |
| Tyr-Tyr-Tyr | 0.14 | 39 | 453 |
| cyc[INPYLYP] | 2.15 × 10−4 | 3.4 × 104 | 792 |
See SI Appendix, Table S3 for full parameters and SI Appendix, Fig. S10 for curves used to generate the parameters. N/A, not determined.
Measured at an enzyme concentration of 1 mM PagF.
Although prenylagaramide B contains two Tyr residues, only Tyr4 is prenylated easily by recombinant PagF (see below), as is also found in the natural product isolated from the cyanobacteria O. aghardii. Moreover, the Tyr residue in all substrates was modified regioselectively only in the forward direction on the phenolic oxygen. This is similar to what was found for LynF, where only phenolic reverse O-prenylation was observed in a large series of substrates (23). Thus, the cyanobacterial F family enzymes are distinct from canonical small-molecule prenyltransferases for their preference of amino acid substrates only within linear or cyclic peptides, as well as for their strict regioselectivity.
Cocrystal Structures of Substrates with PagF.
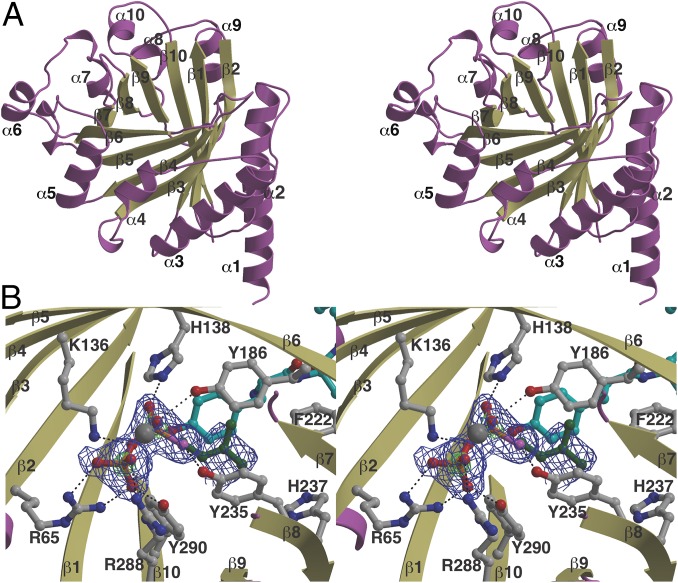
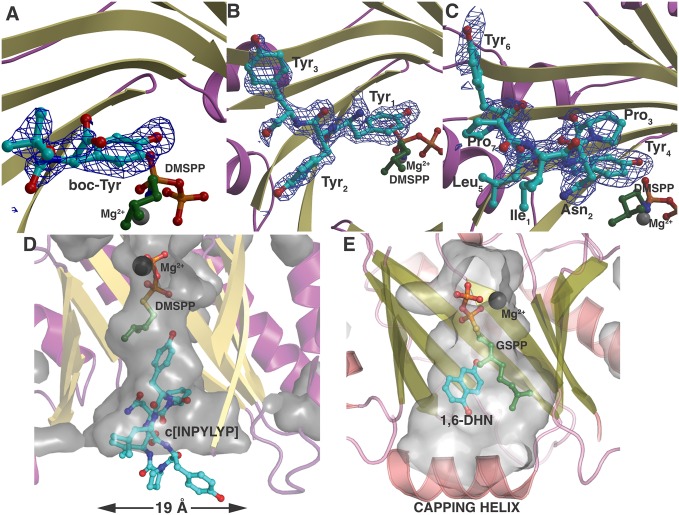
To delineate the determinants for substrate recognition, we solved the crystal structure of native PagF (to 1.8 Å resolution), as well as ternary complex structures with the the prenyl donor analog dimethylallyl S-thiolodiphosphate (DMSPP) and boc-l-Tyr (2.1 Å resolution), a Tyr-Tyr-Tyr tripeptide (1.9 Å resolution), and cyclic[INPYLYP] peptide (2.2 Å resolution), the latter of which is one of the physiological substrates for this enzyme. Relevant data collection and refinement statistics are provided in SI Appendix, Table S3. The overall structure of PagF consists of 10 antiparallel β-strands surrounded by 10 α-helices to form a barrel architecture reminiscent of that observed for aromatic prenyltransferases of the ABBA and indole PTase families of enzymes (Fig. 2A) (14, 16). The DMSPP analog is bound at the center of the barrel, where residues Arg65, Lys136, His138, Tyr186, Tyr235, Arg288, and Tyr290 coordinate the disphosphate (Fig. 2B). A DALI search against the Protein Data Bank (PDB) reveals close structural homology to the indole prenyltransferase (FgPT2) from Aspergillus fumigatus (PDB ID code 3I4Z; rmsd of 3.5 over 250 aligned Cα atoms) (29) and the bacterial ABBA family members NphB (PDB ID code 1ZCW; rmsd of 3.3 over 236 aligned Cα atoms) (28) and CloQ (PDB ID code 2XLQ; rmsd of 3.5 over 216 aligned Cα atoms) (30). Notably, the bottom base of PagF is open, resulting in a solvent-exposed channel that transverses the β barrel core. In contrast, the base of canonical α/β prenyltransferases is occluded by either the presence of a long α helix (in NphB and CloQ) or an extended aromatic residue loop (FgPT2) (SI Appendix, Fig. S2).
Fig. 2.
(A) Stereoview showing the overall structure of PagF with the secondary structural elements labeled and numbered. (B) Stereoview of the active site of PagF in complex with DMSPP (green) and substrate Tyr (cyan). The secondary structural elements are named as in A, and residues that interact with DMSPP are colored in gray. The requisite magnesium ion is shown as a gray sphere. A simulated annealing omit (|Fobs − Fcalc|) difference Fourier electron density map, calculated with the coordinates for DMSPP omitted, is superimposed at 2.4σ (blue) and 10σ (green) above background.
Implications for the Binding of Peptide Substrates.
A comparison of the unliganded PagF structure with those of canonical α/β prenyltransferases suggests that the solvent-exposed channel must be closed before catalysis to prevent nonproductive quenching of the DMAPP-derived allylic carbocation. Each of the PagF cocrystal structures reveals that the substrate acceptor peptide is bound at the base of the active site barrel, resulting in closure of the solvent-exposed channel (Fig. 3 A–C). This substrate-mediated plugging of the base results in the formation of a deep, encapsulated hydrophobic cavity where the DMSPP analog of the substrate donor binds, which would shield the allylic carbocation from solvent (Fig. 3D). In contrast, prenyltransferases that function on small-molecule substrates, such as NphB, which uses indole substrates, are occluded at the base of the barrel by structural elements from the protein (Fig. 3E). A cluster of hydrophobic residues lines the circumference of the active site tunnel, providing a “hydrophobic shield” that further protects reactive intermediates.
Fig. 3.
(A–C) Cocrystal structure of PagF and substrate DMSPP with boc-Tyr (A), Tyr-Tyr-Tyr (B), and cyclic[INPYLYP] (C). A simulated annealing omit (|Fobs − Fcalc|) difference Fourier electron density map is superimposed at 2.4σ (blue). The requisite magnesium ion is shown as a gray sphere. (D) Cutaway diagram showing the volume of the PagF active site viewed parallel to the orientation of the prenyl donor DMSPP and the acceptor cyclic[INPYLYP]. The dimensions of the base of the active site roughly correlate with the size of the natural substrate. (E) Cutaway diagram of the small-molecule prenyltransferase NphB with donor GSPP and acceptor 1,6-dihydroxynapthalene (1,6-DHN) showing a completely occluded active site chamber.
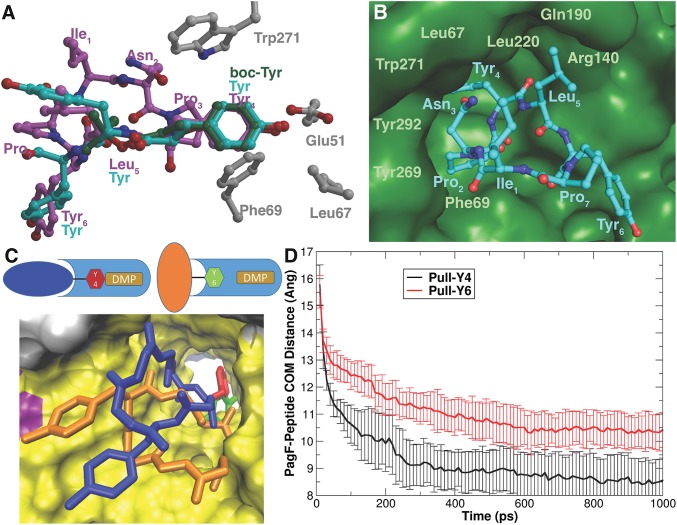
Remarkably, superposition of the cocrystal structures with the cyclic and two linear substrates results in a nearly identical placement of the isoprene acceptor Tyr, which is sandwiched between a wall composed of Trp271 on one side and Leu67 and Phe69 on the other side (Fig. 4A). Glu51 is located roughly 2.6 Å away from the phenolic oxygen of the substrate and likely serves to deprotonate the Tyr to facilitate the electrophilic addition (Fig. 4A). The C-1 atom of the dimethylallyl group of DMSPP is positioned directly above the phenolic oxygen of the substrate at distances of 2.8–3.1 Å (depending on the structure), which would favor the reaction on this atom of the substrate. The distance from the phenolic oxygen to C-3 is nearly 1 Å greater, explaining why PagF catalyzes forward, rather than reverse, prenylation. Similar distances and orientations are observed for the dimethylallyl C-1 and the reactive substrate atom in farnesyl diphosphate synthase (3.2 Å) (31), NphB (4 Å) (28), and the indole prenyltransferase DMATS (3.1 Å) (29). The inability for free amino acids to plug the bottom of the PagF active site provides a rationale for why TruF family enzymes can function only on peptide substrates or amino acids containing bulky N-terminal substituents. The Km values for the tested substrates correlate roughly with their volumes, and the macrocyclic substrate that can best enclose the active site has a Km value four orders of magnitude lower than that for boc-l-Tyr (Table 1).
Fig. 4.
(A) Alignment of linear substrates boc-Tyr (green) and Tyr-Tyr-Tyr (cyan) with the cyclic[INPYLYP] structure (magenta) showing that for all substrates, Tyr is aligned identically among Trp271, Leu67, and Phe69. Glu51 likely serves to deprotonate the phenol to serve in acid/base catalysis. (B) Close-up view of substrate Tyr awaiting isoprene transfer, surrounded by residues that form a hydrophobic chamber. (C) Cartoon illustrating the differences in approach of Tyr4 of cyclic[INPYLYP] vs. Tyr6 to the active site containing DMSPP (DMS; in tan), along with final structures from umbrella sampling. The Tyr4 approach is colored blue/red for cyclic[INPYLYP]/Tyr4 respectively, and the Tyr6 approach is colored orange/green. PagF is colored light blue, and in the structure is shown using a solvent-accessible surface representation looking down the beta barrel toward the active site, colored by secondary structure. Tyr6 cannot penetrate as deeply into the pocket as Tyr4 because of steric interactions of cyclic[INPYLYP] with PagF. (D) Average and SD of the center of mass distance between PagF alpha carbons and cyclic[INPYLYP] alpha carbons during molecular dynamics simulations where either Tyr4 (black line) or Tyr6 (red line) of cyclic[INPYLYP] is guided into the active site of PagF. This figure was generated with Grace 5.1.23.
The PagF-cyclic[INPYLYP] structure also provides a rationale for understanding why cyclic peptides are preferred substrates. Arg140 is within hydrogen-bonding distance to the carbonyl following the substrate Tyr, and the substrate residue immediately carboxyl-terminal is anchored in place through van der Waals contact with Gln190 and Leu220, suggesting that any aliphatic/aromatic residue can be accommodated at this position (Fig. 4B). The residues amino-terminal to the Tyr are supported against a hydrophobic wall composed of Tyr269, Trp271, and Tyr292, and this wall constricts the binding site to favor cyclic peptide substrates over their linear counterparts (Fig. 4B). As with other F family enzymes, PagF requires divalent cations for activity, but lacks the (N/D)DxxD signature motif used in metal ion coordination in other isoprenoid-using enzymes (32). In the cocrystal structure, the diphosphate oxygens of DMSPP coordinate a Mg2+ ion, consistent with the role of the metal in lowering the energy barrier for ionization of the prenyl donor to initiate carbocation formation. The Mg2+ concentration needed for half-maximal activity (45 ± 9 mM; SI Appendix, Fig. S4) is much higher than the KD value for formation of a free DMAPP-Mg2+ complex (120 μM) (33).
Structure-based assignments for the roles of several PagF residues were confirmed through kinetic analysis of site-directed variants using the tri-Tyr peptide as a substrate (SI Appendix, Fig. S12). Mutations H237L and F222V resulted in relatively fast enzymes for which kinetic values could be estimated and compared with wild type, whereas other mutations resulted in much slower enzymes that were only roughly compared using initial activity estimates at high substrate concentrations (Table 2). Mutation of R65 or H138, which engages the diphosphate of DMAPP, decreased activity. Mutations at residues F222 and Y235 that compose the hydrophobic shield, which is proposed to encapsulate and protect the carbocation from unwanted nucleophilic attack, result in decreases in activity that correlate with the volume created by the mutation. The Y290A mutation results in the greatest loss in activity, possibly reflecting the multiple roles that Tyr290 likely plays in solvent shielding, interacting with the pyrophosphate of the isoprene donor, and possibly stabilizing the allylic carbocation through cation–π interactions, as has been proposed for the mechanism of prenylation by NphB (34). Adjacent to the active site, His237 acts as a linchpin in supporting the orientation of several residues along one side of the hydrophobic shield. Intriguingly, mutation of His237 into Leu results in a 1.5-fold increase in catalytic efficiency, possibly because of greater accessibility of the active site, especially for the slower (interior) Tyr residue.
Table 2.
Representative kinetic parameters and specific activities of PagF variants using Tyr3 tripeptide as a substrate
| Mutant | [S], mM | Vmax, min−1* | Km, mM | V/K, min−1 mM−1 | Activity, min−1† |
| WT | 1 | 5.7 ± 0.1 | 0.15 ± 0.01 | 38.0 ± 1.0 | 4.99 ± 0.01 |
| H237L | 1 | 17.6 ± 0.4 | 0.32 ± 0.02 | 55.0 ± 2.2 | — |
| F222V | 1 | 8.15 ± 2.05 | 0.68 ± 0.22 | 12.45 ± 0.85 | — |
| H138A | 10 | — | — | — | 3.70 ± 0.09 |
| R65A | 10 | — | — | — | 1.15 ± 0.07 |
| Y235A | 20 | — | — | — | 0.59 ± 0.02 |
| Y290A | 10 | — | — | — | 0.26 ± 0.01 |
Measurements were conducted in duplicate. SI Appendix, Fig. S12 presents progress curves used to calculate the parameters and the activities.
The maximum rate given by the fit depends on the concentration of enzyme in the assay. Vmax was calculated from this maximum rate and the enzyme concentration.
Turnovers per min by a single enzyme molecule. Initial rates were calculated from the linear part of each assay.
Basis of Substrate Specificity.
Biochemical data and the series of crystal structures with various tyrosine substrates suggest that PagF is able to prenylate each substrate with similar kcat values by positioning Tyr in the same conformation in the active site, regardless of the size or other elements in the substrate. Our efforts to understand the specificity of the enzyme have been somewhat hindered by the lack of peptide substrates that are not prenylated. Of the three pag products that are known, prenylagaramides A and C each contain a single tyrosine that is prenylated; however, prenylagaramide B (cyclic[INPYLYP]) has one prenylated tyrosine and one free tyrosine. Assays of PagF with this substrate showed that Tyr4 is prenylated efficiently, whereas prenylation of the Tyr6 is barely detectable even under saturating concentrations of enzyme, except after long incubations. We used molecular dynamics and modeling to further analyze the rationale for this substrate specificity (Fig. 4 C and D).
We conducted two sets of 10 independent pulling molecular dynamics simulations (20 total), in which either Tyr4 or Tyr6 of the peptide was guided into the active site of PagF via distance restraints (SI Appendix, Materials and Methods). Tyr6 required significantly more restraint energy to enter the active site than Tyr4 (SI Appendix, Fig. S14), and in fact Tyr4 was able to penetrate the active site to a greater extent, ∼2.0 Å on average (Fig. 4D). To ensure that artifacts introduced by the nonequilibrium pulling simulations did not influence these results, we conducted umbrella sampling to estimate the free energy of each approach (SI Appendix, Figs. S13 and S15). Again, approach of Tyr6 was much less favorable than that of Tyr4. Closer examination of the simulations reveal that the environment around Tyr6 was sterically crowded compared with that around Tyr4, leading to physically unfavorable interactions (Fig. 4C). Tyr4 is able to enter the active site to a greater extent then Tyr6 and to form many favorable interactions with PagF residues, as observed in the crystal structure.
Biotechnological Application of Broad Substrate Specificity.
Superposition of the cocrystal structures of PagF with each ligand (boc-l-Tyr, Tyr-Tyr-Tyr tripeptide, and cyclic[INPYLYP] peptide) revealed a nearly identical pose for the substrate Tyr, regardless of the length or complexity of the peptide (Fig. 4A). This unexpected convergence suggests that F family prenyltransferases can modify Tyr residues in the context of any substrate, as long as the geometric and orientation constraints imposed by the active site can be fulfilled. Consequently, the structural data suggest that PagF can accept any linear peptide substrate with the primary sequence of N-Tyr-ψ-R (where ψ is any aliphatic/aromatic residue).
To test this hypothesis, we made two peptides beginning with the sequence Tyr-Leu-Tyr and monitored O-prenylation of these synthetic substrates using PagF. Tyr-Leu-Tyr-Glu-Ile-Ala-Arg is an angiotensin I-converting enzyme inhibitor (35), and Tyr-Leu-Tyr-Gln-Trp-Leu-Gly-Ala-Pro-Val is the N terminus of osteocalcin. The N-terminal Tyr of each peptide was fully prenylated in overnight assays with a 100-fold excess of peptide over enzyme. Over the same time scale, the second Tyr of Tyr-Leu-Tyr-Glu-Ile-Ala-Arg also was almost completely prenylated, whereas the second Tyr of the longer peptide, Tyr-Leu-Tyr-Gln-Trp-Leu-Gly-Ala-Pro-Val, was partly modified (Fig. 5). Prenylation of each of these peptides results in an increase in their hydrophobicity, which increases with successive prenylation [Fig. 5; 1-h trace (red) vs. 5-h trace (green)]. Although the biological consequences of prenylation of these peptides have yet to be established, we recently demonstrated that prenylation can affect the bioactivity profile of the RiPP cyanobactin patellin 2; the prenylated analog of patellin 2 demonstrates neuroactivity, whereas the unmodified analog does not show such activity (36). These data demonstrate that PagF, and the F family enzymes in general, may be used as a biotechnological tool for enzymatic isoprenylation of peptide substrates.
Fig. 5.
Extracted ion chromatograms from LC/MS analysis of PagF assays, showing substrates (blue), singly prenylated products (orange), and doubly prenylated products (green). Extracted ion chromatograms for the substrate mass (blue) are shown at each time point for comparison. Some peaks in the blue chromatograms at each time point come from fragmentation of the prenylated products in the MS. The drawing of the YLY portion of the peptides shows where the first and second prenyl groups are attached. MS/MS fragmentation shows that the first prenylation of the linear peptides is on the N-terminal tyrosine (SI Appendix, Fig. S9), whereas the position of the first prenyl group in the cyclic peptide is inferred from the natural product prenylagaramide B. The cyclic[INPYLYP] assay contained DMAPP (5 mM), peptide substrate (40 μM), and PagF (0.2 μM). Linear peptide assays contained YLYEIAR (0.5 mM) or YLYQWLGAPV (0.5 mM), DMAPP (5 mM), and PagF (3 μM). Singly prenylated YLYQWLGAPV eluted from HPLC as a double peak. All assays' components are described in SI Appendix, Materials and Methods.
Basis for Site Specificity of TruF1 Prenyltransferase.
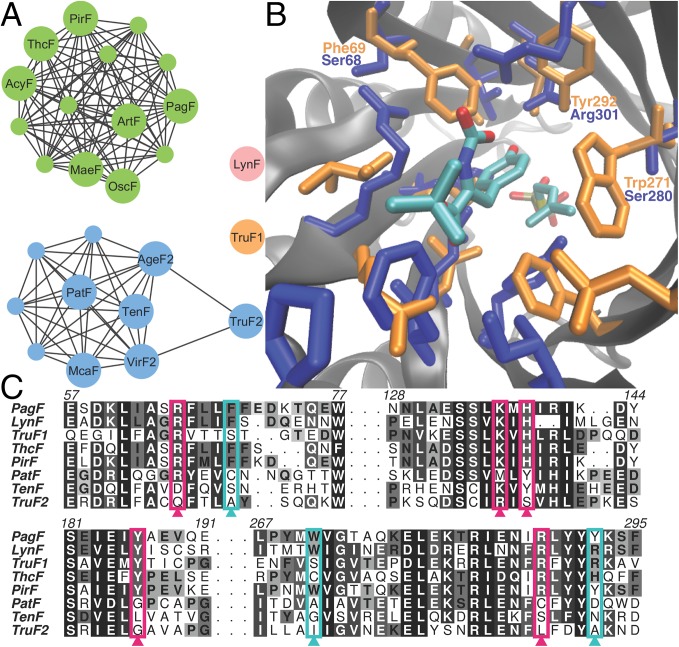
Enzymes of the F family show specificities for a diverse range of substrates, with PagF catalyzing the forward prenylation on Tyr, LynF catalyzing the reverse prenylation on Tyr, and TruF1 catalyzing the reverse prenylation on Ser/Thr. To determine whether the F enzymes could be clustered by substrate specificity based solely only on primary sequence, we used the Enzyme Similarity Tool (EST) of the Enzyme Function Initiative (EFI) to generate a sequence similarity network, using the PagF sequence to query the UniProtKB database for sequences with an E-value threshold of 10−5. Surprisingly, analysis of the 26 retrieved sequences at a cutoff E-value of 10−80 revealed a clustering of sequences with similar substrate specificities (Fig. 6A); e.g., PagF that prenylates Tyr clusters with other enzymes that have been experimentally verified to function on a similar substrate, as well as others with the same presumed activity.
Fig. 6.
(A) Sequence similarity network showing the clustering of F family enzymes into groupings that reflect their cognate substrate preferences. Using a threshold cutoff E value of 10−80, PagF clusters with other enzymes that prenylate on Tyr (shown as green circles). Members that do not demonstrate activity, such as PatF, cluster and are shown in blue spheres. LynF, which catalyzes reverse prenylation on Tyr (in pink), and TruF1, which catalyzes reverse prenylation on Ser/Thr (in orange), do not cluster with any other members. (B) Structure-based homology model of TruF1 (in blue), superimposed on the structure of PagF (in orange), showing changes in the active site composition that would favor the smaller, polar Ser/Thr over Tyr as a substrate. (C) Multiple sequence alignment of F family enzymes illustrating that inactive members lack at least one residue involved in DMAPP binding (shown in pink), and that TruF1 contains polar amino acids at conserved hydrophobic residues (in cyan).
A second cluster consists of orthologs found in biosynthetic clusters that do not produce a prenylated compound. Biochemical analysis demonstrates that some of these enzymes are not functional, and neither PagF nor TruF2 catalyzes isoprenylation on any known peptidic substrate with a range of different prenyl donors. Likewise, nearly all of the proteins that cluster within this group are from clusters that produce nonprenylated cyanobactins. A closer inspection of the primary sequences reveals that each of these enzymes lacks one or more of the residues necessary for engaging the prenyl donor in PagF (Fig. 6B). The lack of activity from these proteins is likely due to the inability to bind DMAPP (or other isoprene donor).
There are two sequences that do not cluster with either of the aforementioned groups, and these two enzymes demonstrate substrate preferences distinct from those of PagF homologs that isoprenylate on Tyr. For example, LynF (40% sequence-identical to PagF) also catalyzes O-prenylation on Tyr, but does so in a “reverse” manner (i.e., via addition of dimethylallylic carbocation at C3 rather than at C1). Similarly, TruF1 (32% sequence-identical to PagF) catalyzes reverse prenylation, but does so on Ser/Thr residues rather than on Tyr. To understand a possible rationale for this change in specificity, we generated a structure-based model for TruF1 using the coordinates of PagF and superimposed the two structures. An inspection of the two active sites suggests that many of the hydrophobic residues that are necessary for stabilizing the Tyr substrate in PagF are altered to either small aliphatic residues (i.e., Phe69 in PagF is Ser68 in TruF1, and Trp271 in PagF is Ser280 in TruF1) or basic residues (i.e., Tyr292 in PagF is Arg301 in TruF1) (Fig. 6C). Other residues that define the PagF active site contours (Fig. 4B) are altered in the TruF1 sequence as well. These changes likely alter both the contour and the charge of the active site in TruF1 to accommodate the smaller, polar Ser residue in the substrate.
Discussion
Biochemical analysis of a TruF family member extends the architectural repertoire of α/β prenyltransferase to a new clade of enzymes that O-prenylate Tyr, Ser, and Thr on linear or cyclic peptide substrates. Biochemical characterization of related F enzymes reveals that a number of members are inactive in their ability to catalyze peptide alkylation, both in vivo and in vitro. The trunkamide cluster contains two F enzymes (TruF1 and TruF2), but only TruF1 is active in reverse prenylation Ser/Thr. (24). Likewise, the patellamide cluster contains PatF, but the products of this cluster are not prenylated, and the enzyme lacks prenyl transfer activity (36). A closer examination of the primary sequences of these inactive enzymes, in the context of the biochemical and structural data presented here, demonstrates that these enzymes lack at least one of the conserved residues that are required to engage the DMAPP substrate.
In the FgPT2, the regiochemistry of the Friedel–Crafts type alkylation at C4 of the indole substrate is dictated by steric constraints that preclude σ-complex formation at the more reactive C7 atom. For PagF, peptide binding at the base of the active site orients the Tyr hydroxyl in the vicinity of the prenyl donor at a sufficient distance from the allylic carbocation to favor the direct electrophilic attack on the Tyr oxygen. This geometric positioning of reactive atoms and the greater reactivity of the hydroxyl group dictate direct O-alkylation over aromatic substitution of Tyr substrates in cyclic peptides.
The structural data enable elucidation of spatial and volume considerations for substrate suitability, which we have used to expand the substrate scope to encompass any peptide that contains an amino terminal N-Tyr-ψ motif. These enzymes catalyze a diverse array of reactions, including forward and reverse Tyr O-prenylation, Ser and Thr reverse O-prenylation, N-prenylation at the N terminus (7), and potentially other modifications, such as addition of geranyl and Trp prenylation (37). Many of these enzymes act on large numbers of very chemically disparate substrates without compromising chemoselectivity. For example, so far PagF has been seen to prenylate diverse substrates only in the forward direction on Tyr, whereas LynF performs only reverse prenylation of Tyr. Likewise, TruF1 catalyzes reverse prenylation, but has a substrate preference for Ser/Thr over Tyr. Although sequence analysis suggests a possible rationale for the specificity of TruF1 for Ser/Thr, the basis of forward vs. reverse prenylation is not as obvious. However, given that this preference is not dependent on the structure of the acceptor peptide, it is likely that LynF simply orients the two reactive species differently relative to PagF. This is a rather unusual feature, in that in some indole prenyltransferases the regioselectivity of reaction appears to be substrate-driven (38). As more cyanobactin prenyltransferase family enzymes are described and cataloged, general rules underlying broad substrate specificity will be uncovered, leading to better control over biology to direct specific synthetic organic reactions.
Materials and Methods
Methodology for the cloning, expression, purification, biochemical analysis, molecular modeling, crystallization, and structure determination of PagF and ligand complexes is described in detail, chemical data are provided, and analytical methods are reported in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Greg Bulaj for stimulating discussions, Scott Endicott for cyclic peptide synthesis, Amy Barrios and Elena Shuangyu Ma for use of the Aapptek peptide synthesizer and help with linear peptide synthesis, Chris M. Ireland and Thomas E. Smith for use of the Waters Micromass Q-Tof mass spectrometer and assistance with mass spectrometry, and Jay Olsen and Zhenjian Lin for assistance with nuclear magnetic resonance. This work was supported by NIH Grants GM102602 (to E.W.S. and S.K.N.) and GM103219 (to E.P.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 5TTY (native PagF), 5TU4 (PagF-Boc-Tyr), 5TU5 (PagF-(Tyr)3), and 5TU6 (PagF-cyclic[INPYLYP])].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609869113/-/DCSupplemental.
References
- 1.Ruffner DE, Schmidt EW, Heemstra JR. Assessing the combinatorial potential of the RiPP cyanobactin tru pathway. ACS Synth Biol. 2015;4(4):482–492. doi: 10.1021/sb500267d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donia MS, et al. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat Chem Biol. 2006;2(12):729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 3.Koehnke J, et al. The structural biology of patellamide biosynthesis. Curr Opin Struct Biol. 2014;29:112–121. doi: 10.1016/j.sbi.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sivonen K, Leikoski N, Fewer DP, Jokela J. Cyanobactins—ribosomal cyclic peptides produced by cyanobacteria. Appl Microbiol Biotechnol. 2010;86(5):1213–1225. doi: 10.1007/s00253-010-2482-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donia MS, Ravel J, Schmidt EW. A global assembly line for cyanobactins. Nat Chem Biol. 2008;4(6):341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30(1):108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leikoski N, et al. Genome mining expands the chemical diversity of the cyanobactin family to include highly modified linear peptides. Chem Biol. 2013;20(8):1033–1043. doi: 10.1016/j.chembiol.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 8.Lee J, McIntosh J, Hathaway BJ, Schmidt EW. Using marine natural products to discover a protease that catalyzes peptide macrocyclization of diverse substrates. J Am Chem Soc. 2009;131(6):2122–2124. doi: 10.1021/ja8092168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McIntosh JA, et al. Circular logic: Nonribosomal peptide-like macrocyclization with a ribosomal peptide catalyst. J Am Chem Soc. 2010;132(44):15499–15501. doi: 10.1021/ja1067806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oman TJ, van der Donk WA. Follow the leader: The use of leader peptides to guide natural product biosynthesis. Nat Chem Biol. 2010;6(1):9–18. doi: 10.1038/nchembio.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sardar D, Pierce E, McIntosh JA, Schmidt EW. Recognition sequences and substrate evolution in cyanobactin biosynthesis. ACS Synth Biol. 2015;4(2):167–176. doi: 10.1021/sb500019b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang FL, Casey PJ. Protein prenylation: Molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 13.Hannoush RN, Sun J. The chemical toolbox for monitoring protein fatty acylation and prenylation. Nat Chem Biol. 2010;6(7):498–506. doi: 10.1038/nchembio.388. [DOI] [PubMed] [Google Scholar]
- 14.Heide L. Prenyl transfer to aromatic substrates: Genetics and enzymology. Curr Opin Chem Biol. 2009;13(2):171–179. doi: 10.1016/j.cbpa.2009.02.020. [DOI] [PubMed] [Google Scholar]
- 15.Tanner ME. Mechanistic studies on the indole prenyltransferases. Nat Prod Rep. 2015;32(1):88–101. doi: 10.1039/c4np00099d. [DOI] [PubMed] [Google Scholar]
- 16.Tello M, Kuzuyama T, Heide L, Noel JP, Richard SB. The ABBA family of aromatic prenyltransferases: Broadening natural product diversity. Cell Mol Life Sci. 2008;65(10):1459–1463. doi: 10.1007/s00018-008-7579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zverina EA, Lamphear CL, Wright EN, Fierke CA. Recent advances in protein prenyltransferases: Substrate identification, regulation, and disease interventions. Curr Opin Chem Biol. 2012;16(5-6):544–552. doi: 10.1016/j.cbpa.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palsuledesai CC, Distefano MD. Protein prenylation: Enzymes, therapeutics, and biotechnology applications. ACS Chem Biol. 2015;10(1):51–62. doi: 10.1021/cb500791f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wollack JW, et al. Multifunctional prenylated peptides for live cell analysis. J Am Chem Soc. 2009;131(21):7293–7303. doi: 10.1021/ja805174z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Bulaj G. Converting peptides into drug leads by lipidation. Curr Med Chem. 2012;19(11):1602–1618. doi: 10.2174/092986712799945003. [DOI] [PubMed] [Google Scholar]
- 21.Marahiel MA, Nakano MM, Zuber P. Regulation of peptide antibiotic production in Bacillus. Mol Microbiol. 1993;7(5):631–636. doi: 10.1111/j.1365-2958.1993.tb01154.x. [DOI] [PubMed] [Google Scholar]
- 22.Okada M. Post-translational isoprenylation of tryptophan. Biosci Biotechnol Biochem. 2011;75(8):1413–1417. doi: 10.1271/bbb.110087. [DOI] [PubMed] [Google Scholar]
- 23.McIntosh JA, Donia MS, Nair SK, Schmidt EW. Enzymatic basis of ribosomal peptide prenylation in cyanobacteria. J Am Chem Soc. 2011;133(34):13698–13705. doi: 10.1021/ja205458h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt EW, Donia MS. Chapter 23. Cyanobactin ribosomally synthesized peptides—a case of deep metagenome mining. Methods Enzymol. 2009;458:575–596. doi: 10.1016/S0076-6879(09)04823-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt EW, et al. Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci USA. 2005;102(20):7315–7320. doi: 10.1073/pnas.0501424102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bent AF, et al. Structure of PatF from Prochloron didemni. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013;69(Pt 6):618–623. doi: 10.1107/S1744309113012931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Donia MS, Schmidt EW. Linking chemistry and genetics in the growing cyanobactin natural products family. Chem Biol. 2011;18(4):508–519. doi: 10.1016/j.chembiol.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuzuyama T, Noel JP, Richard SB. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature. 2005;435(7044):983–987. doi: 10.1038/nature03668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Metzger U, et al. The structure of dimethylallyl tryptophan synthase reveals a common architecture of aromatic prenyltransferases in fungi and bacteria. Proc Natl Acad Sci USA. 2009;106(34):14309–14314. doi: 10.1073/pnas.0904897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Metzger U, Keller S, Stevenson CE, Heide L, Lawson DM. Structure and mechanism of the magnesium-independent aromatic prenyltransferase CloQ from the clorobiocin biosynthetic pathway. J Mol Biol. 2010;404(4):611–626. doi: 10.1016/j.jmb.2010.09.067. [DOI] [PubMed] [Google Scholar]
- 31.Tarshis LC, Yan M, Poulter CD, Sacchettini JC. Crystal structure of recombinant farnesyl diphosphate synthase at 2.6-A resolution. Biochemistry. 1994;33(36):10871–10877. doi: 10.1021/bi00202a004. [DOI] [PubMed] [Google Scholar]
- 32.Koyama T, et al. Identification of significant residues in the substrate binding site of Bacillus stearothermophilus farnesyl diphosphate synthase. Biochemistry. 1996;35(29):9533–9538. doi: 10.1021/bi960137v. [DOI] [PubMed] [Google Scholar]
- 33.Pickett JS, Bowers KE, Fierke CA. Mutagenesis studies of protein farnesyltransferase implicate aspartate beta 352 as a magnesium ligand. J Biol Chem. 2003;278(51):51243–51250. doi: 10.1074/jbc.M309226200. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Miao Y, Wang B, Cui G, Merz KM., Jr Catalytic mechanism of aromatic prenylation by NphB. Biochemistry. 2012;51(12):2606–2618. doi: 10.1021/bi201800m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nakagomi K, et al. Acein-1, a novel angiotensin-I-converting enzyme inhibitory peptide isolated from tryptic hydrolysate of human plasma. FEBS Lett. 1998;438(3):255–257. doi: 10.1016/s0014-5793(98)01311-8. [DOI] [PubMed] [Google Scholar]
- 36.Tianero MD, et al. Metabolic model for diversity-generating biosynthesis. Proc Natl Acad Sci USA. 2016;113(7):1772–1777. doi: 10.1073/pnas.1525438113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leikoski N, et al. Highly diverse cyanobactins in strains of the genus Anabaena. Appl Environ Microbiol. 2010;76(3):701–709. doi: 10.1128/AEM.01061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu X, et al. Catalytic mechanism of stereospecific formation of cis-configured prenylated pyrroloindoline diketopiperazines by indole prenyltransferases. Chem Biol. 2013;20(12):1492–1501. doi: 10.1016/j.chembiol.2013.10.007. [DOI] [PubMed] [Google Scholar]
- 39.Gerlt JA, et al. Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks. Biochim Biophys Acta. 2015;1854(8):1019–1037. doi: 10.1016/j.bbapap.2015.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shannon P, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.