Significance
The proteostasis pathway may be a source of new drug targets in Mycobacterium tuberculosis (Mtb). The conserved protein chaperone DnaK is essential in Mycobacterium smegmatis and predicted to be essential in Mtb. DnaK is regulated by cofactors, J proteins and nucleotide exchange factor GrpE. In contrast to most bacterial pathogens, Mtb has two J proteins, DnaJ1 and DnaJ2. Here, we characterize in vitro activities of Mtb DnaK, DnaJ1, DnaJ2, and GrpE, the disaggregase ClpB, and the small heat shock protein Hsp20, in reactivation of a protein aggregate. We found that DnaJ1 and DnaJ2 are individually dispensable, but collectively essential and mutations in a conserved motif of each result in cellular loss of function. These findings will help in identifying and characterizing inhibitors of Mtb’s proteostasis network.
Keywords: DnaK, proteostasis, DnaJ proteins, M. tuberculosis, chaperones
Abstract
During host infection, Mycobacterium tuberculosis (Mtb) encounters several types of stress that impair protein integrity, including reactive oxygen and nitrogen species and chemotherapy. The resulting protein aggregates can be resolved or degraded by molecular machinery conserved from bacteria to eukaryotes. Eukaryotic Hsp104/Hsp70 and their bacterial homologs ClpB/DnaK are ATP-powered chaperones that restore toxic protein aggregates to a native folded state. DnaK is essential in Mycobacterium smegmatis, and ClpB is involved in asymmetrically distributing damaged proteins during cell division as a mechanism of survival in Mtb, commending both proteins as potential drug targets. However, their molecular partners in protein reactivation have not been characterized in mycobacteria. Here, we reconstituted the activities of the Mtb ClpB/DnaK bichaperone system with the cofactors DnaJ1, DnaJ2, and GrpE and the small heat shock protein Hsp20. We found that DnaJ1 and DnaJ2 activate the ATPase activity of DnaK differently. A point mutation in the highly conserved HPD motif of the DnaJ proteins abrogates their ability to activate DnaK, although the DnaJ2 mutant still binds to DnaK. The purified Mtb ClpB/DnaK system reactivated a heat-denatured model substrate, but the DnaJ HPD mutants inhibited the reaction. Finally, either DnaJ1 or DnaJ2 is required for mycobacterial viability, as is the DnaK-activating activity of a DnaJ protein. These studies lay the groundwork for strategies to target essential chaperone–protein interactions in Mtb, the leading cause of death from a bacterial infection.
The pathogen Mycobacterium tuberculosis (Mtb) infects an estimated one-third of the world’s population in a latent state and can persist for the life of the host despite an active immune response (1). In patients with active tuberculosis (TB), Mtb survives months of chemotherapy with drugs to which the infecting strain is sensitive. Many of the stresses that Mtb encounters, such as reactive oxygen and nitrogen species, heat encountered from fever, changes in pH, or mistranslation caused by antibiotics that target the bacterial ribosome, can cause reversible or irreversible damage to proteins, often resulting in aggregation (2–5). Like all organisms, Mtb deploys a suite of protein chaperones that unfold and refold aggregated proteins to their native structures to restore function, or that shuttle irreversibly damaged proteins to degradation machinery for recycling. Hsp70 proteins are one of the most widely expressed chaperone families, because they are found in both prokaryotes and eukaryotes, and act as a hub for protein folding, refolding, and shuttling (6–9). The Hsp-70 homolog in bacteria is the chaperone DnaK. Where studied, DnaK has been found to resolve protein aggregates in concert with a conserved disaggregase called ClpB, the bacterial homolog of eukaryotic Hsp-104 (10–12).
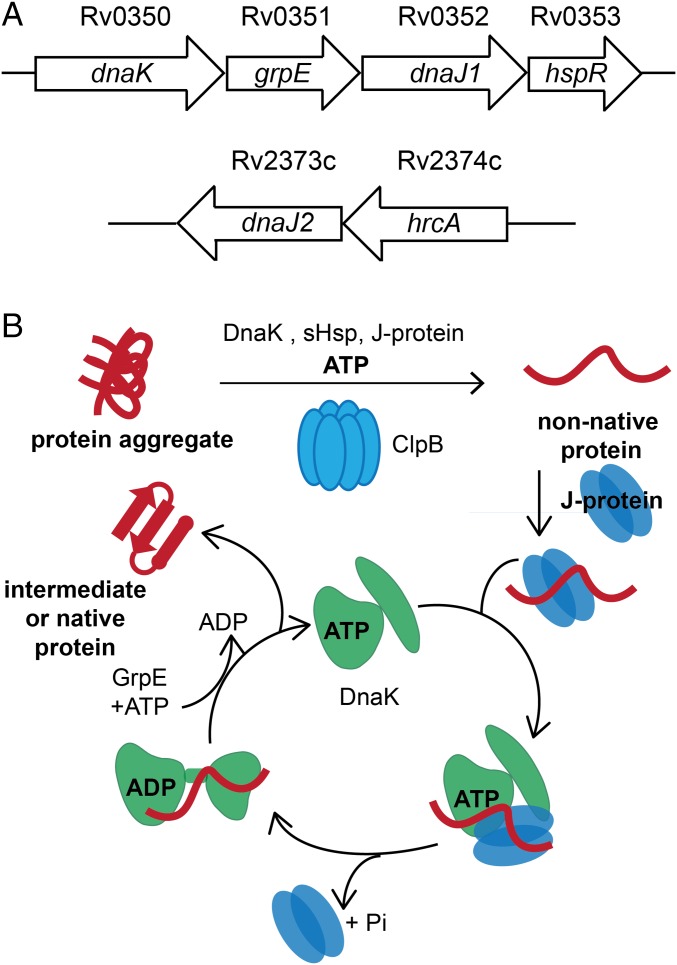
Recent work suggests that DnaK and ClpB play unique roles in mycobacteria. In M. smegmatis (Msm), a saprophytic Mycobacterium used as a model for pathogenic species, DnaK is essential for cell growth and native protein folding (13). Transposon mutagenesis analysis predicts that dnaK is also essential for growth in Mtb (14, 15). In contrast, DnaK is nonessential in other bacteria studied, such as Escherichia coli, where its functions are redundant with those of trigger factor (encoded by tig), such that dnaK and tig form a synthetic lethal pair (16–18). In mycobacteria, the repressor HspR negatively regulates the heat shock response and is encoded in the same operon as dnaK (Fig. 1A) (19). Upon heat shock, the C-terminal tail of HspR unfolds and interacts with DnaK, relieving repression of the operon. Deletion of hspR leads to overexpression of other chaperones in addition to DnaK, such as ClpB, and results in decreased persistence after initial infection of a mouse by Mtb (20). However, we reported that deletion of clpB in Mtb leads to defects in growth and persistence in mice (21). When cells are stressed, ClpB asymmetrically sequesters irreversibly damaged proteins in dividing Mtb as a mechanism for survival. These studies suggest that proper regulation of the dnaK operon and clpB are important for sustained host infection. Given their importance in Mtb physiology and persistence in the host, the proteostasis pathway governed by DnaK and ClpB may represent a class of targets for TB chemotherapy.
Fig. 1.
The conserved ClpB/DnaK bichaperone network resolves protein aggregates in mycobacteria. (A) Schematic of Mtb dnaK operon containing grpE, dnaJ1 and the regulator hspR. Another gene annotated as dnaJ2 is also present in the Mtb genome with the proposed regulator hrcA. ClpB is encoded by the gene Rv0384c. (B) Schematic of aggregated protein unfolding by the hexamer ClpB and refolding by DnaK with the assistance of the protein cofactors J proteins (DnaJs) and GrpE. DnaK and ClpB are ATPases. J proteins bring aggregated substrates to ClpB/DnaK and enhance ATP hydrolysis by DnaK. GrpE is a nucleotide exchange factor that stimulates release of ADP by DnaK. Small heat shock proteins (sHsps) are proposed to help solubilize aggregates for reactivation. Modified from ref. 8 with permission from the Annual Review of Biochemistry, Volume 82, © by Annual Reviews, www.annualreviews.org.
Despite its importance, aggregated protein reactivation by mycobacterial DnaK, ClpB, and accessory proteins has not been reconstituted in vitro, although the general mechanism of the ClpB/DnaK bichaperone system has been worked out in other organisms (Fig. 1B) (8, 22, 23). For example, in E. coli, reactivation is powered by the ATPase activity of ClpB and DnaK. Low molecular weight proteins called small heat shock proteins (sHsps) are believed to solubilize aggregates for unfolding by ClpB before refolding by DnaK (24). The reaction cycle of DnaK is regulated by cofactors called J proteins (DnaJ in bacteria, Hsp40 in eukaryotes) and the nucleotide exchange factor (NEF) GrpE (25, 26). Bacteria have only one NEF, and GrpE is essential in Msm (13). J proteins are thought to recruit aggregated client proteins to DnaK and ClpB bound to ATP, and to stimulate the ATPase activity of DnaK upon binding with an unfolded protein substrate. GrpE enhances the release of ADP by DnaK so that ATP can bind to restart the cycle. DnaK exhibits an “open” conformation when bound to ATP and shows high on/off rates for substrate, whereas it exhibits slow on/off rates and a closed conformation when bound to ADP (27–30).
However, unlike most bacteria, but like eukaryotes, Mtb contains more than one DnaJ. A gene annotated dnaJ2 is found distant from the dnaK operon, which contains dnaJ1 and grpE (Fig. 1A). The multiple J proteins of eukaryotes are believed to increase the ability of Hsp70 to recognize diverse substrates, albeit with some redundancy (31). Different eukaryotic J proteins produce varying effects on in vitro Hsp70 ATPase and refolding activities (32–34), or can act as holdases that prevent protein aggregation (35, 36). Unique mixtures of J proteins can function as disaggregases (37, 38). The function of the J proteins of mycobacteria has not been examined.
To better understand the mycobacterial ClpB/DnaK bichaperone network and to explore the function of the two mycobacterial J proteins, we reconstituted the activities of purified Mtb DnaK, ClpB, DnaJ1, DnaJ2, GrpE, and a sHsp, Hsp20. Using mass spectrometry-based ATPase assays, binding assays and a model aggregate reactivation assay, we characterized the chaperone network biochemically and distinguished the function of each J protein. We constructed defective DnaJ point mutants and demonstrated that they can inhibit aggregated protein reactivation by Mtb chaperones in a concentration-dependent manner in vitro. Finally, we showed that Mtb wild-type J proteins are functional in mycobacterial cells, but their HPD point mutants are not, as predicted by their behavior in vitro, and that deletion of dnaJ1 and dnaJ2 is synthetic lethal in Msm.
Results
Mtb DnaK ATPase activity Is Stimulated by Cofactors DnaJ1 or J2 and GrpE.
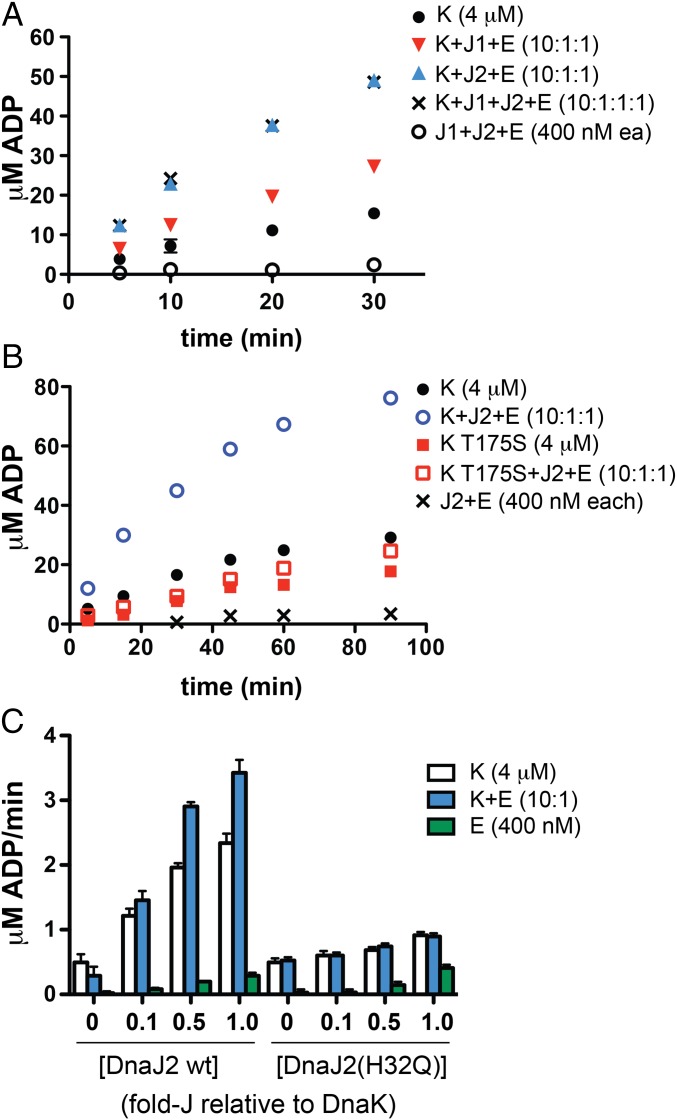
We purified recombinant Mtb DnaK, DnaJ1, DnaJ2, and GrpE as N-terminal His-SUMO fusions to promote the solubility of the overexpressed proteins in E. coli before cleavage of the tag (SI Appendix, Fig. S1A) (39). A mass spectrometry (MS)-based assay was developed to measure ATP turnover directly from changes in the ratio of ATP and ADP concentrations (SI Appendix, Fig. S2). DnaK alone had only modest ATPase activity (Fig. 2A). However, addition of substoichiometric DnaJ1 or DnaJ2 and GrpE to the same concentration of DnaK led to increases in the initial rate of approximately twofold and fivefold, respectively. Addition of DnaJ1 to the mix of DnaK, DnaJ2, and GrpE did not result in further enhancement. To verify that the increase in ATP hydrolysis was due to stimulation of DnaK, we purified a mutant of DnaK at threonine-175 (T175S), a conserved residue that is autophosphorylated in homologs and critical for activity (40–42). We compared the ATPase activity of DnaK(T175S) and wild-type DnaK alone and with substoichiometric DnaJ2 and GrpE over a longer time course (Fig. 2B). Whereas wild-type DnaK ATPase activity was stimulated by DnaJ2 and GrpE, DnaK(T175S) was not activated by the cofactors, although it showed basal ATPase activity. Hence, the ATP hydrolysis observed in these reactions is due to the activity of Mtb DnaK.
Fig. 2.
Mass-spectrometry analysis of the ATPase activity of Mtb DnaK shows that addition of substoichiometric DnaJ1 or DnaJ2 and GrpE enhance the initial rate of ATP hydrolysis. (A) Initial time course of DnaK with or without indicated cofactor proteins shows that addition of DnaJ2 and GrpE results in the highest initial rate, which is not further enhanced by added DnaJ1. (B) Time course of wild-type DnaK compared with the catalytically defective point mutant DnaK(T175S). Addition of DnaJ2 and GrpE stimulates ATP hydrolysis by wild-type DnaK but not by DnaK(T175S). (C) Initial reaction rates of ATP hydrolysis by DnaK ± GrpE titrated with DnaJ2 and a point mutant in the conserved “HPD motif,” DnaJ2(H32Q). Whereas DnaJ2 stimulates turnover in a concentration-dependent manner, addition of DnaJ2(H32Q) shows little effect on activity. Reactions were performed with 100 μM ATP at 37 °C in PBS (pH 7.4), 20 mM MgCl2. E, GrpE; J1, DnaJ1; J2, DnaJ2; K, DnaK. Error bars for all parts represent SEM for n = 2; some cannot be seen due to scale.
To determine the optimal DnaK:cofactor ratios and evaluate cofactor relationships, we performed titration experiments by varying the concentration of each cofactor added to DnaK alone or with another cofactor present, and evaluated the initial rate of ATP hydrolysis (SI Appendix, Fig. S3A). As shown in Fig. 2C, Left, DnaJ2 afforded a concentration-dependent stimulation of DnaK’s ATPase activity. The highest initial rate was seen in reactions containing GrpE, but DnaJ2 was also able to stimulate DnaK in the absence of GrpE (compare blue and white bars, respectively, and see SI Appendix, Fig. S3B). When DnaJ1 was titrated into reactions containing DnaK with or without other cofactors, significant activation was only observed at equimolar DnaJ1 and DnaK regardless of the presence of DnaJ2 or GrpE (SI Appendix, Fig. S3C) and did not exceed the initial rate observed in the presence of substoichiometric DnaJ2. Titration of GrpE in the same manner showed that at equimolar concentrations, GrpE slightly inhibited reactions containing DnaK plus DnaJ2 (SI Appendix, Fig. S3D). This observation may help explain why overexpression of GrpE disrupts DnaK function in cells (13). Taken together, these titrations demonstrated that DnaJ2 and GrpE at low concentrations optimally activate DnaK’s ATPase activity. Kinetic analysis of the DnaK reaction with and without substoichiometric DnaJ2 and GrpE revealed that whereas the KM remained nearly unchanged (∼20 μM), the kcat increased approximately threefold in reactions containing the cofactors (SI Appendix, Fig. S3E).
Analysis of J proteins from species as diverse as bacteria and humans has shown that a highly conserved “HPD” motif in the N terminus is involved in proper function (26, 43). A histidine-to-glutamine mutation in the motif was originally identified as a heat-sensitive mutant in E. coli and has been studied in other homologs (44–46). We identified this motif in Mtb DnaJ1 and DnaJ2 through protein sequence alignments (SI Appendix, Fig. S4). We purified Mtb DnaJ2(H32Q) and tested increasing concentrations for stimulation of DnaK reactions in the presence or absence of GrpE. We saw only modest increases in ATP turnover (on the right side of Fig. 2C and SI Appendix, Fig. S5A). The same behavior was observed with the DnaJ1 His to Gln mutant (SI Appendix, Fig. S5B). We then tested a conservative mutation, DnaJ2(D34E), with GrpE and also observed no activation of DnaK’s ATPase activity (SI Appendix, Fig. S5A). The inability of Mtb DnaJ HPD mutants to activate DnaK was likely not a consequence of misfolding of the mutants, because they behaved the same during gel filtration and anion exchange purification as their respective wild-type proteins.
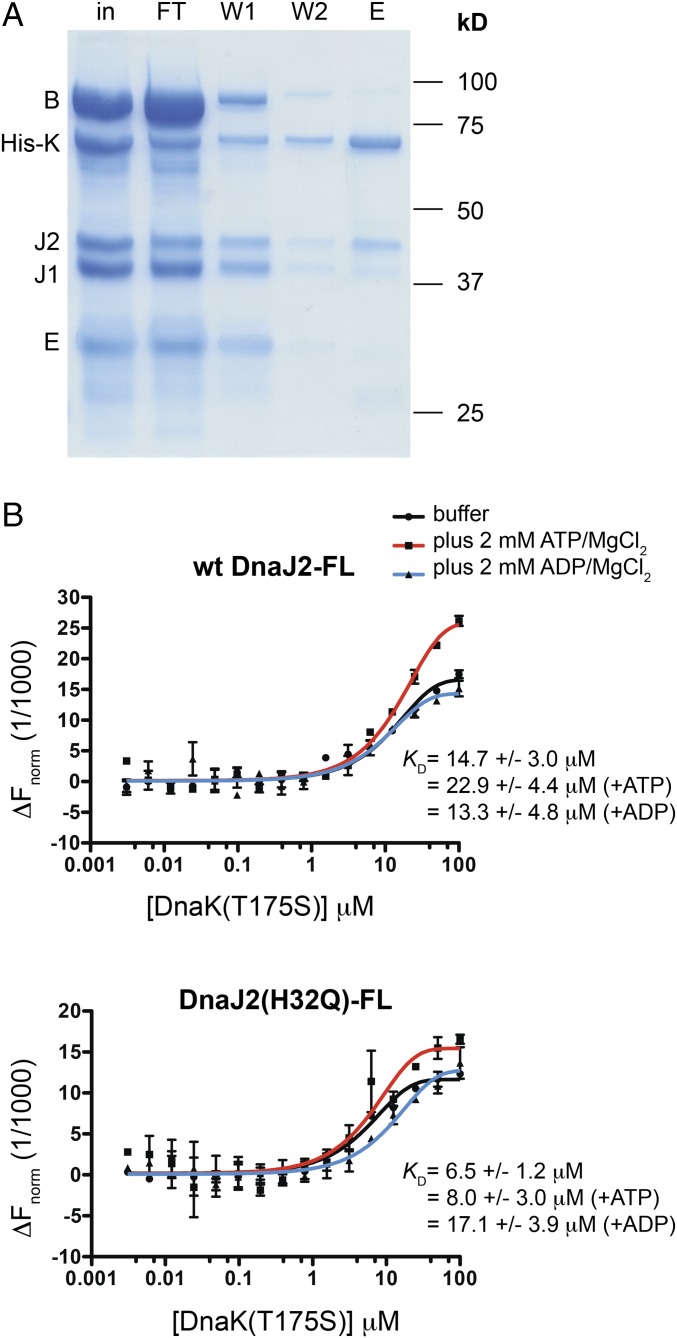
DnaJ2 Binds to DnaK in Vitro Without Aggregated Substrate Present.
To evaluate whether the cofactors physically interact with DnaK to form a stable complex, we performed a protein pull-down by using purified components. ClpB is known to function with DnaK in other organisms (12, 23, 47), and so we also purified Mtb ClpB by using the methods described above. We incubated His-DnaK with untagged ClpB, DnaJ1, DnaJ2, and GrpE in the presence of ATP (Fig. 3A). We optimized the wash steps to minimize nonspecific binding to Ni-NTA beads (SI Appendix, Fig. S6A). DnaJ2 coeluted with His-DnaK, along with faint bands representing DnaJ1 and ClpB, as confirmed by protein band quantification of replicate experiments (SI Appendix, Fig. S6B). Because interactions with DnaK are predicted to be dynamic, especially in the presence of ATP, we did not anticipate that all cofactors would elute with His-DnaK under these conditions. We found DnaJ2 incubated alone with His-DnaK coeluted without additional cofactors (SI Appendix, Fig. S6C), suggesting it formed a stable interaction with DnaK. To quantify the DnaK–DnaJ2 binding interaction, we performed microscale thermophoresis (MST) analysis by using DnaJ2 labeled with a lysine-reactive fluorophore (DnaJ2-FL) (48). Initial experiments titrating DnaK into DnaJ2-FL in the presence of ATP resulted in erratic changes in the initial fluorescence that prevented MST analysis (SI Appendix, Fig. S7A), but which may signify conformational changes of the label due to interaction (perhaps with DnaK reacting with ATP) (49). We then tested DnaK(T175S) as the titrant under the same conditions and found that the initial fluorescence was stable, which allowed us to evaluate the pair with ATP present. As shown in Fig. 3B, Upper, DnaJ2 binds to DnaK(T175S) with nearly equal affinity with and without nucleotide present, although the binding curve with ATP does not reach full saturation because of the limited solubility of DnaK(T175S) at high concentration. To ensure that the wild-type proteins interact as well, DnaK was labeled and varying [DnaJ2] was added with no nucleotide present, and a binding curve was observed. However, the KD was slightly lower (3.7 ± 1.0 μM) than observed with DnaJ2-FL (SI Appendix, Fig. S7B). We confirmed that labeled DnaK and DnaJ2 were still active by using the ATPase activity assay (SI Appendix, Fig. S7 C and D). The variation in KD values may be a consequence of the potential of the fluorophore to modify binding sites on the surface of DnaJ2 or DnaK.
Fig. 3.
DnaJ2 binds to DnaK in vitro. (A) SDS/PAGE analysis of pull-down using purified untagged ClpB (B), DnaJ1 (J1), DnaJ2 (J2), and GrpE (E) with His-DnaK (H-K) shows that DnaJ2 coelutes with His-DnaK (E, eluate; FT, flow-through; in, input; W, wash). (B) MST analysis using wild-type DnaJ2-FL (Upper) and DnaJ2(H32Q)-FL (Lower) (∼20 nM each) with varying concentration of DnaK(T175S) in the presence and absence of nucleotide. Both J proteins interact with DnaK(T175S) under all conditions tested; however, DnaJ2(H32Q) binds DnaK(T175S) with slightly higher affinity than wild-type DnaJ2 except when ADP is present. Error bars represent SD for n = 2. DnaK mutant was used to avoid effects due to ATP turnover.
His-DnaK could also pull-down DnaJ2(H32Q) (SI Appendix, Fig. S6D), indicating that the mutant binds to DnaK although it does not activate the protein. MST analysis using DnaJ2(H32Q)-FL under the same conditions as described above showed binding to DnaK(T175S) with slightly higher affinity than calculated for wild-type DnaJ2, except when ADP was present (Fig. 3B, Lower). Studies in yeast and E. coli have also demonstrated that HPD J-protein point mutants interact with DnaK homologs (46, 50).
Refolding of a Model Aggregated Substrate by Mtb Chaperones Is Inhibited by J-Protein HPD Motif Mutants.
A well-accepted model for protein aggregate resolution proposes that DnaK and J proteins recruit substrates to ClpB (or Hsp-104 homologs), which unfolds aggregates and releases them for refolding by DnaK and cofactors (Fig. 1B) (12, 23, 51, 52). In its active form, ClpB from other organisms is known to form hexamers in an ATP- and concentration-dependent manner (53, 54). We performed gel filtration analysis on Mtb ClpB and observed hexamer formation only when ATP was added (SI Appendix, Fig. S8A) (53). When the concentration of ClpB was doubled and analyzed in a buffer lacking ATP, we observed a similar elution profile by gel filtration (SI Appendix, Fig. S8B).We used the fractions that lacked ATP for enzymatic assays and confirmed that the fractions had ATPase activity by using the MS-based assay described above (SI Appendix, Fig. S8C).
Additional molecular players in the ClpB/DnaK bichaperone network include sHsps that bind and help solubilize aggregates for unfolding by ClpB (24, 55). Based on analysis of >500 reported Mtb transcriptomic experiments, the most highly correlated gene for coexpression with clpB is Rv0251c, which encodes the annotated sHsp protein Hsp20 (56). We purified Mtb Hsp20 (SI Appendix, Fig. S1B) and evaluated its ability to act as a sHsp with the heat denatured model substrate luciferase (24, 38, 57). Briefly, we heated luciferase at 42 °C with and without excess Hsp20 for different times and monitored changes in solubility by ultracentrifugation to separate aggregated and native protein fractions, followed by Western blot analysis (13, 58). After 10 min of heating, the soluble fraction contained almost no detectable luciferase in samples lacking Hsp20 (SI Appendix, Fig. S9A). Whereas heating luciferase significantly diminished activity in samples with or without Hsp20 by the same amount, repeated experiments showed that only samples with Hsp20 had detectable luciferase remaining in the soluble fraction (SI Appendix, Fig. S9B).
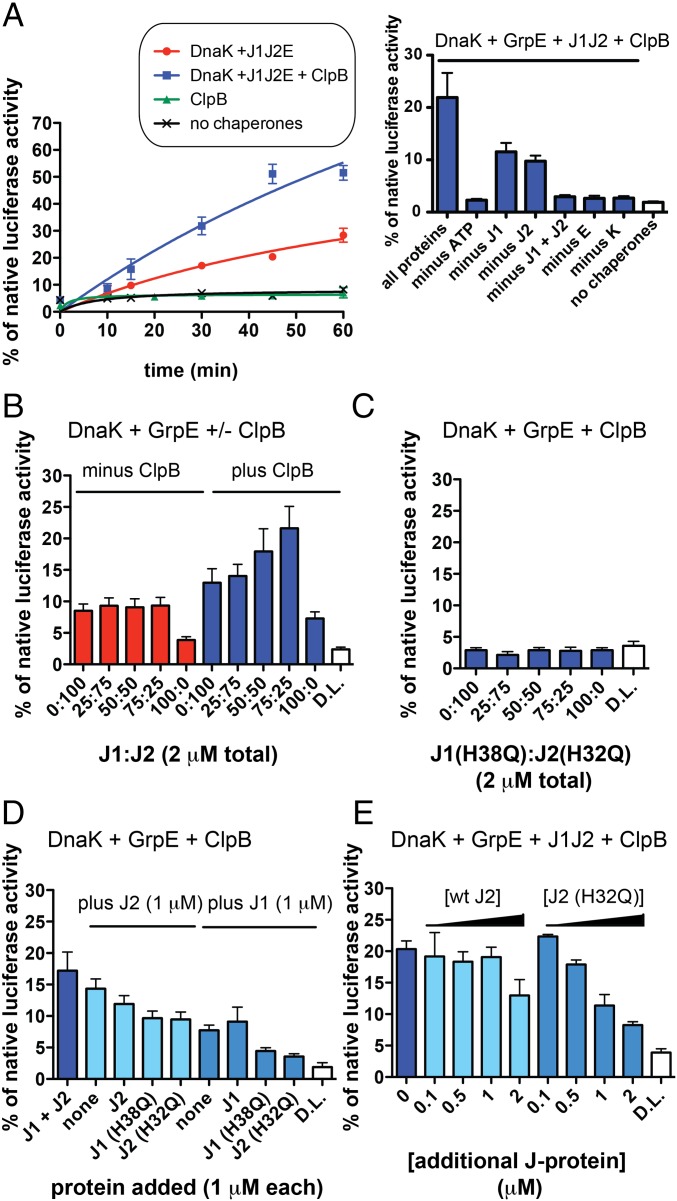
We next sought to reactivate luciferase heat-denatured in the presence of Hsp20 by using purified Mtb chaperones and cofactors. We added DnaK, DnaJ1, DnaJ2, and GrpE with or without ClpB, or ClpB alone, to denatured luciferase with excess Hsp20 and measured the percent recovered luciferase activity relative to a native luciferase control over a time course (Fig. 4A). ClpB alone could not restore luciferase activity, but DnaK and cofactors gave a time-dependent increase in luciferase reactivation that was enhanced when ClpB was added to the reaction. We further evaluated the function of Hsp20 in reactivation by adding different concentrations of Hsp20 to luciferase before heating and compared chaperone reaction yields with and without ClpB (SI Appendix, Fig. S9C) (55). We saw a slight increase in the yield of refolding reactions with increasing [Hsp20]; however, at 40-fold Hsp20 relative to luciferase, chaperone reactions with and without ClpB exhibited almost the same yield. These data suggest that Hsp20 helps to solubilize heat-formed aggregates, but to detect the activity of ClpB, only fourfold excess Hsp20 was used in remaining refolding reactions.
Fig. 4.
Luciferase heat-denatured in the presence of Hsp20 can be reactivated by Mtb ClpB/DnaK with at least one J protein and GrpE, whereas J-protein HPD mutants inhibit refolding. (A) Time course of luciferase activation by different chaperone mixtures containing 2 mM ATP. DnaK and cofactors reactivate aggregated luciferase, and the addition of ClpB to reactions increases the yield. (Right) t = 30 min time point under the same conditions with indicated components removed. Removal of ATP, GrpE, DnaK, or both J proteins prevents refolding. (B) Reactivation of heat-denatured luciferase (D.L.) by DnaK and GrpE with varying ratios of DnaJ1 and DnaJ2 with or without ClpB suggests that J proteins play specific roles in unfolding and/or refolding mechanisms (t = 30 min). (C) Same experiment as described in B with ClpB using DnaJ1 and DnaJ2 point mutants in the HPD motif. Even under optimal J1:J2 ratios, there is no reactivation (t = 30 min). (D) DnaK, GrpE, and ClpB mixtures containing only one J protein with additional DnaJ or mutants added show that DnaJ point mutants cause a decrease in recovery (t = 30 min). (E) Titration of wild-type DnaJ2 or DnaJ2(H32Q) into reactions containing all chaperones and cofactors shows a concentration-dependent inhibition of refolding by DnaJ2(H32Q) (t = 30 min). For all reactions, 25 nM luciferase was denatured with fourfold Hsp20 at 42 °C (SI Appendix, Fig. S9). Reactions were conducted at 25 °C with the following protein concentrations: 4 μM DnaK, 2 μM each ClpB, GrpE, each J protein in A, and 1 μM each J protein in remaining parts unless otherwise noted. Error bars represent SEM for n = 2. For an example of normalized results, see SI Appendix, Fig. S10.
To study the role of each Mtb chaperone and cofactor shown in the time course in Fig. 4A, we varied the concentrations of different components. As shown by the graph (Fig. 4A, Right), when ATP, GrpE, DnaK, and both J proteins were removed, we no longer saw reactivation at t = 30 min. When one J protein was removed, there was approximately half the amount of refolding compared with the reaction with both J proteins. To test whether this loss in activity depended on total [J protein], we added increasing [DnaJ2] to the reaction, but saw no effect on yield (SI Appendix, Fig. S11A). Because DnaJ1 and DnaJ2 stimulate DnaK’s ATPase activity differently, and because varying ratios of J proteins from other organisms have been shown to affect their combined function (38), we then varied the ratio of DnaJ1:DnaJ2 in chaperone reactions with and without ClpB. Although changing the J-protein ratio did not significantly change the amount of refolding in reactions lacking ClpB, it impacted reactions containing ClpB (Fig. 4B). Maximal refolding was seen with a slight excess of DnaJ1 to DnaJ2, although there was low refolding activity when only DnaJ1 was added. To evaluate the potential of DnaK to form a stable complex with both DnaJ1 and DnaJ2 in the presence of aggregated substrate, we performed the pulldown described in Fig. 3A with added denatured luciferase plus Hsp20 (SI Appendix, Fig. S6E). We did not see a significant change in the elution composition compared with experiments lacking substrate, as judged by band quantification (SI Appendix, Fig. S6F compared with SI Appendix, Fig. S6B, respectively). We also did not see a change in DnaJ-mediated activation of DnaK’s ATPase activity in the presence of aggregated substrate (SI Appendix, Fig. S12). Taken together, these observations suggest that there is coordination between DnaJ1 and DnaJ2 in the reactivation of heat-treated substrate and confirm that DnaJ1 and DnaJ2 interact differently with DnaK.
To evaluate the effect of J-protein HPD mutants on reactivation, we used different ratios of DnaJ1(H38Q) and DnaJ2(H32Q) in reactions containing DnaK, ClpB, and GrpE as was done for wild-type proteins. We did not observe denatured luciferase reactivation with DnaJ mutant proteins at any ratio tested (Fig. 4C). Next, we tested whether J-protein mutants could cooperate with wild-type J proteins by adding each mutant to refolding reactions containing only one wild-type J protein. As shown in Fig. 4D, the addition of equimolar DnaJ1(H38Q) or DnaJ2(H32Q) to wild-type J-protein reactions decreased the amount of refolding. To evaluate the possibility that J-protein mutants act as chaperone inhibitors, we titrated DnaJ2(H32Q) into chaperone reactions where only DnaJ2 was present (SI Appendix, Fig. S11B) or both J proteins were present (Fig. 4E). The mutant inhibited all reactions in a concentration-dependent manner, whereas the wild-type had an inhibitory effect only at high concentration. The same experiment was performed by using wild-type and mutant DnaJ1, and we observed the same inhibitory effect of DnaJ1(H38Q) but not wild-type (SI Appendix, Fig. S11C). Because DnaJ2(H32Q) binds DnaK (Fig. 3B), we examined whether DnaJ2(H32Q) competes with wild-type DnaJ2 by binding to DnaK, which would stall aggregate reactivation. To do so, we analyzed the ATPase activity of DnaK stimulated by substoichiometric GrpE and DnaJ2 in the presence of similar concentrations of DnaJ2(H32Q). We observed partial inhibition of DnaK activation (SI Appendix, Fig. S13). This inhibition suggests that DnaJ point mutants can disrupt Mtb DnaK-DnaJ interactions.
At Least One Functional J Protein Is Required for Growth in M. smegmatis.
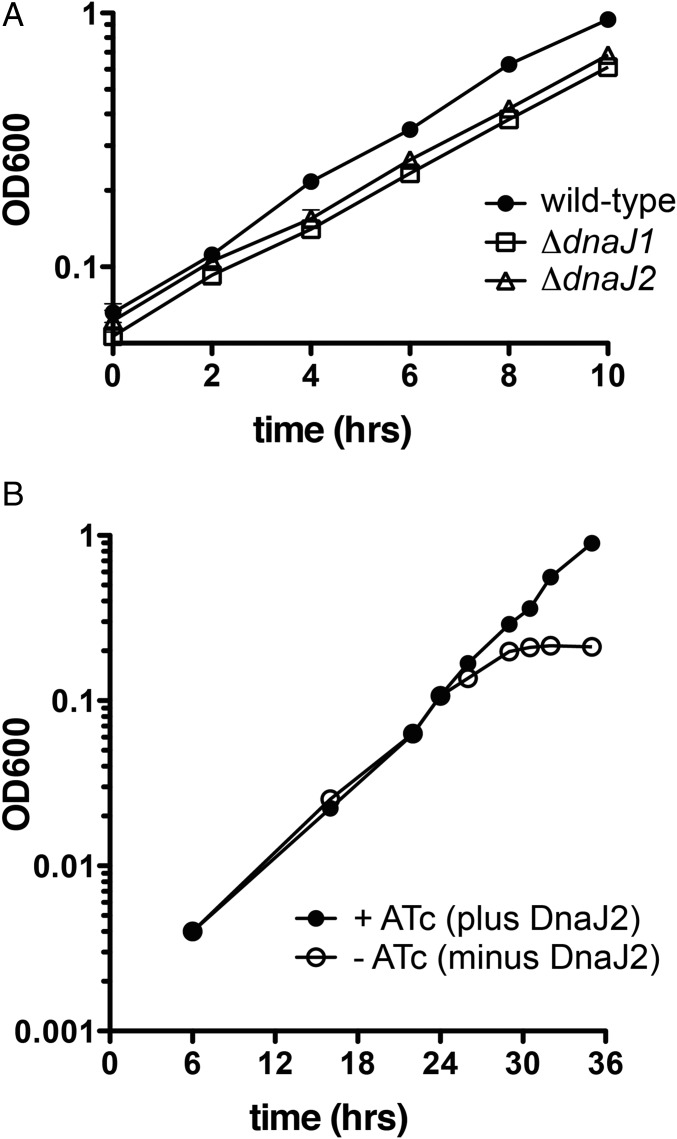
We recently reported that DnaK and GrpE are required for growth of Msm (13). To determine whether DnaJ proteins are individually essential, we deleted each dnaJ individually and in combination. We were able to delete either dnaJ1 or dnaJ2 from Msm (SI Appendix, Fig. S14) and did not observe major growth defects in these strains (Fig. 5A), nor did we see differences from wild-type in recovery from heat shock or from exposure to antibiotics known to promote formation of aggregates (SI Appendix, Fig. S15) (21). Initial attempts to create a ΔdnaJ1 ΔdnaJ2 double mutant were unsuccessful, suggesting synthetic lethality. We were able to delete dnaJ2 from ΔdnaJ1 when a second copy of dnaJ2 was supplied, supporting synthetic lethality. To study the effect of loss of both J proteins, we developed a depletion strain in the ΔdnaJ1 ΔdnaJ2 background that encodes an anhydrotetracycline (ATc)-inducible allele of DnaJ2. The strain was depleted of DnaJ2 by removing ATc and waiting 6 h before dilution, and then its subsequent growth was compared over time to cells that remained in the presence of ATc. As observed previously with depletion of DnaK (13), depletion of DnaJ2 in cells lacking DnaJ1 resulted in arrested growth at a time when replication continued in cells that expressed DnaJ2 (Fig. 5B). These data indicate that dnaJ1 and dnaJ2 are individually nonessential but in combination are required for viability and cell growth, as observed for DnaK and GrpE (13).
Fig. 5.
Single dnaJ deletion mutants do not show strong growth defects, but depletion of DnaJ2 in cells lacking DnaJ1 stalls growth of Msm. (A) Growth curves of wild-type Msm and single dnaJ deletion mutants. The doubling times (in h) of each are calculated as 2.80 ± 0.02 (WT), 2.88 ± 0.12 (ΔdnaJ1), and 2.89 ± 0.17 (ΔdnaJ2). (B) Growth curves of a depletion strain in the ΔdnaJ1 ΔdnaJ2 background that encodes an anhydrotetracycline (ATc)-inducible allele of dnaJ2 in the presence and absence of ATc. Cells were washed and grown ± ATc for 6 h and then diluted back to an OD600 of 0.004, and optical densities were measured. Error bars represent SD for n = 3.
Msm DnaJ1 and DnaJ2 share approximately 87% and 81% sequence identity, respectively, with their Mtb homologs (Clustal Omega). To determine whether the Mtb DnaJ proteins were functional in Msm, we complemented Msm lacking one or both J proteins by marker exchange of attB integrated plasmids containing wild-type or mutant Mtb J proteins. We transformed each strain with different attB:Pmop-dnaJ strepR constructs encoding mycobacterial J proteins and selected for colonies that were sensitive to kanamycin but resistant to streptomycin. We found that Msm dnaJ1 or dnaJ2 or Mtb dnaJ1 or dnaJ2 were each able to support viability as the only J protein, as indicated by successful allelic replacement (Table 1 and SI Appendix, Fig. S16). We analyzed cell lysates by Western blot to ensure that dnaJ constructs expressed the desired protein (SI Appendix, Fig. S17). These results reinforced the conclusion that only one J protein is needed for growth of mycobacteria, and demonstrated that both Mtb DnaJ1 and DnaJ2 are functional in Msm. In contrast, Msm and Mtb dnaJ1 and dnaJ2 sequences encoding point mutations in the His and Asp residues of the HPD motif (which are unable to activate DnaK; Fig. 2C and SI Appendix, Fig. S5) were unable to support viability at either low or high temperature (Table 1 and SI Appendix, Fig. S16), indicating that the mycobacterial DnaJ HPD point mutants are not functional in vivo.
Table 1.
Viability of different M. smegmatis (Msm) background strains expressing mycobacterial DnaJs and HPD motif point mutants
| dnaJ allele | M. smegmatis transformation strain background | |||
| attB:Pmop- (strepR) | Wild type | ΔdnaJ1 | ΔdnaJ2:hyg | ΔdnaJ1 ΔdnaJ2:hyg |
| Msm dnaJ1 | + | + | + | + |
| Msm dnaJ2 | + | + | + | + |
| Mtb dnaJ1 | ND | + | ND | + |
| Mtb dnaJ2 | + | + | + | + |
| Msm dnaJ1 H38Q | + | + | + | − |
| Msm dnaJ2 H32Q | + | + | + | − |
| Mtb dnaJ1 H38Q | + | + | + | − |
| Mtb dnaJ2 H32Q | + | + | + | − |
| Mtb dnaJ1 D40E | + | ND | ND | − |
| Mtb dnaJ2 D34E | + | ND | ND | − |
+, yielded >105 KanS/StrepR transformants; −, yielded no KanS/StrepR transformants; ND, not determined.
Discussion
In this work, we have identified the minimal components required for in vitro reactivation of protein aggregates by the Mtb ClpB/DnaK-bichaperone system. Our observations agree with a well-accepted mechanism in which ClpB disaggregates client proteins bound to sHps to provide nonnative substrate for refolding by DnaK (Fig. 1B) (12, 23, 57). In the cell, mycobacterial dnaK, but not clpB, is necessary for growth in conditions that support logarithmic replication (13, 21). However, under stress conditions, such as exposure to otherwise sublethal concentrations of antibiotics, stationary phase, or host infection, the presence of functional ClpB is important for responding to accumulation of denatured protein (21). We have previously shown that in stressed Mtb, ClpB mediates the asymmetric distribution of irreversibly damaged proteins during cell division. Here, we showed that Mtb ClpB also participates in recovery of reversibly damaged proteins with DnaK and its cofactors, as anticipated from studies in live mycobacterial cells (13).
The cofactors GrpE, DnaJ1, and DnaJ2 play important roles in DnaK activation and protein refolding. In agreement with our report that grpE is essential in mycobacteria (13), we found that denatured protein reactivation required GrpE in vitro (Fig. 4A). Although the two mycobacterial J proteins are individually but not collectively dispensable for Msm viability (Table 1) and for aggregate reactivation (Fig. 4A), we found that DnaJ1 and J2 are not identical in their behavior. DnaK activation by DnaJ1 was not affected by the addition of GrpE, although all three proteins are encoded by genes in the same operon. DnaJ2, however, maximally activated ATPase activity in the presence of GrpE. These cofactor relationships bear some resemblance to the mycobacterial chaperonin/cochaperonin GroEL/ES system, in which GroEL2 cooperates with the cochaperonin GroES encoded in the same operon as groEL1, but GroEL1 does not function with GroES (59). Our pulldowns and MST analyses suggest that Mtb DnaK and DnaJ2 form a stable interaction (Fig. 3). There is a wealth of data on E. coli DnaK–DnaJ interactions that supports this observation, although the binding affinity that we measured (∼15 μM) is more similar to that of E. coli DnaK with the truncated N-terminal J domain of DnaJ (50, 60, 61) than with the full-length protein (∼70 nM) (62, 63). There are differing reports on the dependence of the DnaK–DnaJ interaction on the presence of nucleotides (50, 60–62, 64, 65). Our MST results suggest that the interaction is not affected by binding of ADP to DnaK. However, we used the catalytically defective DnaK point mutant for our studies, which may affect structural changes. It is likely that inconsistencies arise because this interaction is dynamic, and different techniques are thought to measure different binding states (63).
Our results suggest that each J protein may play a predominant role at different stages in the process of aggregate reactivation. Overexpression studies on Mtb DnaJ1 and DnaJ2 suggested that each has a different cellular function (66). Only high-GC content Gram-positive actinobacteria such as mycobacteria have more than one annotated dnaJ gene, each of which is thought to have evolved separately (67). We found that each Mtb DnaJ activated DnaK’s ATPase activity (Fig. 2A and SI Appendix, Fig. S3 A–D) and contributed to refolding (Fig. 4A, Right) to different extents. In recruiting aggregates to DnaK and its homologs, J proteins are proposed to act upstream of ClpB homologs (Hsp104) (Fig. 1B) (51, 52). Higher eukaryotes lack cytosolic ClpB homologs (37), and it was shown that when purified metazoan J proteins from two different protein classes were mixed, they could disaggregate denatured proteins in vitro (38). Because both Mtb J proteins are in the same class (class A, so categorized for their hallmark C-terminal zinc-finger domain), we predicted they could not together perform this function. Indeed, we saw that in chaperone reactions lacking ClpB, there was no difference in luciferase reactivation regardless of the ratio of DnaJ1:DnaJ2 present (Fig. 4B). However, when ClpB was added, a clear pattern emerged, demonstrating that the optimal reactivation reaction contains both J proteins but required ClpB. Because DnaJ1 did not further enhance stimulation of DnaK’s ATPase activity by DnaJ2 and GrpE (Fig. 2A and SI Appendix, Fig. S3 B and C), it is unlikely that DnaJ1 and DnaJ2 bind DnaK simultaneously. Perhaps each DnaJ preferentially assists DnaK at different stages in substrate reactivation relative to the disaggregation step by ClpB. We speculate that DnaJ1 predominantly delivers substrates with DnaK to ClpB, because clpB and dnaj1 are often coexpressed in Mtb (56) and ClpB-containing reactions prefer an excess of DnaJ1.
The biochemical and cellular consequences of mutating the conserved HPD motif of J domains offer insight into the importance of functional chaperone–DnaJ interactions in mycobacteria. Typically, mutation of any residue in the motif will abrogate the ability of a given J domain to activate its cognate DnaK homolog (45, 46, 68, 69), as we observed (Fig. 2C and SI Appendix, Fig. S5). Point mutation in the HPD motif does not always result in complete loss of DnaJ function (26, 43, 70). However, we observed that mutation of His to Gln in the HPD motif of either DnaJ1 or DnaJ2 not only resulted in loss of activity in aggregate reactivation assays, but DnaJ point mutants inhibited the reaction even when wild-type J proteins were present (Fig. 4 D and E and SI Appendix, Fig. S11). Our binding studies suggest that this mutation does not affect affinity to DnaK, and may even improve it compared with wild-type J protein (Fig. 3B). This observation is similar to published binding measurements of E. coli DnaK and a J-domain HPD mutant (50). We found that the HPD point mutant can prevent J protein from fully activating DnaK when aggregates are absent (SI Appendix, Fig. S13). When aggregate is present, as in the refolding reactions, the HPD mutants may titrate substrate away from wild-type J proteins and/or bind DnaK to form an inactive complex with substrate. It has been shown that unlike the wild-type yeast J protein, YDJ1, YDJ1 HPD point mutant added to Hsp70 does not stimulate release of bound substrate (46). In addition, excess YDJ1 mutant prevented wild type from competing substrate off of Hsp70. Taken together, these data suggest that DnaJ HPD point mutants can interfere with functional DnaK–DnaJ–substrate interactions and in doing so inhibit the reactivation process. DnaJ HPD point mutants cannot act as the sole J proteins in Msm (Table 1), which can be explained by our observations that they cannot catalyze protein folding (Fig. 4C). When DnaJ point mutants are expressed with one or both wild-type J proteins, Msm strains are viable, suggesting that wild-type DnaJ function is not inhibited by mutants in the cell. The reported excess of Hsp70/DnaK relative to its cofactors provides a rationalization for this phenotype, because it is unlikely the expressed mutants can bind all of the DnaK present (6, 31). Nonetheless, we showed that a mutation that is temperature sensitive in some organisms can be lethal in mycobacteria because of the essentiality of the DnaK chaperone system, a finding which supports that the essentiality of this system could be phenocopied by small molecules.
The relationship among ClpB, DnaK, and cochaperones must be properly concerted to maintain proteostasis in the cell. Because there is an urgent need to validate new targets in Mtb, the ability to cripple essential functions by targeting chaperone–cofactor interactions represents one approach. This work also sets the stage for additional drug discovery strategies by using the tools that we have developed, such as mass spectrometry-based high-throughput screening for small molecule inhibitors of DnaK’s ATPase activity. Reports of molecules that target Mtb enzymes selectively over human homologs (71–73) or show specificity for single isoforms of human Hsp70 (74) lend support to the feasibility of targeting Mtb DnaK while sparing its homologs in the host. Because this chaperone network plays an important role not only in growth but also in the stress response, molecules that disrupt chaperone function are likely to be synergistic with host immune chemistries and chemotherapeutic agents.
Methods
Materials.
E. coli were grown in Luria–Bertani (LB) medium (BD Biosciences). Msm strains were derivatives of mc2155 (75). Msm was cultured in LB with 0.5% glycerol, 0.5% dextrose (LBsmeg), and 0.05% Tween-80 was added to all liquid media. Antibiotic concentrations used for selection of Msm strains were as follows: kanamycin 20 µg/mL, hygromycin 50 µg/mL, streptomycin 20 µg/mL, zeocin 12.5 µg/mL. Standard procedures were used to manipulate recombinant DNA and to transform E. coli. Mycobacterial chromosomal DNA was purified by using a reported protocol (76). Primers were purchased from Invitrogen, and DNA sequencing was performed by Macrogen. Vectors and expression hosts were obtained from Novagen and Addgene. pET His6 Sumo TEV LIC cloning vector (2S-T) was a gift from Scott Gradia, University of California, Berkeley, CA, (Addgene plasmid 29711) and pHYRS52 was a gift from Hideo Iwai, University of Helsinki, Helsinki, (Addgene plasmid 31122) (77). Phusion DNA polymerase was from Thermo, and restriction endonucleases were purchased from New England Biolabs. All strains used in this study are listed in SI Appendix, Table S1, along with details for construction. Plasmids including relevant features, and primers are listed in SI Appendix, Tables S2 and S3, respectively. PBS buffer (calcium chloride and magnesium chloride free) was from Gibco. Nonstick centrifuge tubes were from VWR. Polyvinylidene fluoride (PVDF) membranes (Bio-Rad) and ECL Western blotting substrate (Pierce) were used for immunoblot experiments. All other reagents were purchased from Sigma-Aldrich unless otherwise specified. FPLC analyses were performed using an AKTA pure 15 L instrument (GE Healthcare).
Cloning of Expression Vectors for Recombinant Chaperones/Cofactors and Mutant Overexpression in E. coli.
Overexpression plasmids are listed in SI Appendix, Table S2 and were constructed by using overlap extension PCR cloning techniques with indicated plasmid and primer pairs (SI Appendix, Tables S2 and S3) (78). Following PCR steps and DpnI treatment, DNA samples were purified by using a PCR purification kit (Qiagen) and transformed into Mach1 competent cells (Invitrogen). For point mutagenesis, QuikChange Site-Directed Mutagenesis Kit, (Stratagene) along with the indicated primers (SI Appendix, Table S3), were used. Following confirmation of gene insertion by DNA sequencing, selected plasmids were transformed into Rosetta2 competent cells (Novagen) for overexpression.
Overexpression, Purification, and FPLC Analysis of Recombinant Mtb Proteins from E. coli.
For N-terminal His-SUMO tagged DnaK, DnaJ1, DnaJ2, GrpE, and ClpB, and point mutants (strains EcTL02-TL06, EcTL09-13, SI Appendix, Table S1): E. coli Rosetta2 cultures in LB medium supplemented with 50 μg/mL carbenicillin, 30 μg/mL chloramphenicol, and 0.1% glucose containing corresponding plasmids (SI Appendix, Table S2) were used to inoculate 0.5 L of LB medium (1:100) supplemented with 50 μg/mL carbenicillin and 30 μg/mL chloramphenicol at 37 °C and grown to OD600 = 0.3–0.4 with shaking. Cells were cooled to 25 °C and grown to OD600 ∼ 0.5 before induction with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 5 h with shaking (DnaK-expressing cells were induced for 3.5 h). Cells were harvested by centrifugation (3,100 × g, 10 min, 4 °C) and all pellets, except DnaJ1-expressing cells, were each resuspended on ice with 15 mL of buffer A [25 mM Tris(hydroxymethyl)aminomethane (Tris) (pH = 8.0), 400 mM NaCl, 10% (vol/vol) glycerol] supplemented with 100 μg/mL lysozyme and 3 μg/mL DNaseI. For DnaJ1-expressing cells, buffer A′ {25 mM 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid (Hepes), (pH 7.5), 400 mM NaCl, 10% (vol/vol) glycerol} was used with the same enzymes added. Cells were rocked for at least 30 min at 4 °C and were lysed by sonication on ice by using a 30-s interval program at an amplitude of 5 for 5 min total. Samples were then ultracentrifuged at 39,191 × g for 30 min at 4 °C. Resulting supernatants of each were added to 1.5 mL, washed Ni-NTA agarose resin (Qiagen) with 2 mM added imidazole, and rocked at 4 °C for 30 min. Resin was then washed with 30 mL of wash buffer (30 mM imidazole in buffer A or A′) and His6-SUMO–tagged proteins were eluted with 10 mL of elution buffer (200 mM imidazole in buffer A or A′). The eluate fractions were dialyzed against 2 L of buffer A or A′ overnight by using a 10-kDa molecular mass cutoff (MWCO) Slide-A-Lyzer dialysis cassette (Pierce). His6-SUMO protease (His-Ulp1) (0.04 mg/mL) was added to the dialysis sample to cleave the His6-SUMO tag from each protein, as has been described (39). To separate His-tagged and nontagged proteins, dialysis samples were incubated with 1.5 mL of washed Ni-NTA resin for 1 h at 4 °C. The flow-through and 3 mL of buffer A or A′ passed through the resin were collected, which contained the desired untagged protein without nonnative residues. For DnaK, DnaJ1, and DnaJ2, an additional step using 5 mL of appropriate wash buffers was also collected and combined with the other fractions. Protein samples were concentrated to <1 mL by using 10-kDa MWCO Amicon Ultra Centrifugal Filter Device (Millipore) at 4 °C. GrpE was stored at this point with a final yield of ∼10 mg/L.
The remaining proteins were subjected to different purification steps. DnaK and point mutant were transferred into buffer A by using 30-kDa MWCO Amicon Ultra Centrifugal Filter Devices following the manufacturer’s instructions and concentrated to <500 μL. DnaJ1 and point mutant were purified by gel filtration using a Superdex 200 10/300 GL column (GE Healthcare) in 25 mM Hepes (pH 7.5), 400 mM NaCl, 10% (vol/vol) glycerol. Fractions containing main elution peaks were concentrated as described above to ∼1 mL. DnaJ2, and point mutants were purified by anion exchange using a HiTrap Q XL 1-mL column (GE Healthcare) with the following buffers: buffer B: 20 mM Tris (pH 8.0), 50 mM NaCl, 10% (vol/vol) glycerol and buffer C: 20 mM Tris (pH 8.0), 1 M NaCl, 10% (vol/vol) glycerol. DnaJ2 protein was transferred into buffer B by using a 10 kDa MWCO Amicon Ultra Centrifugal Filter Device following the manufacturer’s instructions and concentrated to 500 μL. After equilibration of the anion exchange column in buffer B, DnaJ2 protein was injected and eluted over a linear gradient of 0–100% C over 20 column volumes. Fractions containing main elution peaks (∼35% buffer C) were concentrated as described above to ∼100 μL. Final protein yields were ∼1 mg/L for DnaJ1, ∼1.5 mg/L for DnaJ2, and ∼10 mg/L for DnaK.
ClpB was purified by gel filtration using a Superdex 200 10/300 GL column in the buffers indicated in SI Appendix, Fig. S8 A and B. For enzymatic assays, fractions containing main elution peaks from SI Appendix, Fig. S8B were concentrated as described above to <500 μL with a final yield ∼10 mg/L. High molecular mass (HMW) protein standards (GE Healthcare) were injected according to manufacturer’s instructions by using the same protocol used for ClpB in each buffer. The molecular mass of eluted ClpB was calculated by using the manufacturer’s protocol, in which the partition coefficient (Kav) was calculated for each elution peak by using the equation Kav = (Ve − Vo)/(Vc − Vo) where Ve = elution volume, Vo = column void volume = 8.09 mL, and Vc = geometric column volume = 23.6 mL. A standard curve was generated from the HMW protein data by plotting Kav vs. log(MW), which was used to calculate the Kav of ClpB.
For His-DnaK (EcTL07), the same protocol as described above was followed up to the dialysis step and instead the sample was purified by anion exchange as detailed for DnaJ2 above. The buffers used were as follows: buffer D: 50 mM Hepes (pH 7.5), 50 mM NaCl and buffer E: 50 mM Hepes (pH 7.5), 1 M NaCl. Fractions containing main elution peaks (∼30% buffer D) were concentrated as described above to ∼150 μL with a final yield of ∼3 mg/L, and 50% (vol/vol) glycerol in 50 mM Hepes (pH 7.5), 100 mM NaCl was added to a final concentration of 10% (vol/vol) glycerol.
For N-terminal His-tagged Hsp20 (EcTL08), the growth step was carried out as described above except in LB supplemented with 25 μg/mL kanamycin and 30 μg/mL chloramphenicol. The purification protocol was followed as described above except at the dialysis step, the eluate was dialyzed against 2 L of buffer A overnight (no protease was added). The next day, the sample was concentrated by using a 10-kDa MWCO Amicon Ultra Centrifugal Filter Device to 500 μL with a final yield of ∼8 mg/L.
The yield of each purified protein was found by the DC protein assay (Bio-Rad). Proteins were each flash frozen by using N2(l) and stored at −80 °C.
Construction of dnaJ1 and dnaJ2 Deletion Mutants in Msm and Expression Strains.
Gene deletion of dnaJ1 in Msm was achieved by homologous recombination and double negative selection (79). Gene deletion of dnaJ2 in Msm was achieved by specialized transduction by using a temperature-sensitive mycobacteriophage, phAE87 (80). Alleleic replacement were confirmed by Southern blotting (SI Appendix, Fig. S14). Additional strains expressing mycobacterial J proteins were constructed as described in SI Appendix, Table S2. For all experiments, Msm was transformed by electroporation (2500 V, 2.5 µF, 1,000 Ω).
DnaJ1 and DnaJ2 Antibodies for Immunoblotting.
Polyclonal antibodies from rabbits were generated against DnaJ1 by using the peptide KEYDETRRLFAGGGF (EZbiolab), and against DnaJ2 using the peptide GGGLFSRLRETFTGR (Pocono Rabbit Farms). Peptide-specific antibodies were affinity purified by using the respective peptides and SulfoLink Immobilization Kit for Peptides (Pierce). For immunoblotting, affinity purified antibodies were used at 1:5,000 with anti-Rabbit IgG-HRP at 1:10,000 (Zymed).
Growth Curves of Msm Strains and DnaJ Depletion from Msm Lacking Native DnaJ1 and DnaJ2.
For growth curves, overnight log phase cultures for each strain, mc2155, MGM6069, and MGM6160, were diluted in triplicate in 50 mL of LBsmeg to an OD600 of 0.002. After 15 h of growth with shaking at 37 °C, OD600 was taken every 2 h for 10 h. For depletion growth curve, triplicate MGM6282 cultures were grown in LBsmeg supplemented with hygromycin, kanamycin, and anhydrotetracycline (ATc, 50 ng/mL). Log phase cultures were collected by centrifugation, washed once in media, and diluted twofold and split into two replicate cultures with and without ATc. After 6 h, the cultures were diluted to an OD600 of 0.004 and growth was continued in the absence or presence of ATc. At the indicated time points OD600 was taken for each of the six cultures for a total of 35 h.
Analysis of Mtb Chaperone ATPase Activity by Mass Spectrometry.
ATPase reactions were conducted with indicated amounts of protein in PBS buffer (pH 7.4) with 2 mM MgCl2 containing 100 μM ATP, unless otherwise indicated, at 37 °C. In a typical reaction, all components were added except ATP, prewarmed to 37 °C for 10 min before initiation by addition of ATP. At each time point, 40 μL of reaction was quenched into 40 μL of 0.05% formic acid. For kinetic analysis, reactions were carried out at different concentrations of ATP and initial rates were measured within the initial linear phase of the reaction. For ClpB ATPase activity measurements, indicated concentrations of ClpB were added to 50 mM Tris (pH 8.0), 100 mM NaCl, 20 mM MgCl2, and reactions were performed as described above.
For mass-spectrometry analysis, quenched reaction mixtures were transferred to 384-well polypropylene plates (Greiner). In each plate, triplicate 12-point twofold dilution series of ATP or ADP in 40 μL of reaction buffer with 40 μL of 0.05% formic acid were added. Plates were centrifuged at 180 × g for 2 min. Samples were analyzed by using a RapidFire 365 high-throughput solid phase extraction system interfaced with a 6230 TOF mass spectrometer (Agilent Technologies). The load/wash solvent A was aqueous 5 mM ammonium acetate, pH 10. The elution solvent B was 50% (vol/vol) water, 25% (vol/vol) acetone, and 25% (vol/vol) acetonitrile containing 5 mM ammonium acetate (pH 10). Samples (35 μL each) were separated on a Graphitic Carbon Type D cartridge. Once the sample was loaded onto the cartridge, it was washed for 4 s at 1.5 mL/min by using solvent A. ATP and ADP were eluted for 5 s by using solvent B at a flow rate of 1.5 mL/min. Thereafter, the cartridge was reequilibrated with solvent A for 5 s at a flow rate of 1.5 mL/min. The mass spectrometer was operated in negative mode, with a gas temperature of 350 °C, nebulizer pressure of 35 psig, and gas flow rate of 15 L/min. The acquisition range was 300–800 m/z. The molecular masses of the peaks detected were ATP: 505.9928 and ADP: 426.0264. For both analytes, the area under the curve (AUC) of the extracted ion counts was integrated by using RapidFire Integrator software. To determine the concentration of ADP formed and the concentration of ATP remaining, we generated standard curves (AUC vs. concentration) of ATP and ADP for each plate. Data points are represented as a concentration of ADP, which was obtained by multiplying the percent product formation by the initial nucleoside triphosphate concentration present. The formula that was used is as follows: product formation (%) = (µmoles of ADP formed × 100)/(µmoles of ADP formed + µmoles of ATP remaining). ADP background present in ATP stock was subtracted in Prism software before graphing. For Michaelis–Menten kinetics, data were analyzed by using Prism software using the kcat equation with 4 µM DnaK.
Protein Labeling with Fluorophore and Analysis of Binding Interactions by MST.
MST binding experiments were performed in a Monolith NT.115 (Nanotemper Technologies). Labeling of 100 μL of 20 μM DnaJ or DnaK proteins at lysine side chains was performed by using a NT-647 Nanotemper N-hydroxy succinimide ester according to the Nanotemper labeling kit protocol following buffer exchange (48). For experiments using labeled DnaJ2 protein, aliquots of labeled protein at ∼180 and 470 nM (DnaJ2 and mutant, respectively) were used for analysis (samples were not frozen). Each labeled protein was stored in 25 mM Tris (pH 8.0), 400 mM NaCl, 5% (vol/vol) glycerol (buffer E). Before each experiment, labeled protein was diluted to 40 nM in 20 mM Tris (pH 8.0), 150 mM NaCl, 0.05% Tween-20 (buffer F) ± 4 mM MgCl2 and 4 mM indicated nucleoside triphosphate. DnaK protein was serial diluted in manufacturer’s tubes (10 μL in each) in 25 mM Tris (pH 8.0), 400 mM NaCl, 10% (vol/vol) glycerol, 0.05% Tween-20 (buffer G). Diluted labeled protein and DnaK stock were centrifuged for 5 min at 20,000 × g before use. Ten microliters of the labeled protein supernatant were added to the tubes containing DnaK protein and mixed by pipetting. MST premium-coated capillaries were filled with protein mixture. The experiments were run at LED = 60% at 20, 40, and 80% MST powers, and 40% results were chosen for thermophoresis analysis because change in thermophoresis was >10 units. Manufacturer’s software was used for analysis. The change in thermophoresis on the y axis is expressed as the change in the normalized fluorescence (∆Fnorm), which is defined as Fhot/Fcold (bound/unbound), vs. DnaK mutant concentration. For experiments using labeled DnaK protein, aliquots of labeled protein at ∼1.5 µM were used for analysis. Labeled DnaK was stored in buffer E. Before each experiment, DnaK-FL was diluted to 30 nM in buffer F. DnaJ2 was serial diluted in manufacturer’s tubes (10 μL in each) in buffer G. Samples were prepared as above, and the experiments were run at LED = 30% at 20, 40, and 80% MST powers, and 20% results were chosen for thermophoresis analysis as described above.
Purified Protein Pull-Down Using His-DnaK and Untagged Proteins.
For in vitro pull-downs, 4 μM of each indicated “input” protein, except 8 μM GrpE, was incubated with or without His-DnaK (4 μM) in PBS (pH 7.4), 2 mM MgCl2, plus 1 mM ATP for 1 h at 37 °C with shaking. A 20-μL sample was saved as the “input.” Protein mixtures were then added to 25 μL of washed Ni-NTA agarose beads and incubated for 30 min at 37 °C with shaking. Samples were spun at 1,500 × g for 5 min to pellet Ni-NTA beads. A 40-μL sample of the supernatant was saved as the flow-through (FT). After removing the supernatant, 500 μL of wash buffer [30 mM imidazole in PBS (pH 7.4), 2 mM MgCl2] was added to Ni-NTA beads and incubated for 5 min at room temperature (RT) with shaking. Samples were spun as above, a 20-μL sample was saved as wash 1 (W1), and this step was repeated to obtain wash 2 (W2). Finally, all supernatant was removed, 75 μL of elution buffer [250 mM imidazole in PBS (pH 7.4), 2 mM MgCl2] was added to Ni-NTA beads and incubated for 5 min before centrifuging as described above. A 40-μL sample of the supernatant was saved as the elution (E). Samples were mixed with 10 μL of laemmli solution and analyzed on 10% (wt/vol) polyacrylamide gels (Bio-Rad) prepared according to manufacturer’s instructions.
Separation of Soluble and Insoluble Luciferase Fractions and Immunoblotting.
Luciferase (QuantiLum, Promega) (100 nM) was incubated with or without Hsp20 (400 nM, unless otherwise noted) in 10-μL aliquots for 10 min at 42 °C (denatured) or RT (native) in buffer H [50 mM Tris (pH 7.5), 150 mM KCl, 20 mM MgCl2, 2 mM DTT] in nonstick tubes (24). Samples were diluted to 40 μL in buffer H plus 1 mg/mL BSA. Luminescence was measured by using 2-μL diluted samples in black 96-well polystyrene plates (Costar) and adding 100 μL of luciferase reagent (Promega). Light emission was measured by using a SpectraMax L microplate reader luminometer (Molecular Devices) within the linear range of the instrument, and analysis was performed with SoftMax Pro software. An integration time of 10 s was used for all measurements. To separate soluble and insoluble luciferase fractions, diluted samples were placed in polypropylene ultracentrifuge tubes (Beckman, 1.5 mL) and spun at 166,000 × g for 1 h at 4 °C, similar to a published protocol (58). Resulting supernatants were transferred to a new tube and an equal volume of resuspension buffer [buffer H plus 0.5% Triton X-100 and 10% (vol/vol) glycerol] was added to the pellet (13). Pellet samples were vortexed and sonicated for 5 min to help resuspend. To all samples, 10 μL of laemmli solution was added before heating at 90 °C and analysis on 12% (wt/vol) polyacrylamide gels (Bio-Rad) prepared according to manufacturer’s instructions. Immunoblotting was performed by using firefly luciferase antibody-HRP (Thermo) at 1:3,000 dilution.
Protein Reactivation Assay Using Denatured Firefly Luciferase.
Protocol was modified from other reports (24, 38, 57). Luciferase (100 nM) was heated with Hsp20 (400 nM, unless otherwise noted) in 5-μL aliquots for 5 min at 42 °C (at this time point, no luciferase could be recovered from the supernatant and the relative fluorescence units were similar to the value measured for the denaturation protocol above) in buffer H in nonstick tubes. Protein disaggregation/refolding reactions were performed by adding indicated amounts of chaperones and/or cofactors in buffer H plus 1 mg/mL BSA, and initiated by adding 2 mM ATP to a final reaction volume of 20 μL and placing tubes at 25 °C. Luminescence was measured by using 2-μL reaction aliquots at indicated time points as described above. One hundred percent native luciferase activity was measured by using luciferase ± Hsp20 prepared in the same manner without the denaturation step, diluted in buffer H plus 1 mg/mL BSA and incubated for the same period at the same temperature. Time course was performed over stable native luciferase activity measurements. Percent luciferase activity was determined as percent of reaction luminescence relative to native luminescence for duplicate samples of each (native and nonnative) by using Prism software. Separate reactions were set up for each time point to prevent loss of aggregated luciferase substrate because of pipetting.
Supplementary Material
Acknowledgments
We thank Stanislav Dikiy and Hongjun Yu (Brookhaven) for experimental help and Dinorah Levya (Nanotemper Technologies), Jason Young (McGill University), and Kristin Burns-Huang for technical help and scientific discussion. Mass spectrometry and binding experiments were performed at the Rockefeller High-Throughput Screening and Spectroscopy Resource Center, and we thank J. Fraser Glickman for guidance on these projects. The Agilent Rapid-Fire Instrument was placed in the Rockefeller High-Throughput and Spectroscopy Resource Center with grant funds from The Leona M. and Harry B. Helmsley Charitable Trust. Work at Weill Cornell Medicine was supported by a grant from the Bill & Melinda Gates Foundation (TB Drug Accelerator) and by the Milstein Program in Chemical Biology and Translational Medicine. The Department of Microbiology & Immunology is supported by the William Randolph Hearst Trust. Work in the M.S.G. laboratory was supported by NIH Grant P30CA008748, and A.F. was supported by T32 CA009149. The C.F.N. and M.S.G. laboratories were both supported by NIH Grant U19 AI111143-01, Tri-Institutional TB Research Unit. T.J.L. was supported by a Helen Hay Whitney and Simons Foundation Postdoctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617644113/-/DCSupplemental.
References
- 1.WHO . Global Tuberculosis Report 2013. WHO Press; Geneva: 2013. [Google Scholar]
- 2.Kaufmann SH, Cole ST, Mizrahi V, Rubin E, Nathan C. Mycobacterium tuberculosis and the host response. J Exp Med. 2005;201(11):1693–1697. doi: 10.1084/jem.20050842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Russell DG. Mycobacterium tuberculosis and the intimate discourse of a chronic infection. Immunol Rev. 2011;240(1):252–268. doi: 10.1111/j.1600-065X.2010.00984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dahl JU, Gray MJ, Jakob U. Protein quality control under oxidative stress conditions. J Mol Biol. 2015;427(7):1549–1563. doi: 10.1016/j.jmb.2015.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehrt S, Schnappinger D. Mycobacterial survival strategies in the phagosome: Defence against host stresses. Cell Microbiol. 2009;11(8):1170–1178. doi: 10.1111/j.1462-5822.2009.01335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calloni G, et al. DnaK functions as a central hub in the E. coli chaperone network. Cell Reports. 2012;1(3):251–264. doi: 10.1016/j.celrep.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 7.Zuiderweg ER, et al. Allostery in the Hsp70 chaperone proteins. Top Curr Chem. 2013;328:99–153. doi: 10.1007/128_2012_323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Hartl FU. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem. 2013;82:323–355. doi: 10.1146/annurev-biochem-060208-092442. [DOI] [PubMed] [Google Scholar]
- 9.Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science. 2016;353(6294):aac4354. doi: 10.1126/science.aac4354. [DOI] [PubMed] [Google Scholar]
- 10.Mogk A, et al. Identification of thermolabile Escherichia coli proteins: Prevention and reversion of aggregation by DnaK and ClpB. EMBO J. 1999;18(24):6934–6949. doi: 10.1093/emboj/18.24.6934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zietkiewicz S, Krzewska J, Liberek K. Successive and synergistic action of the Hsp70 and Hsp100 chaperones in protein disaggregation. J Biol Chem. 2004;279(43):44376–44383. doi: 10.1074/jbc.M402405200. [DOI] [PubMed] [Google Scholar]
- 12.Doyle SM, Hoskins JR, Wickner S. Collaboration between the ClpB AAA+ remodeling protein and the DnaK chaperone system. Proc Natl Acad Sci USA. 2007;104(27):11138–11144. doi: 10.1073/pnas.0703980104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fay A, Glickman MS. An essential nonredundant role for mycobacterial DnaK in native protein folding. PLoS Genet. 2014;10(7):e1004516. doi: 10.1371/journal.pgen.1004516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48(1):77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 15.Griffin JE, et al. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog. 2011;7(9):e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature. 1999;400(6745):693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- 17.Genevaux P, et al. In vivo analysis of the overlapping functions of DnaK and trigger factor. EMBO Rep. 2004;5(2):195–200. doi: 10.1038/sj.embor.7400067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Teter SA, et al. Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell. 1999;97(6):755–765. doi: 10.1016/s0092-8674(00)80787-4. [DOI] [PubMed] [Google Scholar]
- 19.Bandyopadhyay B, Das Gupta T, Roy D, Das Gupta SK. DnaK dependence of the mycobacterial stress-responsive regulator HspR is mediated through its hydrophobic C-terminal tail. J Bacteriol. 2012;194(17):4688–4697. doi: 10.1128/JB.00415-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart GR, et al. Overexpression of heat-shock proteins reduces survival of Mycobacterium tuberculosis in the chronic phase of infection. Nat Med. 2001;7(6):732–737. doi: 10.1038/89113. [DOI] [PubMed] [Google Scholar]
- 21.Vaubourgeix J, et al. Stressed mycobacteria use the chaperone ClpB to sequester irreversibly oxidized proteins asymmetrically within and between cells. Cell Host Microbe. 2015;17(2):178–190. doi: 10.1016/j.chom.2014.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young JC, Agashe VR, Siegers K, Hartl FU. Pathways of chaperone-mediated protein folding in the cytosol. Nat Rev Mol Cell Biol. 2004;5(10):781–791. doi: 10.1038/nrm1492. [DOI] [PubMed] [Google Scholar]
- 23.Rosenzweig R, Moradi S, Zarrine-Afsar A, Glover JR, Kay LE. Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science. 2013;339(6123):1080–1083. doi: 10.1126/science.1233066. [DOI] [PubMed] [Google Scholar]
- 24.Mogk A, Deuerling E, Vorderwülbecke S, Vierling E, Bukau B. Small heat shock proteins, ClpB and the DnaK system form a functional triade in reversing protein aggregation. Mol Microbiol. 2003;50(2):585–595. doi: 10.1046/j.1365-2958.2003.03710.x. [DOI] [PubMed] [Google Scholar]
- 25.Bracher A, Verghese J. The nucleotide exchange factors of Hsp70 molecular chaperones. Front Mol Biosci. 2015;2:10. doi: 10.3389/fmolb.2015.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL. Not all J domains are created equal: Implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 2005;14(7):1697–1709. doi: 10.1110/ps.051406805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kityk R, Kopp J, Sinning I, Mayer MP. Structure and dynamics of the ATP-bound open conformation of Hsp70 chaperones. Mol Cell. 2012;48(6):863–874. doi: 10.1016/j.molcel.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 28.Qi R, et al. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat Struct Mol Biol. 2013;20(7):900–907. doi: 10.1038/nsmb.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kityk R, Vogel M, Schlecht R, Bukau B, Mayer MP. Pathways of allosteric regulation in Hsp70 chaperones. Nat Commun. 2015;6:8308. doi: 10.1038/ncomms9308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhuravleva A, Gierasch LM. Substrate-binding domain conformational dynamics mediate Hsp70 allostery. Proc Natl Acad Sci USA. 2015;112(22):E2865–E2873. doi: 10.1073/pnas.1506692112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11(8):579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bhangoo MK, et al. Multiple 40-kDa heat-shock protein chaperones function in Tom70-dependent mitochondrial import. Mol Biol Cell. 2007;18(9):3414–3428. doi: 10.1091/mbc.E07-01-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tzankov S, Wong MJ, Shi K, Nassif C, Young JC. Functional divergence between co-chaperones of Hsc70. J Biol Chem. 2008;283(40):27100–27109. doi: 10.1074/jbc.M803923200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baaklini I, et al. The DNAJA2 substrate release mechanism is essential for chaperone-mediated folding. J Biol Chem. 2012;287(50):41939–41954. doi: 10.1074/jbc.M112.413278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gillis J, et al. The DNAJB6 and DNAJB8 protein chaperones prevent intracellular aggregation of polyglutamine peptides. J Biol Chem. 2013;288(24):17225–17237. doi: 10.1074/jbc.M112.421685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nunes JM, Mayer-Hartl M, Hartl FU, Müller DJ. Action of the Hsp70 chaperone system observed with single proteins. Nat Commun. 2015;6:6307. doi: 10.1038/ncomms7307. [DOI] [PubMed] [Google Scholar]
- 37.Nillegoda NB, Bukau B. Metazoan Hsp70-based protein disaggregases: Emergence and mechanisms. Front Mol Biosci. 2015;2:57. doi: 10.3389/fmolb.2015.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nillegoda NB, et al. Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature. 2015;524(7564):247–251. doi: 10.1038/nature14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uehara T, Parzych KR, Dinh T, Bernhardt TG. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 2010;29(8):1412–1422. doi: 10.1038/emboj.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCarty JS, Walker GC. DnaK as a thermometer: Threonine-199 is site of autophosphorylation and is critical for ATPase activity. Proc Natl Acad Sci USA. 1991;88(21):9513–9517. doi: 10.1073/pnas.88.21.9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barthel TK, Zhang J, Walker GC. ATPase-defective derivatives of Escherichia coli DnaK that behave differently with respect to ATP-induced conformational change and peptide release. J Bacteriol. 2001;183(19):5482–5490. doi: 10.1128/JB.183.19.5482-5490.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang L, Thompson AD, Ung P, Carlson HA, Gestwicki JE. Mutagenesis reveals the complex relationships between ATPase rate and the chaperone activities of Escherichia coli heat shock protein 70 (Hsp70/DnaK) J Biol Chem. 2010;285(28):21282–21291. doi: 10.1074/jbc.M110.124149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ajit Tamadaddi C, Sahi C. J domain independent functions of J proteins. Cell Stress Chaperones. 2016;21(4):563–570. doi: 10.1007/s12192-016-0697-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sell SM, Eisen C, Ang D, Zylicz M, Georgopoulos C. Isolation and characterization of dnaJ null mutants of Escherichia coli. J Bacteriol. 1990;172(9):4827–4835. doi: 10.1128/jb.172.9.4827-4835.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wall D, Zylicz M, Georgopoulos C. The NH2-terminal 108 amino acids of the Escherichia coli DnaJ protein stimulate the ATPase activity of DnaK and are sufficient for lambda replication. J Biol Chem. 1994;269(7):5446–5451. [PubMed] [Google Scholar]
- 46.Tsai J, Douglas MG. A conserved HPD sequence of the J-domain is necessary for YDJ1 stimulation of Hsp70 ATPase activity at a site distinct from substrate binding. J Biol Chem. 1996;271(16):9347–9354. doi: 10.1074/jbc.271.16.9347. [DOI] [PubMed] [Google Scholar]
- 47.Schlee S, Beinker P, Akhrymuk A, Reinstein J. A chaperone network for the resolubilization of protein aggregates: Direct interaction of ClpB and DnaK. J Mol Biol. 2004;336(1):275–285. doi: 10.1016/j.jmb.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 48.Jerabek-Willemsen M, et al. Microscale thermophoresis: Interaction analysis and beyond. J Mol Struct. 2014;1077:101–113. [Google Scholar]
- 49.Jerabek-Willemsen M, Wienken CJ, Braun D, Baaske P, Duhr S. Molecular interaction studies using microscale thermophoresis. Assay Drug Dev Technol. 2011;9(4):342–353. doi: 10.1089/adt.2011.0380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wittung-Stafshede P, Guidry J, Horne BE, Landry SJ. The J-domain of Hsp40 couples ATP hydrolysis to substrate capture in Hsp70. Biochemistry. 2003;42(17):4937–4944. doi: 10.1021/bi027333o. [DOI] [PubMed] [Google Scholar]
- 51.Schlieker C, Tews I, Bukau B, Mogk A. Solubilization of aggregated proteins by ClpB/DnaK relies on the continuous extraction of unfolded polypeptides. FEBS Lett. 2004;578(3):351–356. doi: 10.1016/j.febslet.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 52.Reidy M, et al. Hsp40s specify functions of Hsp104 and Hsp90 protein chaperone machines. PLoS Genet. 2014;10(10):e1004720. doi: 10.1371/journal.pgen.1004720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zolkiewski M, Kessel M, Ginsburg A, Maurizi MR. Nucleotide-dependent oligomerization of ClpB from Escherichia coli. Protein Sci. 1999;8(9):1899–1903. doi: 10.1110/ps.8.9.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weibezahn J, et al. Thermotolerance requires refolding of aggregated proteins by substrate translocation through the central pore of ClpB. Cell. 2004;119(5):653–665. doi: 10.1016/j.cell.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 55.Cashikar AG, Duennwald M, Lindquist SL. A chaperone pathway in protein disaggregation. Hsp26 alters the nature of protein aggregates to facilitate reactivation by Hsp104. J Biol Chem. 2005;280(25):23869–23875. doi: 10.1074/jbc.M502854200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wattam AR, et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42(Database issue):D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Glover JR, Lindquist S. Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell. 1998;94(1):73–82. doi: 10.1016/s0092-8674(00)81223-4. [DOI] [PubMed] [Google Scholar]
- 58.Rampelt H, et al. Metazoan Hsp70 machines use Hsp110 to power protein disaggregation. EMBO J. 2012;31(21):4221–4235. doi: 10.1038/emboj.2012.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fan M, et al. The unusual mycobacterial chaperonins: Evidence for in vivo oligomerization and specialization of function. Mol Microbiol. 2012;85(5):934–944. doi: 10.1111/j.1365-2958.2012.08150.x. [DOI] [PubMed] [Google Scholar]
- 60.Greene MK, Maskos K, Landry SJ. Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci USA. 1998;95(11):6108–6113. doi: 10.1073/pnas.95.11.6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ahmad A, et al. Heat shock protein 70 kDa chaperone/DnaJ cochaperone complex employs an unusual dynamic interface. Proc Natl Acad Sci USA. 2011;108(47):18966–18971. doi: 10.1073/pnas.1111220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Suh WC, et al. Interaction of the Hsp70 molecular chaperone, DnaK, with its cochaperone DnaJ. Proc Natl Acad Sci USA. 1998;95(26):15223–15228. doi: 10.1073/pnas.95.26.15223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suh WC, Lu CZ, Gross CA. Structural features required for the interaction of the Hsp70 molecular chaperone DnaK with its cochaperone DnaJ. J Biol Chem. 1999;274(43):30534–30539. doi: 10.1074/jbc.274.43.30534. [DOI] [PubMed] [Google Scholar]
- 64.Wawrzynów A, Zylicz M. Divergent effects of ATP on the binding of the DnaK and DnaJ chaperones to each other, or to their various native and denatured protein substrates. J Biol Chem. 1995;270(33):19300–19306. doi: 10.1074/jbc.270.33.19300. [DOI] [PubMed] [Google Scholar]
- 65.Mayer MP, Laufen T, Paal K, McCarty JS, Bukau B. Investigation of the interaction between DnaK and DnaJ by surface plasmon resonance spectroscopy. J Mol Biol. 1999;289(4):1131–1144. doi: 10.1006/jmbi.1999.2844. [DOI] [PubMed] [Google Scholar]
- 66.Stewart GR, Robertson BD, Young DB. Analysis of the function of mycobacterial DnaJ proteins by overexpression and microarray profiling. Tuberculosis (Edinb) 2004;84(3-4):180–187. doi: 10.1016/j.tube.2003.12.009. [DOI] [PubMed] [Google Scholar]
- 67.Ventura M, et al. Genetic characterization of the Bifidobacterium breve UCC 2003 hrcA locus. Appl Environ Microbiol. 2005;71(12):8998–9007. doi: 10.1128/AEM.71.12.8998-9007.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yan W, Gale MJ, Jr, Tan SL, Katze MG. Inactivation of the PKR protein kinase and stimulation of mRNA translation by the cellular co-chaperone P58(IPK) does not require J domain function. Biochemistry. 2002;41(15):4938–4945. doi: 10.1021/bi0121499. [DOI] [PubMed] [Google Scholar]
- 69.Shen Y, Hendershot LM. ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP’s interactions with unfolded substrates. Mol Biol Cell. 2005;16(1):40–50. doi: 10.1091/mbc.E04-05-0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Michels AA, Kanon B, Bensaude O, Kampinga HH. Heat shock protein (Hsp) 40 mutants inhibit Hsp70 in mammalian cells. J Biol Chem. 1999;274(51):36757–36763. doi: 10.1074/jbc.274.51.36757. [DOI] [PubMed] [Google Scholar]
- 71.Bryk R, et al. Selective killing of nonreplicating mycobacteria. Cell Host Microbe. 2008;3(3):137–145. doi: 10.1016/j.chom.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin G, et al. Inhibitors selective for mycobacterial versus human proteasomes. Nature. 2009;461(7264):621–626. doi: 10.1038/nature08357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bryk R, et al. Triazaspirodimethoxybenzoyls as selective inhibitors of mycobacterial lipoamide dehydrogenase. Biochemistry. 2010;49(8):1616–1627. doi: 10.1021/bi9016186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rodina A, et al. Identification of an allosteric pocket on human hsp70 reveals a mode of inhibition of this therapeutically important protein. Chem Biol. 2013;20(12):1469–1480. doi: 10.1016/j.chembiol.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR., Jr Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol Microbiol. 1990;4(11):1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 76.van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: Evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29(11):2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Muona M, Aranko AS, Iwai H. Segmental isotopic labelling of a multidomain protein by protein ligation by protein trans-splicing. ChemBioChem. 2008;9(18):2958–2961. doi: 10.1002/cbic.200800604. [DOI] [PubMed] [Google Scholar]
- 78.Bryksin AV, Matsumura I. Overlap extension PCR cloning: A simple and reliable way to create recombinant plasmids. Biotechniques. 2010;48(6):463–465. doi: 10.2144/000113418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barkan D, Stallings CL, Glickman MS. An improved counterselectable marker system for mycobacterial recombination using galK and 2-deoxy-galactose. Gene. 2011;470(1-2):31–36. doi: 10.1016/j.gene.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bardarov S, et al. Conditionally replicating mycobacteriophages: A system for transposon delivery to Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 1997;94(20):10961–10966. doi: 10.1073/pnas.94.20.10961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.