Significance
Asn-linked glycosylation of newly synthesized polypeptides occurs in the endoplasmic reticulum of eukaryotic cells. Glycan structures are trimmed and remodeled as they transit the secretory pathway, and processing intermediates play various roles as ligands for folding chaperones and signals for quality control and intracellular transport. Key steps for the generation of these trimmed intermediates are catalyzed by glycoside hydrolase family 47 (GH47) α-mannosidases that selectively cleave α1,2-linked mannose residues. Despite the sequence and structural similarities among the GH47 enzymes, the molecular basis for residue-specific cleavage remains obscure. The present studies reveal enzyme–substrate complex structures for two related GH47 α-mannosidases and provide insights into how these enzymes recognize the same substrates differently and catalyze the complementary glycan trimming reactions necessary for glycan maturation.
Keywords: Asn-linked glycan processing, α-mannosidase, glycosidase mechanism, glycoside hydrolase, bioinorganic chemistry
Abstract
Maturation of Asn-linked oligosaccharides in the eukaryotic secretory pathway requires the trimming of nascent glycan chains to remove all glucose and several mannose residues before extension into complex-type structures on the cell surface and secreted glycoproteins. Multiple glycoside hydrolase family 47 (GH47) α-mannosidases, including endoplasmic reticulum (ER) α-mannosidase I (ERManI) and Golgi α-mannosidase IA (GMIA), are responsible for cleavage of terminal α1,2-linked mannose residues to produce uniquely trimmed oligomannose isomers that are necessary for ER glycoprotein quality control and glycan maturation. ERManI and GMIA have similar catalytic domain structures, but each enzyme cleaves distinct residues from tribranched oligomannose glycan substrates. The structural basis for branch-specific cleavage by ERManI and GMIA was explored by replacing an essential enzyme-bound Ca2+ ion with a lanthanum (La3+) ion. This ion swap led to enzyme inactivation while retaining high-affinity substrate interactions. Cocrystallization of La3+-bound enzymes with Man9GlcNAc2 substrate analogs revealed enzyme–substrate complexes with distinct modes of glycan branch insertion into the respective enzyme active-site clefts. Both enzymes had glycan interactions that extended across the entire glycan structure, but each enzyme engaged a different glycan branch and used different sets of glycan interactions. Additional mutagenesis and time-course studies of glycan cleavage probed the structural basis of enzyme specificity. The results provide insights into the enzyme catalytic mechanisms and reveal structural snapshots of the sequential glycan cleavage events. The data also indicate that full steric access to glycan substrates determines the efficiency of mannose-trimming reactions that control the conversion to complex-type structures in mammalian cells.
Mammalian Asn-linked glycoproteins are initially synthesized by cotranslational transfer of a Glc3Man9GlcNAc2 glycan precursor to nascent polypeptide chains on the luminal face of the endoplasmic reticulum (ER) (1). During transit through the secretory pathway, all three glucose residues and six of nine mannose residues are trimmed, and the structures are extended further by Golgi glycosyltransferases to generate the diverse collection of complex-type glycans found on cell-surface and secreted glycoproteins (2). The transiently formed glycan-processing intermediates resulting from glucose and mannose cleavage also play critical roles in the early secretory pathway, including acting as ligands for ER chaperones, signals for ER quality control and ER-associated degradation (ERAD), and targeting signals during intracellular transport (3–6).
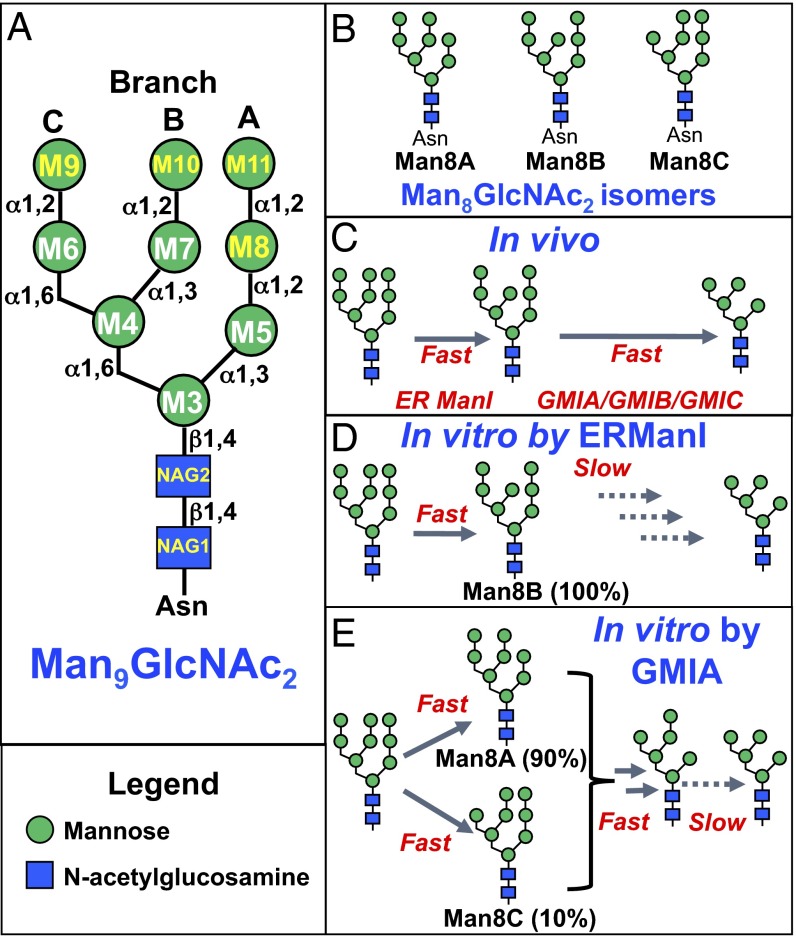
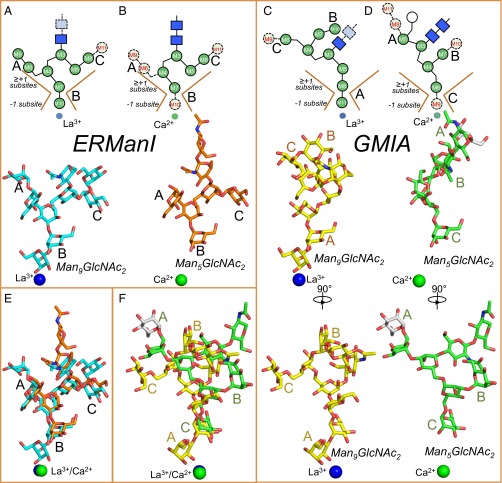
Key steps in the generation of the trimmed oligomannose glycans are catalyzed by a collection of α-mannosidases, termed “class 1” or “Carbohydrate Active Enzymes Database (CAZy) glycoside hydrolase family 47” (GH47) α-mannosidases (7, 8), that selectively cleave α1,2-mannose residues from the Man9GlcNAc2 precursor. In mammals, there are seven GH47 members (6), including ER α1,2-mannosidase I (ERManI), three Golgi α1,2-mannosidases (GMIA, GMIB, and GMIC), which play important roles in N-glycan trimming in the ER and Golgi complex, and three ER degradation-enhancing α-mannosidase-like (EDEM) proteins (EDEM1, EDEM2, and EDEM3) that play roles in ERAD (3, 6). In classical models for mammalian N-glycan maturation, Man9GlcNAc2 structures are acted upon first by ERManI to cleave a single α1,2-mannose residue (M10) from the glycan B branch to generate a Man8GlcNAc2 B isomer (Man8B) (Fig. 1) (6, 9, 10). This cleavage has been proposed to contribute to the mannose-timer mechanism for ER quality control and ERAD (1, 3, 5). Following completion of protein folding and release from ER chaperones, N-glycoproteins with Man8B isomer glycans are transported to the Golgi complex where the remaining three α1,2-mannosyl residues are cleaved by GMIA, GMIB, or GMIC to generate Man5GlcNAc2 (Fig. 1C) (11–13). In vitro, ERManI and the Golgi α-mannosidases exhibit complementary and nonoverlapping specificities for cleavage of Man9GlcNAc2 substrates. ERManI rapidly cleaves the single M10 (branch B) residue (9, 14) but cleaves the remaining mannose residues on other branches with greatly reduced efficiency (Fig. 1D). In contrast, GMIA preferentially cleaves residue M11 to generate the Man8A isomer (90%) with a minor cleavage of residue M9 (10% Man8C) followed by the ordered and sequential cleavage of residues M9 and M8 (branches C and A) to generate Man6GlcNAc2 (Fig. 1E) (15). Residue M10 (branch B), the preferred residue for cleavage by ERManI, is poorly hydrolyzed by GMIA. Thus, both in vivo and in vitro studies of ERManI and GMIA indicate that the two enzymes cleave distinct subsets of terminal α1,2-linked mannose residues during glycan maturation in the secretory pathway.
Fig. 1.
Diagrams of glycan cleavage specificity for ERManI and GMIA in vivo and in vitro. (A) Schematic diagram of the branched Man9GlcNAc2 Asn-linked glycan including the residue nomenclature and glycosidic linkages for mannose (M) and N-acetylglucosamine (NAG) residues as indicated by the symbol nomenclature shown in the legend. (B) Three Man8GlcNAc2 isomers can be generated by α-mannosidase action as indicated by the Man8A, Man8B, and Man8C nomenclature. (C) Cleavage of Man9GlcNAc2 in vivo is initiated by ERManI action to produce the Man8B isomer, and further digestion by GMIA/GMIB/GMIC produces Man5GlcNAc2. (D) In vitro cleavage of Man9GlcNAc2 by ERManI results in the rapid formation of the Man8B isomer and slow progression to smaller structures. (E) In vitro cleavage of Man9GlcNAc2 by GMIA results in the predominant cleavage of residue M11 (90%) and further cleavage to a Man6GlcNAc2 isomer that retains the M10 α1,2-linked mannose residue. Further cleavage to Man5GlcNAc2 occurs at a >10-fold slower rate.
In addition to studies on glycan cleavage, insights into substrate recognition and cleavage by GH47 α-mannosidases also have come from crystal structures of their respective catalytic domains in the absence and presence of various inhibitors and substrate analogs. The enzyme catalytic domains are comprised of a conserved (αα)7 barrel structure with a β-hairpin plugging one end of the barrel; the other end is composed of a broad cleft that leads to the active site in the barrel core (10, 16). A Ca2+ ion is bound at the apex of the β-hairpin through direct coordination to eight oxygen atoms: two from carbonyl and γ oxygen atoms of a Thr residue and six from water molecules. Substrate interactions displace two of the water molecules through coordination with the C2 and C3 hydroxyls of the terminal α1,2-mannose residue within the catalytic −1 subsite (where bond cleavage occurs between the terminal catalytic −1 subsite and the subterminal +1 subsites) (Fig. S1B). Previous studies characterized interactions with glycone-mimicking inhibitors within the −1 subsite (14, 16–19) and a thiodisaccharide substrate analog that bridged the −1 and the adjoining +1 subsites (14, 19) through a highly conserved collection of hydrophobic and H-bonding interactions as well as direct coordination with the Ca2+ ion.
Fig. S1.
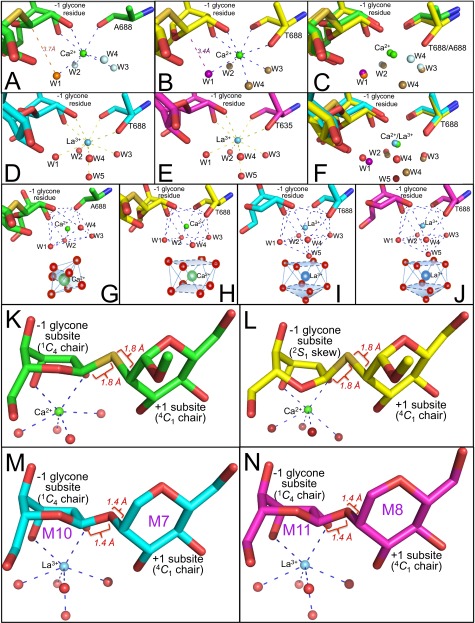
Ion coordination for wild-type and mutant forms of ERManI and conformations of the mannose residues in the −1 and +1 subsites for the ERManI– and GMIA–glycan complexes. (A and B) The structures of bound glycans and ion coordination for the ERManI T688A mutant (A) and wild-type ERManI (1X9D) (14) (B) are presented for complexes with Ca2+ and a thiodisaccharide substrate analog. The Ca2+ ion is shown as a green sphere, and the thiodisaccharide is in stick form (green for the T688A mutant and yellow for wild-type ERManI). Coordinating water molecules are displayed as light blue spheres for the T688A mutant and as orange spheres for the pronucleophile water molecule (A) and as brown spheres for wild-type ERManI and purple spheres for the pronucleophile water molecule (B). The coordinating Thr/Ala residue is shown in stick form. The distance from the pronucleophile water molecule to the C1 of the thiodisaccharide glycone (3.7 Å) in the T688A mutant (A) is greater than the distance for the wild-type enzyme (3.4 Å) (B). (C) An overlay of the structures from A and B. (D and E) The structures of the nine-coordinate enzyme–La3+–Man9GlcNAc2–PA complexes for ERManI (D) and GMIA (E) are presented with the La3+ ion shown as a blue sphere and the glycan as cyan sticks in the ERManI–La3+–Man9GlcNAc2–PA complex and as magenta sticks in the ERManI–La3+–Man9GlcNAc2–PA complex. Coordinating water molecules are displayed as red spheres, and the coordinating Thr residue is shown in stick form. (F) An overlay of the structures from B and D. (G–J) The region of cation coordination in the structures of the ERManI(T688A) mutant (G) and wild-type ERManI (H) as Ca2+–thiodisaccharide complexes and wild-type ERManI (I) and wild-type GMIA (J) as La3+-Man9GlcNAc2–PA complexes. Coordinating water molecules are displayed as red spheres. Below each structure is a diagram representing the coordination geometry of each complex: G, Ca2+-centered pentagonal bipyramid; H, Ca2+-centered eightfold square antiprismatic; I and J, La3+-centered ninefold trigonal prismatic (square face tricapped). The structure of the Man-α (1, 2)-Man-O-methyl thiodisaccharide in the −1 and +1 subsites is shown for the ERManI(T688A) –Ca2+–thiodisaccharide complex (shown as green sticks in G and K) and the for wild-type ERManI–Ca2+–thiodisaccharide complex (1X9D) (14) (shown as yellow sticks in H and L). The equivalent Man-α (1, 2)-Man disaccharide in the −1 and +1 subsites for the ERManI–La3+–Man9GlcNAc2 complex (cyan sticks in I and M) and the GMIA–La3+–Man9GlcNAc2 complex (magenta sticks in J and N) are shown also. In K–N the glycans are displayed in a similar position based on the alignment of the corresponding protein structures. The conformations of the residues in the −1 and +1 subsites and the glycosidic bond lengths between the respective residues are indicated in each panel. The Ca2+ ion is shown in green space fill representation (A–C, G, H, K, and L) and the La3+ ion is shown in blue space fill (D–F, I, J, M, and N). Coordinating water residues are shown in red space fill (D, E, and G–N).
In contrast to the highly conserved −1 and +1 subsites, the topologies of the broad clefts (≥+1 subsites) leading to the active-site cores are distinct among the GH47 family members (10, 16–20); these distinct topologies have been proposed to account for the differences in branch specificities (20). For example, protein–glycan complexes were obtained for both mouse GMIA (20) and the yeast ERManI (10), in which the equivalent of Man5GlcNAc2 enzymatic products occupied the ≥+1 subsites as ligands, but the −1 glycone subsites remained unoccupied. The orientations of the resulting glycans were distinct, and different branch termini were docked in the respective active-site clefts. However, these structures did not reveal interactions with the intact Man9GlcNAc2 substrate or the determinants of initial glycan branch specificity.
A challenge for obtaining structural insights into GH47 α-mannosidase glycan cleavage intermediates has been the progressive digestion of glycan substrates by even partially inactivated mutant enzymes (21). As an alternative approach for enzyme inactivation, we examined the role of the enzyme-bound Ca2+ ion in catalysis. The role of ion coordination was probed by determining the structure of a mutant form of ERManI (T688A), which reduced the Ca2+ ion coordination number and compromised catalysis while partially enhancing substrate-binding affinity. We further tested replacement of the Ca2+ ion with other ions that could potentially eliminate enzymatic activity but maintain substrate-binding affinity. These studies revealed that exchanging the enzyme-bound Ca2+ ion with a lanthanum (La3+) ion resulted in complete enzyme inactivation while enhancing substrate-binding affinity. Cocrystallization of La3+-inactivated GMIA and ERManI with natural Man9GlcNAc2 substrate analogs led to the formation of uncleaved enzyme–glycan complexes and provided detailed insights into initial enzyme–substrate interactions. Binding-cleft residues were probed by mutagenesis, time-course studies on the kinetics of glycan cleavage, and binding-affinity studies. These data demonstrated that the enzyme–glycan complexes reflected substrate-bound Michaelis complexes and provided insights into both the catalytic mechanism and mapping of the molecular determinants for branch-specific substrate recognition for this family of enzymes. The resulting models also highlight the essential roles of steric restrictions for glycan cleavage and maturation of N-linked glycans, a fundamental process for glycoprotein biosynthesis in the eukaryotic secretory pathway.
Results
Structure and Ion Coordination of the ERManI T688A Mutant.
Substrate interactions with the GH47 α-mannosidases are stabilized in part by the direct association of the α1,2-linked mannose glycone residue with the enzyme-bound Ca2+ ion in the −1 subsite. Chelation of the Ca2+ ion leads to enzyme inactivation, whereas its replacement with alternative cations can lead to enzyme inhibition (9, 22). Mutation of the Ca2+-coordinating Thr residue (T688A for ERManI) reduced catalysis but also increased substrate-binding affinity, whereas the Ca2+-binding affinity was essentially unchanged (21). Because the T688A mutant retains some catalytic activity toward α-mannoside substrates, we determined the crystal structure of the T688A mutant in complex with an uncleavable thiodisaccharide substrate analog (Table S1). The ERManI(T688A)–thiodisaccharide complex was highly similar in structure to the wild-type ERManI–thiodisaccharide complex [rmsd 0.13 Å versus Protein Data Bank (PDB) ID code 1X9D, hereafter “1X9D”] (14) with the major structural differences resulting from an altered glycone conformation and changes in Ca2+ coordination (Fig. S1). The glycone residue of the thiodisaccharide assumed a 1C4 chair conformation rather than the 3S1 skew boat found in the prior 1X9D structure (Fig. S1) (14). In addition, the Ca2+ ion was seven-coordinate for the T688A mutant rather than eight-coordinate for the wild-type enzyme (Fig. S1). The positions of the ligand for the substrate analog, the carbonyl oxygen of A688, and three of the coordinating water molecules (W1, W2, and W3) were similar to their positions in the wild-type ERManI–thiodisaccharide complex (Fig. S1). No additional ligands replaced the missing T688 side-chain hydroxyl group, but the fourth water ligand (W4) in the T668A mutant complex was repositioned significantly closer to the missing Thr side chain (Fig. S1 A, C, and G). Thus, the structure indicates that the removal of one point of Ca2+ coordination in the T688A mutant likely accounts for the reduced catalytic turnover by the mutant enzyme and demonstrates that ion coordination plays a direct and essential role in catalysis.
Table S1.
Crystal data collection and refinement statistics for the ERManI(T688A)–thiodisaccharide (TDS) complex, ERManI-La3+–Man9GlcNAc2–PA complex, and GMIA–La3+–Man9GlcNAc2–PA complex
| Property | Value | ||
| ERManI(T688A)–TDS | ERManI–La3+–Man9GlcNAc2–PA | GMIA–La3+–Man9GlcNAc2–PA | |
| Data collection | |||
| PDB ID code | 5KK7 | 5KIJ | 5KKB |
| Space group | P 1 | P 1 | P 21 21 2 |
| Cell dimensions | |||
| a, b, c, Å | 53.9, 56.3, 89.6 | 50.6, 53.8, 56.1 | 94.9, 131.5, 87.8 |
| α, β, γ, degrees | 105.7, 94.1, 114.2 | 89.6, 63.6, 62.7 | 90.0 90.0 9.00 |
| Resolution, Å | 42.39–1.73 | 48.69–1.65 | 41.76–1.77 |
| <I/σ(I)> | 2.0 (at 1.75 Å) | 5.35 (at 1.65 Å) | 3.22 (at 1.77 Å) |
| Data completeness,% | 90.60 | 87.20 | 94.80 |
| Refinement | |||
| Resolution, Å | 1.73 | 1.65 | 1.77 |
| No. reflections | 85,409 | 53,095 | 101,615 |
| R, Rfree | 0.176, 0.209 | 0.186, 0.229 | 0.181, 0.219 |
| No. atoms | |||
| Total | 8,930 | 4,549 | 8,518 |
| Protein | 7,592 | 3,778 | 7,635 |
| Ligand (ion) | 153 | 161 | 268 |
| Average B factors | |||
| All atoms, Å | 20.1 | 10.2 | 29.0 |
| Protein, Å | 17.7 | 8.35 | 28.0 |
| Ligand/ion, Å | 48.2 | 16.3 | 38.4 |
| Water, Å | 32.1 | 20.4 | 36.8 |
| Rmsd | |||
| Bond angles, degrees | 1.02 | 0.88 | 0.88 |
| Bond lengths, Å | 0.01 | 0.01 | 0.01 |
Cation Effects on Enzyme Activity and Substrate-Binding Affinity.
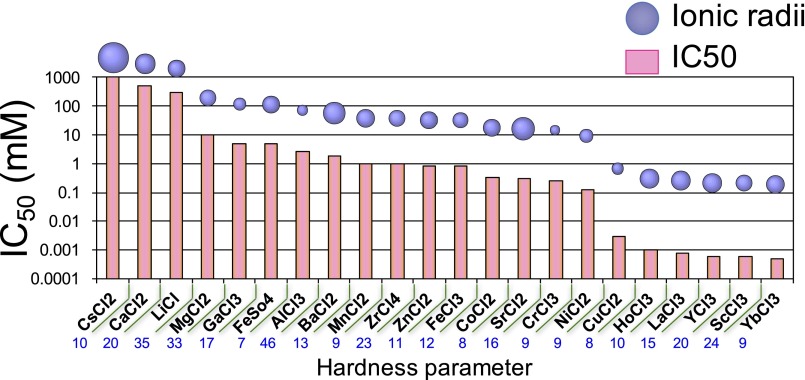
We next tested the effects of exchanging the enzyme-bound Ca2+ ion with alternative metal ions with a goal of identifying conditions in which catalysis would be compromised. Human ERManI was screened with a panel of metal ions to test their ability to rescue enzyme activity following Ca2+ depletion. Significant α-mannosidase activity was rescued in the presence of Cs2+, Ca2+, and Li+, but reduced activities were observed in the presence of other divalent and trivalent cations (Fig. S2). A representative subset of the cations (Ca2+, Sr2+, Ba2+, Mg2+, La3+) was tested further for effects on enzyme kinetics. Replacement of Ca2+ with Sr2+, Ba2+, or Mg2+ decreased enzymatic rescue (Table S2) with a primary effect on kcat but not Km. In contrast, no enzyme activity was detected in the presence of La3+. A similar inactivation of GMIA was seen following La3+ substitution.
Fig. S2.
Effect of cation substitution on the rescue of ERManI activity. ERManI was treated with EGTA and desalted as described in Material and Methods. Enzyme assays were performed with Man9GlcNAc2–PA as substrate in the presence of a range of cation concentrations. IC50 values were calculated for each cation and were plotted on a log scale in descending order of IC50. The ionic radius of the respective ion is indicated by the sphere above the plot, and hardness parameters are indicated in blue below each plot (59).
Table S2.
Effects of various cations on ERManI kinetics
| Ion | kcat, × 0.001 s−1 | Km, μM | kcat/Km, s−1/M | , % |
| Ca2+ | 3,700 | 110 | 34,000 | 1 |
| Sr2+ | 10 | 24 | 420 | 1.2 |
| Ba2+ | 0.9 | 140 | 6 | 0.02 |
| Mg2+ | N.d. | N.d. | 1* | 0.03 |
| La3+ | 0 | 0 | 0 | >0.00001 |
Wild-type ERManI was treated with 20 mM EGTA and 20 mM EDTA for 2 h and desalted over a Superdex 75 column in the absence of Ca2+. The indicated ions were added at 5-mM concentrations and were assayed with Man9GlcNAc2–PA at various concentrations a substrate to determine values for kcat and Km from the initial rates of cleavage of Man9GlcNAc2–PA to Man8GlcNAc2–PA. N.d. not determined.
The kcat/Km was determined by the slope of the Michaelis–Menten plot.
Substrate-binding affinity also was tested by surface plasmon resonance (SPR) using both wild-type ERManI and a mutant with reduced catalytic activity (E330Q) (21) in the presence of the respective cations (Table S3). These studies indicated that substrate-binding affinity was comparable in the presence of Ca2+, Sr2+, or Ba2+ but was significantly reduced in the presence of Mg2+ (Table S3) Surprisingly, high glycan-binding affinity was observed in the presence of La3+, indicating that, despite complete enzyme inactivation by the cation, the enzyme retained high substrate-binding affinity and might provide access to structural studies on enzyme–substrate complexes.
Table S3.
Effects of various cations on the binding affinity of the Man9GlcNA2–PA substrate analog to ERManI by SPR
| Ion | ERManI form | ka, s−1⋅M−1 × 1,000 | kd, s−1 × 0.001 | KD, μM | |
| Ca2+ | Wild type | — | — | 205* | 1 |
| E330Q | 18.5 | 41.5 | 2.24 | 0.01 | |
| Sr2+ | Wild type | 11 | 498 | 45.4 | 0.22 |
| E330Q | 20 | 20.8 | 1.04 | 0.01 | |
| Ba2+ | Wild type | N.d. | N.d. | N.d. | — |
| E330Q | 12.9 | 76.1 | 5.89 | 0.03 | |
| Mg2+ | Wild type | N.d. | N.d. | N.d. | — |
| E330Q | 0.388 | 37.4 | 96.5 | 0.47 | |
| La3+ | Wild type | 9.7 | 114 | 11.8 | 0.06 |
| E330Q | 5.4 | 12.3 | 2.2 | 0.01 | |
| La3+ | Wild type | 17.5 | 197 | 11.3 | 1 |
| La3+ | R461L | 0.453 | 81.7 | 180 | 16.0 |
| La3+ | W389A | 0.299 | 129 | 433 | 38.4 |
| La3+ | Combinatorial mutant | — | — | 2,160* | 191 |
Wild-type and mutant forms of ERManI were isolated and immobilized on the SPR chip surface as described in SI Materials and Methods. Bound cations were chelated by continuous flow of EGTA overnight. The indicated cations then were added in the buffer flow, and binding was examined using Man9GlcNAc2–PA as analyte as described in SI Materials and Methods. Two sets of experiments were performed. One set of experiments examined the effects of various cations on substrate-binding affinity using wild-type ERManI or the ERManI(E330Q) mutant as the immobilized binding protein. The other set of experiments examined the effects of site-directed mutants or a combinatorial mutant in which glycan-interacting residues from ERManI were swapped for equivalent residues from GMIA (SI Materials and Methods). The latter data were collected in the presence of La3+ as the enzyme-bound cation. On- and off-rates (ka and kd) and equilibrium dissociation constants (KD) were determined by fitting to a 1:1 Langmuir binding algorithm as described in SI Materials and Methods. Alternatively, maximal steady-state equilibrium sensorgram values were used to plot a saturation binding curve and calculate values for the equilibrium dissociation constant (KD) directly. N.d., binding not detectable (KD >10 mM).
The KD was determined from the steady-state affinity values and fitting from a plot of the saturation curve.
Structures of Substrate Complexes for GMIA and ERManI.
Bound Ca2+ in both ERManI and GMIA was exchanged for La3+, and substrate interactions were examined for each enzyme by cocrystallization with Man9GlcNAc2 substrate analogs. The resulting enzyme–La3+–glycan complex structures for both ERManI and GMIA were solved by molecular replacement at 1.65-Å and 1.77-Å resolution, respectively. The structure of the ERManI–La3+–glycan complex was highly similar to the prior ERManI–Ca2+–thiodisaccharide complex (rms 0.121 Å versus 1X9D) (14). For the GMIA–La3+–glycan complex, each of the two chains in the asymmetric unit were highly similar (rms 0.07 Å for chain A versus chain B), and the overall protein folds were highly similar to the structure of the GMIA–Ca2+–glycan complex (rms of 0.30 Å for both chains A and B versus PDB ID code 1NXC, hereafter “1NXC”) (20), with the predominant structural changes for both enzymes being found in ion coordination and for the residues within the glycan-binding clefts.
Both the ERManI– and GMIA–La3+–Man9GlcNAc2 complexes contained clear densities in the active-site clefts representing bound glycan structures (Fig. 2). However, the orientations of the glycans were quite different. For both enzymes, the modeled structures unambiguously identified eight of nine mannose residues and the core N-acetylglucosamine (GlcNAc) residue (NAG2) (Figs. 2 and 3). Only the M11 residue was missing from the refined electron density for ERManI, and this residue would have been positioned facing the solvent in the structural model (Figs. 2 and 3). Similarly, residue M9 was missing in the GMIA structure and also would have faced into the solvent. Both complexes were also missing the first GlcNAc residue (NAG1) and the pyridylamine tag on the substrate analogs, which also face into the solvent. For ERManI, the glycan was positioned with the M10 residue (branch B) in the −1 subsite as expected for an enzyme–substrate complex, and the remainder of the glycan structure formed a broad, flattened collection of interactions across the glycan-binding cleft extending from the −1 subsite toward the solvent at the opening of the cleft (Figs. 3 and 4 and Table S4). For GMIA, the glycan was positioned with the terminal residue of branch A (M11) extending into the −1 subsite. The other glycan branches were considerably less flattened across the glycan-binding cleft than the equivalent branches in the corresponding ERManI–glycan complex and instead formed a more compact arrangement with fewer direct interactions with residues in the GMIA glycan-binding cleft (Figs. 3 and 4 and Table S4).
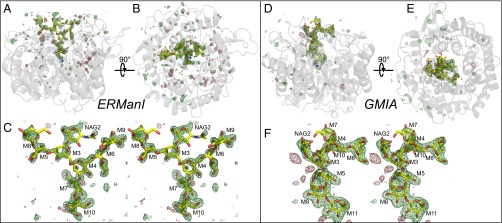
Fig. 2.
Oligosaccharide structures in the glycan-binding cleft of ERManI and GMIA. The crystal structures of ERManI (A–C) and GMIA (D–F) were determined in the presence of La3+ and Man9GlcNAc2–PA, and the electron densities of the bound glycan were revealed in Fo-Fc difference density maps calculated after omitting the ligands and subjecting the models to simulated annealing and contoured to 3σ. Cartoon diagrams of ERManI (A and B) and GMIA (D and E) are shown as side-on (A and D) or end-on (B and E) views with the protein structures in gray cartoon representation and the Fo-Fc map shown in mesh form. The structures of the bound Man9GlcNAc2 glycans are shown as yellow sticks. Wall-eyed stereo diagrams of the Fo-Fc omit maps for ERManI (C) and GMIA (F) in the glycan-binding cleft are also shown in a mesh representation with the structures of the glycans shown as yellow sticks. Monosaccharide residue designations based on the nomenclature shown in Fig.1 are indicated in the stereo figures.
Fig. 3.
Comparison of glycan structural conformations in the active sites of GH47 α-mannosidases. Enzyme-bound glycan conformations are compared for two pairs of GH47 α-mannosidase enzyme–glycan complexes (ERManI–glycan complexes in A, B, and E and GMIA–glycan complexes in C, D, and F) to illustrate the differences in glycan conformations bound in the active sites of the respective enzymes. (A–D, Upper) Simplified cartoon representations of each glycan structure. The respective enzyme active-site clefts are depicted as brown lines representing the sides of the glycan-binding cleft with labeling of the −1 glycone-binding and ≥+1 glycan-binding subsites. Mannose residues are indicated by green circles and GlcNAc residues by blue squares. The monosaccharides that are missing from the respective glycan structures are shown as light green or light blue symbols with a dotted outline for each monosaccharide. (Lower) Stick representations of the glycan associated with the human ERManI–La3+–Man9GlcNAc2 complex (A, cyan sticks), yeast ERManI-Ca2+-Man5GlcNAc2 complex (1DL2) (10) (B, orange sticks), GMIA–La3+–Man9GlcNAc2 complex (C, yellow sticks), and GMIA–Ca2+–Man6GlcNAc2 complex (1NXC) (20) (D, green sticks); the residues in the +1 subsites are positioned identically. The positions of the La3+ or Ca2+ ions in the respective structures are shown as spheres at the bottom of the respective diagrams. The crystal structure of the mouse GMIA–Ca2+–Man6GlcNAc2 glycan complex contains a single additional mannose residue (α1,6-mannose linkage shown as a white circle in upper diagram of D and as white sticks in D and F) that was added during secretion in the yeast recombinant host. Because this residue is not a part of the mammalian glycan structure in vivo and faces into the solvent in the GMIA–Ca2+–glycan complex, the glycan structure is referred to as a “GMIA–Ca2+–Man5GlcNAc2” complex throughout this paper. In the lower diagrams in C and D, two additional sets of glycan structural representations are shown with a 90° rotation to illustrate the differences in glycan structural positions. (E) A structural overlay of the Man9GlcNAc2 glycan from human ERManI in A and the Man5GlcNAc2 glycan from yeast ERManI in B demonstrating a close structural superimposition for the two glycan structures. (F) A structural overlay of the Man9GlcNAc2 glycan from GMIA in C and the Man6GlcNAc2 glycan from GMIA in D demonstrating that the glycan structural conformations in these two complexes are quite different and also are quite different from the corresponding ERManI–glycan complexes. Branch designations for the individual arms of the Man9GlcNAc2–glycan structure, based on the nomenclature shown in Fig. 1, are indicated in the figures.
Fig. 4.
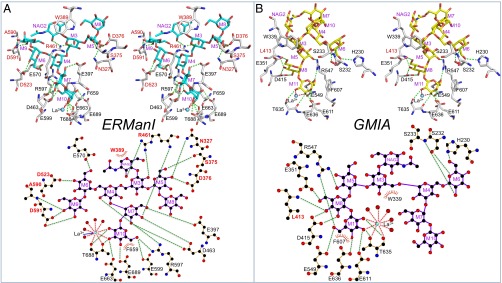
Interactions with the glycan structures in the active sites of ERManI and GMIA. (A and B, Upper) Wall-eyed stereo diagrams of the glycans bound in the active sites of ERManI (A, cyan sticks) and GMIA (B, yellow sticks) are shown along with active-site residues (white sticks) that interact with the glycan structures. (A and B, Lower) LigPlot displays of the residues that interact with the respective glycans. Hydrogen bonds are indicated by green dotted lines in each panel. Monosaccharides in the glycan structures are labeled in purple using the nomenclature described in Fig. 1. Amino acid residues labeled in red are residues that were subjected to mutagenesis and time-course studies for glycan cleavage (Fig. S4 and Table S5). Other amino acid residues in the active-site cleft are labeled in black.
Table S4.
Interactions between glycan ligands and enzyme active-site cleft residues for ERManI and GMIA
| ERManI Protein- and Glycan-interacting residues | GMIA Protein- and Glycan-interacting residues | |||||||||||
| Glycan residue | Branch | Position | Subsite | Yeast ERManI (Man5GlcNAc2) | Human ERManI (Man9GlcNAc2–PA) | Glycan residue | Branch | Position | Subsite | Mouse GMIA (Man5GlcNAc2) | Subsite | Mouse GMIA (Man9GlcNAc2–PA) |
| Man (M10) (glycone) | B | O2 | −1 | Missing | T688 (OG1) | Man (11) (glycone) | A | O2 | >3 | Missing | −1 | T635 (OG1) |
| La3+ | La3+ | |||||||||||
| O3 | Missing | T688 (O) | O3 | Missing | T635 (O) | |||||||
| La3+ | La3+ | |||||||||||
| E663 (OE2) | E611 (OE2) | |||||||||||
| O4 | Missing | E689 (OE1) | O4 | Missing | E636 (OE1) | |||||||
| O6 | Missing | R597 (NH2) | O6 | Missing | R547 (NH1) | |||||||
| E599 (OE2) | E549 (OE2) | |||||||||||
| C4-C5 | F659 | C4-C5 | F607 | |||||||||
| Hydrophobic face | Hydrophobic face | |||||||||||
| Man (7) | B | O3 | +1 | D275 (OD1) | D463 (OD2) | Man (M8) | A | O3 | >3 | Missing | +1 | D415 (OD2) |
| O4 | D275 (OD2) | D463 (OD1) | O4 | Missing | D415 (OD1) | |||||||
| R273 (N) | R461 (N) | L413 (N) | ||||||||||
| O6 | E207 (OE2) | E397 (OE2) | O6 | Missing | E351 (OE1) | |||||||
| Man (M4) | Core | O4 | +2 | R433 (NH1) | R597 (NH2) | Man (M5) | A | O3 | >3 | S233 (OG) | +2 | R547 (NH2) |
| Man (M3) | Core | O2 | +3 | R273 (NH2) | R461 (NH2) | Man (M3) | Core | Hydrophobic | +3 | No interaction | +3 | W339 |
| face | ||||||||||||
| Man (M6) | C | O4 | >3 | D336 (OD1) | D523 (OD2) | |||||||
| S272 (OG) | Man (M4) | Core | O2 | +2 | E524 (OE2) | >3 | No interaction | |||||
| O6 | E399 (OE2) | E570 (OE1) | ||||||||||
| Man (M6) | C | O2 | +1 | No interaction | >3 | S233 (N) | ||||||
| Man (M9) | C | O3 | >3 | Missing | D591 (OD1) | O3 | D415 (OD2) | |||||
| O4 | Missing | D591 (OD1) | O4 | D415 (OD1) | ||||||||
| A590 (O) | L413 (N) | |||||||||||
| O5 | No Interaction | S232 (OG) | ||||||||||
| Man (M5) | A | O4 | >3 | S185 (OG) | D376 (OD1) | O6 | E351 (OE1) | H230 (NE2) | ||||
| N129 (OD1) | N327 (ND2) | |||||||||||
| O6 | N196 (ND2) | S375 (OG) | Man (M9) | C | Missing | Missing | ||||||
| Man (M8) | A | >3 | Missing | No interaction | Man (M7) | B | >3 | No Interaction | >3 | No interaction | ||
| Man (M11) | A | O3 | >3 | Missing | No interaction | Man (M10) | B | O3 | >3 | Missing | >3 | No interaction |
| NAG2 | Core | O3 | >3 | R273 (NH2) | R461 (NH2) | NAG2 | Core | Hydrophobic face | >3 | W341 | >3 | No interaction |
| Hydrophobic face | W389 | |||||||||||
Entries in the table indicate the atoms from the respective amino acids in the indicated GH47 α-mannosidase structures that interact with the corresponding glycan monosaccharide atoms. The compositions of the bound glycans in the enzyme active sites are indicated in parentheses. The structural data for the yeast ERManI–Ca2+–Man5GlcNAc2 complex are from 1DL2 (15), and the data for the GMIA–Ca2+–Man5GlcNAc2 complex are from 1NXC (4). For the GMIA–Ca2+–Man5GlcNAc2 structure, the bound glycan has a Man6GlcNAc2 composition, because the yeast recombinant host used for mouse GMIA production added an additional mannose residue (α1,6-mannose linkage) to the glycan during secretion (see Fig. 3). Because this residue is not a part of the mammalian glycan structure in vivo, and the residue largely faces into the solvent in the GMIA–Ca2+–glycan complex, the complex is referred to as the “GMIA–Ca2+–Man5GlcNAc2” complex throughout the article. All interactions listed are hydrogen bonds except interactions between aromatic side chains and glycan residues indicated as “hydrophobic face.” The monosaccharide and branch designations are based on the nomenclature described in Fig. 1. The “subsite” designation indicates the position of the respective monosaccharide in binding-cleft subsites.
Ion Coordination and Sugar Conformations in the Active Sites.
We hypothesized that replacing Ca2+ with La3+ might result in altered ion coordination and account for the loss of enzyme activity. Comparison of the ERManI–La3+–Man9GlcNAc2 complex with the ERManI–Ca2+–thiodisaccharide complex (14) indicated that the substrate glycone and the T688 residue were coordinated to the cation in identical positions (Fig. S1 D and F). On the opposite side of the La3+ ion, five water molecules were coordinated rather than the four found in the enzyme–Ca2+ complex, resulting in a La3+-centered ninefold trigonal prismatic (square-faced tricapped) coordination (Fig. S1 D, E, I, and J). Three waters in the enzyme–La3+ complex (W1–3La) were located at approximately equivalent positions relative to the enzyme–Ca2+ complex (W1–3Ca) (Fig. S1). However, both W4La and W5La were in positions distinct from W4Ca (Fig. S1F). The ERManI– and GMIA–La3+–Man9GlcNAc2 complexes were virtually superimposable in both their −1 and +1 subsites, indicating that La3+ coordination is the same for both enzymes (Fig. S1 D and E). Thus, the only differences in ion coordination between the enzyme–La3+ structures and the corresponding enzyme–Ca2+ complexes were the altered positions of one water molecule, the appearance of one additional coordinated water molecule in the La3+ complex, and the increased charge of the bound ion.
Prior structures of inhibitors or substrate analogs that occupy the GH47 enzyme −1 subsites as Ca2+ complexes revealed distorted 1C4, 3H4, or 3S1 sugar conformations (14, 16–19) that mimic intermediates in the proposed conformational itinerary during catalysis (14, 19). In the ERManI– and GMIA–La3+–substrate complexes the mannose residue in the −1 subsite assumed a 1C4 chair conformation and also retained normal glycosidic bond lengths to the +1 mannose residue (Fig. S1 M and N). Thus, the two structures represent interactions between the enzymes and uncleaved substrates that presumably resemble Michaelis complexes in which the glycone conformations are approaching the transition states for glycoside bond hydrolysis.
Glycan Interactions in the Enzyme Active-Site Clefts.
Prior studies on yeast ERManI revealed a complex with a Man5GlcNAc2 glycan product in the enzyme-binding cleft (≥+1 subsites) (PDB ID code 1DL2, hereafter “1DL2”) (10). The Man5GlcNAc2 ligand was devoid of α1,2-linked mannose residues, and the −1 subsite was unoccupied, but residue M7 from branch B was found in the enzyme +1 subsite (Fig. 3B). By comparison, the human ERManI–La3+–Man9GlcNAc2 complex reflected an intact substrate analog docked in the active-site cleft with residue M10 of branch B in the −1 subsite and residue M7 in the +1 subsite (Figs. 3A and 4A). Alignment of the yeast ERManI–Ca2+–Man5GlcNAc2 complex with the human ERManI–La3+–Man9GlcNAc2 complex demonstrated virtually identical positions and conformations for the core Man5GlcNAc residues in the respective active-site clefts despite the two glycans being quite different in size and yeast and human ERManI being only 42% identical in primary sequence (Fig. 3E). Interactions with the core Man5GlcNAc glycan residues also were quite similar (Fig. 4 and Table S4), particularly for glycan residues M7, M4, and M3. Differences in H-bonding were found for residues M6, M5, and NAG2, and hydrophobic stacking interactions with NAG2 were found for the human enzyme (with W389) but not for the yeast enzyme.
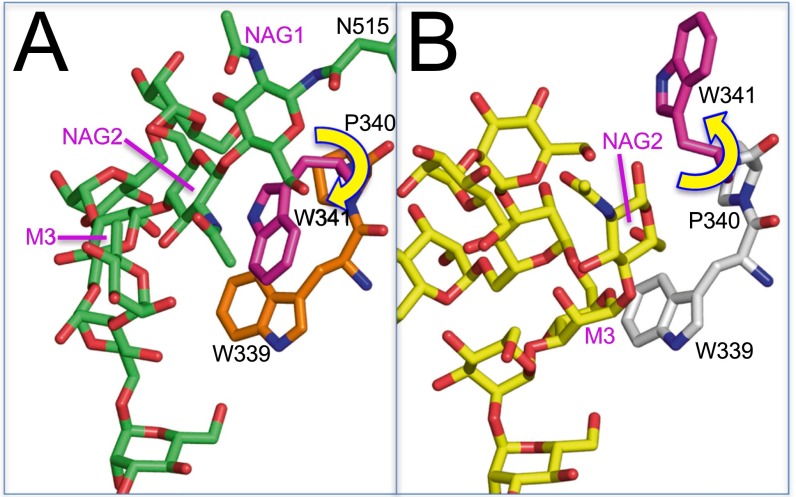
The original crystal structure of mouse GMIA also represented an enzyme–product complex with the equivalent of Man5GlcNAc2 occupying the ≥+1 subsites and the −1 subsite unoccupied (1NXC) (20). Binding of the Man5GlcNAc2 glycan in the GMIA cleft led to the insertion of residue M6 from branch C into the +1 subsite (Fig. 3D). In contrast, the present GMIA–La3+–Man9GlcNAc2 complex reflected the insertion of residue M11 from branch A into the −1 subsite, and residue M8 occupied the +1 subsite (Figs. 3C and 4B). As expected from the distinct glycan branches occupying the −1/+1 subsites for the two GMIA–glycan complexes, each complex had different interactions between the glycan and residues in the binding cleft (Fig. 4B and Table S4). Interacting residues in the +1 subsite were identical, but all other interactions in the >+1 subsites for the GMIA–La3+–Man9GlcNAc2 and GMIA–Ca2+–Man5GlcNAc2 complexes were entirely different. For both glycan complexes the interactions extended all the way from the +1 subsite to the core Man-β1,4–NAG linkage at the cleft opening (Fig. 4 and Table S4). These interactions combine both direct and indirect (through bridging water molecules) H-bonding and hydrophobic stacking to either M3 (with W339 for the Man9GlcNAc2 complex) (Fig. 4) or NAG2 (with W341 for the Man5GlcNAc2 complex). Interestingly, W341 is rotated by ∼180° in the Man9GlcNAc2 complex to avoid a steric clash with NAG2, whereas an alternative hydrophobic anchoring interaction with W339 was used with the C5–C6 region of the M3 residue (Fig. S3). Overall, there were fewer direct H-bonding interactions to fewer glycan residues in the GMIA–La3+–Man9GlcNAc2 complex (14 total H-bonds to five glycan residues) than in the ERManI–La3+–Man9GlcNAc2 complex (21 total H-bonds to nine glycan residues) (Table S4).
Fig. S3.
Hydrophobic interactions with bound glycans in the active site of GMIA. Hydrophobic stacking of W341 with NAG2 is indicated for the GMIA–Ca2+–Man5GlcNAc2 complex (1NXC) (20) (A) and between W339 and mannose residue M3 in the GMIA–La3+–Man9GlcNAc2 complex (B). The two protein structures were aligned, and the positions of the W339 and P340 residues were found to be essentially identical in the two complexes. W341 exhibits hydrophobic stacking interactions with NAG2 in the GMIA–Ca2+–Man5GlcNAc2 complex. However, W341 has rotated ∼180° in the GMIA–La3+–Man9GlcNAc2 complex to alleviate a steric clash with NAG2, and W339 stacks over the C5–C6 region of mannose residue M3 in the latter complex.
Kinetic Analysis of Wild-Type and Mutant Forms of ERManI and GMIA.
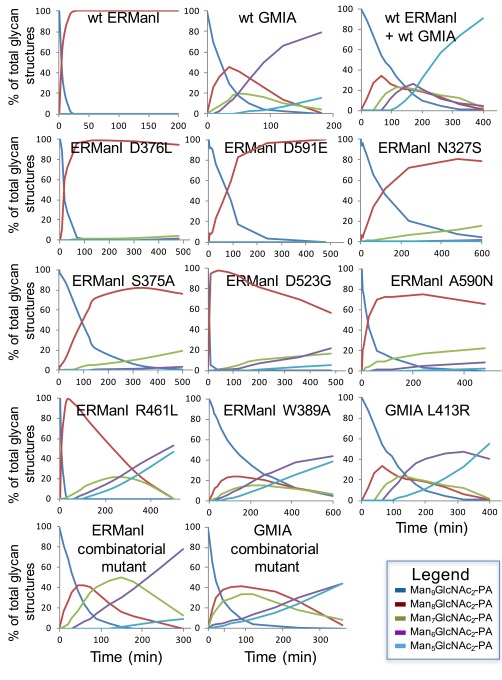
The differences in substrate specificity between ERManI and GMIA were seen most readily through in vitro time-course studies of Man9GlcNAc2 substrate cleavage. ERManI readily converted Man9GlcNAc2 to Man8B by removal of the M10 residue, but further digestion to Man7–5GlcNAc2 occurred comparatively slowly (Fig. 1 and Fig. S4). In contrast, GMIA readily cleaved Man9GlcNAc2 to Man6GlcNAc2 with transient accumulations of each intermediate structure (Man8GlcNAc2–Man6GlcNAc2) in progression through the time course and slow cleavage to Man5GlcNAc2 (Fig. 1 and Fig. S4). To test the roles of the individual amino acids in the substrate-binding cleft in substrate cleavage, residues in ERManI were mutagenized to the equivalently positioned residues in GMIA, and time-course studies of substrate cleavage were performed.
Fig. S4.
Time course of glycan digestion for wild-type and mutant forms of ERManI and GMIA. Wild-type and mutant forms of ERManI or GMIA were isolated and used in glycan digestion time-course studies with Man9GlcNAc2–PA as substrate. Individual time points in the digestion series were removed and subjected to NH2-HPLC as described in SI Materials and Methods. The relative abundance of Man9GlcNAc2–PA, Man8GlcNAc2–PA, Man7GlcNAc2–PA, Man6GlcNAc2–PA, and Man5GlcNAc2–PA structures in each time point was determined from the HPLC profiles, and glycan relative abundance was plotted versus digestion time. Enzyme sources are indicated in each time-course plot. Wild-type ERManI, wild-type GMIA, and a mixture of the two enzymes are shown in the three plots in the top row. Eight ERManI point mutants and a single-point mutant of GMIA are shown in the next three rows. The two plots in the bottom row show glycan digestion time-course plots for an ERManI combinatorial mutant containing equivalent residues swapped from GMIA and a combinatorial mutant of GMIA in which equivalent residues from ERManI were swapped into the GMIA sequence. Initial rate kinetic values for the enzymes are shown in Table S5. Details of the generation of the enzyme mutants and the glycan digestion time course are described in SI Materials and Methods.
Mutagenesis of several of the individual glycan-interacting residues in the ERManI–La3+–Man9GlcNAc2 complex (Fig. 4 and Table S4) resulted in minor alterations in glycan cleavage including, in some cases, low levels of cleavage beyond Man8GlcNAc2 (i.e., in N327S, S375A, D523G, D376L, A590N, and D591E) (Fig. S4). All the latter mutants showed some degree of compromised catalytic activity (Table S5).
Table S5.
Effects of ERManI mutations on enzyme kinetics
| Enzyme form | kcat, s−1 × 0.001 | Km, μM | kcat/Km, s−1/M | |
| ERManI (wild type) | 4,120 ± 60 | 104 ± 9 | 39,600 | 100 |
| N327S | 1,260 ± 10 | 359 ± 31 | 3,510 | 8.8 |
| S375A | 1,800 ± 30 | 455 ± 62 | 3,960 | 10.0 |
| D376L | 790 ± 10 | 371 ± 22 | 2,130 | 5.4 |
| W389A | 280 ± 90 | 970 ± 45 | 289 | 0.73 |
| R461L | 520 ± 190 | 680 ± 80 | 764 | 1.9 |
| D523G | 830 ± 20 | 204 ± 10 | 4,070 | 10.3 |
| D591E | 2,580 ± 50 | 277 ± 14 | 9,310 | 23.5 |
| Combinatorial mutant | 350 ± 80 | 2,260 ± 390 | 154 | 0.39 |
| GMIA (wild type) | 1,600 ± 300 | 142 ± 15 | 11,300 | 100 |
| L413R | 211 ± 10 | 309 ± 30 | 683 | 0.60 |
| Combinatorial mutant | 26 ± 14 | 1,985 ± 202 | 13.1 | 0.033 |
Wild-type ERManI, wild-type GMIA, and a collection of site-directed mutants were assayed with Man9GlcNAc2–PA as substrate at varied substrate concentrations to determine values for kcat and Km from the initial rates of cleavage to Man8GlcNAc2–PA. Values in the first 10 rows reflect kinetic data for wild-type and ERManI mutants (including the “combinatorial mutant” in which the collection of residues interacting with the glycan was swapped for the equivalent residues in GMIA). The final column shows kcat/Km values for each mutant relative to wild-type ERManI. Values in the last three rows reflect wild-type and mutant forms of GMIA, including a combinatorial GMAI mutant in which the collection of residues interacting with the glycan was swapped for the equivalent residues in ERManI. Values of kcat/Km for the mutant are calculated relative to wild-type GMIA.
In contrast, mutagenesis of Arg461 in ERManI, which interacts with residues M3, M7, and NAG2 in the ERManI–La3+–Man9GlcNAc2 complex (Fig. 4 and Table S4), to the topologically equivalent Leu residue in GMIA resulted in a hybrid activity between ERManI and GMIA. Glycan cleavage progressed beyond Man8GlcNAc2 and eventually produced a mixture of Man6GlcNAc2 and Man5GlcNAc2 (Fig. S4). However, the efficiency of cleavage and glycan-binding affinity were significantly reduced (Tables S3 and S5). In contrast, the opposite mutation in GMIA (L413R) resulted in an enzyme that still resembled GMIA (Fig. S4), albeit with significantly reduced catalytic activity (Table S5).
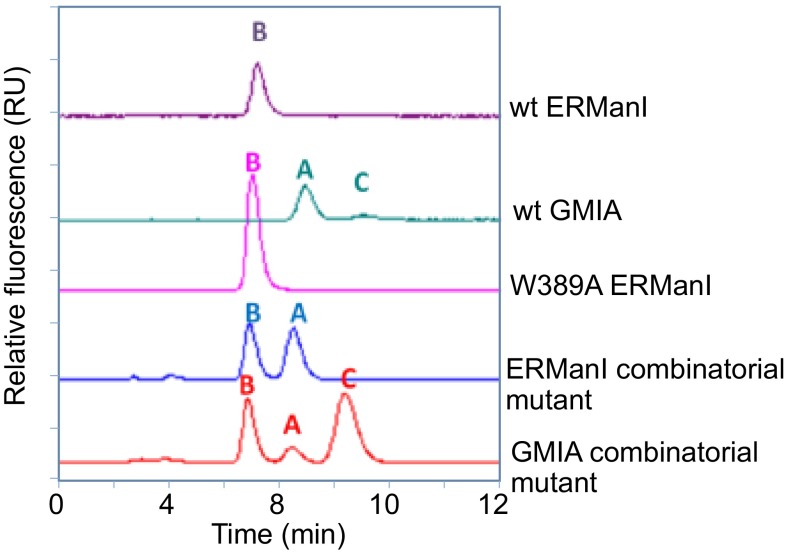
At the core of the bound glycan for both the ERManI–La3+–Man9GlcNAc2 and GMIA–Ca2+–Man5GlcNAc2 complexes (20) a Trp residue (W389 in ERManI and W341 in GMIA) provides hydrophobic stacking interactions with NAG2 in the substrate analog. An ERManI W389A mutant exhibited broader hybrid specificity for cleavage beyond Man8GlcNAc2 (Fig. S4) to produce a mixture of Man6GlcNAc2 and Man5GlcNAc2. This altered specificity presumably results from a reduction in anchoring interactions at the core of the glycan structure and more flexible glycan interactions within the cleft. Further analysis revealed reduced catalytic efficiency and glycan-binding affinity for the W389A mutant (Tables S3 and S5). In addition, the isomer structure generated as a transient cleavage intermediate resembled ERManI specificity rather than the Man8A product that is generated by GMIA (Fig. S5).
Fig. S5.
HPLC separations of Man8GlcNAc2–PA isomers generated by wild-type and mutant forms of ERManI and GMIA. The peaks corresponding to Man8GlcNAc2–PA in the time-course studies were pooled and resolved by C18-HPLC as described in SI Materials and Methods. Man8GlcNAc2–PA isomer standards were used to identify the elution positions of the Man8A (A), Man8B (B), and Man8C (C) as indicated in the figure. Profiles are shown for wild-type ERManI, wild-type GMIA, the ERManI W389A mutant, the ERManI combinatorial mutant in which binding-cleft residues from ERManI were swapped for equivalent residues from GMIA, and an equivalent combinatorial mutant of GMIA in which equivalent residues from ERManI were swapped into the GMIA sequence. Details of generation of the enzyme mutants and the glycan digestion time course are described in SI Materials and Methods. The x axis is the elution time (in min) from the C18-HPLC column.
Additional efforts to swap the specificities of ERManI and GMIA were pursued by the mutagenic conversion of the full collection of glycan-interacting residues in ERManI to the equivalent residues in GMIA (N327S/S375A/D376L/R461L/D523G/A590N/D591E). The combinatorial mutant enzyme resembled wild-type GMIA in glycan cleavage (Fig. S4) with broadened specificity for cleavage beyond Man8GlcNAc2 and accumulation of Man6GlcNAc2 before slowly generating Man5GlcNAc2. However, the catalytic efficiency and glycan-binding affinity of the mutant was reduced considerably compared with wild-type GMIA (Tables S3 and S5), and analysis of the Man8GlcNAc2 structure demonstrated a mixture of Man8B and Man8A isomers (60%:40%, Fig. S5). These results indicate that the mutant gained a broader substrate specificity that more closely resembled GMIA but still retained an initial glycan-cleavage specificity reflecting a hybrid between ERManI and GMIA.
An equivalent combinatorial mutant of GMIA was also generated in which the full collection of glycan-interacting residues in GMIA was swapped with the equivalent residues from ERManI (S279N/A327S/L328D/P340R/L413R/G474D/N540A/E541D). This latter combinatorial mutant did not resemble either ERManI or GMIA in its cleavage profile (Fig. S4). Compared with wild-type GMIA, the combinatorial mutant exhibited a slower cleavage of Man8GlcNAc2 (Table S5), slower progression to Man6GlcNAc2, and more rapid conversion to Man5GlcNAc2 (Fig. S4). Analysis of the Man8GlcNAc2 isomer intermediates demonstrated a mixture of all three Man8GlcNAc2 isomers (Fig. S5) compared with GMIA (90% Man8A) or ERManI (100% Man8B), suggesting a promiscuous specificity unlike either of the wild-type enzymes.
Discussion
The present findings provide an unprecedented level of insight into the mechanism of α1,2-mannosidase reactions, which was made possible by the parallel comparison of two distinct enzymes, trapping of Michaelis complexes by a metal substitution, and mutagenesis to direct the specificity of one enzyme toward that of the other. The studies also advance our understanding of the bioinorganic chemistry of cations in enzymatic catalysis, the conformational itineraries in glycoside bond hydrolysis, the evolution of active-site structures that provide novel enzyme specificities within the constraints of a common catalytic mechanism, and the contributions of steric factors to control the efficiency of metabolic processing during mammalian glycan maturation. Key findings and their implications are summarized below.
Glycan Distortion Precedes Hydrolysis.
Most glycosidases use a pair of amino acid carboxylate side chains in acid/base chemistry or as transient covalent intermediates to achieve bond hydrolysis. Rarely are cations used for either substrate interactions or catalysis. However, the cleavage of α-mannosidic linkages presents an unusual enzymatic challenge because the axially positioned hydroxyl group at C2 sterically hinders nucleophilic attack at the C1 position of the glycone residue. Exo-α-mannosidases generally alleviate this steric barrier by distorting the substrate from its ground state 4C1 chair conformation to reposition the C2 hydroxyl through coordination with an enzyme-bound divalent cation (Fig. 5B) (23). Three families of exo-α-mannosidases (CAZy GH38, GH47, and GH92) (24) are unique among the glycoside hydrolases in their use of divalent cations for substrate binding and catalysis (10, 14, 23, 25). Each enzyme family uses a distinct combination of divalent cation interactions, glycone distortion, transition state structures, and catalytic mechanisms. For the GH47 enzymes, a Ca2+ ion is used both for substrate interactions and for glycone distortion into a proposed 3,OB/3S1⇒3H4⇒1C4 conformational pathway that transiently produces the 3H4 structure as the ring-flattened transition state (Fig. 5B) (14, 19). Both GH47 and GH92 α-mannosidases also use a water pronucleophile that is directly coordinated to the enzyme-bound Ca2+ ion during their inverting catalytic mechanisms (14, 23). Because the water pronucleophile molecule also interacts with a catalytic Brønsted base (Glu in GH47 and Asp in GH92) to achieve deprotonation, it is anticipated that the resulting nucleophilic hydroxide anion will depart from the Ca2+ ion coordination sphere during attack on the glycosidic C1. A consequence of this attack would be the transient change from an eight to seven coordinated Ca2+ ion during catalysis (Fig. 5B).
Fig. 5.
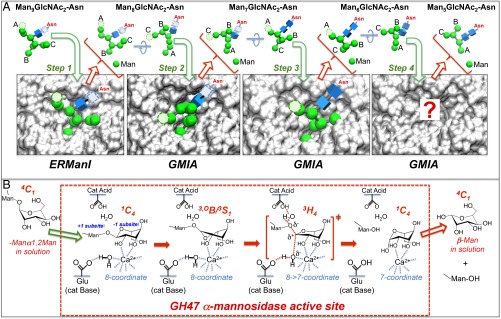
Summary of structural and mechanistic studies of mammalian GH47 α-mannosidases. (A) The sequential cleavage of Man9GlcNAc2-Asn to Man5GlcNAc2-Asn by ERManI and GMIA, in which each cleavage step requires the binding, cleavage, release, and subsequent conformational rearrangement of the glycan before binding to the enzyme in the next cleavage step. The structures of the glycan substrates are derived from the respective enzyme–glycan complexes and are depicted in 3D cartoon representations (50) in which Man residues (green spheres) and GlcNAc residues (blue cubes) are connected by glycosidic linkages (gray lines). Initial substrate interactions with a specific Man9GlcNAc2-Asn conformation in the ERManI glycan-binding cleft (step 1) result in the insertion of the terminal Manα1,2-Man disaccharide from the B branch into the −1/+1 catalytic subsites (enzymes are shown in A as gray surface representations). Residues that were not resolved in the respective enzyme–glycan complexes are shown in their anticipated positions in the structures (Man residues are shown as light green circles with dotted outlines and GlcNAc residues as light blue cubes with dotted outline). Following glycoside bond hydrolysis, the Man8GlcNAc2-Asn product and free Man residue must dissociate completely from the enzyme active site before conformational rearrangement and binding for the subsequent cleavage step. Conformational rearrangement leads to the insertion of branch A of the Man8GlcNAc2-Asn substrate in the GMIA glycan-binding cleft (step 2) with subsequent bond hydrolysis and dissociation of the Man7GlcNAc2-Asn product and the free Man residue. Similar conformational rearrangements lead to the binding of branch C of the Man7GlcNAc2-Asn substrate in step 3 and to the binding of branch A of the Man6GlcNAc2-Asn substrate to the GMIA glycan-binding cleft in step 4 (the structure of the bound complex is unknown). Each step in the cleavage series requires a different glycan structural conformation that is complementary to the geometry of the enzyme glycan-binding cleft. (B) The conformational distortion of the terminal mannose glycone residue upon binding to the active site and the enzymatic mechanism for all GH47 α-mannosidases. The dotted red box represents the proposed catalytic mechanism for bond hydrolysis by all GH47 α-mannosidases (including all ERManI and GMIA cleavage steps). Conformation distortion of the low free energy 4C1 ground state for the terminal α1,2-Man residue occurs as it enters the −1 subsite. Initial binding of the glycone in a 1C4 conformation is facilitated by hydrogen bonding, hydrophobic interactions (not shown), and coordination of the enzyme-bound Ca2+ ion with the glycone residue C2 and C3 hydroxyls. A conformational least-motion twist at C1 and the ring oxygen atom leads to a transient 3,0B/3S1 intermediate conformation followed by ring flattening to the 3H4 oxocarbenium ion transition state and release of the glycone in a 1C4 conformation. Deprotonation and attack of the glycosidic C1 by the hydroxide anion nucleophile (coordinated with the Ca2+ ion as a pronucleophile) leads to a change in coordination number from 8 to 7 for the cation. Release of the β-Man product into solution results in a conformational shift back to the low free energy 4C1 ground state.
La3+ Substitution Reveals the Michaelis Complex and the Role of the Divalent Cation in Catalysis.
We found that replacing the enzyme-bound Ca2+ ion with La3+ potently inhibited α-mannosidase activity. Subsequent structural studies demonstrated a conversion in cation geometry from eight to nine coordinated ions for the enzyme–La3+–substrate complexes. No changes in glycone, enzyme, or water pronucleophile positions were detected in the enzyme–La3+ complexes despite the complete inactivation of the enzyme in the La3+ complex. Lanthanides are frequently used to probe Ca2+- and Mg2+-binding sites in proteins (26–37), although the mechanism by which La3+ influences the enzyme activities of ion-bound complexes has not been studied systematically. Among the lanthanide family, La3+ exhibits physical and chemical properties similar to Ca2+, including a similar ionic radius (1.30 and 1.26 Å, respectively, for eight coordinated metal ions) (38), and acts as a hard Lewis acid that prefers to bind hard bases containing oxygen (39). However, La3+ is distinct in acting as a stronger Lewis acid than Ca2+, with a higher affinity toward oxygen-containing biological ligands, and has a 10-fold slower ligand exchange rate than Ca2+ (40).
Our structural studies with ion substitution indicate that altered coordination geometry and electrostatics of the La3+ complex directly impact catalytic efficiency. The conversion to a nine-coordinate geometry for the La3+–enzyme complex, combined with the stronger Lewis acid character and the tighter electrostatic interactions between the trivalent cation and the developing hydroxide anion nucleophile, led to a trapped Michaelis complex that was incapable of glycoside bond hydrolysis. Thus, the normal role of the enzyme-bound Ca2+ ion in catalysis by the GH47 α-mannosidases arises from an eight-coordinate cation complex that presumably provides an optimal balance of coordination geometry, electrostatics for high-affinity substrate interactions, and the effective release of the forming hydroxide anion for nucleophilic attack on the glycoside bond with a subsequent transient transition to a seven-coordinate Ca2+ ion configuration associated with the enzymatic product.
The Michaelis Complex Demonstrates the Glycone Conformational Itinerary During Bond Hydrolysis.
Rarely is it possible to trap intact, uncleaved natural substrates in glycoside hydrolase active sites as Michaelis complexes without making significant alterations to either the catalytic site residues or the substrate structure. Both enzyme–La3+–Man9GlcNAc2 complexes represent intact enzyme-bound substrates with glycone distortion into a 1C4 chair conformation and normal glycosidic bond lengths. By contrast, prior studies on GH47 enzymes used glycone-mimicking inhibitors or poorly cleaved thiodisaccharide substrate analogs as surrogates for bound natural substrates. The thiodisaccharide complexes displayed glycone residues bound in 3S1 skew boat conformations (14, 19) which may have been induced by the longer C-S-C bond lengths (∼1.8 Å) (Fig. S1). However, the presence of a 1C4 chair conformation for the seven-coordinate ERManI(T688A)–Ca2+–thiodisaccharide complex indicates that the longer bond lengths also can be accommodated in a 1C4 chair conformation for the latter complex. These data demonstrate that ion coordination can influence the conformation of the bound glycone residue in the respective structures. By comparison, several inhibitors bound to GH47 enzymes in 1C4 chair conformations (16, 17, 19), whereas mannoimidazole bound in the proposed 3H4 half-chair transition state conformation (19). These structures led to models suggesting a 3,OB/3S1⇒3H4⇒1C4 conformational itinerary during catalysis by GH47 α-mannosidases (14, 19). Further computational studies demonstrated that the 3,OB/3S1 and 1C4 conformations had similar energetic minima (19), but the 3,OB/3S1 conformation was more favored for catalysis. Our data indicate that initial glycone substrate interactions can be accommodated by a 1C4 chair (Fig. 5B) and that nucleophilic substitution then presumably could occur through a transient conformational progression of 1C4⇒3,OB/3S1⇒3H4⇒1C4 to produce the enzymatic product (Fig. 5B). Thus, the trapped uncleaved substrate complexes provide a direct structural snapshot of a presumed Michaelis complex during glycoside bond hydrolysis without requiring alterations in catalytic residues or the substrate structure.
Cleavage Specificity Depends on Global Substrate Contacts to Orient the Scissile Bond.
Comparison of the GH47 enzyme–glycan complexes (Figs. 3 and 5A) shows how two highly related enzyme structures can bind to the same substrates via completely distinct glycan termini using different binding modes and unique glycan substrate conformations. For the ERManI–La3+–Man9GlcNAc2 complex, it is the geometry of the core Man5GlcNAc2 glycan residues and their interactions with the active-site cleft that specifies the selective recognition and cleavage of the substrate branch B rather than contributions from the extended terminal α1,2-linked mannose residues (Fig. 5B). Presumably the other glycan branch termini are not compatible with the geometry of the ERManI glycan-binding cleft.
In contrast, GMIA may appear to be more promiscuous in its cleavage of three mannose residues from Man9GlcNAc2 (branches A and C) as compared with the single cleavage of residue M10 (branch B) by ERManI. However, the GMIA cleavage series is surprisingly restricted and sequential, with initial cleavage of residue M11 (branch A) followed by residue M9 (branch C) and residue M8 (branch A) and considerably slower cleavage of residue M10 (branch B) (Fig. 5A). In this cleavage series the GMIA–La3+–Man9GlcNAc2 complex would reflect the first cleavage step with the insertion of M11 (branch A) into the −1 subsite. The mode of substrate binding in this complex and the glycan conformation are entirely different from those in the ERManI–La3+–Man9GlcNAc2 complex except for the identical recognition of the terminal Man-α1,2-Man disaccharide in the −1/+1 subsites. By comparison, the GMIA–Ca2+–Man5GlcNAc2 product complex has an insertion of branch C into the +1 subsite and does not reflect the enzymatic product for either the initial or final cleavage of Man9GlcNAc2 by GMIA. Instead, insertion of residue M6 from branch C into the +1 subsite represents the product of the second step in the GMIA cleavage hierarchy (Fig. 5A). The clear differences in glycan geometry and interacting residues between the two GMIA–glycan substrate and –product complexes, with virtually no overlap in glycan positions other than the residue in the +1 subsite, indicates that the patterns of binding interactions for the sequential cleavage reactions by GMIA change substantially at each stage of glycan hydrolysis (Fig. 5A). Because the glycan-binding cleft for GMIA is considerably more constricted than that of ERManI (20), only a restricted set of glycan geometries is available for insertion into the active site. These restraints in cleft geometry, along with the flexible collection of indirect H-bonding substrate interactions in the glycan-binding cleft, allow GMIA to bind unique glycan branch termini for each step in the selective, ordered, and sequential series of glycan-cleavage events. Each cleavage event is presumably followed by the complete dissociation of the glycan product, exit of the cleaved glycone residue from the buried −1 subsite, and reorientation of the resulting glycan structure, before binding as the substrate for the next cleavage step (Fig. 5A).
The collection of GH47 enzyme–glycan complex structures as substrate or product complexes now reveals a series of snapshots for three distinct cleavage events (Fig. 5A). The similarities in overall protein fold for all the GH47 α-mannosidases suggest that these enzymes use a common structural scaffold and catalytic core −1/+1 subsites, but each enzyme employs a distinct cleft geometry to achieve branch-specific glycan recognition. Efforts to swap the specificities of ERManI and GMIA by interchanging individual cleft residues led to limited success, suggesting that substrate recognition is more complicated than the contributions of individual residues. The combinatorial exchange of all glycan-interacting residues in ERManI for equivalent residues from GMIA resulted in hybrid activity that more closely resembled GMIA, including a partial conversion in the initial branch cleavage by the mutant enzyme. However, the inverse combinatorial mutant (a multisite GMIA mutant harboring equivalent ERManI residues) produced an enzyme with a specificity unlike either parent enzyme. These data suggest that it is the broader collection of residues within the overall cleft topology that contributes to efficient and selective glycan cleavage.
Finally, both GH47 α-mannosidases engage their substrates by remarkably large contact interfaces for protein–sugar recognition, which is important for presenting the correct glycan termini to the respective enzyme active sites. A hallmark of the bound substrate complexes for both enzymes is hydrophobic stacking interactions between Trp residues and the glycan core (either NAG2 or M3 residues) that anchors the base of the glycan structure to the enzyme cleft. Mutation of this residue in ERManI (W389A) allowed a broadened cleavage to Man6–5GlcNAc2 structures partially resembling a GMIA-like specificity that presumably results from altered interactions within the glycan-binding cleft. Thus, alternative hydrophobic interactions are used at multiple cleavage steps and combine with a broad H-bonding network to establish an unusual paradigm for protein–glycan interactions that extend across the entire glycan structure.
Implications for Processing of Oligomannose Glycans in the Secretory Pathway.
The extended interactions between the GH47 α-mannosidases and the full glycan substrate structures have significant implications for glycan processing in the secretory pathway. The potential for local protein-specific steric barriers to impair glycan cleavage and maturation has been described previously (41–45), but the structural basis for these observations has remained obscure. Recent studies demonstrated that glycan interactions with an adjacent protein domain in a soluble Pdi1 reporter glycoprotein reduced α1,2-mannose trimming efficiency (45). Removal of the respective domain rescued glycan trimming and maturation in this model system. In HIV gp120, a regional high-density clustering of glycosylation sites results in the formation of an under-processed “mannose patch” comprising a network of interlocking oligomannose structures that protects the virus from antibody-mediated neutralization (46, 47). In contrast, glycan structures outside the mannose patch were converted to complex-type structures (46), indicating that normal glycan maturation would occur if there were adequate steric access to the glycan substrates. Thus, the high-density glycan clustering within the mannose patch presumably impaired steric access to cleavage by the GH47 α-mannosidases. Finally, the impact of protein–glycan interactions on glycan maturation was examined as a part of a larger study on residues that flank glycosylation site acceptor sequons (48). Aromatic residues at the n-2 positions relative to glycosylation sites on the polypeptide backbone resulted in increased glycan occupancy but also reduced the efficiency of glycan trimming. The authors suggested that the presence of the n-2 aromatic residue resulted in the formation of a reverse turn as a result of hydrophobic stacking between the aromatic residue and the nonpolar face of the adjoining glycan core GlcNAc residue (49). These interactions presumably would compete with steric access for binding to the cleft in the GH47 α-mannosidases and would lead to less efficient mannose cleavage. Thus, complete steric access to the core glycan residues contributes to the efficiency of glycan cleavage by the GH47 α-mannosidases in vivo and influences the conversion to complex-type structures at individual glycan sites on cell-surface and secreted glycoproteins. By extension, these steric issues also would impact glycan maturation on many secreted recombinant products including commercially important biomolecules such as glycosylated therapeutics. Similar issues of steric access to substrate structures presumably also occur for many other glycan-processing enzymes in the secretory pathway and are likely contributors to the diversity and heterogeneity of glycan structures in eukaryotic cells.
Materials and Methods
Detailed discussions of the materials and methods used in this study are provided in SI Materials and Methods.
Protein Expression and Purification.
The catalytic domains of wild-type or site-directed mutants of human ERManI and mouse GMIA were expressed in Pichia pastoris and purified as described SI Materials and Methods.
Generation of Mutants.
Site-directed mutagenesis was performed using the QuikChange mutagenesis kit in the respective pPICZα or pHIL-S1 expression vectors as described in SI Materials and Methods.
Preparation of the Man9GlcNAc2–PA Substrate Analog.
Man9GlcNAc2 was isolated from soybean agglutinin (SBA) and was reductively aminated with 2-amino pyridine (PA) as described in SI Materials and Methods.
Crystallization and X-Ray Diffraction.
Crystals of wild-type and mutant forms of GMIA and ERManI were generated from purified enzyme preparations and premixed with Man9GlcNAc2–PA or thiodisaccharide substrate analogs as described in SI Materials and Methods. Crystals were grown using hanging-drop vapor-diffusion methods, and diffraction data were collected at the Southeast Regional Collaborative Team/Advanced Photon Source (SER-CAT/APS) at Argonne National Laboratory and processed as described in SI Materials and Methods. The structures were solved by molecular replacement as described in SI Materials and Methods.
Enzyme Assays and Time Course of Glycan Digestion.
Enzyme assays were performed using Man9GlcNAc2–PA as substrate, and enzymatic products were resolved at individual time points by NH2-HPLC as described in SI Materials and Methods.
Binding Studies by SPR.
SPR analyses were conducted using a Biacore 3000 apparatus (Biacore AB) with recombinant enzymes immobilized on the CM5 SPR chip surface and Man9GlcNAc2–PA as the analyte as described in SI Materials and Methods.
SI Materials and Methods
Recombinant Enzyme Expression, Mutagenesis, and Extraction of Divalent Cations.
Recombinant expression of wild-type and mutant forms of human ERManI and mouse GMIA was performed in P. pastoris. Construction of the wild-type ERManI expression vector (16) in pPICZαA (Invitrogen) and the GMIA expression vector (15) in pHIL-S1 (Invitrogen), transformation, culture growth, protein expression, and purification were carried out as previously described (14, 20). For ion inhibition and structural studies the enzymes were treated with 20 mM EGTA and 20 mM EDTA for 2 h and were desalted over a Superdex 75 column in the absence of Ca2+. The desired cations were added at 5-mM concentrations for structural studies. Site-directed mutagenesis was performed using the QuikChange mutagenesis kit (Stratagene) in the respective expression vectors as previously described (21). The full coding region of each mutant was sequenced to confirm that only the desired mutation was generated.
Deglycosylation of GMIA.
Prior structural studies on murine GMIA used a glycosylated form of the enzyme (1NXC) (20). In this latter GMIA structure, the Man6GlcNAc2 glycan attached to one protein unit in the crystal lattice was bound in the active site of an adjoining monomer in the crystal lattice, resulting in extensive cross-lattice contacts. To eliminate this competition between cross-lattice contacts and the binding of an exogenous soluble Man9GlcNAc2–PA glycan ligand in the enzyme active-site cleft, the single glycan structure associated with recombinant GMIA was removed by digestion with recombinant endoglycosidase F1 (endoF1) produced as a cellulose-binding module fusion (51) and bound to cellulose beads. Following digestion, the immobilized endoF1 was removed by centrifugation, and the detached glycans were separated from the deglycosylated protein by dialysis. The enzyme was purified further by Superdex-75 gel filtration chromatography in 20 mM Hepes (pH 6.0) and was concentrated in 20 mM Hepes, 5 mM LaCl3, 200 mM NDSB-201 (pH 6.5), using a stirred concentration pressure cell (Amicon, Inc.) to a final concentration of 20 mg/mL for crystallization studies.
Preparation of Substrate Analog Man9GlcNAc2–PA.
Man9GlcNAc2 glycan structures were isolated from crude soybean paste by a modification of prior procedures (14, 52). Briefly, crude SBA was extracted from soybean paste at pH 4.5, subjected to (NH4)2SO4 precipitation, and digested with trypsin to generate glycopeptides that were further separated by Sephadex G50 size-exclusion chromatography and on a C18 SPE cartridge as described previously (14). Glycans were cleaved from the glycopeptides by PNGaseF digestion as previously described (53). Phenol/chloroform extraction then was used to remove most of the peptides, and the free oligosaccharides were reductively aminated with 2-PA as previously described (15). Phenol/chloroform extraction was used to remove the free PA, and Bio-Gel P-2 size-exclusion chromatography was used to desalt the tagged glycans from the remaining pyridylamine. The fluorescently tagged oligosaccharides were separated further on a preparative Hypersil APS-2 amine column to generate highly enriched substrate structures for enzymatic analysis (15).
Enzyme Assays, Time-Course Studies of Glycan Digestion, and Man8GlcNAc2–PA Isomer Separations.
Enzyme assays on the respective wild-type and mutant α-mannosidase forms were performed using Man9GlcNAc2–PA as a substrate as previously described (15, 21). Briefly, the enzyme reactions (20 μL) were carried out in reaction buffer [20 mM Mes (pH 6.5), 150 mM NaCl, and 5 mM of the indicated cation] at 37 °C for the indicated times, stopped by the addition of 50 μL of acetonitrile, and resolved and quantitated using Hypersil APS-2 NH2-HPLC column (4.6 × 250 mm) (Alltech) chromatography (15) using an isocratic mobile-phase elution in 40% acetonitrile, 50 mM Na phosphate (pH 4.0) and a flow rate of 1 mL/min. Elution of the Man9–5GlcNAc2–PA glycans was monitored by in-line fluorescence detection (excitation wavelength, 320 nm; emission wavelength, 400 nm). Pure Man9–5GlcNAc2–PA standards previously confirmed by 1H-NMR (15) were used under identical separation conditions to identify the oligosaccharides from the enzymatic cleavage reactions. The areas under the resolved peaks were integrated to determine the quantities of the corresponding oligosaccharides in the reaction product mixtures and to determine the decreasing abundance of the Man9GlcNAc2–PA substrate and the appearance of Man8–5GlcNAc2–PA products. The decreasing abundance of Man9GlcNAc2–PA was used in a time point before the appearance of Man7–5GlcNAc2–PA to determine the initial reaction rates for both ERManI and GMIA. Initial rates (ν) for the enzyme reactions were determined at various substrate concentrations ranging from 10 to 1,600 µM. Catalytic coefficient (kcat) and Michaelis constant (Km) values were determined by fitting initial rates to the Michaelis–Menten function by nonlinear regression analysis using SigmaPlot (Jandel Scientific). One unit of enzyme activity is defined as the amount of enzyme that generates 1 μmol of Man8GlcNAc2 from Man9GlcNAc2 in 1 min at 37 °C.
To determine the rates of conversion among trimmed glycan products, time-course assays were performed by mixing 1 nmol Man9GlcNAc2–PA in 50 μL of 20 mM Mes (pH 6.5), 150 mM NaCl, 5 mM CaCl2, and 2–20 μL of enzyme stock solution (10 μg/mL in the same buffer), depending on the activity of the enzymes, in a total volume of 100 μL of reaction buffer. The digestion was incubated at 37 °C, and 5-μL aliquots of the reaction mix were removed at 5-min intervals and mixed immediately with 5 μL acetonitrile to stop the reaction. The 10-μL samples were separated by HPLC using the same methods as in the kinetic analyses. The peak areas of the Man9–5GlcNAc2–PA structures were integrated, and the percentage ratio of each oligosaccharide in the reaction mix was calculated. Curves of Man9–5GlcNAc2–PA glycan percentages versus time for all of the oligosaccharides in the reaction mixture were generated in the same plot for a single-enzyme reaction.
Man8GlcNAc2–PA isomers generated as cleavage intermediates in the time-course studies were characterized by pooling the fractions containing Man8GlcNAc2–PA from the Hypersil APS-2 NH2-HPLC column separations, drying in a Speed-Vac, dissolving in water, and separation by reverse-phase HPLC using a Cosmosil C18 column (6 × 0.5 cm) (JM Science, Inc.). Isocratic elution was used in a mobile phase containing 20 μm NH4 acetate and 1% butanol (pH 4.0) and a flow rate of 0.8 μL/min.
Protein concentration was determined using the BCA protein assay reagent (Pierce) as described by the manufacturer. Oligosaccharide concentrations were determined by phenol-sulfuric acid assays (54).
Crystallization and X-Ray Diffraction.
For the ERManI T688A mutant–thiodisaccharide complex, protein crystallization was initiated from an enzyme preparation (10 mg/mL final concentration) in 20 mM Mes (pH 7.0), 150 mM NaCl, 5 mM CaCl2, 750 mM NDSB-201, and a 50-mM final concentration of thiodisaccharide. The enzyme–thiodisaccharide mixture was preincubated for 2 h at room temperature before crystallization. For the ERManI–La3+–Man9GlcNAc2–PA complex, the crystallization conditions were identical except that 5 mM LaCl3 was used in place of CaCl2, and a final concentration of 5 mM Man9GlcNAc2–PA was used in place of the thiodisaccharide. Hanging drops for ERManI crystals were prepared by mixing the enzyme–glycan preparation with an equal volume of mother liquor solution containing 24% (wt/vol) polyethylene glycol 4000, 100 mM Mes/NaOH (pH 6.0), 50 mM ammonium sulfate, and 10% (vol/vol) 1,4- butanediol. For the GMIA–La3+–Man9GlcNAc2 structure, the enzyme (10 mg/mL final concentration) in 20 mM Hepes, 5 mM LaCl3, 200 mM NDSB-201 (pH 6.5), was preincubated with Man9GlcNAc2–PA (5 mM final concentration) for 2 h before crystallization. Crystals were prepared by mixing the enzyme–glycan preparation with an equal volume of mother liquor solution containing 100 mM Mes, 50 mM ammonium sulfate, 15% PEG 4000 (pH 6.5). Crystals were mounted and flash-frozen, and diffraction data were collected at SER-CAT/APS by remote data collection from University of Georgia. The data were collected with beam line 22ID using a MAR300 detector. A total of 360 frames of 1° oscillation images were collected. HKL2000 software (55) was used for data indexing, integration, and scaling. The ERManI and GMIA structures were solved by molecular replacement using the PHENIX software suite (56) and the ERManI–Ca2+–thiodisaccharide complex (1X9D) (14) or GMIA (1NXC) (20), respectively, as probes. The structure models were built with COOT software (57) and were refined repeatedly using the PHENIX software suite. All figures of protein structures were generated using PyMOL v1.8.2 software (Schrodinger LLC) except for the cartoon representations of the glycan structures in Fig. 5, which were prepared using VMD version 1.9.3, and the 3D symbol nomenclature for glycans (50).
Binding Studies by SPR.
SPR analyses were conducted using either a Biacore 3000 or T-100 apparatus (Biacore AB) with recombinant ERManI or GMIA enzyme forms immobilized on the CM5 sensor chip surfaces at 25 °C by the amine-coupling method as described previously (14, 21). Mock-derivatized flow cells served as reference surfaces. The binding analyses were performed at 10 °C with continuous flow (30 μL/min) of running buffer [10 mM Mes (pH 7.0), 300 mM NaCl, and 5 mM of the indicated cation]. For studies using ions other than CaCl2, the chip surface was subjected to overnight treatment with 20 mM Na Mes (pH 7.0), 300 mM NaCl, 5 mM EGTA, at a flow rate of 5 μL/min, before binding analyses using running buffer containing the respective cations. Analytes were prepared in the respective running buffers by twofold serial dilution to obtain an appropriate concentration range. The binding of Man9GlcNAc2–PA glycans was analyzed in a concentration series (from 0.4 to 400 μM or more) over an immobilization surface of recombinant protein (3,000 response units) (14, 21). SPR data for each analyte concentration were collected in duplicate and globally fit to a 1:1 Langmuir binding algorithm model to calculate the on-rate (ka), the off-rate (kd), and the equilibrium dissociation constant (kd/ka = KD) using the BIAevaluation 3.1 software (58). Alternatively, the maximal equilibrium sensorgram values were used to plot a saturation-binding curve and to calculate KD values directly.
Acknowledgments
We thank Dr. Aloysius Siriwardena for supplying the thiodisaccharide; Drs. Z. Wood and W. Lanzilotta for assistance in the protein structural studies and for help in preparation of the Fo-Fc difference maps; Dr. Todd Harrop for helpful discussions on bioinorganic chemistry; David Thieker for assistance with the 3D symbol nomenclature for glycans; Drs. R. Haltiwanger, C. West, C. Glover, L. Wells, and M. Tiemeyer for their helpful reading of the manuscript; and Dr. L. Chen and B. C. Wang for assisting with access to the SER-CAT/APS beamline. This research was supported by NIH Grants P41GM103390, R01DK075322, R01GM047533, and P01GM107012 (to K.W.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors reported in this paper have been deposited in the Protein Data Bank database (ID codes 5KK7, 5KIJ, and 5KKB).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611213113/-/DCSupplemental.
References
- 1.Moremen KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: Diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13(7):448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- 3.Tannous A, Pisoni GB, Hebert DN, Molinari M. N-linked sugar-regulated protein folding and quality control in the ER. Semin Cell Dev Biol. 2015;41:79–89. doi: 10.1016/j.semcdb.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamriben L, Graham JB, Adams BM, Hebert DN. N-glycan-based ER molecular chaperone and protein quality control system: The calnexin binding cycle. Traffic. 2016;17(4):308–326. doi: 10.1111/tra.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pisoni GB, Molinari M. Five questions (with their answers) on ER-associated degradation. Traffic. 2016;17(4):341–350. doi: 10.1111/tra.12373. [DOI] [PubMed] [Google Scholar]
- 6.Moremen KW, Molinari M. N-linked glycan recognition and processing: The molecular basis of endoplasmic reticulum quality control. Curr Opin Struct Biol. 2006;16(5):592–599. doi: 10.1016/j.sbi.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mast SW, Moremen KW. Family 47 alpha-mannosidases in N-glycan processing. Methods Enzymol. 2006;415:31–46. doi: 10.1016/S0076-6879(06)15003-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henrissat B, Davies G. Structural and sequence-based classification of glycoside hydrolases. Curr Opin Struct Biol. 1997;7(5):637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez DS, Karaveg K, Vandersall-Nairn AS, Lal A, Moremen KW. Identification, expression, and characterization of a cDNA encoding human endoplasmic reticulum mannosidase I, the enzyme that catalyzes the first mannose trimming step in mammalian Asn-linked oligosaccharide biosynthesis. J Biol Chem. 1999;274(30):21375–21386. doi: 10.1074/jbc.274.30.21375. [DOI] [PubMed] [Google Scholar]
- 10.Vallée F, et al. Crystal structure of a class I alpha1,2-mannosidase involved in N-glycan processing and endoplasmic reticulum quality control. EMBO J. 2000;19(4):581–588. doi: 10.1093/emboj/19.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moremen K. 2000. α-Mannosidases in asparagine-linked oligosaccharide processing and catabolism. Oligosaccharides in Chemistry and Biology: A Comprehensive Handbook, eds Ernst B, Hart G, Sinay P (Wiley and Sons, New York), Vol II, Part 1, pp 81–117.
- 12.Lal A, Schutzbach JS, Forsee WT, Neame PJ, Moremen KW. Isolation and expression of murine and rabbit cDNAs encoding an alpha 1,2-mannosidase involved in the processing of asparagine-linked oligosaccharides. J Biol Chem. 1994;269(13):9872–9881. [PubMed] [Google Scholar]
- 13.Tremblay LO, Herscovics A. Characterization of a cDNA encoding a novel human Golgi alpha 1, 2-mannosidase (IC) involved in N-glycan biosynthesis. J Biol Chem. 2000;275(41):31655–31660. doi: 10.1074/jbc.M004935200. [DOI] [PubMed] [Google Scholar]
- 14.Karaveg K, et al. Mechanism of class 1 (glycosylhydrolase family 47) α-mannosidases involved in N-glycan processing and endoplasmic reticulum quality control. J Biol Chem. 2005;280(16):16197–16207. doi: 10.1074/jbc.M500119200. [DOI] [PubMed] [Google Scholar]
- 15.Lal A, et al. Substrate specificities of recombinant murine Golgi alpha1, 2-mannosidases IA and IB and comparison with endoplasmic reticulum and Golgi processing alpha1,2-mannosidases. Glycobiology. 1998;8(10):981–995. doi: 10.1093/glycob/8.10.981. [DOI] [PubMed] [Google Scholar]
- 16.Vallee F, Karaveg K, Herscovics A, Moremen KW, Howell PL. Structural basis for catalysis and inhibition of N-glycan processing class I α 1,2-mannosidases. J Biol Chem. 2000;275(52):41287–41298. doi: 10.1074/jbc.M006927200. [DOI] [PubMed] [Google Scholar]
- 17.Lobsanov YD, et al. Structure of Penicillium citrinum alpha 1,2-mannosidase reveals the basis for differences in specificity of the endoplasmic reticulum and Golgi class I enzymes. J Biol Chem. 2002;277(7):5620–5630. doi: 10.1074/jbc.M110243200. [DOI] [PubMed] [Google Scholar]
- 18.Van Petegem F, Contreras H, Contreras R, Van Beeumen J. Trichoderma reesei alpha-1,2-mannosidase: Structural basis for the cleavage of four consecutive mannose residues. J Mol Biol. 2001;312(1):157–165. doi: 10.1006/jmbi.2001.4946. [DOI] [PubMed] [Google Scholar]
- 19.Thompson AJ, et al. The reaction coordinate of a bacterial GH47 α-mannosidase: A combined quantum mechanical and structural approach. Angew Chem Int Ed Engl. 2012;51(44):10997–11001. doi: 10.1002/anie.201205338. [DOI] [PubMed] [Google Scholar]
- 20.Tempel W, et al. Structure of mouse Golgi α-mannosidase IA reveals the molecular basis for substrate specificity among class 1 (family 47 glycosylhydrolase) α1,2-mannosidases. J Biol Chem. 2004;279(28):29774–29786. doi: 10.1074/jbc.M403065200. [DOI] [PubMed] [Google Scholar]
- 21.Karaveg K, Moremen KW. Energetics of substrate binding and catalysis by class 1 (glycosylhydrolase family 47) α-mannosidases involved in N-glycan processing and endoplasmic reticulum quality control. J Biol Chem. 2005;280(33):29837–29848. doi: 10.1074/jbc.M505130200. [DOI] [PubMed] [Google Scholar]
- 22.Schutzbach JS, Forsee WT. Calcium ion activation of rabbit liver alpha 1,2-mannosidase. J Biol Chem. 1990;265(5):2546–2549. [PubMed] [Google Scholar]
- 23.Zhu Y, et al. Mechanistic insights into a Ca2+-dependent family of alpha-mannosidases in a human gut symbiont. Nat Chem Biol. 2010;6(2):125–132. doi: 10.1038/nchembio.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coutinho PM, Henrissat B. 1999. Carbohydrate-active enzymes: An integrated database approach. Recent Advances in Carbohydrate Bioengineering, eds Gilbert HJ, Davies G, Henrissat B, Svensson B (The Royal Society of Chemistry, Cambridge, UK), pp 3–12.
- 25.Numao S, Kuntz DA, Withers SG, Rose DR. Insights into the mechanism of Drosophila melanogaster Golgi alpha-mannosidase II through the structural analysis of covalent reaction intermediates. J Biol Chem. 2003;278(48):48074–48083. doi: 10.1074/jbc.M309249200. [DOI] [PubMed] [Google Scholar]
- 26.Evans CH. Interesting and useful biochemical properties of lanthanides. Trends Biochem Sci. 1983;8:445–449. [Google Scholar]
- 27.Dimicoli JL, Bieth J. Location of the calcium ion binding site in porcine pancreatic elastase using a lanthanide ion probe. Biochemistry. 1977;16(25):5532–5537. doi: 10.1021/bi00644a022. [DOI] [PubMed] [Google Scholar]
- 28.Dux L, Taylor KA, Ting-Beall HP, Martonosi A. Crystallization of the Ca2+-ATPase of sarcoplasmic reticulum by calcium and lanthanide ions. J Biol Chem. 1985;260(21):11730–11743. [PubMed] [Google Scholar]
- 29.Fitzpatrick LA. Differences in the actions of calcium versus lanthanum to influence parathyroid hormone release. Endocrinology. 1990;127(2):711–715. doi: 10.1210/endo-127-2-711. [DOI] [PubMed] [Google Scholar]
- 30.Gomez JE, Birnbaum ER, Darnall DW. The metal ion acceleration of the conversion of trypsinogen to trypsin. Lanthanide ions as calcium ion substitutes. Biochemistry. 1974;13(18):3745–3750. doi: 10.1021/bi00715a020. [DOI] [PubMed] [Google Scholar]
- 31.Guo S, et al. Inhibition mechanism of lanthanum ion on the activity of horseradish peroxidase in vitro. Spectrochim Acta A Mol Biomol Spectrosc. 2010;75(2):936–940. doi: 10.1016/j.saa.2009.11.033. [DOI] [PubMed] [Google Scholar]
- 32.Switzer ME. The lanthanide ions as probes of calcium ion binding sites in biological systems. Sci Prog. 1978;65(257):19–30. [PubMed] [Google Scholar]
- 33.Wilkins AL, et al. Metal-binding studies for a de novo designed calcium-binding protein. Protein Eng. 2002;15(7):571–574. doi: 10.1093/protein/15.7.571. [DOI] [PubMed] [Google Scholar]
- 34.Xue J, et al. Sugar-metal ion interactions: The coordination behaviors of lanthanum with erythritol. Carbohydr Res. 2012;361:12–18. doi: 10.1016/j.carres.2012.07.026. [DOI] [PubMed] [Google Scholar]
- 35.Furie BC, Furie B, Gottlieb AJ, Williams WJ. Activation of bovine factor X by the venom coagulant protein of Vipera russelli: Complex formation of the activation fragments. Biochim Biophys Acta. 1974;365(1):121–132. doi: 10.1016/0005-2795(74)90256-6. [DOI] [PubMed] [Google Scholar]
- 36.Marquis JK, Black EE. Activation and inactivation of bovine caudate acetylcholinesterase by trivalent cations. Biochem Pharmacol. 1985;34(4):533–538. doi: 10.1016/0006-2952(85)90186-8. [DOI] [PubMed] [Google Scholar]
- 37.Wang L, Lu A, Lu T, Ding X, Huang X. Interaction between lanthanum ion and horseradish peroxidase in vitro. Biochimie. 2010;92(1):41–50. doi: 10.1016/j.biochi.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 38.Shannon RD. Revised effective ionic radii and systematic studies of interatomic distances in halides and chalcogenides. Acta Crystallogr A. 1976;32:751–767. [Google Scholar]
- 39.Terrier C, Vitorge P, Gaigeot MP, Spezia R, Vuilleumier R. Density functional theory based molecular dynamics study of hydration and electronic properties of aqueous La(3+) J Chem Phys. 2010;133(4):044509. doi: 10.1063/1.3460813. [DOI] [PubMed] [Google Scholar]
- 40.Evans CH. Biochemistry of the Lanthanides. Plenum; New York, NY: 1990. [Google Scholar]
- 41.Hsieh P, Rosner MR, Robbins PW. Selective cleavage by endo-beta-N-acetylglucosaminidase H at individual glycosylation sites of Sindbis virion envelope glycoproteins. J Biol Chem. 1983;258(4):2555–2561. [PubMed] [Google Scholar]
- 42.Hubbard SC. Regulation of glycosylation. The influence of protein structure on N-linked oligosaccharide processing. J Biol Chem. 1988;263(36):19303–19317. [PubMed] [Google Scholar]
- 43.Thaysen-Andersen M, Packer NH. Site-specific glycoproteomics confirms that protein structure dictates formation of N-glycan type, core fucosylation and branching. Glycobiology. 2012;22(11):1440–1452. doi: 10.1093/glycob/cws110. [DOI] [PubMed] [Google Scholar]
- 44.Trimble RB, Maley F, Chu FK. GlycoProtein biosynthesis in yeast. Protein conformation affects processing of high mannose oligosaccharides on carboxypeptidase Y and invertase. J Biol Chem. 1983;258(4):2562–2567. [PubMed] [Google Scholar]
- 45.Hang I, et al. Analysis of site-specific N-glycan remodeling in the endoplasmic reticulum and the Golgi. Glycobiology. 2015;25(12):1335–1349. doi: 10.1093/glycob/cwv058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pritchard LK, et al. Glycan clustering stabilizes the mannose patch of HIV-1 and preserves vulnerability to broadly neutralizing antibodies. Nat Commun. 2015;6:7479. doi: 10.1038/ncomms8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stewart-Jones GB, et al. Trimeric HIV-1-Env structures define glycan shields from clades A, B, and G. Cell. 2016;165(4):813–826. doi: 10.1016/j.cell.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murray AN, et al. Enhanced aromatic sequons increase oligosaccharyltransferase glycosylation efficiency and glycan homogeneity. Chem Biol. 2015;22(8):1052–1062. doi: 10.1016/j.chembiol.2015.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen W, et al. Structural and energetic basis of carbohydrate-aromatic packing interactions in proteins. J Am Chem Soc. 2013;135(26):9877–9884. doi: 10.1021/ja4040472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thieker DF, Hadden JA, Schulten K, Woods RJ. 3D implementation of the symbol nomenclature for graphical representation of glycans. Glycobiology. 2016;26(8):786–787. doi: 10.1093/glycob/cww076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kwan EM, Boraston AB, McLean BW, Kilburn DG, Warren RA. N-Glycosidase-carbohydrate-binding module fusion proteins as immobilized enzymes for protein deglycosylation. Protein Eng Des Sel. 2005;18(10):497–501. doi: 10.1093/protein/gzi055. [DOI] [PubMed] [Google Scholar]
- 52.Franco-Fraguas L, et al. Preparative purification of soybean agglutinin by affinity chromatography and its immobilization for polysaccharide isolation. J Chromatogr B Analyt Technol Biomed Life Sci. 2003;790(1-2):365–372. doi: 10.1016/s1570-0232(03)00086-2. [DOI] [PubMed] [Google Scholar]
- 53.Sutton CW, O’Neill JA. Preparation of glycopeptides. Methods Mol Biol. 1997;64:73–79. doi: 10.1385/0-89603-353-8:73. [DOI] [PubMed] [Google Scholar]
- 54.Saha SK, Brewer CF. Determination of the concentrations of oligosaccharides, complex type carbohydrates, and glycoproteins using the phenol-sulfuric acid method. Carbohydr Res. 1994;254:157–167. doi: 10.1016/0008-6215(94)84249-3. [DOI] [PubMed] [Google Scholar]
- 55.Otwinowski Z, et al. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 56.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 58.Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12(5):279–284. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 59.Haynes WM. CRC Handbook of Chemistry and Physics. 95th Ed CRC; Boca Raton, FL: 2015. [Google Scholar]