Abstract
The neuregulins (NRGs) are a family of four structurally related growth factors that are expressed in the developing and adult brain. NRG-1 is essential for normal heart formation and has been implicated in the development and maintenance of both neurons and glia. NRG-2 was identified on the basis of its homology to NRG-1 and, like NRG-1, is expressed predominantly by neurons in the central nervous system. We have generated mice with the active domain of NRG-2 deleted in an effort to characterize the biological function of NRG-2 in vivo. In contrast to the NRG-1 knockout animals, NRG-2 knockouts have no apparent heart defects and survive embryogenesis. Mutant mice display early growth retardation and reduced reproductive capacity. No obvious histological differences were observed in the major sites of NRG-2 expression. Our results indicate that in vivo NRG-2 activity differs substantially from that of NRG-1 and that it is not essential for normal development in utero.
The neuregulin (NRG) family of growth factors comprises numerous glycoproteins that arise via alternative splicing from four distinct genes (NRG-1, NRG-2, NRG-3, and NRG-4). These proteins have a similar mosaic-like structure with a number of recognizable protein domains. The prototypic member of this family, NRG-1, was independently identified as a Schwann cell mitogen (12, 19), an activator of erbB2 (15, 29), and an inducer of acetylcholine receptor synthesis at the neuromuscular junction (7). The NRG-1 gene gives rise to as many as 15 different splice forms, generated by alternative splicing and differential promoter usage. These variants are also known as heregulin, acetylcholine receptor-inducing activity (ARIA), sensory and motor neuron-derived differentiation factor, Neu differentiation factor, and glial growth factor. More recently, a number of genes encoding proteins related to NRG-1 have been identified. These genes, named NRG-2 (also identified as Don-1 and NTAK) (2-4, 14), NRG-3 (32), and NRG-4 (13), were discovered by using sequences from NRG-1 to either search nucleotide databases or perform low-stringency hybridization and/or PCR.
Of the four NRG genes, NRG-1 and NRG-2 are the most closely related, although all members possess a 50-amino-acid region with homology to epidermal growth factor. This domain alone is sufficient for receptor binding and activation (2-4, 15, 32). The epidermal growth factor-like domains of NRGs 1 to 4 are 40 to 48% similar to each other in pairwise comparisons. Despite this low homology, all the NRG epidermal growth factor-like domains bind to either ErbB4 or ErbB3, albeit with different affinities (16). The epidermal growth factor-like domains of NRG-1 and NRG-2 each have two variants, designated α and β, arising from alternative exon usage, which also exhibit differential receptor binding affinities (16).
The majority of studies examining NRG activity have focused on NRG-1, and indeed numerous in vitro activities have been ascribed to the various forms of NRG-1 (for a review, see reference 1). Targeted disruptions of the NRG-1, erbB2, and erbB4 genes have also revealed new roles for NRGs. These mutants died in utero between embryonic day 10 (E10) and E11 as a result of almost identical heart defects (8, 17, 20). In contrast, the majority of mice deficient in ErbB3 died at E13.5 (20% survive to birth), also exhibiting lethal heart defects (6, 24). Although all of these mutant mice died midgestation, abnormalities within the nervous system were still apparent. NRG-1 and erbB2 null mice have reduced numbers of cranial sensory neurons (17, 20), and erbB4 mutants exhibit aberrant targeting of cranial sensory and motor axons (8, 11). In addition, NRG-1 knockout mice have reduced numbers of Schwann cell precursors and spinal cord oligodendrocytes (27).
Genetic rescue of the heart phenotype of erbB2 knockout mice has also revealed lethal peripheral nervous system defects (22, 30). While these mice have provided valuable information on the functions of NRG-ErbB interactions in early development, the early lethality has prevented the study of these factors both in later neural development and in the adult nervous system. Interestingly, the different phenotypes observed in the hindbrain of NRG-1 and erbB4 knockouts is highly suggestive of distinct activities for other ErbB receptor ligands. Unfortunately, very little is known about the function of NRG-2, -3, and -4. NRG-2 is expressed throughout the developing nervous system and is also found in the embryonic heart, lung, and bladder (2, 3). In the adult brain, however, NRG-2 expression is confined to three areas, the cerebellum (granule cells and purkinje cells), dentate gyrus (granule cells), and olfactory bulb (granule cells) (2-4). The restricted expression pattern of NRG-2 in the adult differs substantially from that of NRG-1, suggesting specific biological roles for each gene.
In the present study, we have begun to examine the in vivo functions of NRG-2 via the generation of a mouse strain with a targeted deletion of part of the NRG-2 gene. The resultant homozygous mice are viable and do not display any overlapping phenotypes with previously reported NRG-1/erbB knockout mice. The work reported here details the first characterization of NRG-2 knockout mice.
MATERIALS AND METHODS
Construction of the NRG-2 targeting vector and generation of mutant mice.
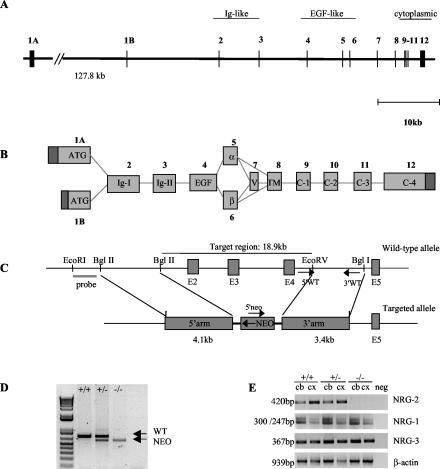
Genomic clones containing exons 2 to 7 were obtained from a mouse E129 bacterial artificial chromosome library (Stratagene). Sequencing and restriction mapping were used to identify suitable regions for subcloning into the targeting vector. These sequences have since been confirmed by the mouse genome sequencing project (28). A 4.1-kb BglII fragment from intron 2 was blunt-end ligated into the NotI site of the pBluescript-neo-tk vector as the 5′ arm. EcoRV and BglI were used to generate a 3.4-kb 3′ arm from intron 5, which was then blunt-end ligated into the ClaI site of the targeting vector. Homologous recombination of the targeting construct with the wild-type gene results in deletion of exons 2 to 4, as depicted in Fig. 1C.
FIG. 1.
Targeted disruption of the mouse NRG-2 gene. (A) Schematic representation of the genomic structure of the mouse NRG-2 gene. Exons are numbered 1 to 12 and are represented by vertical lines. The two alternative start exons are designated 1A and 1B. (B) Numerous transcripts of NRG-2 are generated via alternative splicing events. Exons are numbered as above and labeled according to the structural region that they encode. (C) Targeting strategy. The structures of the wild-type allele and the disrupted allele are shown. Exon numbering is consistent with A. The primers used for routine genotyping are represented by arrows. (D) PCR genotyping with primers for wild-type and recombinant alleles. A 0.75-kb fragment was generated from the wild-type allele, and a 0.6-kb fragment was generated from the mutant allele. (E) Total RNA prepared from adult cerebellum and cortex/hippocampus was reverse transcribed and then used for PCR amplification with primers specific for NRG-2, NRG-1, and NRG-3. No amplification product for NRG-2 was observed in the −/− mice. ATG, start methionine; Ig, immunoglobulin-like domain; EGF, epidermal growth factor-like domain; α and β, alternative epidermal growth factor sequence; V, variable region; TM, transmembrane domain; C1 to C4, cytoplasmic region; +/+, wild-type; +/−, heterozygous; −/−, homozygous knockout; cb, cerebellum; cx, cortex/hippocampus.
The targeting construct was linearized with SacII and electroporated into 129/SvJ embryonic stem (ES) cells. Resistant cells were selected in the presence of G418 and ganciclovir as described previously (5). A total of 384 clones were picked and DNA was isolated. A 1.8-kb EcoRI/BglII fragment generated from sequence immediately upstream of the 5′ arm was used to screen ES cell genomic DNA for correct homologous recombination by Southern blotting and hybridization. Two independent ES cell lines were injected into C57BL/6 blastocysts, which were subsequently transferred into pseudo-pregnant females to generate chimeric offspring. Chimeras were bred with C57BL/6 females to generate heterozygotes.
Mice were housed in a 12-h light-dark cycle facility with free access to food and water. All procedures were carried out in accordance with institutional policies following granting of ethics approval from the Royal Perth Hospital Ethics Committee.
Genotyping of mutant mice.
The genotypes of mutant mice were determined by PCR and confirmed by Southern blot analysis of genomic DNA prepared from tail biopsy samples. Briefly, tail samples were incubated for 16 h in lysis buffer (50 mM Tris [pH 8.0], 100 mM EDTA, 100 mM NaCl, 1% sodium dodecyl sulfate, 1 mg of proteinase K per ml), followed by isopropanol precipitation. Routine genotyping through PCR was performed with ≈0.3 μg of genomic DNA with specific oligonucleotide primers for the NRG-2 wild-type allele designed from intron 5 (F1, 5′-CCTCACAGAGATGTCCTAG-3′, and R1, 5′-CAAATGGTAGACGTGAGTCC-3′). For the targeted allele, a primer was designed from within the neomycin resistance gene (F2, 5′-CCAGTCATAGCCGAATAGC-3′). The three primers were used in a multiplex PCR with the following amplification conditions: 95°C for 5 min, and 30 cycles of 95°C for 1 min, 55°C for 2 min, and 72°C for 2 min. Amplification products were resolved on a 2% agarose gel.
Reverse transcription-PCR analysis.
Total cellular RNA was isolated from adult cerebellum and cortex with RNAzol reagent (Invitrogen). Three micrograms of this RNA was transcribed into cDNA (Superscript II; Invitrogen) before being amplified with primers specific for NRG-1 (forward, 5′-CTGACTCTGGAGAATATATG-3′, and reverse, 5′-CTTGCACAAGTATCTTGAGG-3′). NRG-2 (forward, 5′-ATGAAGAGCCAGACAGGAGAGG-3′, and reverse, 5′-GATTCTTCGGACAGAGATGT-3′), NRG-3 (forward, 5′-CTACCAAGGAGTCCGTTGTGA-3′, and reverse, 5′-TTGACTCCATTATTTTCTTCA-3′), and β-actin (forward, 5′-GTGACGAGGCCCAGAGCAAGAG-3′, and reverse, 5′ AGGGGCCGGACTCATCGTACTC-3′). The following amplification protocol was used: 95°C for 2 min, and 30 cycles of 95°C for 30 s, 58°C for 1 min, and 72°C for 1 min. Products were electrophoresed through a 2% agarose gel.
Histopathology.
Embryos were removed from pregnant dams 13 days after fertilization (E13), fixed in 4% paraformaldehyde, dehydrated, and embedded in paraffin. Serial 10-μm sections were stained with hematoxylin-eosin according to standard protocols. Dissected brains from 7-day-old pups were fixed in 4% paraformaldehyde and processed as above.
Prior to collection of adult tissue, animals were deeply anaesthetized, and cardiac perfusion with 4% paraformaldehyde was performed. Whole brains were removed, postfixed overnight, dehydrated, and embedded in paraffin. Serial 10-μm sections were stained with cresyl violet according to standard protocols. Immunostaining of sections was performed with primary antibodies specific for neuroD (G-20) (Santa Cruz Biotechnology) or calbindin D-28 (Sigma) following antigen unmasking (Vector Laboratories). For neuroD staining, slides were incubated overnight with primary antibody (1:400 dilution) in phosphate-buffered saline with 3% bovine serum albumin and 0.1% Triton X-100. For calbindin staining, slides were incubated for 1 h with primary antibody (1:3,000 dilution) in phosphate-buffered saline with 1.5% fetal calf serum. Appropriate secondary antibodies were used in conjunction with the ABC staining system (Santa Cruz Biotechnology) according to the manufacturer's instructions.
RESULTS AND DISCUSSION
Characterization of the mouse NRG-2 gene and generation of NRG-2-deficient mice.
The mouse NRG-2 gene spans ≈180 kb on the proximal end of chromosome 18 and contains 12 exons, with two alternatively spliced first exons, 1A and 1B (Fig. 1A). Exon 1A is located approximately 128 kb upstream of the alternative 5′exon 1B, itself 15.5 kb away from exon 2. The remaining exons are spread over 33 kb. We isolated and sequenced bacterial artificial chromosome clones that contained exons 2 to 7. The full sequence of this region of mouse chromosome 18 has since been confirmed by NCBI and the Mouse Genome Sequencing Consortium (accession no. NT_078847) (28). The gene structure is similar to that reported for the human NRG-2 gene (25, 31). Exons 1A and 1B account for all reported N-terminal sequence variants of NRG-2 (2-4, 14), although it cannot be ruled out that additional exons may be found further upstream, similar to the NRG-1 gene, where a 5′ ATG-containing exon is found 5 Mb away from other coding exons (26).
To determine the physiological role of NRG-2 in vivo, we used homologous recombination in ES cells to disrupt part of the NRG-2 gene (Fig. 1C). Because of the size of the mouse NRG-2 gene and the presence of alternative start exons more than 127 kb apart, we chose to target a region of the gene essential for receptor binding. A similar strategy was successfully used to eliminate the NRG-1 gene in mice (20). The targeting vector was designed to replace exons 2 to 4 with the neomycin resistance gene. The deleted region (≈19 kb) encodes the immunoglobulin domain and most of the epidermal growth factor-like domain. Independent ES cell clones with confirmed homologous recombination were injected into blastocysts to generate chimeras. Male chimeras from two different ES cell clones were crossed with C57BL/6 females to establish strains with a mixed genetic background (129/SvJ × C57BL/6) heterozygous for the mutated allele. The data presented here are representative of one chimeric line, although both showed a similar phenotype. Homologous recombination in offspring was confirmed by both Southern blot analysis (data not shown) and PCR (Fig. 1D).
To confirm that NRG-2 mRNA was absent in the knockout mice, reverse transcription-PCR was performed with cDNA generated from cerebellum and cortex/hippocampus RNA. As can be seen in Fig. 1E, no NRG-2 product was amplified from knockout tissue. Primers specific for NRG-1 and NRG-3 were also used to ensure that expression of these genes was not inadvertently dysregulated. Two amplification products were obtained with the NRG-1 primers. Sequencing confirmed that these products correspond to alternative splice variants between the immunoglobulin-like domain and epidermal growth factor-like domain.
Genotyping of 117 offspring at birth from heterozygous intercrosses demonstrated Mendelian inheritance of the mutated allele (Table 1), indicating that disruption of NRG-2 did not result in embryonic lethality. Examination of genotype frequencies following weaning, however, revealed a skewing in the Mendelian ratio, caused by the death of ≈34% of NRG-2-deficient pups derived from heterozygous matings (Table 1). These deaths coincided with unexplained growth retardation during the first 6 weeks of life.
TABLE 1.
Numbers of wild-type, heterozygous and knockout micea
| Time | No. (%) of mice with genotype:
|
||
|---|---|---|---|
| +/+ | +/− | −/− | |
| Birth | 31 (27) | 53 (45) | 33 (28) |
| Weaning | 60 (28) | 112 (53) | 39 (18) |
At birth, a Mendelian distribution of wild-type, heterozygous, and homozygous knockout mice was observed following mating of heterozygous animals. This ratio was dramatically skewed at weaning (P28 to P35) due to the death of ∼34% of the knockout mice.
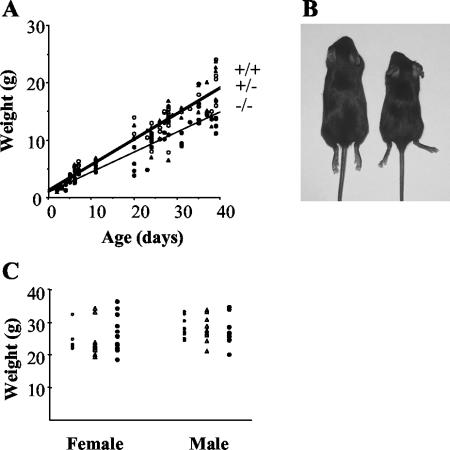
In the first few days of postnatal development, NRG-2-deficient pups were indistinguishable from their wild-type and heterozygous littermates in size, weight, and external appearance. By day 6 postbirth (P6), however, it became apparent that NRG-2 knockout pups were significantly smaller than their littermates (Fig. 2A and B). At P6, NRG-2−/− pups were ≈35% lighter than both wild-type and heterozygous littermates. Visual inspection of the pups revealed no obvious suckling or motor coordination abnormalities, and milk was present in the stomachs of the knockout animals. This growth retardation was observed throughout the weaning period (P28 to P35), and at P39 knockout animals were on average 7 g (35%) lighter than their littermates. Mutant mice that survived past weaning displayed “catch-up” growth and by 4 months were indistinguishable from littermate controls on the basis of weight (Fig. 2C). Adult mice have been monitored for over 18 months, and no differences in mortality compared to littermate controls have been observed.
FIG. 2.
Growth of NRG-2−/− mice. (A) The weights of wild-type (open circles, n = 51), heterozygous (triangles, n = 77), and knockout (solid circles, n = 51) mice at various ages were plotted. Each data point is representative of an individual mouse. Lines of best fit were drawn to better represent the growth retardation exhibited by knockout mice. The correlation coefficient of each line was 0.86 or greater. (B) Photograph of wild-type (left) and knockout (right) male littermates at P28, demonstrating the difference in size. (C) The weights of 4-month-old male and female mice revealed no significant difference between knockouts and wild-types or heterozygotes.
It has been reported that NRG-ErbB interactions are essential for normal sexual maturation in female mice (18, 23). This effect is thought to be due to signaling between neurons and astrocytes in the hypothalamus. To investigate the effect of NRG-2 on both male and female fertility, a number of cross-matings with wild-type mates were performed. These confirmed that both knockout males and females were fertile. Twenty-one litters from four NRG-2−/− females (mated with wild-type males) revealed that the interval of time between the addition of a wild-type male and the first litter was comparable between wild-type and knockout females, suggesting normal mating behavior. Similar observations were made when knockout males were mated with wild-type females. However, when knockout pairs were mated, there was a tendency towards smaller litters, and a significant reduction in pup survival was evident (P = 7 × 10−6, Table 2). The results of wild-type and homozygous mixed matings showed that both the litter size and average number of pups surviving per pregnancy were not significantly different from that observed in wild-type-only matings. These results do not point to a specific maternal or paternal effect but rather to a synergistic negative effect of two knockout parents.
TABLE 2.
Litter size and pup survival from breeding NRG-2−/− micea
| Mating (male × female) | No. of matings | Mean litter size (no. of pups) ± SE | Mean survival (%) ± SE |
|---|---|---|---|
| +/+ × +/+ | 15 | 7.6 ± 0.7 | 82 ± 7.2 |
| +/− × +/− | 109 | 6.2 ± 0.2 | 59 ± 4.0 |
| −/− × +/+ | 11 | 6.3 ± 0.6 | 74 ± 10.2 |
| +/+ × −/− | 21 | 7.4 ± 0.6 | 61 ± 8.5 |
| −/− × −/− | 15 | 4.6 ± 0.7* | 18 ± 9.0** |
Statistically significant (t test) difference between knockout matings and wild-type matings: *, P = 0.007; **, P = 7 × 10−6.
Histopathological analysis of NRG-2-deficient mice.
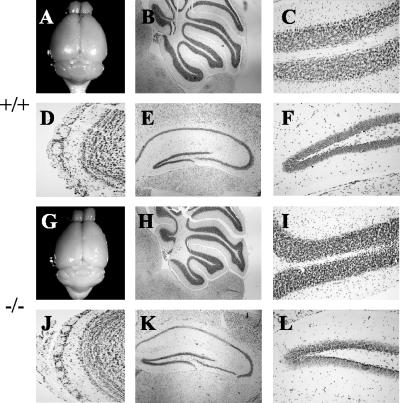
A macroscopic analysis of major organs revealed no obvious changes in the mutant mice compared to wild-type controls. We examined histological sections of heart, muscle, kidney, spleen, liver, lung, and gonads of adult mice of both sexes and saw no overt differences (data not shown). In the adult, NRG-2 is expressed primarily in the central nervous system. The gross morphology of brains taken from NRG-2 knockout animals of both sexes did not differ from that of wild-type or heterozygous brains (Fig. 3A and G). A closer examination of the regions of the brain where NRG-2 is expressed (cerebellum, hippocampus, and olfactory bulb) also revealed no major abnormalities (Fig. 3). From the cresyl violet-stained sections of the cerebellum, it is clear that foliation is normal and that the molecular layer, purkinje cell layer, and granule cell layer are all present. The magnitude of cells in the granule cell layer made it impractical to quantify cell numbers, but the thickness and density of the granule cell layer are commensurate with wild-type controls. These data show that deletion of the NRG-2 gene does not result in any major degeneration of cerebellar structure.
FIG. 3.
Gross histology of adult brains. Macroscopic analysis of brains from adult wild-type (A) and knockout (G) mice revealed no major abnormalities. Sagittal sections (stained with cresyl violet) of the cerebellum show normal foliation and cell layering in both wild-type (B and C) and knockout (H and I) mice. Sagittal sections of the olfactory bulb also showed no major abnormality (D and J). Coronal sections through the hippocampus revealed that the characteristic cytoarchitecture was unaltered (E, F, K, and L). Magnification: ×4 (B and H); ×10 (E and K); ×20 (C, D, F, I, J, and L).
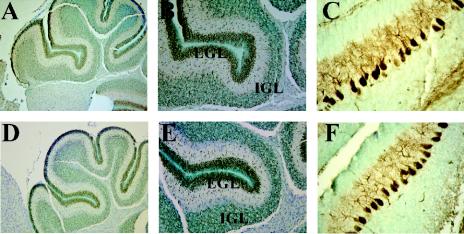
We also examined the cerebellum at P7, when granule cells migrate from the external germinal layer to form the internal granule cell layer. The cerebella of NRG-2−/− and control mice at this age were indistinguishable in size and morphology, and immunohistochemical staining with neuroD, a marker for migrating granule cells (21) indicated normal migration patterns of granule cell neurons (Fig. 4). Calbindin staining of adjacent sections revealed the presence of a defined purkinje cell layer with dendritic extensions similar to that observed in control animals.
FIG. 4.
Immunohistochemistry of P7 brain sections. Immunoperoxidase staining of P7 cerebellum from wild-type (A to C) and knockout (D to F) animals with neuroD (A, B, D, and E) and calbindin (C and F). Note the typical staining patterns of neuroD in granule cells in both the external germinal layer (EGL) and internal granule cell layer (IGL). Calbindin staining was specific to purkinje cells, and no obvious abnormalities were detected in NRG-2-deficient mice at this age. Magnification: ×10 (A and D); ×20 (B and E); ×40 (C and F).
Sections of the hippocampus revealed that the characteristic cytoarchitecture was unaltered in NRG-2 knockout mice and that the dentate gyrus was fully formed (Fig. 3). No obvious changes in granule cell numbers or density were observed. Similarly, layers within the olfactory bulb of adult NRG-2−/− mice appeared normal, with no noticeable changes to the granule cell population (Fig. 3). Taken together, these results show that NRG-2 is dispensable for the normal development of these regions.
Comparison of NRG-2-deficient mice with NRG-1 and erbB knockouts.
It has been speculated that NRG-2 and NRG-3 may be ErbB4 ligands in the developing hindbrain (8), where their activity may be essential for appropriate innervation of craniofacial ganglia from the rhombomeres. Whole-mount immunohistochemistry of E11 embryos with antibodies against neurofilament revealed that the organization and innervation of craniofacial ganglia V, VII, and VIII were unaltered in NRG-2-deficient embryos (data not shown). Specific defects in innervation of the hindbrain had previously been observed in erbB4 receptor knockouts that were not mimicked in NRG-1 knockouts (8, 11). Given our observations, it is unlikely that NRG-2 is a ligand for ErbB4 in this region of the hindbrain at that developmental stage. This is not surprising, as in situ hybridization studies have not detected NRG-2 transcripts in rhombomeres of E9 to E10.5 embryos (3).
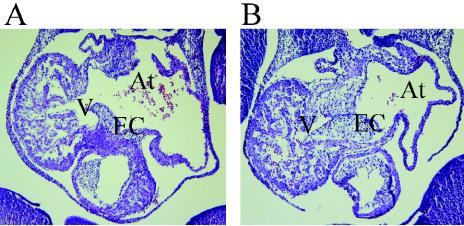
ErbB2, -3, and -4 signaling is also critical for normal heart development (6, 8, 17, 24). It has been reported that NRG-2 is expressed in the endothelial lining of the atrium during development (3). Examination of the heart at E12.5 revealed normal morphology of both the ventricle and atrium (Fig. 5). Ventricular trabeculae were present, and the endocardial cushion appeared normal. The atrial chambers of NRG-2−/− mice also appeared normal.
FIG. 5.
Histological analysis of developing heart in NRG-2−/− mice. Sagittal sections of wild-type (A) and knockout (B) E12.5 embryos illustrate normal ventricular trabeculation (V) and atrium formation (At). EC, endocardial cushion. Magnification, ×10.
These observations suggest that NRG-2 is not a significant ligand for the ErbB receptors in the heart or hindbrain during development of these tissues. However, it is possible that NRG-1 and NRG-3 are able to functionally compensate for the lack of NRG-2 during this period.
We have described the generation of NRG-2-deficient mice. In contrast to NRG-1, NRG-2 is not essential for normal development of the heart or craniofacial innervation, and knockout mice survive embryogenesis. However, following birth, NRG-2−/− pups display severe growth retardation and increased morbidity. Intriguingly, the chances of pup survival are further reduced when they are derived from a homozygous null mating. This phenomenon is suggestive of a more complex behavioral defect which warrants further study.
Deletion of NRG-2 also appears to have no major effects on cell specification and patterning in regions of the brain where it is normally expressed. This may in part be due to functional redundancy with other NRG ligands. During the later stages of embryogenesis (E16 to E18) NRG-1, NRG-2, and NRG-3 have overlapping patterns of expression. In the adult, however, patterns of expression become more restricted, especially in the cerebellum, where NRG-2 is the predominant NRG. ErbB receptors are expressed widely throughout the adult brain (9), including regions that also express NRG-2. It is therefore likely that NRG-2/ErbB signaling has an important role in the mature brain. Although no overt anatomical abnormalities were detected in these mice, it is possible that more subtle defects are present that may have important consequences for behavior and pharmacological responses. Indeed, it has been shown previously that heterozygous NRG-1 mutant mice and mice expressing a dominant negative form of ErbB4 exhibit a number of behavioral deficits (10, 26). Further studies will focus on a detailed examination of behavioral traits, including the reduced reproductive capacity of these mice, and on the analysis of potentially compensatory effects of other ErbB ligands.
Acknowledgments
We thank the following people for their assistance with this work: Carol Bartlett, Shane Meakins, Nora Murray, Anne Storrie, and the staff of the Royal Perth Hospital Research Centre.
This work is supported by the National Health and Medical Research Council (grants 254650 and 139167 to S.J.B. and 139165 to J.M.B.), Neurotrauma Research Program (Western Australia), and the Royal Perth Hospital Medical Research Foundation.
REFERENCES
- 1.Buonanno, A., and G. D. Fischbach. 2001. Neuregulin and ErbB receptor signaling pathways in the nervous system. Curr. Opin. Neurobiol. 11:287-296. [DOI] [PubMed] [Google Scholar]
- 2.Busfield, S. J., D. A. Michnick, T. W. Chickering, T. L. Revett, J. Ma, E. A. Woolf, C. A. Comrack, B. J. Dussault, J. Woolf, A. D. Goodearl, and D. P. Gearing. 1997. Characterization of a neuregulin-related gene, Don-1, that is highly expressed in restricted regions of the cerebellum and hippocampus. Mol. Cell. Biol. 17:4007-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carraway, K. L., J. L. Weber, M. J. Unger, J. Ledesma, N. Yu, M. Gassmann, and C. Lai. 1997. Neuregulin-2, a new ligand of ErbB3/ErbB4-receptor tyrosine kinases. Nature 387:512-516. [DOI] [PubMed] [Google Scholar]
- 4.Chang, H., D. J. Riese, W. Gilbert, D. F. Stern, and U. J. McMahan. 1997. Ligands for ErbB-family receptors encoded by a neuregulin-like gene. Nature 387:509-512. [DOI] [PubMed] [Google Scholar]
- 5.de Haan, J. B., C. Bladier, P. Griffiths, M. Kelner, R. D. O'Shea, N. S. Cheung, R. T. Bronson, M. J. Silvestro, S. Wild, S. S. Zheng, P. M. Beart, P. J. Hertzog, and I. Kola. 1998. Mice with a homozygous null mutation for the most abundant glutathione peroxidase, Gpx1, show increased susceptibility to the oxidative stress-inducing agents paraquat and hydrogen peroxide. J. Biol. Chem. 273:22528-22536. [DOI] [PubMed] [Google Scholar]
- 6.Erickson, S. L., K. S. O'Shea, N. Ghaboosi, L. Loverro, G. Frantz, M. Bauer, L. H. Lu, and M. W. Moore. 1997. ErbB3 is required for normal cerebellar and cardiac development: a comparison with ErbB2-and heregulin-deficient mice. Development 124:4999-5011. [DOI] [PubMed] [Google Scholar]
- 7.Falls, D. L., K. M. Rosen, G. Corfas, W. S. Lane, and G. D. Fischbach. 1993. ARIA, a protein that stimulates acetylcholine receptor synthesis, is a member of the neu ligand family. Cell 72:801-815. [DOI] [PubMed] [Google Scholar]
- 8.Gassmann, M., F. Casagranda, D. Orioli, H. Simon, C. Lai, R. Klein, and G. Lemke. 1995. Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature 378:390-394. [DOI] [PubMed] [Google Scholar]
- 9.Gerecke, K. M., J. M. Wyss, I. Karavanova, A. Buonanno, and S. L. Carroll. 2001. ErbB transmembrane tyrosine kinase receptors are differentially expressed throughout the adult rat central nervous system. J. Comp. Neurol. 433:86-100. [DOI] [PubMed] [Google Scholar]
- 10.Gerlai, R., P. Pisacane, and S. Erickson. 2000. Heregulin, but not ErbB2 or ErbB3, heterozygous mutant mice exhibit hyperactivity in multiple behavioral tasks. Behav. Brain Res. 109:219-227. [DOI] [PubMed] [Google Scholar]
- 11.Golding, J. P., P. Trainor, R. Krumlauf, and M. Gassmann. 2000. Defects in pathfinding by cranial neural crest cells in mice lacking the neuregulin receptor ErbB4. Nat. Cell Biol. 2:103-109. [DOI] [PubMed] [Google Scholar]
- 12.Goodearl, A. D., J. B. Davis, K. Mistry, L. Minghetti, M. Otsu, M. D. Waterfield, and P. Stroobant. 1993. Purification of multiple forms of glial growth factor. J. Biol. Chem. 268:18095-18102. [PubMed] [Google Scholar]
- 13.Harari, D., E. Tzahar, J. Romano, M. Shelly, J. H. Pierce, G. C. Andrews, and Y. Yarden. 1999. Neuregulin-4: a novel growth factor that acts through the ErbB-4 receptor tyrosine kinase. Oncogene 18:2681-2689. [DOI] [PubMed] [Google Scholar]
- 14.Higashiyama, S., M. Horikawa, K. Yamada, N. Ichino, N. Nakano, T. Nakagawa, J. Miyagawa, N. Matsushita, T. Nagatsu, N. Taniguchi, and H. Ishiguro. 1997. A novel brain-derived member of the epidermal growth factor family that interacts with ErbB3 and ErbB4. J. Biochem. 122:675-680. [DOI] [PubMed] [Google Scholar]
- 15.Holmes, W. E., M. X. Sliwkowski, R. W. Akita, W. J. Henzel, J. Lee, J. W. Park, D. Yansura, N. Abadi, H. Raab, G. D. Lewis, et al. 1992. Identification of heregulin, a specific activator of p185erbB2. Science 256:1205-1210. [DOI] [PubMed] [Google Scholar]
- 16.Jones, J. T., R. W. Akita, and M. X. Sliwkowski. 1999. Binding specificities and affinities of egf domains for ErbB receptors. FEBS Lett. 447:227-231. [DOI] [PubMed] [Google Scholar]
- 17.Lee, K. F., H. Simon, H. Chen, B. Bates, M. C. Hung, and C. Hauser. 1995. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature 378:394-398. [DOI] [PubMed] [Google Scholar]
- 18.Ma, Y. J., D. F. Hill, K. E. Creswick, M. E. Costa, A. Cornea, M. N. Lioubin, G. D. Plowman, and S. R. Ojeda. 1999. Neuregulins signaling via a glial erbB-2-erbB-4 receptor complex contribute to the neuroendocrine control of mammalian sexual development. J. Neurosci. 19:9913-9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchionni, M. A., A. D. Goodearl, M. S. Chen, O. Bermingham-McDonogh, C. Kirk, M. Hendricks, F. Danehy, D. Misumi, J. Sudhalter, K. Kobayashi, D. Wroblewski, C. Lynch, M. Baldassare, I. Hiles, J. B. Davis, J. J. Hsuan, N. F. Totty, M. Otsu, R. N. McBurney, M. D. Waterfield, P. Stroobant, and D. Gwynne. 1993. Glial growth factors are alternatively spliced erbB2 ligands expressed in the nervous system. Nature 362:312-318. [DOI] [PubMed] [Google Scholar]
- 20.Meyer, D., and C. Birchmeier. 1995. Multiple essential functions of neuregulin in development. Nature 378:386-390. [DOI] [PubMed] [Google Scholar]
- 21.Miyata, T., T. Maeda, and J. E. Lee. 1999. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 13:1647-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris, J. K., W. Lin, C. Hauser, Y. Marchuk, D. Getman, and K. F. Lee. 1999. Rescue of the cardiac defect in ErbB2 mutant mice reveals essential roles of ErbB2 in peripheral nervous system development. Neuron 23:273-283. [DOI] [PubMed] [Google Scholar]
- 23.Prevot, V., C. Rio, G. J. Cho, A. Lomniczi, S. Heger, C. M. Neville, N. A. Rosenthal, S. R. Ojeda, and G. Corfas. 2003. Normal female sexual development requires neuregulin-erbB receptor signaling in hypothalamic astrocytes. J. Neurosci. 23:230-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riethmacher, D., E. Sonnenberg-Riethmacher, V. Brinkmann, T. Yamaai, G. R. Lewin, and C. Birchmeier. 1997. Severe neuropathies in mice with targeted mutations in the ErbB3 receptor. Nature 389:725-730. [DOI] [PubMed] [Google Scholar]
- 25.Ring, H. Z., H. Chang, A. Guilbot, A. Brice, E. LeGuern, and U. Francke. 1999. The human neuregulin-2 (NRG2) gene: cloning, mapping and evaluation as a candidate for the autosomal recessive form of Charcot-Marie-Tooth disease linked to 5q. Hum. Genet. 104:326-332. [DOI] [PubMed] [Google Scholar]
- 26.Stefansson, H., E. Sigurdsson, V. Steinthorsdottir, S. Bjornsdottir, T. Sigmundsson, S. Ghosh, J. Brynjolfsson, S. Gunnarsdottir, O. Ivarsson, T. T. Chou, O. Hjaltason, B. Birgisdottir, H. Jonsson, V. G. Gudnadottir, E. Gudmundsdottir, A. Bjornsson, B. Ingvarsson, A. Ingason, S. Sigfusson, H. Hardardottir, R. P. Harvey, D. Lai, M. Zhou, D. Brunner, V. Mutel, A. Gonzalo, G. Lemke, J. Sainz, G. Johannesson, T. Andresson, D. Gudbjartsson, A. Manolescu, M. L. Frigge, M. E. Gurney, A. Kong, J. R. Gulcher, H. Petursson, and K. Stefansson. 2002. Neuregulin 1 and susceptibility to schizophrenia. Am. J. Hum. Genet. 71:877-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vartanian, T., G. Fischbach, and R. Miller. 1999. Failure of spinal cord oligodendrocyte development in mice lacking neuregulin. Proc. Natl. Acad. Sci. USA 96:731-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril, P. Agarwal, R. Agarwala, R. Ainscough, M. Alexandersson, P. An, S. E. Antonarakis, J. Attwood, R. Baertsch, J. Bailey, K. Barlow, S. Beck, E. Berry, B. Birren, T. Bloom, P. Bork, M. Botcherby, N. Bray, M. R. Brent, D. G. Brown, S. D. Brown, C. Bult, J. Burton, J. Butler, R. D. Campbell, P. Carninci, S. Cawley, F. Chiaromonte, A. T. Chinwalla, D. M. Church, M. Clamp, C. Clee, F. S. Collins, L. L. Cook, R. R. Copley, A. Coulson, O. Couronne, J. Cuff, V. Curwen, T. Cutts, M. Daly, R. David, J. Davies, K. D. Delehaunty, J. Deri, E. T. Dermitzakis, C. Dewey, N. J. Dickens, M. Diekhans, S. Dodge, I. Dubchak, D. M. Dunn, S. R. Eddy, L. Elnitski, R. D. Emes, P. Eswara, E. Eyras, A. Felsenfeld, G. A. Fewell, P. Flicek, K. Foley, W. N. Frankel, L. A. Fulton, R. S. Fulton, T. S. Furey, D. Gage, R. A. Gibbs, G. Glusman, S. Gnerre, N. Goldman, L. Goodstadt, D. Grafham, T. A. Graves, E. D. Green, S. Gregory, R. Guigo, M. Guyer, R. C. Hardison, D. Haussler, Y. Hayashizaki, L. W. Hillier, A. Hinrichs, W. Hlavina, T. Holzer, F. Hsu, A. Hua, T. Hubbard, A. Hunt, I. Jackson, D. B. Jaffe, L. S. Johnson, M. Jones, T. A. Jones, A. Joy, M. Kamal, E. K. Karlsson, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 29.Wen, D., E. Peles, R. Cupples, S. V. Suggs, S. S. Bacus, Y. Luo, G. Trail, S. Hu, S. M. Silbiger, R. B. Levy, et al. 1992. Neu differentiation factor: a transmembrane glycoprotein containing an EGF domain and an immunoglobulin homology unit. Cell 69:559-572. [DOI] [PubMed] [Google Scholar]
- 30.Woldeyesus, M. T., S. Britsch, D. Riethmacher, L. Xu, E. Sonnenberg-Riethmacher, F. Abou-Rebyeh, R. Harvey, P. Caroni, and C. Birchmeier. 1999. Peripheral nervous system defects in erbB2 mutants following genetic rescue of heart development. Genes Dev. 13:2538-2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamada, K., N. Ichino, K. Nishii, H. Sawada, S. Higashiyama, H. Ishiguro, and T. Nagatsu. 2000. Characterization of the human NTAK gene structure and distribution of the isoforms for rat NTAK mRNA. Gene 255:15-24. [DOI] [PubMed] [Google Scholar]
- 32.Zhang, D., M. X. Sliwkowski, M. Mark, G. Frantz, R. Akita, Y. Sun, K. Hillan, C. Crowley, J. Brush, and P. J. Godowski. 1997. Neuregulin-3 (NRG3): a novel neural tissue-enriched protein that binds and activates ErbB4. Proc. Natl. Acad. Sci. USA 94:9562-9567. [DOI] [PMC free article] [PubMed] [Google Scholar]