Significance
The liver is a common site for metastatic disease, and liver metastasis is strongly correlated with poor prognosis. Therefore, an understanding of how liver metastasis is regulated by the immune system is one of the most important issues in cancer immunology. Liver-resident immune cells may either suppress or promote liver metastasis. In this study, we show that Dectin-2 and macrophage C-type lectin, both of which belong to the C-type lectin family of innate receptors, is expressed on resident liver macrophages known as Kupffer cells and play critical roles in the suppression of liver metastasis by enhancing the cells’ phagocytotic activity against cancer cells. Our study sheds light on the protective role of Kupffer cells in liver metastasis with therapeutic implications.
Keywords: liver metastasis, C-type lectin receptor, Dectin-2, Kupffer cell, phagocytosis
Abstract
Tumor metastasis is the cause of most cancer deaths. Although metastases can form in multiple end organs, the liver is recognized as a highly permissive organ. Nevertheless, there is evidence for immune cell-mediated mechanisms that function to suppress liver metastasis by certain tumors, although the underlying mechanisms for the suppression of metastasis remain elusive. Here, we show that Dectin-2, a C-type lectin receptor (CLR) family of innate receptors, is critical for the suppression of liver metastasis of cancer cells. We provide evidence that Dectin-2 functions in resident macrophages in the liver, known as Kupffer cells, to mediate the uptake and clearance of cancer cells. Interestingly, Kupffer cells are selectively endowed with Dectin-2–dependent phagocytotic activity, with neither bone marrow-derived macrophages nor alveolar macrophages showing this potential. Concordantly, subcutaneous primary tumor growth and lung metastasis are not affected by the absence of Dectin-2. In addition, macrophage C-type lectin, a CLR known to be complex with Dectin-2, also contributes to the suppression of liver metastasis. Collectively, these results highlight the hitherto poorly understood mechanism of Kupffer cell-mediated control of metastasis that is mediated by the CLR innate receptor family, with implications for the development of anticancer therapy targeting CLRs.
Metastasis to distal organs is a critical pathological feature of cancer malignancies. Among the types of metastatic disease, liver metastasis occurs in many cancer types and is strongly correlated with poor prognosis (1). Colon cancer is notable, in that ∼80% of metastasis is confined to the liver (2). Therefore, the understanding on how liver metastasis is controlled is of general interest to both basic and clinical tumor immunology.
Innate immune cells, such as Kupffer cells and natural killer (NK) cells, are known to play critical roles in the regulation of liver metastasis (3). Numerous highly specialized cell types are distributed within the sinusoidal structure of the liver, with hepatocytes composing a major proportion of the total number (4). The cells of the innate immune system, including Kupffer cells, NK cells, NKT cells, and dendritic cells (DCs), also reside within the sinusoid, where they play a role in immunity (5). Kupffer cells are particularly critical to the maintenance of homeostasis, with their absence resulting in pathogen invasion and/or systemic inflammation (6). On the other hand, Kupffer cells also can contribute to pathogenies, as has been reported in such conditions as nonalcoholic fatty liver disease, in which the innate receptor Toll-like receptor (TLR) 4 plays a critical role (7). Thus, like many other components of the innate immune system, the appropriate functional activity of Kupffer cells is critical to the maintenance of a healthy organism.
Compared with other tissue macrophages, Kupffer cells have some unique features, such as their high phagocytotic ability (8). It also has been reported that Kupffer cells can directly kill cancer cells through the secretion of cytotoxic molecules, such as tumor necrosis factor (TNF)-α and reactive oxygen species, and Kupffer cells enhance antitumor responses mediated by other immune cells, such as NK cells (9). On the other hand, several reports have argued that Kupffer cells also have a protumorigenic effect through the production of inflammatory cytokines and chemokines, which contribute to extracellular matrix remodeling and angiogenesis (9). Thus, the actual role of Kupffer cells in liver metastasis has warranted further investigation.
Previously we showed that Dectin-1, a C-type lectin receptor (CLR) family member, plays a critical role in the suppression of tumor growth and metastasis by NK cells. This suppression is indirect, in that cancer cell recognition by Dectin-1 results in the receptor activation in DCs and macrophages, which in turn can enhance the tumoricidal activity of NK cells (10). That study prompted us to study whether other CLR family receptors manifest a similar or distinct antitumor function in the innate antitumor responses, particularly in the context of the regulation of metastasis. Dectin-2 was of particular interest because of its high sequence homology to Dectin-1 (11) and because, similarly if not identical to Dectin-1, it recognizes high-mannose carbohydrate structures presented on bacteria and fungi (12). Also of interest was the fact that stimulation of Dectin-2 induces phagocytosis through signaling by Fc receptor γ chain (FcRγ) (13).
Here we provide evidence for the antimetastatic function of Dectin-2 in Kupffer cells in the liver. We first show the enhancement of cancer cell metastasis in the livers of mice deficient in the Dectin-2 gene. We also report that among the liver-resident cells, Dectin-2 is dominantly expressed in Kupffer cells, and that the removal of these cells also results in enhanced metastasis. Interestingly, Kupffer cells engulf cancer cells, a process that is impaired by the absence of Dectin-2. Such Dectin-2–mediated activity is specific to Kupffer cells, because neither bone marrow-derived macrophages (BMDMs) nor alveolar macrophages engulf the same cancer cells in Dectin-2–dependent manner.
Furthermore, we also present evidence that macrophage C-type lectin (MCL; also known as Dectin-3), which is known to form a heterodimer with Dectin-2 (14), contributes to the suppression of liver metastasis by enhancing the phagocytotic activity of Kupffer cells, indicating that Dectin-2 and MCL cooperatively suppress liver metastasis. These findings shed light on the hitherto poorly understood mechanism of Kupffer cell-mediated control of liver metastasis and suggest the promising prospect of the manipulation of CLR-mediated antitumor responses for controlling liver metastasis.
Results
Selective Contribution of Dectin-2 to the Suppression of Liver Metastasis.
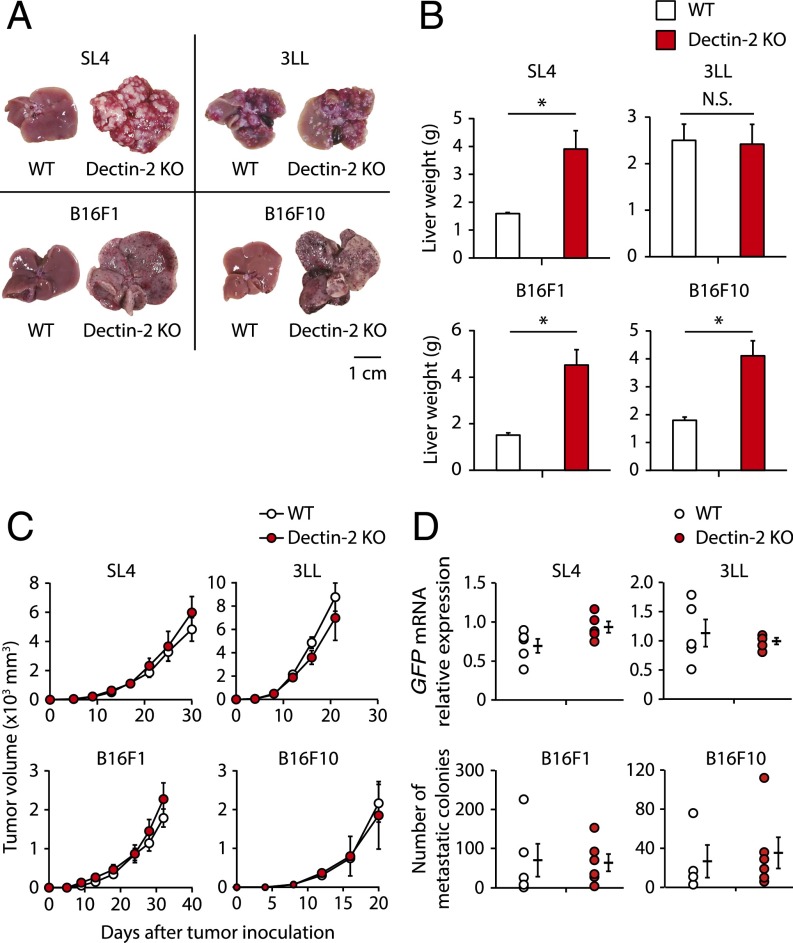
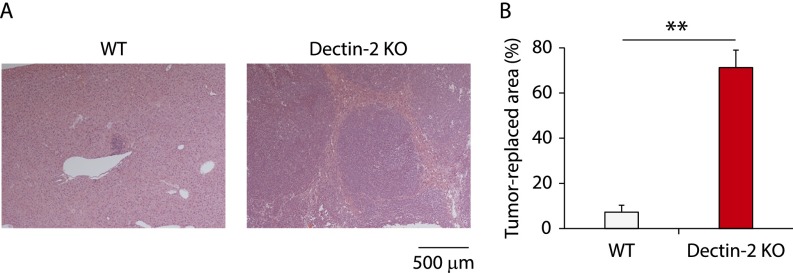
We first examined the contribution of Dectin-2 to antitumor immunity by evaluating wild-type (WT) and Dectin-2–deficient (Dectin-2 KO) mice for subcutaneous tumor growth, lung metastasis, and liver metastasis of colon carcinoma cell line SL4, Lewis lung carcinoma cell line 3LL, and melanoma cell line B16F1 and B16F10, all of which have been demonstrated to undergo metastasis in mice (15–18). We found that Dectin-2 KO mice showed more metastatic nodules in the liver compared with WT mice at 14 d after the intrasplenic inoculation with SL4, B16F1, or B16F10 cells, but not after inoculation with 3LL cells (Fig. 1A). Consistently, liver weight was significantly increased in the mice inoculated with these three cell lines (Fig. 1B). In the case of SL4 cell liver metastasis, the tumor-replaced area was approximately 10-fold larger in Dectin-2 KO mice compared with WT mice (Fig. S1).
Fig. 1.
Selective contribution of Dectin-2 to the suppression of liver metastasis. (A and B) SL4 cells (2 × 105 cells), 3LL cells (3 × 105 cells), B16F1 cells (1 × 106 cells), or B16F10 cells (2 × 105 cells) were inoculated into the spleens of WT and Dectin-2 KO mice. Fourteen days later, the livers were observed macroscopically (A) and liver weights were measured (B). (Scale bar: 1 cm.) (C) SL4 cells (2 × 105 cells), 3LL cells (5 × 105 cells), B16F1 cells (5 × 105 cells), or B16F10 cells (1 × 105 cells) were inoculated s.c. into WT and Dectin-2 KO mice, and tumor volumes were measured every 3 or 4 d. (D) SL4-GFP cells (3 × 105 cells), 3LL-GFP cells (1 × 106 cells), B16F1 cells (1 × 106 cells), or B16F10 cells (5 × 105 cells) were inoculated i.v. into WT and Dectin-2 KO mice, and the metastatic levels of SL4-GFP and 3LL-GFP cells were evaluated by quantifying GFP mRNA in the lung on day 12. The numbers of B16F1 and B16F10 colonies in the lung were counted at 14 d after inoculation. Data are shown as mean ± SEM. *P < 0.05. N.S., not significant.
Fig. S1.
Histological analysis for Dectin-2–mediated suppression of liver metastasis. At 14 d after WT and Dectin-2 KO mice were inoculated intrasplenically with SL4 cells (2 × 105 cells), liver sections were analyzed by H&E staining. Representative staining images (A) and tumor-replaced areas (B) are shown. (Scale bar: 500 μm.) Data are displayed as mean ± SEM. **P < 0.01.
Interestingly, notable differences between WT and Dectin-2 KO mice were not observed for subcutaneous tumor growth and lung metastasis of SL4, 3LL, B16F1, or B16F10 cancer cell lines (Fig. 1 C and D). These observations are quite different from what we previously reported for another CLR family member, Dectin-1, the absence of which affects the subcutaneous growth and lung metastasis of some cancer cells (10), and indicate that Dectin-2 is selectively involved in the suppression of liver metastasis.
Essential Role of Kupffer Cells in Dectin-2–Mediated Suppression of Liver Metastasis.
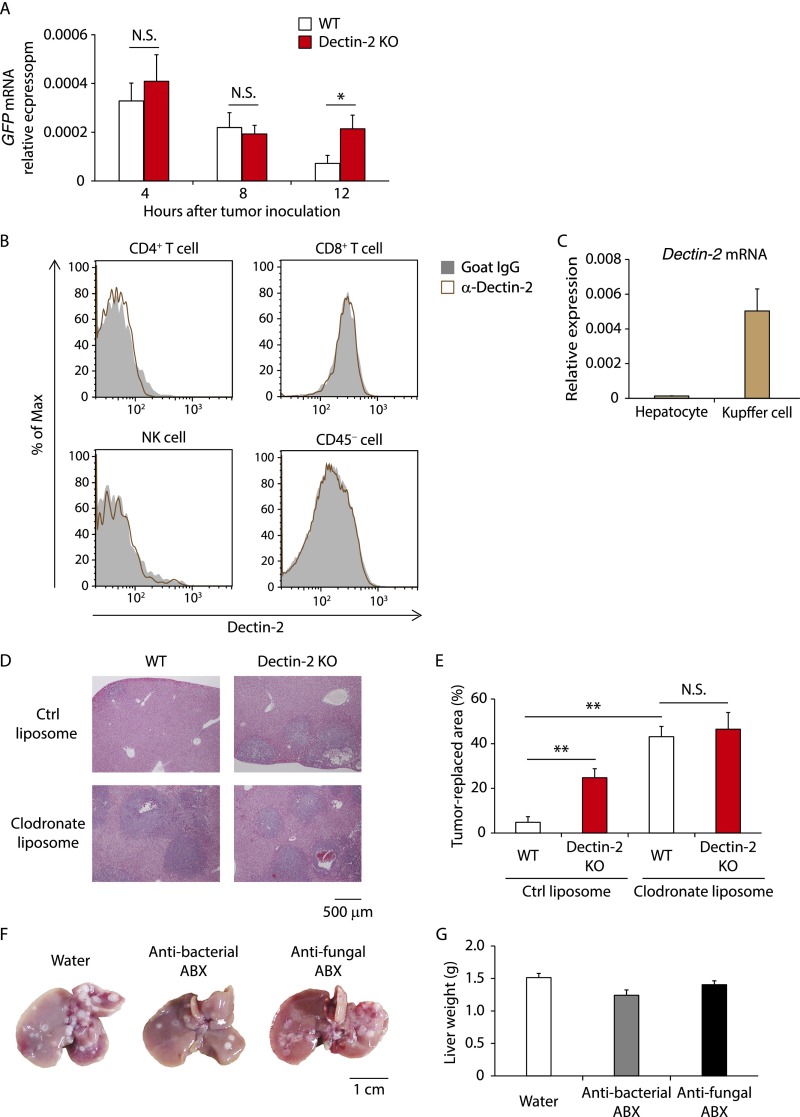
Cancer cell metastasis, which is initiated by cells leaving the primary site and entering the circulation, proceeds in a stepwise manner that includes, for example, cell adhesion to the endothelial wall of the target organ, extravasation, establishment of micrometastatic colonies, and subsequent tumor growth (1). Previous studies have shown that when cancer cells are intrasplenically inoculated, approximately one-half of the cells are extravasated by around 24 h after inoculation and undergo micrometastasis 4 d later (19, 20). To determine the stage of the metastatic event that is suppressed by Dectin-2, we inoculated SL4 cells expressing green fluorescent protein (GFP) into mice and monitored GFP mRNA in the liver at various time points thereafter. We found a marked increase in GFP mRNA levels in the livers of Dectin-2 KO mice compared with the livers of WT mice as early as 12 h after cancer cell inoculation (Fig. S2A). These results suggest that Dectin-2 mediates antitumor responses during an early phase of liver metastasis, perhaps before or during the extravasation step.
Fig. S2.
Critical role of Dectin-2–expressing Kupffer cells in cancer rejection at the liver. (A) SL4-GFP cells (2 × 105 cells) were inoculated into the spleen of WT and Dectin-2 KO mice. GFP mRNA levels in the livers were quantified at the indicated time points. (B) Dectin-2 expression on liver-residing cells was analyzed by flow cytometry. Representative histograms gated with NK cells (CD45+ NK1.1+ CD3ε− cells), CD4+ T cells (CD45+ NK1.1− CD3ε+ CD4+ cells), CD8+ T cells (CD45+ NK1.1− CD3ε+ CD8+ cells), and CD45− cells are shown. (C) Dectin-2 mRNA expression levels in isolated hepatocytes and Kupffer cells were measured by qRT-PCR. (D and E) WT and Dectin-2 KO mice were treated with control (ctrl) liposomes or clodronate liposomes on days −2 and 2 of intrasplenic inoculation of SL4 cells (2 × 105 cells). On day 10, liver sections were analyzed histologically with H&E staining. Representative staining images (D) and tumor-replaced areas (E) are shown. (Scale bar: 500 μm.) (F and G) Mice were treated with or without an antibacterial or antifungal antibiotic mixture via drinking water for 3 wk. SL4 cells (2 × 105 cells) were inoculated intrasplenically into the mice, and 14 d later, liver metastases were evaluated by macroscopic observation (F) and liver weights (G). The antibiotic treatments were continued until the livers were collected. (Scale bar: 1 cm.) Data are shown as mean ± SEM. *P < 0.05; **P < 0.01. N.S., not significant.
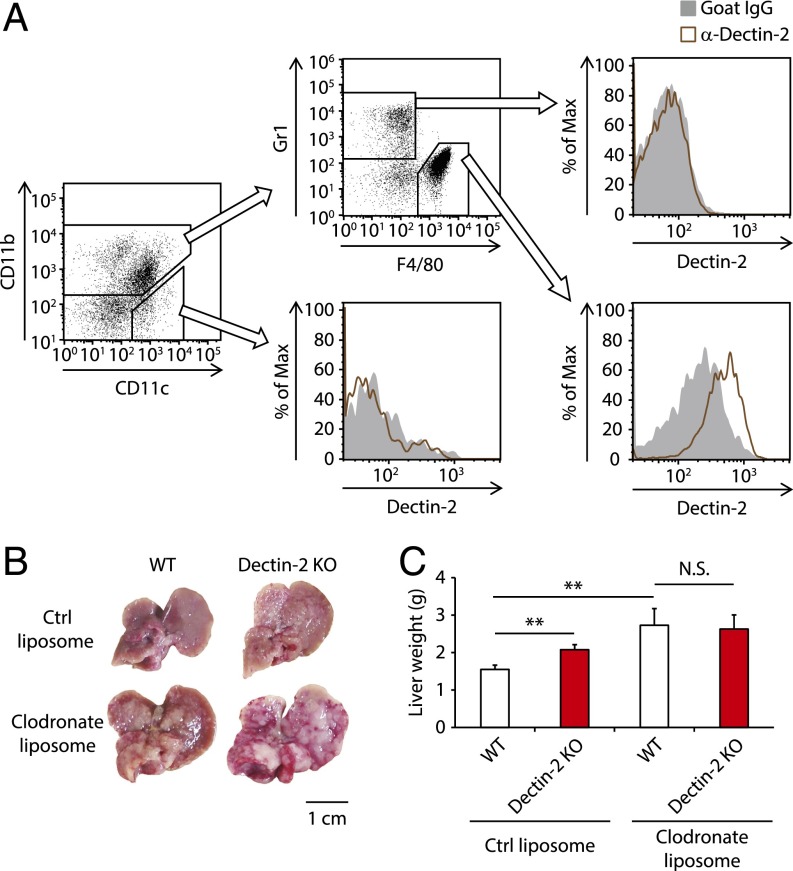
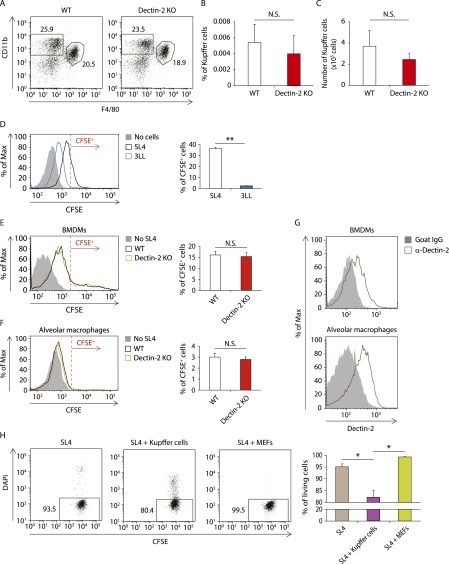
We next asked which cell types use Dectin-2 for the antitumor response. To address this question, we assessed cells residing in the liver for Dectin-2 expression by flow cytometry analysis of the cellular populations. As shown in Fig. 2A, CD11b+ F4/80+ cells expressed Dectin-2 at high levels, whereas neither CD11c+ cells nor CD11b+ Gr1+ cells expressed Dectin-2. In addition, Dectin-2 expression was not observed on NK cells, CD4+ T cells, CD8+ T cells, or CD45− T cells (Fig. S2B). Perhaps expectedly, hepatocytes showed little if any expression of Dectin-2 mRNA (Fig. S2C). These observations suggest that CD11b+ F4/80+ Kupffer cells are critically involved in the Dectin-2–mediated suppression of liver metastasis.
Fig. 2.
Requirement of Kupffer cells for the Dectin-2–mediated antitumor system against liver metastasis. (A) Dectin-2 expression on liver-residing cells was analyzed with flow cytometry. Plots gated on CD45+ cells are shown. (B and C) WT and Dectin-2 KO mice were treated with control (ctrl) liposomes or clodronate liposomes 2 d before and after intrasplenic inoculation of SL4 cells (2 × 105 cells). On day 10, the livers were collected. Macroscopic images of the liver (B) and liver weights (C) are shown. (Scale bar: 1 cm.) Data are displayed as mean ± SEM. **P < 0.01. N.S., not significant.
To further address the role of Dectin-2 in Kupffer cells in the suppression of metastasis, we treated WT and Dectin-2 KO mice with clodronate liposomes to deplete macrophages during the early phase of liver metastasis. We found that the administration of clodronate liposomes markedly enhanced SL4 metastasis in both WT and Dectin-2 KO mice, with no significant difference in tumor burden between them (Fig. 2 B and C and Fig. S2 D and E). This result indicates that Kupffer cells play a central role in Dectin-2–triggered antitumor immunity against liver metastasis.
It has been reported that gut commensal microbiota can influence tumor development in tissues distal from the intestine (21, 22). Given that Dectin-2 expression in intestinal tissues has been reported (23), and Dectin-2 is known to recognize high-mannose carbohydrate structures present on bacteria and fungi (12), we next examined whether commensal bacteria and fungi are involved in the suppression of liver metastasis by Dectin-2. We treated WT mice with antibacterial or antifungal antibiotics and then inoculated the mice with SL4 cells to examine liver metastasis. We found no significant increase in SL4 cell metastatic levels in the livers of mice treated with these antibiotics (Fig. S2 F and G), further supporting the idea that Dectin-2 in Kupffer cells acts directly on cancer cells.
Dectin-2–Dependent Phagocytotic Activity of Kupffer Cells Against Cancer Cells.
Flow cytometry analysis of the cell composition in livers revealed similar proportions and numbers of Kupffer cells in WT and Dectin-2 KO mice (Fig. S3 A–C). These data implicate Dectin-2 in the regulation of Kupffer cell function, but not in Kupffer cell expansion, in the suppression of liver metastasis.
Fig. S3.
The role of Dectin-2 in Kupffer cell differentiation and phagocytosis against cancer cells. (A–C) The composition of liver-residing cells in WT and Dectin-2 KO mice was analyzed by flow cytometry. (A) Representative plots gated with CD45+ cells. (B and C) The proportion (B) and number (C) of CD45+ CD11b+ F4/80+ Kupffer cells. (D) Kupffer cells (1 × 105 cells) were cocultured with or without CFSE-labeled SL4 cells or 3LL cells (0.25 × 105 cells) for 2 h, and the CFSE intensity in CD45+ CD11b+ F4/80+ cells was analyzed by flow cytometry. (Left) Representative histograms of CFSE level. Cells with a CFSE level exceeding the red line are identified as CFSE+ cells. (Right) Proportion of CFSE+ cells. The experiments were performed three times, with high reproducibility. (E and F) BMDMs (1 × 105 cells) (E) or alveolar macrophages (1 × 105 cells) (F) isolated from WT and Dectin-2 KO mice were cocultured with or without CFSE-labeled SL4 cells (0.25 × 105 cells) for 2 h. The CFSE intensity in CD45+ CD11b+ F4/80+ cells (E) or CD45+ CD11c+ F4/80+ cells (F) was analyzed by flow cytometry. (Left) Representative histograms of CFSE level. Cells with a CFSE level exceeding the red line are identified as CFSE+ cells. (Right) Proportion of CFSE+ cells. (G) Dectin-2 expression on BMDMs and alveolar macrophages were analyzed by flow cytometry. Representative histograms are shown. (H) CFSE-labeled SL4 cells (0.25 × 105 cells) were cocultured with Kupffer cells (1 × 105 cells) or MEF-mCherry cells (1 × 105 cells) for 12 h. The proportion of DAPI− cells among the CD11b− CFSE+ cells or mCherry− CFSE+ cells was analyzed by flow cytometry when SL4 cells were cultured in the presence of Kupffer cells or MEF-mCherry cells, respectively. Representative plots are shown. Data are shown as mean ± SEM. *P < 0.05; **P < 0.01. N.S., not significant.
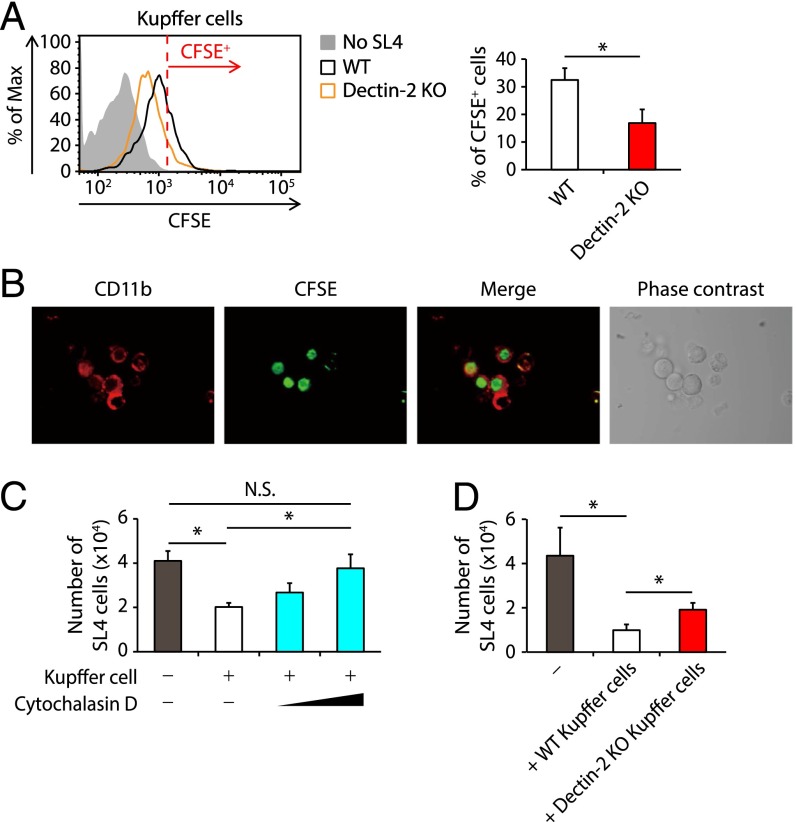
How does Dectin-2 function in Kupffer cells? Because Dectin-2 can trigger the engulfment of its ligand into cells (24, 25), we first asked whether Kupffer cells engulf cancer cells in a Dectin-2–dependent manner. Kupffer cells sorted from WT or Dectin-2 KO mice were cocultured with carboxyfluorescein diacetate succinimidyl ester (CFSE)-labeled SL4 cells, and then subjected to flow cytometry analysis. As shown in Fig. 3A, this coculturing of SL4 and Kupffer cells resulted in a significant increase in CFSE intensity in WT Kupffer cells, indicating engulfment of the SL4 cancer cells by Kupffer cells. Consistent with this, we observed SL4 cells engulfed by Kupffer cells by confocal microscopy and time-lapse imaging (Fig. 3B and Movie S1). Notably, the intensity of CFSE was significantly lower in the Dectin-2–deficient Kupffer cells (Fig. 3A). These results indicate that Dectin-2 enhances the phagocytotic activity of Kupffer cells against SL4 cancer cells. However, the fact that Dectin-2 deficiency did not completely abrogate this phagocytic activity suggests the involvement of additional molecule(s) in this process (Fig. 3A). Interestingly, 3LL cells, which underwent liver metastasis independently of Dectin-2 (Fig. 1 A and B), showed more marked resistance to the engulfment by Kupffer cells compared with SL4 cells (Fig. S3D).
Fig. 3.
Dectin-2–dependent engulfment and clearance of cancer cells by Kupffer cells. (A) Kupffer cells (1 × 105 cells) isolated from WT and Dectin-2 KO mice were cocultured with or without CFSE-labeled SL4 cells (0.25 × 105 cells) for 2 h. The CFSE intensity in CD45+ CD11b+ F4/80+ cells was analyzed by flow cytometry. (Left) Representative histograms of CFSE level. The cells with a CFSE level exceeding the red line were identified as CFSE+ cells. (Right) Proportion of CFSE+ cells. (B) Kupffer cells (1 × 105 cells) were cocultured with CFSE-labeled SL4 cells (0.25 × 105 cells) and after 2 h, the cells were observed by confocal microscopy. (C) CFSE-labeled SL4 cells (0.25 × 105 cells) were cultured in the presence or absence of Kupffer cells (1 × 105 cells) pretreated with DMSO or cytochalasin D, and the number of PI− CD45− CFSE+ cells was determined at 24 h after culturing. (D) CFSE-labeled SL4 cells (0.25 × 105 cells) were cultured in the presence or absence of Kupffer cells (1 × 105 cells) derived from WT and Dectin-2 KO mice, and the number of PI− CD45− CFSE+ cells was determined at 24 h after culturing. Data are shown as mean ± SEM. *P < 0.05. N.S., not significant.
We also analyzed the phagocytotic potential of BMDMs and alveolar macrophages, and found that neither of these cells showed any significant dependence on Dectin-2 for phagocytotic activity against SL4 cells (Fig. S3 E and F), even though both exhibited substantial levels of Dectin-2 expression (Fig. S3G). This observation is consistent with the foregoing data showing that subcutaneous tumor growth and lung metastasis are not affected by Dectin-2 deficiency (Fig. 1 C and D).
Interestingly, after SL4 cells were cocultured with Kupffer cells, we found a decreased proportion of cells negative for DAPI, an indicator of dead cells, among the unphagocytosed SL4 cells (Fig. S3H). This finding suggests that Kupffer cells may actively engulf living cancer cells. Indeed, the number of SL4 cells was reduced after coculturing with Kupffer cells in vitro, which was inhibited by the treatment of Kupffer cells with cytochalasin D, a phagocytosis inhibitor (Fig. 3C). Furthermore, a significantly higher number of SL4 cells was observed after cancer cells were cocultured with Dectin-2–deficient Kupffer cells (Fig. 3D), further supporting the role of Dectin-2 in the phagocytosis of cancer cells by Kupffer cells (26).
We next asked whether Dectin-2 can recognize a particular molecular structure on cancer cells. We previously showed that soluble Dectin-1 conjugated to human IgG1 Fc (Dectin-1–Fc) can bind to the surface of cancer cells (10). We then generated Dectin-2–Fc (soluble Dectin-2; sDectin-2) and evaluated its binding to SL4 cells. We were unable to detect any significant binding of sDectin-2 to SL4 cells, however (Fig. S4A). This finding may indicate either that the affinity of sDectin-2 is too low to allow detection of its binding or that Dectin-2 does not directly bind to the cells. In addition, because Dectin-2 recognizes carbohydrate structures on bacteria and fungi (12), we also examined the effects of N-glycosidase or O-glycosidase treatment of SL4 cells on the differential phagocytotic activity between WT and Dectin-2–deficient Kupffer cells. We found that the activity remained essentially unchanged after treatment with either of these glycosidases (Fig. S4B). Therefore, unlike the previous study showing the Dectin-1 recognition of N-glycan structures on cancer cells (10), N/O-glycan structures on SL4 cells are dispensable for the Dectin-2–mediated engulfment by Kupffer cells. As such, the nature of the Dectin-2–mediated recognition of cancer cells requires future investigation.
Fig. S4.
Analysis of cancer cell recognition by Dectin-2 and Dectin-2–mediated gene induction in response to cancer cells. (A) Binding of sDectin-2 to SL4 cells and zymosan particles was analyzed by flow cytometry. Representative histograms are shown. (B) Kupffer cells (1 × 105 cells) isolated from WT and Dectin-2 KO mice were cocultured with or without 0.25 × 105 CFSE-labeled SL4 cells pretreated with N-glycosidase or a combination of O-glycosidase and neuraminidase (10), and 2 h later, CFSE intensity in CD45+ CD11b+ F4/80+ cells was analyzed by flow cytometry. Representative histograms of CFSE levels and proportion of CFSE+ cells are shown. (C) Kupffer cells (1 × 105 cells) and SL4 cells (1 × 105 cells) were cultured together or separately, and mRNA levels of Il6, Il23a, Cxcl1, and Ccl2 at the indicated time points were quantified by qRT-PCR analysis. (D and E) Kupffer cells (1 × 105 cells) were isolated from WT and Dectin-2 KO mice and cultured together with or separately from SL4 cells (1 × 105 cells). Eight hours later, mRNA expression levels of Il6, Il23a, Cxcl1, and Ccl2 (D) or Tnf, Arg1, and Cd206 (E) were analyzed by qRT-PCR. Data are shown as mean ± SEM. *P < 0.05; **P < 0.01. N.S., not significant.
The foregoing observations raise the interesting question of whether Dectin-2 signaling is involved in the phagocytotic activity of Kupffer cells. Because Dectin-2 signaling is known to induce the expression of inflammatory cytokines (24, 25), we examined whether the interaction between Kupffer cells and cancer cells results in Dectin-2–dependent gene expression for various cytokine mRNAs by coculturing these cells. Although the expression levels of interleukin 6 (Il6), Il23a, chemokine (C-X-C motif) ligand 1 (Cxcl1), and chemokine (C-C motif) ligand 2 (Ccl2) mRNA were up-regulated by the coculture, Dectin-2 deficiency did not affect the mRNA expression levels (Fig. S4 C and D). We also examined the mRNA expression profile by microarray analysis (GEO accession no. GSE88809), and found no Dectin-2–dependent induction of any mRNA measured. These results lend support to the notion that Dectin-2 participates in the engulfment of cancer cells by Kupffer cells independently of gene induction.
Of final note, we also found no evidence of Dectin-2–dependent skewing of Kupffer cell polarization toward an M1 or M2 type in this experimental setting. In WT and Dectin-2–deficient Kupffer cells cocultured with SL4 cells, the mRNA expression signatures for M1 polarization (Il6, Il23a, and Tnf) and those for M2 polarization [arginases, liver (Arg1) and mannose receptor, C-type 1 (Mrc1; Cd206)] remained unchanged (Fig. S4 D and E). Although further clarification may be necessary, these observations suggest that Kupffer cells exert their antimetastatic function in vivo without their polarization.
Contribution of CLR Family Member MCL (Dectin-3) to the Suppression of Liver Metastasis.
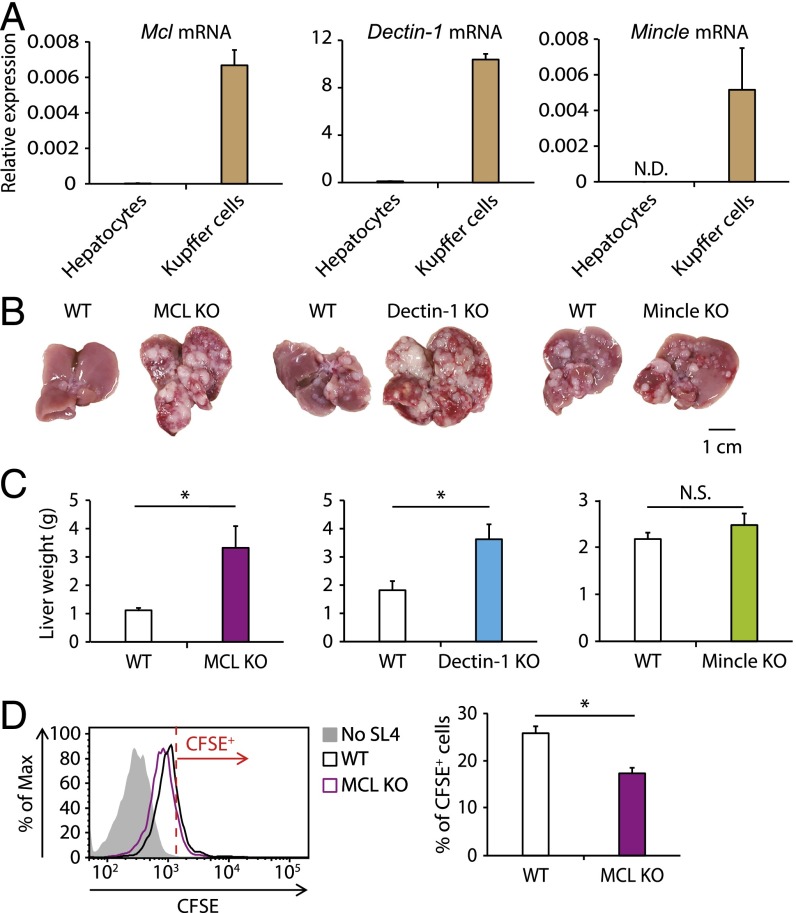
Previous studies have shown that Dectin-2’s ability to protect the host from fungal infection requires that it form a complex with another CLR member, MCL (or Dectin-3) (14). MCL regulates another CLR, Mincle, by enhancing its cell surface expression (27). Mincle signaling inhibits signal transduction downstream of Dectin-1 (28). Given our finding of mRNA expression for these three CLRs in Kupffer cells (Fig. 4A), we examined whether Dectin-1, MCL, and Mincle play roles in liver metastasis. Intrasplenic inoculation of SL4 cells into mice deficient in any of these CLR members revealed that a deficiency of MCL or Dectin-1, but not of Mincle, aggravated liver metastasis, indicating that MCL and Dectin-1 are also involved in the antimetastatic immune response in the liver (Fig. 4 B and C).
Fig. 4.
MCL-mediated uptake of cancer cells by Kupffer cells and suppression of liver metastasis. (A) Expression levels of Mcl, Dectin-1, and Mincle mRNAs in hepatocytes and Kupffer cells were analyzed by qRT-PCR. (B and C) SL4 cells (2 × 105 cells) were inoculated into the spleens of WT, MCL KO, Dectin-1 KO, and Mincle KO mice. On day 14, macroscopic images of livers were obtained (B), and livers were weighed (C). (Scale bar: 1 cm.) (D) Kupffer cells collected from WT and MCL KO mice were cocultured with or without CFSE-labeled SL4 cells (0.25 × 105 cells) for 2 h. The CFSE intensity in CD45+ CD11b+ F4/80+ cells was analyzed by flow cytometry. (Left) Representative histograms of CFSE levels. Cells with a CFSE level exceeding the red line were identified as CFSE+ cells. (Right) Proportion of CFSE+ cells. Data are displayed as mean ± SEM. *P < 0.05. N.S., not significant; N.D., not detected.
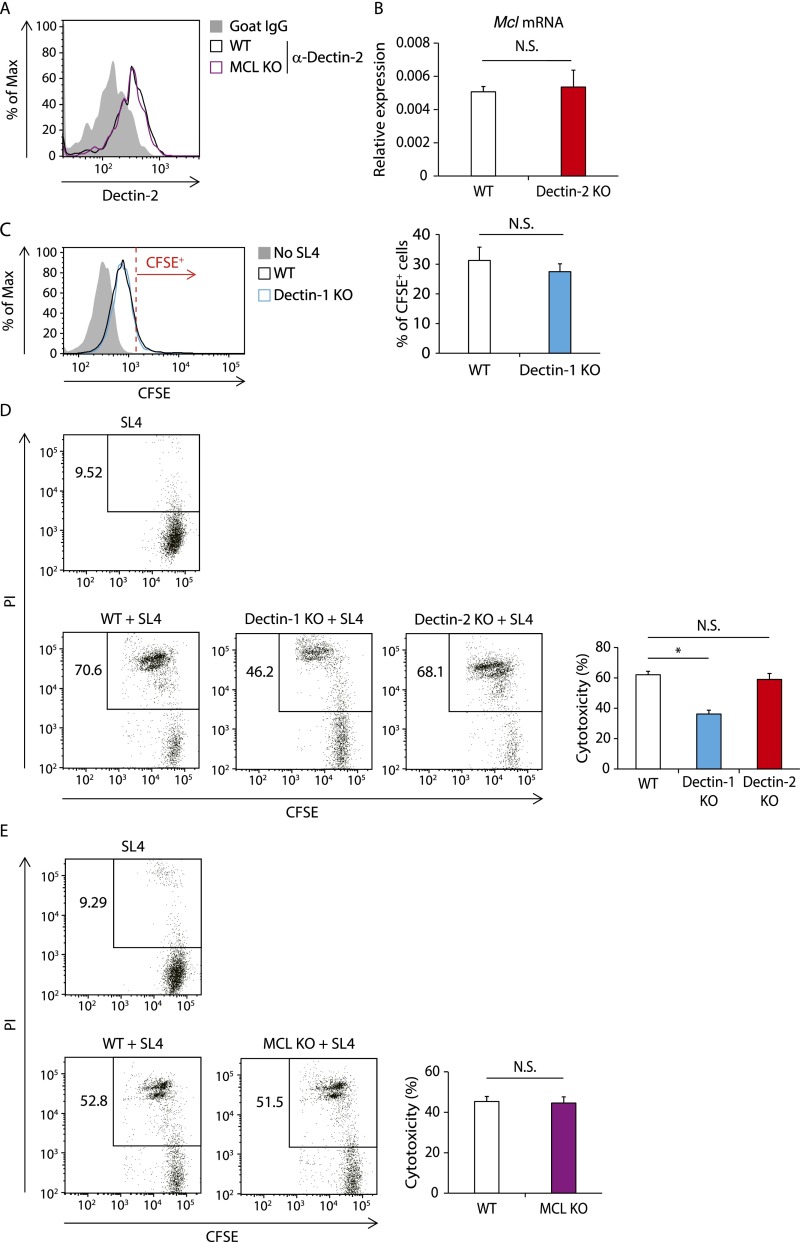
Consistent with the foregoing in vivo data, MCL-deficient Kupffer cells showed weaker engulfing activity for SL4 cells compared with WT cells (Fig. 4D). The decrease in SL4 cell uptake in MCL-deficient Kupffer cells was not due to the down-regulation of Dectin-2, given that Dectin-2 expression on MCL KO mouse-derived Kupffer cells was similar to that on WT Kupffer cells (Fig. S5A). It is unlikely that the impaired phagocytotic activity of Dectin-2–deficient Kupffer cells is caused by MCL down-regulation, because Mcl mRNA expression remained unaffected in Dectin-2–deficient Kupffer cells (Fig. S5B). Taken together, these results suggest that, similar to the antifungal innate response, MCL contributes to the engulfment of cancer cells by Kupffer cells by pairing with Dectin-2.
Fig. S5.
The roles of MCL and Dectin-1 in the regulation of Dectin-2 expression, phagocytotic activity of Kupffer cells, and cytotoxic activity of liver NPCs against cancer cells. (A) Dectin-2 expression on Kupffer cells isolated from WT and MCL KO mice were analyzed by flow cytometry. Representative histograms are shown. (B) Mcl mRNA expression levels in Kupffer cells isolated from WT and Dectin-2 KO mice were measured with qRT-PCR analysis. (C) Kupffer cells collected from WT and Dectin-1 KO mice were cocultured with or without CFSE-labeled SL4 cells (0.25 × 105 cells) for 2 h. The CFSE intensity in CD45+ CD11b+ F4/80+ cells was analyzed by flow cytometry. (Left) Representative histograms of CFSE level. Cells with a CFSE level exceeding the red line are identified as CFSE+ cells. (Right) Proportion of CFSE+ cells. (D and E) CFSE-labeled SL4 cells (1 × 105 cells) were cocultured with or without liver NPCs (5 × 106 cells) derived from WT, Dectin-1 KO, and Dectin-2 KO mice (D) or from WT and MCL KO mice (E). The proportion of PI+ population in SL4 cells was analyzed by flow cytometry at 4 h after the coculture. (Left) Representative plots gated with CD45− CFSE+ cells. (Right) Calculated cytotoxicity of SL4 cells. Data are displayed as mean ± SEM. *P < 0.05. N.S., not significant.
In contrast to MCL, although Kupffer cells expressed Dectin-1 mRNA at high levels (Fig. 4A), the phagocytotic activity of Kupffer cells against SL4 cells was not affected by Dectin-1 deficiency (Fig. S5C). Instead, Dectin-1 induced antitumor killing mediated by liver nonparenchymal cells (NPCs) (Fig. S5D). Indeed, NK cells are mainly responsible for the cytotoxic activity of liver NPCs (29), and this observation is consistent with our previous report showing that Dectin-1 signaling in DCs and macrophages enhances NK cell-mediated tumoricidal activity (10). Of note, neither Dectin-2 nor MCL contributed to this cytotoxic response of the NPCs (Fig. S5 D and E), further supporting the idea that, unlike Dectin-1–mediated antitumor responses, Dectin-2– and MCL-mediated antimetastatic responses are mediated by enhanced phagocytotic activity of Kupffer cells.
Discussion
Liver metastasis is a feature of many types of malignant cancers and is correlated with poor prognosis. Although innate immune cells have been identified as key players in the control of liver metastasis, innate immune receptors, such as TLRs, can promote tumor metastasis (30, 31). The role of CLRs in mediating antimetastatic responses has remained largely elusive. Here we provide evidence that Dectin-2, a CLR family member, promotes the engulfment and clearance of cancer cells by Kupffer cells and suppresses liver metastasis. This suppression mechanism is not operational for all cancer cells, however; the liver metastasis level for the 3LL cancer cell line remained unaffected in the absence of Dectin-2 (Fig. 1 A and B). It is worth recalling that Dectin-1 plays a critical role in controlling the metastasis of 3LL cancer cells through activation of NK cells (10). Thus, these results indicate the differential contribution of CLR members in the control of metastasis; that is, Dectin-2, together with MCL, contributes to the phagocytosis of cancer cells by Kupffer cells, whereas Dectin-1 contributes to the NK cell-mediated killing of cancer cells.
Interestingly, the Dectin-2–mediated antitumor response is selective for Kupffer cells, as demonstrated by our finding that Dectin-2 deficiency did not affect cancer cell engulfment by BMDMs and alveolar macrophages. Nor is Dectin-2 involved in the suppression of lung metastasis or subcutaneous tumor growth. Moreover, our results show that MCL, a heterodimeric counterpart of Dectin-2, is also critical for the suppression of liver metastasis, consistent with the idea that Dectin-2 and MCL function through heterodimeric complex formation (14). These findings provide insights into the mechanism of the antimetastatic response mediated by the CLR innate receptor family, i.e., the enhanced phagocytosis of cancer cells by Kupffer cells.
Analysis of the mechanism of Dectin-2–mediated suppression of metastasis revealed that Dectin-2 facilitates the engulfment of SL4 cells by Kupffer cells, but not by BMDMs or alveolar macrophages (Fig. 3A and Fig. S3 E and F). These results suggest that Dectin-2 functions as a phagocytotic receptor against cancer cells selectively in Kupffer cells. Kupffer cells exhibit greater phagocytotic ability than other types of macrophages, such as alveolar and peritoneal macrophages (8); thus, Kupffer cells may have some specific molecular features that enhance phagocytotic activity, with Dectin-2 involved in their unique phagocytotic function against cancer cells. Previous studies have shown that several CLR family members, including CLEC4G, CD207, and CLEC4F, are more highly expressed in Kupffer cells compared with other macrophages (32); therefore, it is possible that the Dectin-2–MCL complex cooperates with such CLRs, leading to the selective antitumor function in Kupffer cells. This is an interesting issue that will be addressed in future studies.
We further examined the antitumor potential of Kupffer cells and found data suggesting that the ability of living cell-engulfing Kupffer cells to eliminate cancer cells depends on Dectin-2 (Fig. 3 C and D and Fig. S3H). Previous studies have shown that macrophages engulf dead cells to trigger adaptive immune responses through antigen presentation to T cells, and regulate the tumor microenvironment by producing immune mediators in response to cancer (33). It was recently suggested that macrophages phagocytose living cancer cells and suppress tumor development (26, 34), although the molecular mechanism for this remains largely unknown. In view of our present study, it will be interesting to examine the contributions of CLR family members other than Dectin-2 and MCL to these antitumor responses.
Dectin-2–mediated phagocytosis against fungi is associated with the up-regulation of inflammatory cytokines, such as IL-6 and CXCL1 (24, 25). Nevertheless, our analysis of Dectin-2–dependent gene induction in response to cancer cells revealed that Dectin-2 did not regulate the mRNA expression of any tested genes, even Il6 and Cxcl1 (Fig. S4D). These observations suggest the interesting idea that the Dectin-2–triggered response in Kupffer cells against cancer cells is distinct from the responses reported previously. Supporting this notion, CARD9, a signaling molecule downstream of Dectin-2, was found to promote the liver metastasis of SL4 cells by manipulating the tumor microenvironment (35), which is inconsistent with our findings. Moreover, NLRP3, which is known to be activated dependently on Dectin-2 (13), enhances NK cell-mediated antitumor killing to suppress liver metastasis (36), although we found that Dectin-2 was dispensable for NK cell-mediated cytotoxicity against SL4 cells (Fig. S5D). Therefore, Dectin-2–triggered phagocytosis of cancer cells may be mediated by a unique signaling pathway.
The molecular nature of how Kupffer cells recognize some, but not all, cancer cells merits further examination. We could not identify the binding of sDectin-2 to cancer cells, and it is possible that, similar to the antifungal response (14), Dectin-2 may need to associate with MCL for the recognition of structure(s) associated with cancer cells. Alternatively, ligand recognition by the Dectin-2–MCL complex might not be required, in that the complex may augment the phagocytotic activity induced by other receptor molecules that recognize cancer cell-associated ligands. Clearly, this is an interesting issue for future studies.
At present, however, we cannot exclude the possibility that conventional signal transduction mediated by FcRγ downstream of Dectin-2 contributes to the Kupffer cell-mediated phagocytosis. FcRγ activation induces endocytosis through the phosphorylation of its own ITAM motif (37). Such posttranslational modifications of the FcRγ are also observed when an anti–Dectin-2 agonistic antibody is taken up by macrophages (25). Previous studies have shown that Mincle transduces FcRγ-mediated signaling pathways (13); however, our data indicate that Mincle is not involved in the control of liver metastasis (Fig. 4 B and C). Therefore, Dectin-2 may possess some selectivity in sensing cancer cells to induce antitumor responses.
Of note, it has been reported that treatment with an anti-CD47 antibody effectively suppresses in vivo tumor growth without coadministration of other chemotherapeutic agents (38, 39). Given that CD47 is a well-known “don’t eat me” signal for inhibiting phagocytosis (40), the uptake of cancer cells by phagocytes is a promising target for anticancer therapy, particularly when combined with a method that accelerates the cancer cell phagocytosis. As such, our findings may reveal a way to enhance the phagocytotic activity of macrophages by developing an agonist for Dectin-2 and/or other CLR family members for controlling metastasis in the liver and other organs.
Materials and Methods
Mice.
C57BL/6 mice were purchased from CLEA Japan. Clec7a−/− mice (Dectin-1 KO mice), Clec4n−/− mice (Dectin-2 KO mice), Clec4e−/− mice (Mincle KO mice), and Clec4d−/− mice (MCL KO mice) on a C57BL/6 background were generated as described previously (41–44). All animal experiments were approved and performed in accordance with guidelines of The University of Tokyo’s Animal Research Committee.
Cells.
Mouse colon carcinoma cell line SL4 was kindly provided by Dr. T. Irimura (The University of Tokyo). Mouse melanoma cell lines B16F1 and B16F10 and Lewis lung carcinoma cell line 3LL were maintained as described previously (10). GFP-transduced SL4 cells (SL4-GFP) and 3LL cells (3LL-GFP) were prepared as described previously (10). Mouse embryonic fibroblasts (MEFs) were retrovirally transfected with pmCherry-N1 vector (Clontech) and used as MEF-mCherry cells after selection with puromycin.
Liver Metastasis Model.
The liver metastasis model has been described previously (36). In brief, after the mouse was anesthetized, the spleen was exposed from a small incision in the left flank, and 2 × 105 SL4 cells, 3 × 105 3LL cells, 1 × 106 B16F1 cells, or 2 × 105 B16F10 cells were inoculated into the spleen. Five minutes later, the spleen was excised, and the incision was closed by clip. The mouse was killed on day 14, followed by the macroscopic observation of the liver and the measurement of liver weight. For the evaluation of tumor burden at early stage of liver metastasis, 2 × 105 SL4-GFP cells were inoculated, and liver specimens were collected 4, 8, and 12 h later. The GFP mRNA level in the liver was measured by quantitative RT-PCR (qRT-PCR).
Additional information is provided in SI Materials and Methods.
SI Materials and Methods
In Vivo Tumor Growth Model.
In this model, 2 × 105 SL4 cells, 5 × 105 3LL cells, 5 × 105 B16F1 cells, or 1 × 105 B16F10 cells were inoculated s.c. into mice. Tumor volume was calculated as ab2/2 by measuring longer axes (a) and shorter axes (b) of the tumor every 3 or 4 d.
Lung Metastasis Model.
Mice were inoculated i.v. with 3 × 105 SL4-GFP cells, 1 × 106 3LL-GFP cells, 1 × 106 B16F1 cells, or 5 × 105 B16F10 cells. The metastatic levels of 3LL-GFP or SL4-GFP cells were evaluated by quantifying GFP mRNA in the lung on day 12. In the mice bearing B16F1 or B16F10 cells, the number of metastatic colonies in the lung was counted at 14 d after inoculation.
Evaluation of Tumor-Replaced Area in Liver.
Metastasized livers were stored in PBS containing 4% paraformaldehyde. Hematoxylin and eosin (H&E) staining of the liver section was performed by Biopathology Institute Co., Ltd. Staining images obtained microscopically were analyzed with ImageJ, and the tumor-replaced area was calculated as tumor-replaced area (%) = 100 × (tumor area)/(whole liver area).
Preparation of Liver-Residing Cells.
For flow cytometry analysis of liver-residing cells, minced liver was digested by 750 μg/mL collagenase D (Roche Life Science), 40 μg/mL DNase I (Roche Life Science), and 500 μg/mL Dispase (Gibco) in RPMI 1640 for 1 h at 37 °C. In other experiments, liver was perfused with HBSS containing 0.2 mM EDTA and then with 1 mM CaCl2 and 750 μg/mL collagenase D in HBSS. After enzymatic digestion, liver was mashed on a 70-μm cell strainer (BD Biosciences).
Isolation of Hepatocytes, Liver NPCs, and Kupffer Cells.
Hepatocytes were isolated from liver-residing cells by centrifugation at 50 × g for 2 min. The supernatant was centrifuged twice more to remove hepatocytes. NPCs were obtained by centrifugation of the supernatant at 500 × g for 3 min, after which CD45+ CD11b+ F4/80+ Kupffer cells were isolated by cell sorting.
Flow Cytometry Analysis.
Anti-CD16/CD32 (93), -CD45.2 (104), -CD11b (M1/70), -CD11c (N418), –Gr-1 (RB6-8C5), -F4/80 (BM8), -CD49b (DX5), -CD3ε (145-2C11), -CD4 (RM4-5), and -CD8a (53-6.7) antibodies were purchased from Biolegend. Goat IgG and anti–Dectin-2a antibodies were obtained from Santa Cruz Biotechnology and R&D Systems, respectively. To inhibit binding of the antibodies to the Fc receptor, cells were incubated with anti-CD16/CD32 antibody before staining. Biotin-labeled antibodies were reacted with streptavidin-FITC (BD Pharmingen). Flow cytometry analysis was performed with an LSR Fortessa flow cytometer (BD Biosciences), and data were analyzed with FlowJo software (Tree Star). Kupffer cells, BMDMs, and alveolar macrophages were sorted using a FACSAria cell sorter (BD Biosciences) in the FACS core laboratory at the Center for Stem Cell Biology and Regenerative Medicine, Institute of Medical Science, The University of Tokyo.
Quantification of mRNA Expression.
Total RNA was extracted from tissue or cells with NucleoSpin RNA II (Macherey-Nagel) and reverse-transcribed into cDNA using PrimeScript RT Master Mix (Takara) according to the manufacturer’s instructions. qRT-PCR analysis was performed with a LightCycler 480 (Roche Life Science) by detecting the cDNA amplified with SYBR Premix Ex Taq (Takara). mRNA expression was normalized to the Gapdh mRNA level. Primer sequences were as follows: GFP forward, 5′-CTTCTTCAAGTCCGCCATGC-3′; GFP reverse, 5′-GTGTCGCCCTCGAACTTCAC-3′; Dectin-2 forward, 5′-TTCTTACTTCCTGGGTCTTTCG-3′; Dectin-2 reverse, 5′-AACACACCGCTCTTCTGGA-3′; Il6 forward, 5′-ACGATGATGCACTTGCAGAA-3′; Il6 reverse, 5′-GTAGCTATGGTACTCCAGAAGAC-3′; Il23a forward, 5′-TGGTTGTGACCCACAAGGAC-3′; Il23a reverse, 5′-CAGGCTCCCCTTTGAAGATG-3′; Cxcl1 forward, 5′-TGCAGACCATGGCTGGGATTCA-3′; Cxcl1 reverse, 5′-GTGTGGCTATGACTTCGGTTTGGG-3′; Ccl2 forward, 5′-GTTGGCTCAGCCAGATGCA-3′; Ccl2 reverse, 5′-AGCCTACTCATTGGGATCATCTTG-3′; Arg1 forward, 5′-GCAACCTGTGTCCTTTCTCC-3′; Arg1 reverse, 5′-GCAAGCCAATGTACACGATG-3′; Cd206 forward, 5′-AGAGCCCACAACAACTCCTG-3′; Cd206 reverse, 5′-TCTCGAGATTCAAACCACGTT-3′; Dectin-1 forward, 5′-CATCGTCTCACCGTATTAATGCAT-3′; Dectin-1 reverse, 5′-CCCAGAACCATGGCCCTT-3′; Mincle forward, 5′-CATCCCACCACACAGAGAGA-3′; Mincle reverse, 5′-GGTGATGAAACAGCCACTGA-3′; Mcl forward, 5′-GCTGGAAGAATCCCAAATGA-3′; Mcl reverse, 5′-AAAGCAAGCACTAAGGAACGAG-3′; Gapdh forward, 5′-CTCATGACCACAGTCCATGC-3′; Gapdh reverse, 5′-CACATTGGGGGTAGGAACAC-3′.
Depletion of Macrophages in Vivo.
For this analysis, 200 μL of plain control liposomes for Clophosome-N (neutral) or Clophosome-N clodronate liposomes (Neutral) (FormuMax) were injected i.v. into mice at 2 d before and after SL4 inoculation.
Clearance of Intestinal Bacteria and Fungi.
Drinking water containing antibacterial antibiotics (1 mg/mL ampicillin, 1 mg/mL neomycin, 1 mg/mL metronidazole, and 0.5 mg/mL vancomycin) or antifungal antibiotics (0.25 mg/mL fluconazole, 0.5 mg/mL amphotericin B, and 0.25 mg/mL terbinafine) were administered to C57BL/6 mice for 3 wk, after which the mice were inoculated with SL4 cells. The antibiotic treatments were continued until the mice were killed.
Preparation of Alveolar Macrophages.
Lung was vigorously minced and digested with collagenase D, DNase I, and Dispase as described above. After the lysis of red blood cells, CD45+ CD11c+ F4/80+ alveolar macrophages were isolated by cell sorting.
Evaluation of Macrophage Phagocytotic Activity.
BMDMs were prepared as described previously (45). First, 1 × 105 Kupffer cells, alveolar macrophages, or BMDMs were cocultured with 0.25 × 105 SL4 cells or 3LL cells labeled with CFSE using the CellTrace CFSE Cell Proliferation Kit (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. Then, 2 h later, the CFSE intensity in CD45+ CD11b+ F4/80+ cells was analyzed by flow cytometry when BMDMs or Kupffer cells were used. The phagocytotic activity of alveolar macrophages was evaluated by measuring the CFSE level in CD45+ CD11c+ F4/80+ cells. The engulfment of SL4 cells by Kupffer cells was further visualized on a glass-bottom 96-well plate by confocal microscopy with an Olympus FluoView FV1000 confocal microscope and time-lapse imaging with a Keyence BZ-X700 fluorescence microscope.
Analysis of Kupffer Cell-Mediated Clearance of Cancer Cells.
After labeling with CFSE, 0.25 × 105 SL4 cells were cultured with 1 × 105 Kupffer cells. In the experiment to inhibit phagocytosis, Kupffer cells were pretreated with 0.5 or 1 μM cytochalasin D (Sigma-Aldrich) or DMSO for 1 h. Kupffer cell-mediated clearance of living cancer cells was estimated by staining the cells with DAPI and evaluating the proportion of the DAPI− population among CD11b− CFSE+ SL4 cells. For the examination of cancer cell elimination by Kupffer cells, the number of CD45− CFSE+ cells in the propidium iodide (PI)-negative population was determined at 24 h after culturing.
Binding of sDectin-2 to Cancer Cells.
cDNA encoding the extracellular domain of Dectin-2 (amino acids 50–209) was amplified by PCR using forward primer 5′-GATATCGAGAAGACTATATGAACTTCA-3′ and reverse primer 5′-AGATCTTAGGTAAATCTTCTTCATTT-3′. The DNA fragments were inserted between the EcoRV and BglII sites of pFUSE-hIgG1-Fc2 (Invivogen). These vectors or pFUSE-hIgG1-Fc2 without any cDNA fragments were transfected into HEK293T cells with X-tremeGENE 9 DNA Transfection Reagent (Roche Life Science). On the next day, the cells were cultured in serum-free medium. The supernatants were collected 36 h later, followed by incubation with Protein G Sepharose Fast Flow (GE Healthcare) for 2 h at 4 °C. Soluble hIgG1-Fc (Fc) or hIgG1-Fc–conjugated Dectin-2 (sDectin-2) were eluted by 100 mM Glycine-HCl solution (pH 3.0) and dialyzed into PBS. SL4 cells were incubated with 50 μg/mL of Fc or sDectin-2 in Tris-buffered saline (TBS) (pH 8.0) containing 10 mM CaCl2 for 20 min at 4 °C. After staining with goat anti-human IgG Fc (DyLight 550) (Abcam), the cells were analyzed by flow cytometry.
N- and O-Glycosidase Treatment on Cancer Cells.
Following CFSE labeling, 1 × 105 of SL4 cells were treated for 1 h at 37 °C with 25 U/mL N-glycosidase F (Roche Life Science) or a combination of 25 mU/mL O-glycosidase (Roche Life Science) and 250 mU/mL neuraminidase (Roche Life Science) as described previously (10).
Gene Expression Profiles in Kupffer Cell–Cancer Cell Interaction.
Here 1 × 105 Kupffer cells were cocultured with 1 × 105 of SL4 cells for 0, 6, 8, 12, 18, or 24 h. Total RNA was extracted from whole the cells and analyzed with qRT-PCR or microarray analysis (Takara). As a control, the same number of Kupffer cells and SL4 cells were cultured separately, and the total RNA of each cell type was mixed at a 1:1 ratio. The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE88809).
Analysis for the Cytotoxic Activity of Liver NPCs.
Here 5 × 106 liver NPCs were cocultured with 1 × 105 CFSE-labeled SL4 cells for 4 h. The proportion of PI+ cells among CD45− CFSE+ SL4 cells was measured by flow cytometry. Cytotoxicity (%) was calculated as (percentage of the PI+ cells when SL4 cells were cocultured with liver NPCs) − (percentage of the PI+ cells when SL4 cells were cultured alone).
Statistical Analysis.
Each experiment was performed at least twice for in vivo analysis and three times for in vitro analysis. All of the results were highly reproducible. Data obtained from at least three individual samples were analyzed using a two-tailed unpaired Student’s t test, and P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Y. Miyake, M. Oh-hora, and K. Shibata for helpful comments and advice, and M. Sugahara, M. Taniguchi, T. Mizutani, S. Chiba, and members of the FACS core laboratory of the Institute of Medical Science, The University of Tokyo for technical assistance. This work is supported in part by Grant-In-Aid for Scientific Research (S) 15638461 from the Ministry of Education, Culture, Sports, Science, and Technology and by Grant 15656877 from the Japan Agency for Medical Research and Development. The Department of Molecular Immunology at The University of Tokyo is supported by BONAC Corporation and Kyowa Hakko Kirin Co., Ltd.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE88809).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617903113/-/DCSupplemental.
References
- 1.Valastyan S, Weinberg RA. Tumor metastasis: Molecular insights and evolving paradigms. Cell. 2011;147(2):275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manfredi S, et al. Epidemiology and management of liver metastases from colorectal cancer. Ann Surg. 2006;244(2):254–259. doi: 10.1097/01.sla.0000217629.94941.cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van den Eynden GG, et al. The multifaceted role of the microenvironment in liver metastasis: Biology and clinical implications. Cancer Res. 2013;73(7):2031–2043. doi: 10.1158/0008-5472.CAN-12-3931. [DOI] [PubMed] [Google Scholar]
- 4.Protzer U, Maini MK, Knolle PA. Living in the liver: Hepatic infections. Nat Rev Immunol. 2012;12(3):201–213. doi: 10.1038/nri3169. [DOI] [PubMed] [Google Scholar]
- 5.Heymann F, Tacke F. Immunology in the liver—from homeostasis to disease. Nat Rev Gastroenterol Hepatol. 2016;13(2):88–110. doi: 10.1038/nrgastro.2015.200. [DOI] [PubMed] [Google Scholar]
- 6.Bilzer M, Roggel F, Gerbes AL. Role of Kupffer cells in host defense and liver disease. Liver Int. 2006;26(10):1175–1186. doi: 10.1111/j.1478-3231.2006.01342.x. [DOI] [PubMed] [Google Scholar]
- 7.Bieghs V, Trautwein C. The innate immune response during liver inflammation and metabolic disease. Trends Immunol. 2013;34(9):446–452. doi: 10.1016/j.it.2013.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Laskin DL, Weinberger B, Laskin JD. Functional heterogeneity in liver and lung macrophages. J Leukoc Biol. 2001;70(2):163–170. [PubMed] [Google Scholar]
- 9.Paschos KA, Majeed AW, Bird NC. Role of Kupffer cells in the outgrowth of colorectal cancer liver metastases. Hepatol Res. 2010;40(1):83–94. doi: 10.1111/j.1872-034X.2009.00578.x. [DOI] [PubMed] [Google Scholar]
- 10.Chiba S, et al. Recognition of tumor cells by Dectin-1 orchestrates innate immune cells for anti-tumor responses. eLife. 2014;3:e04177. doi: 10.7554/eLife.04177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ariizumi K, et al. Cloning of a second dendritic cell-associated C-type lectin (dectin-2) and its alternatively spliced isoforms. J Biol Chem. 2000;275(16):11957–11963. doi: 10.1074/jbc.275.16.11957. [DOI] [PubMed] [Google Scholar]
- 12.McGreal EP, et al. The carbohydrate-recognition domain of Dectin-2 is a C-type lectin with specificity for high mannose. Glycobiology. 2006;16(5):422–430. doi: 10.1093/glycob/cwj077. [DOI] [PubMed] [Google Scholar]
- 13.Sancho D, Reis e Sousa C. Signaling by myeloid C-type lectin receptors in immunity and homeostasis. Annu Rev Immunol. 2012;30:491–529. doi: 10.1146/annurev-immunol-031210-101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu LL, et al. C-type lectin receptors Dectin-3 and Dectin-2 form a heterodimeric pattern-recognition receptor for host defense against fungal infection. Immunity. 2013;39(2):324–334. doi: 10.1016/j.immuni.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 15.Morimoto-Tomita M, Ohashi Y, Matsubara A, Tsuiji M, Irimura T. Mouse colon carcinoma cells established for high incidence of experimental hepatic metastasis exhibit accelerated and anchorage-independent growth. Clin Exp Metastasis. 2005;22(6):513–521. doi: 10.1007/s10585-005-3585-0. [DOI] [PubMed] [Google Scholar]
- 16.Cullen R, Germanov E, Shimaoka T, Johnston B. Enhanced tumor metastasis in response to blockade of the chemokine receptor CXCR6 is overcome by NKT cell activation. J Immunol. 2009;183(9):5807–5815. doi: 10.4049/jimmunol.0803520. [DOI] [PubMed] [Google Scholar]
- 17.Qi K, et al. Impact of cirrhosis on the development of experimental hepatic metastases by B16F1 melanoma cells in C57BL/6 mice. Hepatology. 2004;40(5):1144–1150. doi: 10.1002/hep.20421. [DOI] [PubMed] [Google Scholar]
- 18.Bezuhly M, et al. Role of activated protein C and its receptor in inhibition of tumor metastasis. Blood. 2009;113(14):3371–3374. doi: 10.1182/blood-2008-05-159434. [DOI] [PubMed] [Google Scholar]
- 19.Martin MD, et al. Rapid extravasation and establishment of breast cancer micrometastases in the liver microenvironment. Mol Cancer Res. 2010;8(10):1319–1327. doi: 10.1158/1541-7786.MCR-09-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritsma L, et al. Intravital microscopy through an abdominal imaging window reveals a pre-micrometastasis stage during liver metastasis. Sci Transl Med. 2012;4(158):158ra145. doi: 10.1126/scitranslmed.3004394. [DOI] [PubMed] [Google Scholar]
- 21.Rutkowski MR, et al. Microbially driven TLR5-dependent signaling governs distal malignant progression through tumor-promoting inflammation. Cancer Cell. 2015;27(1):27–40. doi: 10.1016/j.ccell.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iida N, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342(6161):967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor PR, et al. Dectin-2 is predominantly myeloid restricted and exhibits unique activation-dependent expression on maturing inflammatory monocytes elicited in vivo. Eur J Immunol. 2005;35(7):2163–2174. doi: 10.1002/eji.200425785. [DOI] [PubMed] [Google Scholar]
- 24.Ifrim DC, et al. Role of Dectin-2 for host defense against systemic infection with Candida glabrata. Infect Immun. 2014;82(3):1064–1073. doi: 10.1128/IAI.01189-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato K, et al. Dectin-2 is a pattern recognition receptor for fungi that couples with the Fc receptor γ chain to induce innate immune responses. J Biol Chem. 2006;281(50):38854–38866. doi: 10.1074/jbc.M606542200. [DOI] [PubMed] [Google Scholar]
- 26.Feng M, et al. Macrophages eat cancer cells using their own calreticulin as a guide: Roles of TLR and Btk. Proc Natl Acad Sci USA. 2015;112(7):2145–2150. doi: 10.1073/pnas.1424907112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyake Y, Masatsugu OH, Yamasaki S. C-type lectin receptor MCL facilitates Mincle expression and signaling through complex formation. J Immunol. 2015;194(11):5366–5374. doi: 10.4049/jimmunol.1402429. [DOI] [PubMed] [Google Scholar]
- 28.Wevers BA, et al. Fungal engagement of the C-type lectin mincle suppresses dectin-1–induced antifungal immunity. Cell Host Microbe. 2014;15(4):494–505. doi: 10.1016/j.chom.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 29.Cohen SA, Tzung SP, Doerr RJ, Goldrosen MH. Role of asialo-GM1–positive liver cells from athymic nude or polyinosinic-polycytidylic acid-treated mice in suppressing colon-derived experimental hepatic metastasis. Cancer Res. 1990;50(6):1834–1840. [PubMed] [Google Scholar]
- 30.Kim S, et al. Carcinoma-produced factors activate myeloid cells through TLR2 to stimulate metastasis. Nature. 2009;457(7225):102–106. doi: 10.1038/nature07623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu LX, et al. Platelets promote tumour metastasis via interaction between TLR4 and tumour cell-released high-mobility group box1 protein. Nat Commun. 2014;5:5256. doi: 10.1038/ncomms6256. [DOI] [PubMed] [Google Scholar]
- 32.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157(4):832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke C, Smyth MJ. Calreticulin exposure increases cancer immunogenicity. Nat Biotechnol. 2007;25(2):192–193. doi: 10.1038/nbt0207-192. [DOI] [PubMed] [Google Scholar]
- 34.Kopatz J, et al. Siglec-h on activated microglia for recognition and engulfment of glioma cells. Glia. 2013;61(7):1122–1133. doi: 10.1002/glia.22501. [DOI] [PubMed] [Google Scholar]
- 35.Yang M, et al. Tumor cell-activated CARD9 signaling contributes to metastasis-associated macrophage polarization. Cell Death Differ. 2014;21(8):1290–1302. doi: 10.1038/cdd.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dupaul-Chicoine J, et al. The Nlrp3 inflammasome suppresses colorectal cancer metastatic growth in the liver by promoting natural killer cell tumoricidal activity. Immunity. 2015;43(4):751–763. doi: 10.1016/j.immuni.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 37.Guilliams M, Bruhns P, Saeys Y, Hammad H, Lambrecht BN. The function of Fcγ receptors in dendritic cells and macrophages. Nat Rev Immunol. 2014;14(2):94–108. doi: 10.1038/nri3582. [DOI] [PubMed] [Google Scholar]
- 38.Weiskopf K, et al. CD47-blocking immunotherapies stimulate macrophage-mediated destruction of small-cell lung cancer. J Clin Invest. 2016;126(7):2610–2620. doi: 10.1172/JCI81603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu X, et al. CD47 blockade triggers T cell-mediated destruction of immunogenic tumors. Nat Med. 2015;21(10):1209–1215. doi: 10.1038/nm.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown GC, Neher JJ. Eaten alive! Cell death by primary phagocytosis: “Phagoptosis”. Trends Biochem Sci. 2012;37(8):325–332. doi: 10.1016/j.tibs.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 41.Saijo S, et al. Dectin-1 is required for host defense against Pneumocystis carinii but not against Candida albicans. Nat Immunol. 2007;8(1):39–46. doi: 10.1038/ni1425. [DOI] [PubMed] [Google Scholar]
- 42.Saijo S, et al. Dectin-2 recognition of α-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32(5):681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 43.Yamasaki S, et al. C-type lectin Mincle is an activating receptor for pathogenic fungus, Malassezia. Proc Natl Acad Sci USA. 2009;106(6):1897–1902. doi: 10.1073/pnas.0805177106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyake Y, et al. C-type lectin MCL is an FcRγ-coupled receptor that mediates the adjuvanticity of mycobacterial cord factor. Immunity. 2013;38(5):1050–1062. doi: 10.1016/j.immuni.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 45.Yanai H, et al. Conditional ablation of HMGB1 in mice reveals its protective function against endotoxemia and bacterial infection. Proc Natl Acad Sci USA. 2013;110(51):20699–20704. doi: 10.1073/pnas.1320808110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.