Significance
Legumes are known as pioneer plants colonizing marginal soils, and as enhancers of the nutritional status in cultivated soils. This beneficial activity has been explained by their capacity to engage in symbiotic relationship with nitrogen-fixing rhizobia. We performed a community profiling analysis of Lotus japonicus wild type and mutants to investigate the role of the nodulation pathway on the structure of the root-associated bacterial microbiota. We found that several bacterial orders were almost entirely depleted from the mutant roots, and that an intact symbiosis is needed for the establishment of taxonomically diverse and distinctive bacterial communities in the root and rhizosphere. Our findings imply that a symbiosis-linked bacterial community, rather than dinitrogen-fixing rhizobia alone, contributes to legume growth and ecological performance.
Keywords: Lotus japonicus, microbiota, symbiosis, 16S, nitrogen fixation
Abstract
Lotus japonicus has been used for decades as a model legume to study the establishment of binary symbiotic relationships with nitrogen-fixing rhizobia that trigger root nodule organogenesis for bacterial accommodation. Using community profiling of 16S rRNA gene amplicons, we reveal that in Lotus, distinctive nodule- and root-inhabiting communities are established by parallel, rather than consecutive, selection of bacteria from the rhizosphere and root compartments. Comparative analyses of wild-type (WT) and symbiotic mutants in Nod factor receptor5 (nfr5), Nodule inception (nin) and Lotus histidine kinase1 (lhk1) genes identified a previously unsuspected role of the nodulation pathway in the establishment of different bacterial assemblages in the root and rhizosphere. We found that the loss of nitrogen-fixing symbiosis dramatically alters community structure in the latter two compartments, affecting at least 14 bacterial orders. The differential plant growth phenotypes seen between WT and the symbiotic mutants in nonsupplemented soil were retained under nitrogen-supplemented conditions that blocked the formation of functional nodules in WT, whereas the symbiosis-impaired mutants maintain an altered community structure in the nitrogen-supplemented soil. This finding provides strong evidence that the root-associated community shift in the symbiotic mutants is a direct consequence of the disabled symbiosis pathway rather than an indirect effect resulting from abolished symbiotic nitrogen fixation. Our findings imply a role of the legume host in selecting a broad taxonomic range of root-associated bacteria that, in addition to rhizobia, likely contribute to plant growth and ecological performance.
The transition from an aquatic to terrestrial lifestyle during plant evolution required the formation of roots as organs for water, macronutrient, and micronutrient retrieval from soil. Nutrient-uptake systems of roots are usually specific for plant-available forms of nutrients, for example, inorganic nitrogen such as nitrate (NO3−) or inorganic orthophosphate (Pi) (1). However, phosphorus, per se, is abundant in soil in plant-inaccessible pools, and, likewise, atmospheric dinitrogen (N2) is abundant in aerobic soil [78% (vol/vol)] but cannot be accessed by plants. Soil-resident microbes play important roles in the solubilization and conversion of mineral nutrients into root-available forms, and a subset of these microbes have acquired the capacity to engage in mutualistic interactions with plant roots to trade soil-derived bioavailable macronutrients for plant-derived photoassimilates (2–4).
Healthy, asymptomatic plants live in association with diverse microbes, including bacteria, fungi, viruses, and protists, collectively called the plant microbiota (3, 5). The bacterial root microbiota is taxonomically structured and characterized by the co-occurrence of three main phyla comprising Actinobacteria, Bacteroidetes, and Proteobacteria across different soil types and divergent plant hosts (6, 7). This root-associated bacterial assemblage is mostly derived from the highly diverse bacterial soil biome surrounding roots and is established rapidly within a few days after seed germination (6, 8). Soil type is the main driver of diversification of the bacterial root microbiota at low taxonomic ranks (i.e., at genus and species level), with less variation detectable at the higher phylum rank (8–11). However, root exudates are thought to play an important role as cues to initiate a substrate-driven competition between, and differential proliferation of, soil-resident microbes for root colonization (3, 12). An estimated 17% of photosynthetically fixed carbon is transferred to the rhizosphere, the thin layer of soil surrounding the root, through root exudation (13). These carbon substrates likely contribute to the bacterial community shifts that are often detected in the rhizosphere. A fraction of the bacterial taxa present in the rhizosphere colonize roots either as epiphytes on the root surface (rhizoplane) or as bacterial endophytes inside roots (3, 8). In particular, members of Proteobacteria are consistently found enriched in root and rhizosphere compartments, and diazotrophs within this phylum have evolved the capacity to establish a sophisticated form of mutualistic interaction with plant roots, designated root nodule symbiosis. Unlike the taxonomically diverse root- and rhizosphere-associated bacterial communities that comprise a network of microbe–microbe and plant–microbe associations, the root nodule symbiosis defines a highly specific binary plant–microbe interaction where the compatible nitrogen-fixing soil bacterium is selected by the host for intracellular infection often via plant-derived infection threads and subsequent accommodation and amplification inside nodule cells.
Decades of bacterial and legume genetics allowed a detailed dissection of the regulatory networks behind the stepwise symbiotic association with diazotrophic Alphaproteobacteria. A two-way signal recognition initiates the interaction. Root-secreted flavonoids are perceived by the compatible soil bacteria, which start the production and secretion of the rhizobial symbiotic signal, the Nod factor. On the host side, lysin motif (LysM) receptor kinases, like Nod factor receptor1 (NFR1) and NFR5 in Lotus japonicus, specifically recognize and bind the compatible Nod factors (14, 15) and initiate the symbiotic signaling cascade. Nodule inception (NIN) was identified as an early key regulator of both nodule organogenesis and infection thread formation (16), whereas cytokinin signaling proteins involving Lotus histidine kinase1 (LHK1) receptor (17) control progression of the signaling events from root epidermis into the cortex (18). Inside nodules, a low-oxygen, carbon-rich environment is established by the host, allowing bacteria, upon endocytosis, to start the nitrogen fixation (19). Symbiotic nitrogen fixation reprograms the whole-root transcriptional and metabolic landscape (20–23). Moreover, the process is reiterative and highly asynchronous, because rhizobia from the rhizosphere recapitulate the infection on newly formed, competent root hairs. Nevertheless, the legume host controls the number of infection events and nodule primordia via shoot-derived signal(s) (24, 25).
Symbiotic nitrogen fixation allows legumes to thrive in habitats with limited nitrogen availability (26–28). The beneficial effect of this symbiosis is not limited to legume hosts, but extends to subsequent or concurrent plantings with nonlegumes as exemplified by ancient agricultural practices with legume cropping sequences or intercropping systems. This symbiosis likely involves a beneficial activity of legume roots and their associated microbes on the nutritional status of the soil as well as the soil biome. However, the mechanisms underpinning these symbiotic interactions in a community context and their impact on the complex microbial assemblages associated with roots remain largely unknown. Integrating these highly specific binary interactions into an ecological community context is critical for understanding the evolution of symbiosis and efficient use of rhizobia inoculum in agricultural systems.
Here, we investigated the role of symbiotic nitrogen fixation on the structure of the root-associated bacterial microbiota of the model legume L. japonicus. We performed bacterial 16S rRNA gene-based community profiling experiments of wild-type (WT) plants, grown in natural soil, and symbiotic mutants impaired at different stages of the symbiotic process. We have found that an intact nitrogen-fixing symbiosis in WT Lotus plants is needed for the establishment of taxonomically diverse and distinctive bacterial communities in root and rhizosphere compartments. This finding raises the possibility that the influence of legumes on soil performance in agricultural and ecological contexts is mediated by the enrichment of a symbiosis-linked bacterial community rather than dinitrogen-fixing rhizobia alone.
Results
Characterization of the L. japonicus Root, Nodule, and Rhizosphere Microbiota.
We established a root fractionation protocol for 10-wk-old L. japonicus plants (accession Gifu, designated WT), grown in three batches of natural Cologne soil (10) to account for batch-to-batch and seasonal variation at the soil sampling site (Fig. 1A and Materials and Methods). This fractionation enabled us to compare the structure of bacterial communities present in nodules, roots without nodules (denoted hereafter as “root compartment”), the rhizosphere, and unplanted soil (Materials and Methods and SI Appendix, Fig. S1). Briefly, the “rhizosphere compartment” defines soil particles tightly adhering to Lotus roots that were collected after the first of two successive washing steps. Macroscopically visible nodules and nodule initials were excised from roots with a scalpel and designated the “nodule compartment.” Pooled nodules and washed roots without nodules were separately subjected to a sonication treatment to deplete epiphytes and enrich for endophytic bacteria. Abundant nodulation (∼20 nodules per plant) of healthy WT plants demonstrates that this soil is conducive for nodule formation and contains Lotus-compatible rhizobia (Fig. 1A, Inset). We subjected a total of 27 unplanted soil, 73 rhizosphere, 75 root, and 27 nodule samples to amplification of the 16S rRNA gene with PCR primers targeting the V5–V7 hypervariable regions (29) (Materials and Methods) and generated ∼1 M high-quality sequencing reads (4,670 reads per sample on average). After removal of low-quality reads, chimeras, and sequences assigned to plant-derived organellar DNA, we clustered the data into 1,834 operational taxonomic units (OTUs) at 97% sequence similarity (Materials and Methods and Dataset S1).
Fig. 1.
Images depicting L. japonicus WT (A) and nodule symbiosis-deficient mutant plants lhk1-1 (B), nfr5-3 (C), and nin-2 (D) following harvest. (A and B, Insets) For nodulating genotypes, close-up views of nodules are shown. (Scale bars: 1 cm.)
To assess the effect of the different compartments on the assembly of bacterial communities, we compared the β-diversity (between-samples diversity) using Bray–Curtis distances and performed a canonical analysis of principal coordinates (CAP) (30) (Materials and Methods). This analysis revealed a clear differentiation of samples belonging to the root, rhizosphere, nodule, and soil compartments that explains as much as 19.97% of the overall variance of the data (Fig. 2A; P < 0.001), whereas the effect attributable to the soil batch was comparatively small (8.01% of the variance, P < 0.001). Analysis of α-diversity (within-samples diversity) using the Shannon index indicated a decreasing gradient of complexity from the soil bacterial communities (highest richness) to the rhizosphere, root, and, finally, the nodule microbiota (SI Appendix, Fig. S2).
Fig. 2.
(A) Constrained PCoA plot of Bray–Curtis distances between samples including only the WT constrained by compartment (19.97% of variance, P > 0.001; n = 94). (B) Constrained PCoA plot of Bray–Curtis distances constrained by genotype (9.82% of variance explained, P < 0.001; n = 164). Each point corresponds to a different sample colored by compartment, and each host genotype is represented by a different shape. The percentage of variation indicated in each axis corresponds to the fraction of the total variance explained by the projection. Corresponding unconstrained PCoA plots for each soil batch are shown in SI Appendix, Fig. S3.
Our finding of a bacterial community shift in the Lotus rhizosphere compared with the bulk soil reservoir is consistent with previous reports from WT pea (31), soybean (32), and peanut (33), in which a similar enrichment of members of Burkholderiales, Flavobacteriales, and Rhizobiales has been shown, whereas information on the community structure of the root microbiota is unavailable for other legumes.
Parallel Selection of Nodule- and Root-Specific Bacteria from the Rhizosphere Compartment.
Legume nodules represent a unique environmental niche derived from differentiated cortical root cells where both symbiotic and nonsymbiotic bacteria are allowed accommodation and proliferation. Laboratory studies with single WT or mutant symbiotic strains demonstrated a stepwise, host-controlled colonization process ensuring symbiont selection (34). By contrast, little is known about the extent or the diversity of nodule and root colonization by nonsymbionts (35).
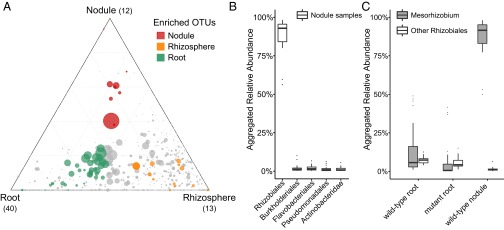
We took advantage of the compatible symbiotic association between Lotus and rhizobia present in Cologne soil and performed an analysis of the bacterial community of epiphyte-depleted, functional nodules of WT plants grown in this soil (Materials and Methods). We found that nodules were inhabited by a distinctive bacterial community compared with those present in the root and rhizosphere (Fig. 2A). Only a small number of the 1,834 OTUs were identified to be nodule-enriched (12 red circles in Fig. 3A) with one dominant member classified as belonging to the Mesorhizobium genus, substantiating that nodules also represent a highly selective bacterial niche for soil-grown Lotus plants. Importantly, the nodule and root communities were similarly divergent from the rhizosphere (separation by second component; Fig. 2A), and nodule-enriched OTUs were found in similar abundances in the root and rhizosphere samples (red circles in Fig. 3A). These findings suggest a parallel rather than consecutive selection of bacterial taxa from the rhizosphere assemblage for enrichment in the two endocompartments, most likely via host-induced infection threads. If there were a sequential selection, then nodule-enriched OTUs would be expected to be more abundant in the root compared with the rhizosphere. Taxonomic assignments at the order level for all OTUs with a relative abundance (RA) >5‰ revealed that the Lotus nodule community is dominated by bacteria belonging to the order Rhizobiales (88.01% average RA; Fig. 3B), which is mainly due to a selective enrichment of Mesorhizobium members (Fig. 3C). This analysis also revealed the presence of Burkholderiales, Flavobacteriales, Pseudomonadales, and Actinobacteridae at detectable abundances (>1% RA; Fig. 3B), showing that nodules of soil-grown Lotus are primarily, but not exclusively, colonized by symbiotic rhizobia.
Fig. 3.
(A) Ternary plot depicting compartment RAs of all OTUs (>5 ‰) for WT root, rhizosphere, and nodule samples (n = 67) across three soil batches (CAS8–CAS10). CAS, Cologne agricultural soil. Each point corresponds to an OTU. Its position represents its RA with respect to each compartment, and its size represents the average across all three compartments. Colored circles represent OTUs enriched in one compartment compared with the others (green in root, orange in rhizosphere, and red in nodule samples), whereas gray circles represent OTUs that are not significantly enriched in a specific compartment. (B) Rank abundance plot depicting RAs aggregated to the order taxonomic level for the top abundant taxa found in the WT nodule samples (n = 21). (C) Comparison of abundances between Mesorhizobium and other Rhizobiales genera in WT roots (n = 48), mutant roots (n = 100), and WT nodule samples (n = 21).
Impairment of Nitrogen-Fixing Symbiosis Dramatically Alters Bacterial Community Structure in the Lotus Root and Rhizosphere Compartments.
Next, we applied the same growth conditions, fractionation protocol, and bacterial community analysis to four symbiotic Lotus mutants (nfr5-2, nfr5-3, nin-2, and lhk1-1) to identify the role of the nodulation signaling pathway on bacterial assemblages. In nfr5 and nin mutants, the infection process is either not initiated or terminated at the microcolony stage, respectively, whereas lhk1 plants develop a large number of root hair infection threads that subsequently fail to infect cortical cells (16, 18, 36). Similar to WT, symbiotic mutant plants appeared healthy, but were smaller and had slightly pale green leaves (Fig. 1 B–D). With the exception of occasional nodules on lhk1 roots (18), no nodules were found on nfr5 or nin root systems (Fig. 1 B–D). Remarkably, we found that communities associated with the roots and rhizospheres of each of the four symbiosis mutants were similar to each other, but significantly different from the communities of WT plants (Fig. 2B and SI Appendix, Fig. S3). This separation between the mutant and WT samples was found to be robust, as indicated by unconstrained principal coordinate analysis (PCoA) performed independently for each soil batch (SI Appendix, Fig. S3). Furthermore, CAP performed on the entire dataset revealed a prominent effect of the host genotype on bacterial communities, explaining 9.82% of the variance (Fig. 2B).
Nodules are root-derived and -anchored structures, and yet the two organs host distinctive bacterial assemblages (Fig. 2A). As a consequence, despite rigorous preparation of root compartments, WT root segments might contain incipient root-concealed nodule primordia and, vice versa, the nodule samples might be contaminated with surrounding root tissue. To clarify whether these potential limitations of our sampling protocol confound the observed host genotype-dependent community differentiation, we performed an in silico depletion of all nodule-enriched OTUs from the WT root dataset and repeated the PCoA and CAP (SI Appendix, Fig. S4). This experiment revealed only a negligible reduction in the portion of the community variance explained by the host genotype (9.82% versus 9.72%), indicating that the differences in the root-associated assemblages caused by the impairment of nitrogen-fixing symbiosis are largely robust against residual cross-contamination between the two compartments.
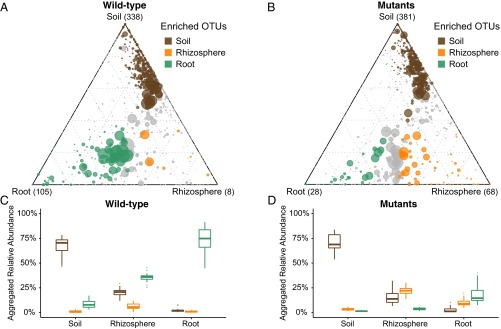
To understand better how the Lotus nodulation pathway influences bacterial community composition, we identified OTUs that are specifically enriched in the root and rhizosphere of WT or mutants compared with unplanted soil (Materials and Methods). Due to the fact that the bacterial assemblages of the tested symbiosis mutants do not significantly differ among each other (Fig. 2B), we performed our analyses using the combined samples from all mutant genotypes across all soil batches (Fig. 4). The Lotus WT root microbiota is characterized by a large number of root-enriched OTUs, mostly belonging to Proteobacteria, Actinobacteria, and Bacteroidetes (105 green circles in Fig. 4A and SI Appendix, Fig. S12). By contrast, only a small number of OTUs were found specifically enriched in the WT rhizosphere samples (8 orange circles in Fig. 4A). Compared with WT, roots of the symbiotic mutants are dramatically depleted of root-enriched OTUs (28 green circles in Fig. 4B), whereas the number of rhizosphere-enriched OTUs increased by a factor of 8 (68 orange circles in Fig. 4B). This pattern was reproducible when we performed the same analysis for each soil batch and mutant genotype independently (SI Appendix, Figs. S5–S9).
Fig. 4.
Ternary plots depicting compartment RA of all OTUs (>5 ‰) for WT samples (A; WT; n = 73) and mutant samples (B; nfr5-2, nfr5-3, nin-2, and lhk1-1; n = 118) across three soil batches (CAS8–CAS10). Each point corresponds to an OTU. Its position represents its RA with respect to each compartment, and its size represents the average across all three compartments. Colored circles represent OTUs enriched in one compartment compared with the others (green in root, orange in rhizosphere, and brown in root samples). Aggregated RAs of each group of enriched OTUs (root-, rhizosphere- and soil-enriched OTUs) in each compartment for the WT samples (C; WT; n = 73) and mutant samples (D; nfr5-2, nfr5-3, nin-2, lhk1-1; n = 118) are shown. In each compartment, the difference from 100% RA is explained by OTUs that are not significantly enriched in a specific compartment.
To characterize the bacterial community shifts further, we calculated separately for WT and symbiotic mutant plants aggregated RAs of OTUs that are specifically enriched in one compartment. As expected, this calculation revealed a decreasing contribution of the soil-enriched OTUs in soil, rhizosphere, and root samples (69.40%, 17.03%, and 2.40% mean aggregated RA, respectively; dark brown box plots in Fig. 4 C and D) in both WT and mutant samples. This finding suggests that impairment of the symbiosis pathway does not affect the capacity of Lotus to exclude colonization by the majority of the detectable bacterial soil biome and the formation of characteristic root-associated microbiota, fully differentiated from the root-associated microbiota present in bulk soil. We observed an inverse pattern for the root-enriched OTUs across the three WT compartments (green box plots in Fig. 4C). The steep increase in the aggregated RAs, from 8.76% in the soil, to 35.72% in rhizosphere, and to 72.34% in roots for WT samples, was almost completely abolished in the mutants (1.49%, 3.63%, and 17.74%, respectively; green box plots in Fig. 4D). Conversely, the aggregated RAs of rhizosphere-specific OTUs are only slightly higher in the rhizosphere samples of WT plants compared with roots and soil, which are influenced by the low number of rhizosphere-specific OTUs (orange box plots in Fig. 4C). However, RAs are significantly higher in the mutant rhizosphere samples with respect to the other compartments (3.29% in soil samples, 22.09% in rhizosphere samples, and 9.94% in root samples; orange box plots in Fig. 4D). Taken together, these data support the hypothesis that the Lotus symbiosis pathway is a key component for the progressive enrichment/selection of specific soil-derived OTUs and the establishment of fully differentiated microbiota in rhizosphere and root compartments.
The Symbiosis Pathway Drives Root and Rhizosphere Differentiation Across Multiple Bacterial Orders.
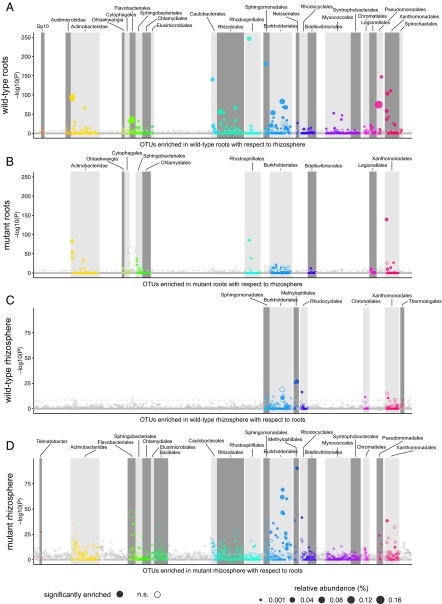
We dissected the observed bacterial community shifts by arranging OTUs according to their taxonomy and displaying their enrichment in the root or rhizosphere of WT and symbiotic mutants in a set of Manhattan plots (Materials and Methods). The results revealed unexpectedly nuanced taxonomic alterations underlying the community shifts in the plant-associated compartments (Fig. 5 A and B). Whereas WT plants host root-enriched OTUs belonging to a wide range of bacterial orders, mutants roots fail to enrich any member of the orders Flavobacteriales, Myxococcales, Pseudomonadales, Rhizobiales, and Sphingomonadales above a threshold of significance (false discovery rate-corrected P values, α = 0.05; Materials and Methods). In addition, a striking enrichment of more than 15 Burkholderiales OTUs in WT roots contrasts with a marginal enrichment of this order in the symbiotic mutants. However, the mutant roots retain the capacity to enrich OTUs belonging to the orders Actinobacteridae, Rhodospirilalles, Sphingobacteriales, and Xanthomonadales (Fig. 5 A and B). Strikingly, we found an almost inverse pattern when we considered the rhizosphere-enriched OTUs in WT and mutant plants: Both the number and the taxonomic diversity of significantly enriched OTUs increased dramatically in the mutants compared with WT (Fig. 5 C and D).
Fig. 5.
Manhattan plots showing root-enriched OTUs in WT (A) or in the mutants (B) with respect to rhizosphere and rhizosphere-enriched OTUs in WT (C) or in the mutants (D) with respect to root. OTUs that are significantly enriched (also with respect to soil) are depicted as full circles. The dashed line corresponds to the false discovery rate-corrected P value threshold of significance (α = 0.05). The color of each dot represents the different taxonomic affiliation of the OTUs (order level), and the size corresponds to their RAs in the respective samples [WT root samples (A), mutant root samples (B), WT rhizosphere samples (C), and mutant rhizosphere samples (D)]. Gray boxes are used to denote the different taxonomic groups (order level).
Next, we compared directly the WT and mutants to identify OTUs differentially abundant in the root or rhizosphere (SI Appendix, Figs. S10 and S11). We found that the community shift that separates host genotypes (Fig. 2B) is largely caused by numerous OTUs that are specifically enriched (n = 45) or depleted (n = 15) in WT roots with respect to mutant root samples (SI Appendix, Fig. S10A) belonging to at least 14 bacterial orders (SI Appendix, Fig. S11 A and B). We observed a parallel effect on OTUs of a similar taxonomic profile when comparing rhizosphere samples across genotypes, and identified numerous OTUs enriched (n = 27) or depleted (n = 6) in the WT rhizosphere (SI Appendix, Figs. S10B and S11 C and D and Dataset S2).
Next, we examined whether the complex community shifts observed at the order taxonomic rank were also detectable at the higher phylum rank. Interestingly, we observed clear differences between the root and rhizosphere samples of WT and mutant Lotus plants for Proteobacteria (66.78% and 46.97%, respectively) and Bacteroidetes (14.57% and 38.10%, respectively), which were largely explained by variation in abundances of OTUs belonging to the Rhizobiales, Burkholderiales, and Flavobacteriales bacterial orders (SI Appendix, Figs. S10–S12). Conversely, we found no significant differences between WT and mutant roots and rhizospheres for Actinobacteria (13.42% and 11.62% average RA, respectively) and Firmicutes (∼1% average RA). These results illustrate that the large shifts observed between the WT and mutant Lotus plants affect root and rhizosphere communities similarly, even at higher taxonomic levels (SI Appendix, Figs. S10–S12).
Comparable Immune- and Symbiosis-Related Metabolic Responses in Soil-Grown WT and Symbiotic Mutant Roots.
The extensive changes of root microbiota structure across multiple bacterial orders in the symbiosis mutants prompted us to investigate whether mutant roots display altered immune- or symbiosis-related metabolic responses that indirectly perturb an orderly microbiota establishment. We quantified relative transcript levels for a panel of defense and symbiotic marker genes using WT and mutant root tissue samples that were processed as for the 16S rRNA gene community profiling (SI Appendix and Dataset S3). Analysis of eight genes induced during pathogen defense in Lotus or likely representing Lotus orthologs of Arabidopsis defense marker genes revealed that WT and mutant roots accumulate similar transcript levels, indicative of a comparable immune status rather than an induced defense in the mutants (SI Appendix, Fig. S13A). We also tested whether WT and mutant roots differed in expression levels of genes that have been reported to contribute to the metabolic state established between host and nitrogen-fixing symbiont (37–40). We found comparable transcript levels of Nodulin26, Nodulin70, Sucrose transporter4, and Invertase1 in the tested genotypes, suggesting similar metabolic responses in the roots of the WT and mutants (SI Appendix, Fig. S13B). On the other hand, early symbiotic genes like Nin, Peroxidase, and Thaumatin were induced in WT or lhk1 and nin-2 mutants, but not in nfr5-2 roots, indicating that soil-grown symbiotic mutants maintain their previously described, gradually impaired root response to nitrogen-fixing rhizobia (16, 18, 22, 36) (SI Appendix, Fig. S13C). Direct measurements of total protein content revealed comparable levels in WT and symbiotic mutants (SI Appendix, Fig. S14), whereas quantification of nitrate levels revealed significant differences between nfr5, nin, and lhk1 or WT, indicating that regulation of nitrate uptake, which has a known inhibitory effect on nodulation (41, 42), operates downstream of Nin (SI Appendix, Fig. S14). Together, these results suggest that a nitrogen-sufficient status is reached in all tested genotypes, but that the nitrogen source, N2 or nitrate, might differ among them.
L. japonicus and Various Brassicaceae Species Assemble Highly Diverged Root-Inhabiting Bacterial Communities.
We have previously shown that Arabidopsis thaliana and three other Brassicaceae species (Cardamine hirsuta, Arabidopsis halleri, and Arabidopsis lyrata), grown in Cologne soil, assemble a highly similar root microbiota, characterized only by small quantitative differences of community profiles (29). We retrieved the corresponding raw 16S sequence reads and performed de novo OTU clustering together with the amplicon data of WT and symbiotic Lotus mutants (Fig. 6). PCoA of Bray–Curtis distances revealed a clear separation of root and soil samples, but also a marked distinction between all Lotus and Brassicaceae samples (Fig. 6A), indicating that both WT and symbiosis-impaired Lotus plants harbor strikingly distinctive root microbiota compared with root microbiota of the four tested Brassicaceae species. Quantitative analysis of WT L. japonicus and A. thaliana root-enriched OTUs revealed significant and contrasting differences already at the phylum level primarily reflected in the abundances of Proteobacteria and Actinobacteria (Fig. 6B). Similar rank abundance analysis performed at the order level identified particular taxonomic lineages contributing to the differences between these two plant species; Rhizobiales, Caulobacterales, Rickettsiales, and Sphingobacteriales were found in larger abundances in Lotus roots, whereas Burkholderiales, Actinomycetales, Myxococcales, and Pseudomonadales were more abundant in Arabidopsis roots (Fig. 6C).
Fig. 6.
(A) PCoA plot of Bray–Curtis distances for root and soil samples showing a clear separation between the roots of all Lotus genotypes (circles) compared with the roots of Arabidopsis and relative species (hollow shapes) grown in Cologne soil and sequenced using the same primer set. RAs aggregated to the phylum (B) and order taxonomic levels (C, 10 most abundant orders) showing a comparison between A. thaliana (n = 26) and L. japonicus root samples (n = 74).
Symbiosis-Impaired Mutants Maintain an Altered Community Structure in Nitrogen-Supplemented Soil.
Nitrogen-fixing symbiosis is nitrogen-sensitive, and already from 2 mM KNO3 concentration, reduced nodulation and infection were observed in Lotus (42). To determine if the community shifts observed in Lotus mutant roots and the rhizosphere were caused by a potentially differential nitrogen status, we performed a similar community analysis using plants grown in Cologne soil (different soil batch) supplemented with 10 mM KNO3. In these conditions, the symbiotic mutants no long had a pale leaf phenotype as observed in nonsupplemented soil (SI Appendix, Fig. S15) and the WT plants developed no functional nodules, based on their low number and small and white appearance at the time of harvest. Despite similar nitrogen content in WT and mutant roots (SI Appendix, Fig. S15), we found that the differential phenotypes (stature and fresh weight) seen in nonsupplemented soil were retained under nitrogen-supplemented conditions (SI Appendix, Fig. S15), indicating that an intact symbiosis pathway promotes plant growth irrespective of the presence of functional nodules. Based on the similar macroscopic phenotypes of the symbiotic mutants in response to the nitrogen supplementation, we then analyzed the composition of bacterial communities in WT and two Nod factor receptor mutants, nfr5-2 and nfr5-3. Remarkably, the PCoA revealed a similar shift in the root and rhizosphere communities of the mutants relative to the corresponding WT compartments for plants grown in nitrogen-supplemented soil (21.20% of the variance, P < 0.001; SI Appendix, Fig. S16A) as in the nonsupplemented Cologne soil (21.80% of the variance, P < 0.001; SI Appendix, Fig. S16B). Finally, no detectable differences in soil biome composition were seen between nitrogen-supplemented and nonsupplemented unplanted soil samples (Dataset S4). Together, these results provide evidence for a direct impact of the disabled symbiosis pathway on the root-associated community structure rather than an indirect effect resulting from abolished symbiotic nitrogen fixation.
Discussion
Here, we have characterized the root microbiota of the model legume L. japonicus using a 16S rRNA amplicon survey. By using a panel of symbiosis-impaired mutants, we have investigated the role of host genes with known functions in the establishment of a highly specific and binary symbiotic plant–microbe association in the context of the root-associated bacterial community. Our study reveals that key symbiotic genes play a major role in the establishment of taxonomically structured bacterial communities in the root and rhizosphere of L. japonicus (Fig. 4), which extends their role beyond the perception and selection of nitrogen-fixing rhizobia for intracellular accommodation in nodules. The observed impact of disenabling symbiosis in Lotus seemingly differs from analogous perturbations in insects and mammals, where impairment of symbiosis in the gut leads to the accumulation of pathogens and dramatic consequences for the health of the host (43–46). Cologne soil-grown Lotus symbiotic mutants showed no signs of disease or altered immune response in comparison to WT (Fig. 1 and SI Appendix, Fig. S13). However, it remains possible that the observed reduced plant growth (Fig. 1) leads to lower reproductive fitness of the mutant plants in a natural ecosystem.
Genetic disruption of the nodulation pathway resulted in depletion of six bacterial orders from the root compartment, including the two most abundant orders identified in WT, Flavobacteriales and Burkholderiales (Fig. 5 A and B). Rhizobium therefore acts as a bacterial “hub” for Lotus roots, in analogy to the Albugo oomycete pathogen that largely affected the phyllosphere microbiome of Arabidopsis after infection (47). Two mutually nonexclusive scenarios could account for the observed dramatic community shift inside roots: (i) cooperative microbe–microbe interactions between symbiotic nitrogen-fixing bacteria and a subset of root microbiota members or (ii) direct use of the nodulation pathway by soil-borne bacteria other than nodulating rhizobia for entry and proliferation inside legume roots. Quantification of selected transcripts in WT and mutant roots provided evidence for a similar immune status and symbiosis-specific metabolic responses, but, as expected, differential expression of the symbiotic genes (SI Appendix, Fig. S13). These results support the hypothesis of a direct rather than indirect engagement of the nitrogen-fixing symbiosis pathway in the selection of specific bacterial taxa other than nodulating rhizobia.
The beneficial association with Burkholderiales is habitual for legumes. For example, Mimosa genera within South America and legumes from the Papilionoideae subfamily in South Africa are known to form an ancient and stable symbiosis with nitrogen-fixing Burkholderia lineages inside nodules (48–50). Members of the order Burkholderiales, which are dramatically depleted in the Lotus mutants, are also known for potent plant growth promotion activities in nonleguminous plants (51). The inclusion of legumes in a cropping rotation sequence in agriculture generally enriches the soil, but the symbiotic nitrogen fixation alone cannot explain the productivity increase in the subsequent crop (26). Thus, our study raises the intriguing possibility that besides the activity of nitrogen-fixing bacteria, the selective enrichment of other symbiosis-linked root microbiota members influences the soil biome and, consequently, soil biofertilization by legumes might involve a much wider taxonomic range than currently thought.
Our statistical analyses revealed a major impact of the host genotype on the Lotus root microbiota, which explains 9.82% of the variance in the data (Fig. 2B). This genotype effect is almost twofold larger than the effect identified when comparing the root-associated communities of WT and severely immunocompromised A. thaliana mutants (52) or between A. thaliana and three Brassicaceae species that diverged up to 35 Mya (29). Interestingly, the community shifts detected in nfr5, nin, and lhk1 mutant roots are highly comparable (Fig. 2B). This similarity is likely related to our stringent root fractionation protocol (10, 11), depleting both epiphytes and epidermal endophytes. However, we cannot rule out differences between the communities of the tested mutants of a magnitude smaller than the effect caused by soil batch variation and technical noise present in the pyrosequencing data.
Recent studies have shown that upon microbiota acquisition, the root-associated bacterial assemblage remains robust against major changes in plant stature and source-sink relationships (9, 11). This microbiota stability is also observed here, where a clear separation between the root and rhizosphere of WT and symbiosis-impaired Lotus plants is retained when plants are grown under different nitrate conditions (SI Appendix, Fig. S16). The latter observation is also strong evidence that the root-associated community shift in the symbiotic mutants is a direct consequence of the disabled symbiosis pathway rather than an indirect effect resulting from abolished symbiotic nitrogen fixation.
Previous controlled coinoculation experiments with Mesorhizobium loti and root endophytes have shown that Lotus can selectively guide endophytic bacteria toward nodule primordia via symbiont-induced infection threads, and that endophytes and symbionts can promote each other’s infection of the host (35). These experiments focused on nodule colonization and were conducted using a gnotobiotic plant system with a limited number of endophytes. Our bacterial community profiling data obtained with soil-grown symbiotic mutants allowed testing the contribution of infection thread-dependent root colonization by natural populations of compatible endophytes present in soil. Indeed, the large number of bacterial taxa found depleted in nfr5, nin, and lhk1 genotypes (SI Appendix, Fig. S11) suggests that infection threads arrested in WT may facilitate root colonization by endophytes. This finding implies additional functions of host genes active in early symbiosis for efficient root colonization by a subset of the root microbiota. Our community profiling data also identified bacterial orders enriched in the mutant roots; thus, their root colonization takes place independent of nitrogen-fixing symbiosis. These endophytic taxa may use alternative entry routes [i.e., crack entry, which occurs at the base of emerging lateral roots and is likely used as an entry portal for root endophytes in nonleguminous plants (53–55)].
Our study revealed that Lotus growth in Cologne soil did not interfere with the host–symbiont compatibility described previously in monoassociations with plants grown in artificial media (56). This high selectivity is evidenced by only 12 nodule-enriched OTUs among a total of 1,834 OTUs in our dataset and by Mesorhizobium members representing the most abundant taxa (Fig. 3A). Remarkably, nodule-enriched OTUs had a similar RA in the root and rhizosphere compartments, suggesting linked selection process(es) in all three compartments. Furthermore, nodule-enriched OTUs were depleted from the mutant root samples, which corroborates our hypothesis that an intact symbiotic pathway is selecting rhizobia during infection in both the root cortex and nodules. Interestingly, nodule-enriched Mesorhizobium OTUs were depleted from both mutant root and rhizosphere compartments, indicating that root-derived diffusible compounds produced by WT exert a role in enriching symbionts in soil that adheres to the legume root surface. Thus, our findings support previous observations that maintenance of highly symbiotic isolates in the soil is not only a function of rhizobia release from decaying nodules but is also dependent on persistent host selective pressure (57, 58). A likely scenario for this enrichment is a positive feedback mechanism in which host-initiated signaling leads to enrichment of symbionts in the root and rhizosphere, a hypothesis that is supported by the markedly similar patterns observed in plants grown under nitrogen-supplemented conditions that impede nitrogen fixation. Legume root-derived flavonoids are candidate diffusible signaling molecules in such a feedback mechanism because their profile was shown to change during root–nodule symbiosis (59, 60) and their broad impact on soil bacterial communities (61, 62), especially on symbiotic rhizobia, has been documented (63, 64).
Our comparative analyses of the root microbiota from L. japonicus and four Brassicaceae species grown in Cologne soil (29) revealed a highly distinctive community composition irrespective of an intact or dysfunctional symbiosis pathway (Fig. 6). Lotus and Arabidopsis ancestors diverged ∼118 Mya (65), and two major evolutionary events took place soon after their separation: loss of arbuscular mycorrhiza (AM) symbiosis in the Brassicaceae (66) and gain of nitrogen-fixation predisposition in the legume predecessor (67). Thus, our findings suggest that the marked distinctiveness of the Lotus-specific root microbiota is not governed by the evolved functions of Nfr5, Nin, or Lhk1, but is possibly linked to the loss of AM symbiosis in the Brassicaceae lineage. This hypothesis can be tested by future experiments with mutants affecting Lotus “common symbiosis genes” that fail to establish both symbiotic relationships with AM fungi and nodulating rhizobia (68–70).
Materials and Methods
A detailed description of the methods used in this study can be found in SI Appendix, Supplementary Materials and Methods.
Soil and Plant Material.
Seeds of L. japonicus WT, ecotype Gifu B-129, and the corresponding symbiosis-deficient mutants (nfr5-2, nfr5-3, lhk1-1, and nin-2) were grown in soil batches collected in the successive seasons. Plants were grown in the greenhouse under long-day conditions (16-h photoperiod), watered with tap water (optionally supplemented with 10 mM KNO3), and harvested after 10 wk.
Sample and 16S Library Preparations.
Fragments of the root systems were washed, and the rhizosphere, root, and nodules were separated. A first wash containing the root-adhering soil layer defined the rhizosphere compartment. Nodules and visible primordia were separated from root fragments of nodulating genotypes (WT and lhk1-1) with a scalpel. Root and nodule samples were ultrasound-treated. DNA extraction was performed using a FastDNA SPIN Kit for Soil (MP Biomedicals). Barcoded primers targeting the variable V5–V7 regions of bacterial 16S rRNA genes (799F and 1193R) (10, 71) were used for amplification. Amplicons were purified (Qiagen), combined, and subjected to 454 sequencing.
Metabolite Analysis.
Root nitrate contents were determined by the ion chromatography method as previously described (72). Proteins were extracted in 10 mM Tris⋅HCl buffer (pH 8) and determined with a Bio-Rad Protein Assay Kit using BSA as the standard.
Computational Analyses.
The 16S rRNA gene sequences were processed using a combination of custom scripts as well as tools from the QIIME (73) and USEARCH (74) pipelines (QIIIME-ready mapping files are provided in Datasets S4 and S5). The resulting OTU table was used in all subsequent statistical analyses of differentially abundant taxa as well as analyses of α- and β-diversity. Indices of α-diversity were calculated after subsampling to an even depth of 1,000 reads. Measures of β-diversity were calculated on a normalized OTU table. The PCoA was done by classical multidimensional scaling of β-diversity distance matrices using the cmdscale function in R. CAP (30) was computed using the capscale function implemented in the vegan R library (75), by constraining for the variable of interest and conditioning for the remaining factors. Statistical analyses of differentially abundant OTUs were performed using the edgeR library (76) by fitting a negative binomial generalized linear model to the OTUs.
Code Availability.
All scripts required for the computational analyses performed in this study as well as the corresponding raw sequencing and intermediate data are available at www.mpipz.mpg.de/R_scripts.
Supplementary Material
Acknowledgments
We thank Diana Kuehn and Bruno Huettel (Max Planck Genome Centre Cologne) for the preparation and sequencing of the 454 libraries and Irene Klinkhammer (Botanical Institute Cologne) for assistance with nitrate analysis. We thank Omri Finkel, Jeffery L. Dangl, Jens Stougaard, Lene H. Madsen, Stig U. Andersen, Ryohei Nakano, Stephane Hacquard, and Thorsten Thiergart for valuable comments on the manuscript. This work was supported by Danish Council for Independent Research Grant 09-070023 and Danish National Research Foundation Grant DNRF79 (to S.R.) and by the Max Planck Society, European Research Council Grant ROOTMICROBIOTA, and the “Cluster of Excellence on Plant Sciences” program funded by the Deutsche Forschungsgemeinschaft (P.S.-L.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequence reported in this paper has been deposited in the European Nucleotide Archive (accession no. PRJEB15623).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1616564113/-/DCSupplemental.
References
- 1.Maathuis FJ. Physiological functions of mineral macronutrients. Curr Opin Plant Biol. 2009;12(3):250–258. doi: 10.1016/j.pbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103(3):626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 4.Parniske M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat Rev Microbiol. 2008;6(10):763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 5.Vorholt JA. Microbial life in the phyllosphere. Nat Rev Microbiol. 2012;10(12):828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 6.Hacquard S, et al. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe. 2015;17(5):603–616. doi: 10.1016/j.chom.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Guttman DS, McHardy AC, Schulze-Lefert P. Microbial genome-enabled insights into plant-microorganism interactions. Nat Rev Genet. 2014;15(12):797–813. doi: 10.1038/nrg3748. [DOI] [PubMed] [Google Scholar]
- 8.Edwards J, et al. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci USA. 2015;112(8):E911–E920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lundberg DS, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488(7409):86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bulgarelli D, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488(7409):91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 11.Dombrowski N, et al. Root microbiota dynamics of perennial Arabis alpina are dependent on soil residence time but independent of flowering time. ISME J. August 2, 2016 doi: 10.1038/ismej.2016.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eilers KG, Lauber CL, Knight R, Fierer N. Shifts in bacterial community structure associated with inputs of low molecular weight carbon compounds to soil. Soil Biol Biochem. 2010;42(6):896–903. [Google Scholar]
- 13.Nguyen C. Rhizodeposition of organic C by plants: Mechanisms and controls. Agronomie. 2003;23(5-6):375–396. [Google Scholar]
- 14.Radutoiu S, et al. LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 2007;26(17):3923–3935. doi: 10.1038/sj.emboj.7601826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broghammer A, et al. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc Natl Acad Sci USA. 2012;109(34):13859–13864. doi: 10.1073/pnas.1205171109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schauser L, Roussis A, Stiller J, Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402(6758):191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- 17.Held M, et al. Lotus japonicus cytokinin receptors work partially redundantly to mediate nodule formation. Plant Cell. 2014;26(2):678–694. doi: 10.1105/tpc.113.119362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray JD, et al. A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science. 2007;315(5808):101–104. doi: 10.1126/science.1132514. [DOI] [PubMed] [Google Scholar]
- 19.Udvardi M, Poole PS. Transport and metabolism in legume-rhizobia symbioses. Annu Rev Plant Biol. 2013;64(64):781–805. doi: 10.1146/annurev-arplant-050312-120235. [DOI] [PubMed] [Google Scholar]
- 20.El Yahyaoui F, et al. Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol. 2004;136(2):3159–3176. doi: 10.1104/pp.104.043612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colebatch G, et al. Global changes in transcription orchestrate metabolic differentiation during symbiotic nitrogen fixation in L. japonicus. Plant J. 2004;39(4):487–512. doi: 10.1111/j.1365-313X.2004.02150.x. [DOI] [PubMed] [Google Scholar]
- 22.Høgslund N, et al. Dissection of symbiosis and organ development by integrated transcriptome analysis of lotus japonicus mutant and wild-type plants. PLoS One. 2009;4(8):e6556. doi: 10.1371/journal.pone.0006556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakagawa T, et al. From defense to symbiosis: Limited alterations in the kinase domain of LysM receptor-like kinases are crucial for evolution of legume-Rhizobium symbiosis. Plant J. 2011;65(2):169–180. doi: 10.1111/j.1365-313X.2010.04411.x. [DOI] [PubMed] [Google Scholar]
- 24.Krusell L, et al. Shoot control of root development and nodulation is mediated by a receptor-like kinase. Nature. 2002;420(6914):422–426. doi: 10.1038/nature01207. [DOI] [PubMed] [Google Scholar]
- 25.Searle IR, et al. Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science. 2003;299(5603):109–112. doi: 10.1126/science.1077937. [DOI] [PubMed] [Google Scholar]
- 26.Peoples MB, et al. The contributions of nitrogen-fixing crop legumes to the productivity of agricultural systems. Symbiosis. 2009;48(1):1–17. [Google Scholar]
- 27.Batterman SA, Wurzburger N, Hedin LO. Nitrogen and phosphorus interact to control tropical symbiotic N2 fixation: A test in Inga punctata. J Ecol. 2013;101(6):1400–1408. [Google Scholar]
- 28.Adams MA, Turnbull TL, Sprent JI, Buchmann N. Legumes are different: Leaf nitrogen, photosynthesis, and water use efficiency. Proc Natl Acad Sci USA. 2016;113(15):4098–4103. doi: 10.1073/pnas.1523936113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlaeppi K, Dombrowski N, Oter RG, Ver Loren van Themaat E, Schulze-Lefert P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci USA. 2014;111(2):585–592. doi: 10.1073/pnas.1321597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson MJ, Willis TJ. Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology. Ecology. 2003;84(2):511–525. [Google Scholar]
- 31.Turner TR, James EK, Poole PS. The plant microbiome. Genome Biol. 2013;14(6):209. doi: 10.1186/gb-2013-14-6-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendes LW, Kuramae EE, Navarrete AA, van Veen JA, Tsai SM. Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 2014;8(8):1577–1587. doi: 10.1038/ismej.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen M, et al. Dynamic succession of soil bacterial community during continuous cropping of peanut (Arachis hypogaea L.) PLoS One. 2014;9(7):e101355. doi: 10.1371/journal.pone.0101355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzaki T, Yoro E, Kawaguchi M. Leguminous plants: Inventors of root nodules to accommodate symbiotic bacteria. Int Rev Cell Mol Biol. 2015;316:111–158. doi: 10.1016/bs.ircmb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 35.Zgadzaj R, et al. A legume genetic framework controls infection of nodules by symbiotic and endophytic bacteria. PLoS Genet. 2015;11(6):e1005280. doi: 10.1371/journal.pgen.1005280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Madsen EB, et al. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425(6958):637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- 37.Szczyglowski K, Kapranov P, Hamburger D, de Bruijn FJ. The Lotus japonicus LjNOD70 nodulin gene encodes a protein with similarities to transporters. Plant Mol Biol. 1998;37(4):651–661. doi: 10.1023/a:1006043428636. [DOI] [PubMed] [Google Scholar]
- 38.Hwang JH, Ellingson SR, Roberts DM. Ammonia permeability of the soybean nodulin 26 channel. FEBS Lett. 2010;584(20):4339–4343. doi: 10.1016/j.febslet.2010.09.033. [DOI] [PubMed] [Google Scholar]
- 39.Flemetakis E, et al. A sucrose transporter, LjSUT4, is up-regulated during Lotus japonicus nodule development. J Exp Bot. 2003;54(388):1789–1791. doi: 10.1093/jxb/erg179. [DOI] [PubMed] [Google Scholar]
- 40.Welham T, et al. A cytosolic invertase is required for normal growth and cell development in the model legume, Lotus japonicus. J Exp Bot. 2009;60(12):3353–3365. doi: 10.1093/jxb/erp169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carroll BJ, Gresshoff PM. Nitrate inhibition of nodulation and nitrogen-fixation in white clover. Z Pflanzenphysiol. 1983;110(1):77–88. [Google Scholar]
- 42.Reid DE, Heckmann AB, Novák O, Kelly S, Stougaard J. Cytokinin oxidase/dehydrogenase3 maintains cytokinin homeostasis during root and nodule development in L. japonicus. Plant Physiol. 2016;170(2):1060–1074. doi: 10.1104/pp.15.00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brandl K, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455(7214):804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dessein R, et al. Toll-like receptor 2 is critical for induction of Reg3 beta expression and intestinal clearance of Yersinia pseudotuberculosis. Gut. 2009;58(6):771–776. doi: 10.1136/gut.2008.168443. [DOI] [PubMed] [Google Scholar]
- 45.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Johnston PR, Rolff J. Host and symbiont jointly control gut microbiota during complete metamorphosis. PLoS Pathog. 2015;11(11):e1005246. doi: 10.1371/journal.ppat.1005246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agler MT, et al. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016;14(1):e1002352. doi: 10.1371/journal.pbio.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bontemps C, et al. Burkholderia species are ancient symbionts of legumes. Mol Ecol. 2010;19(1):44–52. doi: 10.1111/j.1365-294X.2009.04458.x. [DOI] [PubMed] [Google Scholar]
- 49.Garau G, Yates RJ, Deiana P, Howieson JG. Novel strains of nodulating Burkholderia have a role in nitrogen fixation with papilionoid herbaceous legumes adapted to acid, infertile soils. Soil Biol Biochem. 2009;41(1):125–134. [Google Scholar]
- 50.Angus AA, Hirsch AM. Insights into the history of the legume-betaproteobacterial symbiosis. Mol Ecol. 2010;19(1):28–30. doi: 10.1111/j.1365-294X.2009.04459.x. [DOI] [PubMed] [Google Scholar]
- 51.Touceda-Gonzalez M, et al. Combined amendment of immobilizers and the plant growth-promoting strain Burkholderia phytofirmans PsJN favours plant growth and reduces heavy metal uptake. Soil Biol Biochem. 2015;91:140–150. [Google Scholar]
- 52.Lebeis SL, et al. PLANT MICROBIOME. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science. 2015;349(6250):860–864. doi: 10.1126/science.aaa8764. [DOI] [PubMed] [Google Scholar]
- 53.Patriquin DG, Döbereiner J, Jain DK. Sites and processes of association between diazotrophs and grasses. Can J Microbiol. 1983;29(8):900–915. [Google Scholar]
- 54.Egener T, Hurek T, Reinhold-Hurek B. Use of green fluorescent protein to detect expression of nif genes of Azoarcus sp. BH72, a grass-associated diazotroph, on rice roots. Mol Plant Microbe Interact. 1998;11(1):71–75. doi: 10.1094/MPMI.1998.11.1.71. [DOI] [PubMed] [Google Scholar]
- 55.Chi F, et al. Ascending migration of endophytic rhizobia, from roots to leaves, inside rice plants and assessment of benefits to rice growth physiology. Appl Environ Microbiol. 2005;71(11):7271–7278. doi: 10.1128/AEM.71.11.7271-7278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Handberg K, Stougaard J. L. japonicus, an autogamous, diploid legume species for classical and molecular genetics. Plant J. 1992;2(4):487–496. [Google Scholar]
- 57.Triplett EW, Albrecht KA, Oplinger ES. Crop-rotation effects on populations of Bradyrhizobium japonicum and Rhizobium meliloti. Soil Biol Biochem. 1993;25(6):781–784. [Google Scholar]
- 58.Thies JE, Woomer PL, Singleton PW. Enrichment of Bradyrhizobium spp populations in soil due to cropping of the homologous host legume. Soil Biol Biochem. 1995;27(4-5):633–636. [Google Scholar]
- 59.Zuanazzi JAS, et al. Production of Sinorhizobium meliloti nod gene activator and repressor flavonoids from Medicago sativa roots. Mol Plant Microbe Interact. 1998;11(8):784–794. [Google Scholar]
- 60.Rispail N, et al. Secondary metabolite profiling of the model legume L. japonicus during its symbiotic interaction with Mesorhizobium loti. Symbiosis. 2010;50(3):119–128. [Google Scholar]
- 61.White LJ, Jothibasu K, Reese RN, Brözel VS, Subramanian S. Spatio-temporal influence of isoflavonoids on bacterial diversity in the soybean rhizosphere. Molecular plant-microbe interactions. Mol Plant Microbe Interact. 2015;28(1):22–29. doi: 10.1094/MPMI-08-14-0247-R. [DOI] [PubMed] [Google Scholar]
- 62.Szoboszlay M, White-Monsant A, Moe LA. The effect of root exudate 7,4′-dihydroxyflavone and naringenin on soil bacterial community structure. PLoS One. 2016;11(1):e0146555. doi: 10.1371/journal.pone.0146555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hartwig UA, Joseph CM, Phillips DA. Flavonoids released naturally from alfalfa seeds enhance growth rate of Rhizobium meliloti. Plant Physiol. 1991;95(3):797–803. doi: 10.1104/pp.95.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dakora FD, Phillips DA. Diverse functions of isoflavonoids in legumes transcend anti-microbial definitions of phytoalexins. Physiol Mol Plant Pathol. 1996;49(1):1–20. [Google Scholar]
- 65.Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T. A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol. 2015;207(2):437–453. doi: 10.1111/nph.13264. [DOI] [PubMed] [Google Scholar]
- 66.Delaux PM, et al. Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet. 2014;10(7):e1004487. doi: 10.1371/journal.pgen.1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Werner GD, Cornwell WK, Sprent JI, Kattge J, Kiers ET. A single evolutionary innovation drives the deep evolution of symbiotic N2-fixation in angiosperms. Nat Commun. 2014;5:4087. doi: 10.1038/ncomms5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stracke S, et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature. 2002;417(6892):959–962. doi: 10.1038/nature00841. [DOI] [PubMed] [Google Scholar]
- 69.Kanamori N, et al. A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc Natl Acad Sci USA. 2006;103(2):359–364. doi: 10.1073/pnas.0508883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Charpentier M, et al. Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell. 2008;20(12):3467–3479. doi: 10.1105/tpc.108.063255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chelius MK, Triplett EW. The diversity of archaea and bacteria in association with the roots of Zea mays L. Microb Ecol. 2001;41(3):252–263. doi: 10.1007/s002480000087. [DOI] [PubMed] [Google Scholar]
- 72.Koprivova A, Harper AL, Trick M, Bancroft I, Kopriva S. Dissection of the control of anion homeostasis by associative transcriptomics in Brassica napus. Plant Physiol. 2014;166(1):442–450. doi: 10.1104/pp.114.239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 75.Oksanen J, et al. 2015. R Package vegan: Community Ecology Package (R Foundation, Vienna), Version 2.3-3.
- 76.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.