Abstract
The TRAP/Mediator complex serves as a coactivator for many transcriptional activators, including nuclear receptors such as the thyroid hormone receptor (TR) that targets the TRAP220 subunit. The critical but selective function of TRAP220 is evidenced by the embryonic lethal phenotype of Trap220−/− mice and by the observation that Trap220−/− fibroblasts (isolated before embryonic death) are impaired in specific nuclear receptor-dependent pathways. Here we have used a biochemical and genetic approach to understand the basis of specificity in TRAP220 function. We show that Trap220−/− cells possess a TRAP/Mediator complex that is relatively intact and compromised in its ability to support TR-dependent, but not VP16-dependent, transcription in vitro. Transfection studies using TRAP220 mutants revealed that the N terminus of TRAP220 is necessary and sufficient for stable association with the TRAP/Mediator complex and, further, that TRAP220-dependent TR function in transfected cells requires both of the NR boxes that contain the LXXLL motif implicated in nuclear receptor binding. Similarly, an analysis of isolated TRAP/Mediator complexes with mutations in either or both of the two NR boxes confirmed a critical role for them in in vitro coactivator function. The implications of these observations are discussed in terms of our present understanding of coactivator function.
Nuclear receptors constitute a large superfamily of transcription factors that control diverse biological processes such as cell growth, differentiation, and homeostasis (reviewed in references 30 and 45). In the vast majority of cases, nuclear receptor functions are accomplished through a series of molecular events that are triggered by the binding of the cognate ligand. The liganded nuclear receptor, which binds to regulatory elements of target genes, orchestrates the assembly of multiprotein complexes containing various coactivators that directly or indirectly lead to enhanced activity of the preinitiation complex (PIC), which consists of RNA polymerase II (Pol II) and its associated basal transcriptional machinery (reviewed in references 37 and 38).
The various coactivators that have been implicated in the function of nuclear receptors and other transcriptional activators fall into two broad categories. One group is comprised primarily of cofactors that facilitate the initial penetration of chromatin (10, 13, 50) either through ATP-dependent remodeling (SWI/SNF and various I-SWI-containing complexes) or through covalent protein modifications such as histone acetylation (p160 family members, CBP/p300, GCN5-containing SAGA complex) or methylation (CARM1 and PRMT) (41). The other group contains coactivators that were identified, for the most part, by biochemical assays utilizing naked DNA templates and are thus thought to function directly at the level of PIC formation or function (37). This group includes the TATA box binding protein (TBP)-associated factors in TFIID (46); positive cofactors PC1, PC2, PC3, PC4, and PC52 (37) derived from the USA fraction (20); and several multiprotein complexes that are all related to the TRAP/Mediator complex (reviewed in reference 28). TRAP/Mediator was first identified (8) through an intracellular ligand-dependent association with thyroid hormone receptor (TR) and, in turn, is related to Saccharomyces cerevisiae Mediator, the coactivator component of the Pol II holoenzyme (23, 32).
The TRAP/Mediator complex consists of about 25 subunits that are thought to assemble in relatively discrete modules, reflecting both a degree of evolutionary conservation of this coactivator from yeast to metazoans and the coactivator's functional versatility (4, 28). Although additional submodules may be identified, depending on the experimental methods, at least three major subcomplexes have been identified in both the yeast and metazoan complexes. The majority of the subunits are tightly associated and appear to constitute the core of the complex, which likely corresponds to the PC2 and CRSP complexes (25, 28, 43). Nonetheless, some individual subunits that constitute this core, including TRAP220, display variable association with the complex. A second subcomplex consists of polypeptides (SUR2/TRAP150β, TRAP100, and TRAP95) that, as a group, are relatively loosely associated with the core complex (18, 42). Still another module might be composed of CDK8/SRB10, cyclin C/SRB11, TRAP240/SRB9, and TRAP230/SRB8. These subunits are absent in the PC2 and CRSP complexes (25, 28, 43); their respective orthologs in yeast appear to be associated with the complex only under certain cellular states and have been implicated in several negative regulatory transcriptional pathways (3, 5).
Although the precise roles of the various TRAP/Mediator modules and the individual constituent polypeptides are far from clear (either in yeast or metazoan cells), it is believed that they may represent direct physical targets of distinct transcriptional activators (28). They may thereby serve to transduce regulatory signals from activators to Pol II and associated general transcription factors. Thus, many nuclear receptors, including TRα (52), vitamin D receptor (35), peroxisome proliferator-activated receptor γ (PPARγ) (12, 44, 51, 53), estrogen receptor (22, 48), retinoid X receptor (RXR) (51, 53), and hepatocyte nuclear factor 4 (HNF4α) (29), have been demonstrated to physically interact with TRAP220. Consistent with its coactivator role for these receptors, TRAP220 contains two LXXLL motifs (designated NR1 and NR2) (52) that have been shown to mediate nuclear receptor interactions with a number of coactivators (13, 17) that include TRAP220 (22, 36, 48, 51-53). Furthermore, fibroblasts removed from Trap220−/− mouse embryos, which fail to develop beyond 10.5 days postcoitus, do not support efficient TR-mediated activation in transfection assays, whereas activation by nonreceptor activators such as VP16 and p53 is largely unaffected (19). Most strikingly, unlike fibroblasts of wild-type origin, the Trap220−/− fibroblasts display an impaired ability to differentiate into adipocytes, consistent with the defect in the ability of PPARγ to interact with TRAP/Mediator (12). These observations suggest that the TRAP220 subunit plays a specific (specialized) role in receptor function through the TRAP/Mediator complex. They further suggest that, consistent with the modular organization of the TRAP/Mediator complex, the remaining subunits can function largely independently of this subunit.
In this study, we first demonstrate that the residual TRAP/Mediator complex in Trap220−/− fibroblasts is largely intact, except for the absence of the TRAP220 subunit. Further, to establish structure-function relationships for the role of TRAP220 in nuclear receptor function, we report a mutational analysis of this subunit that has identified domains involved in the interaction of TRAP220 with the bulk complex, as well as those involved in physical and functional interactions with the receptors.
MATERIALS AND METHODS
Plasmid constructions.
The vector for generating stable mouse embryo fibroblast (MEF) lines that express the epitope-tagged NUT2 subunit of TRAP/Mediator was derived from a previously described construct (25). Briefly, the NUT2 cDNA fused with sequences encoding FLAG and hemagglutinin (HA) epitopes was amplified by PCR and subcloned into pIRES-hygro (Clontech) to generate pFH-NUT2-hygro. Subcloning of wild-type TRAP220 and its two derivatives (mutants a and b) into expression vector pCIN4 have previously been described (52). The remaining deletion and point mutants of TRAP220 were generated by high-fidelity PCR and FLAG tagged by subcloning into pCIN-neo (Promega). These mutants include the following: a, L607A and L608A located in NR box 1; b, L648A and L649A in NR box 2; 1, V109G and E110S; 2, P213G and R214S; 3, T391G and L392S; Δ1, deletion of residues 108 to 212; Δ2, deletion of residues 215 to 390; AB, region from residue 1 to residue 670; CD, region from residue 624 to residue 1581; and ED, region from residue 1076 to residue 1581.
Epitope-tagged cell lines and affinity purification.
To isolate the residual TRAP/Mediator complex in Trap220−/− MEFs, simian virus 40 (SV40)-immortalized MEFs growing in monolayers were transfected with pFH-NUT2-hygro using Lipofectamine (Invitrogen/GIBCO). After being selected in hygromycin (160 μg/ml; Invitrogen/GIBCO) for about 3 weeks, resistant colonies were picked and expanded for analysis. Clones expressing FLAG- and HA-tagged NUT2 were expanded further. Because the MEFs were not amenable to growth in suspension, Dignam-type nuclear extracts were made directly from monolayers. Nuclear extracts were chromatographed over phosphocellulose (P11) columns, and the combined 0.5 and 0.85 M KCl eluates were subjected to affinity chromatography over M2 agarose as described previously (11, 27).
To generate stable cell lines expressing derivatives of TRAP220, a series of FLAG-tagged TRAP220 mutants were transfected into HeLa-S3 cells by using SuperFect as recommended by the manufacturer (Invitrogen/GIBCO). Stable cell lines were selected with 0.5 mg of G418 (Invitrogen/GIBCO)/ml and, after analysis of clones, expanded into spinner cultures. For each derivative, nuclear extracts were prepared and subjected to affinity chromatography over M2 agarose as described previously (11, 27). Purified preparations were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with a 5 to 15% gradient and visualized with either silver staining (Rapid-AG-Stain; ICN) or immunoblotting using enhanced chemiluminescence (Amersham). In the case of FLAG-tagged proteins, anti-FLAG monoclonal antibodies (Sigma) were used in immunoblotting.
Transient transfections.
MEFs derived from TRAP220−/− embryos were transfected by using Lipofectamine with 0.5 μg of pTRE2-LBK-luc (firefly luciferase), 0.07 μg of pRL-SV40 (Renilla luciferase, internal control), 0.05 μg of pNT7-TRα, and 1 μg of various TRAP220 constructs. Luciferase activity in extracts from transiently transfected cells was determined by the Dual-Luciferase Reporter Assay System (Promega), in which firefly luciferase values were normalized to those of Renilla luciferase. To assess the nuclear protein expression level of a given construct, 10 μg of each construct DNA and 0.3 μg of pRL-SV40 (control for transfection efficiency) were cotransfected into TRAP220−/− MEF cells. Small-scale nuclear extracts from the transfectants were normalized on the basis of the Renilla luciferase activity and analyzed by immunoblotting using either anti-FLAG or anti-HA antibodies.
In vitro transcription.
TRAP/Mediator coactivator activity was assayed in highly purified, reconstituted in vitro transcription systems, as described previously (14-16, 22, 26).
Protein-protein interaction assays.
For glutathione S-transferase (GST) pull-down assays, immobilized GST fusion proteins (2.5 μg) and about 500 ng of TRAP/Mediator complexes purified from the appropriate cell lines were used. Binding of complexes to the immobilized GST proteins and subsequent washing were carried out in buffer containing 180 mM KCl and 0.1% NP-40. The bound TRAP complexes were eluted with a synthetic NR2 polypeptide (TKNHPMLMNLLKDNPA), resolved by SDS-PAGE, and visualized with silver staining.
RESULTS
Composition analysis of a residual TRAP/Mediator complex in TRAP220−/− MEFs.
A previous study from this laboratory showed that although Trap220−/− mouse embryos fail to survive beyond 10.5 days postcoitus, MEFs harvested prior to this could be relatively efficiently propagated in culture (19). Consistent with this ability to survive in culture, we further demonstrated that while these cells could not fully support TR (and T3-)-dependent transcription, the transcriptional potential of several other activators (including p53 and VP16) was not compromised. This indicated that loss of TRAP220 affects only a subset of the transcriptional functions controlled by the TRAP/Mediator complex. It further implied that TRAP220 loss is not accompanied by large-scale disruption of the structural integrity of the complex, although this has remained a critical unanswered question.
Another previous study showed that stable expression in HeLa cells of an epitope-tagged NUT2 subunit, which is an integral component of the core TRAP/Mediator complex (25), allows efficient isolation of the intact TRAP/Mediator complex from corresponding nuclear extracts. Therefore, in order to isolate and characterize the residual complex that supports transcription regulation in Trap220−/− cells, we constructed stable lines of Trap220−/− MEFs (previously immortalized by stably expressing the SV40 T antigen) that expressed a FLAG- and HA-epitope-tagged version of the NUT2 subunit of the Mediator complex. Nuclear extract from one such line (MN-6) was first fractionated over phosphocellulose (P11). The 0.5 and 0.85 M KCl eluates from the column were pooled (so as to reflect essentially the entire TRAP/Mediator population in the extract [27]) and subjected to affinity chromatography over M2 agarose. The resulting preparation was analyzed by both silver staining and immunoblotting (Fig. 1).
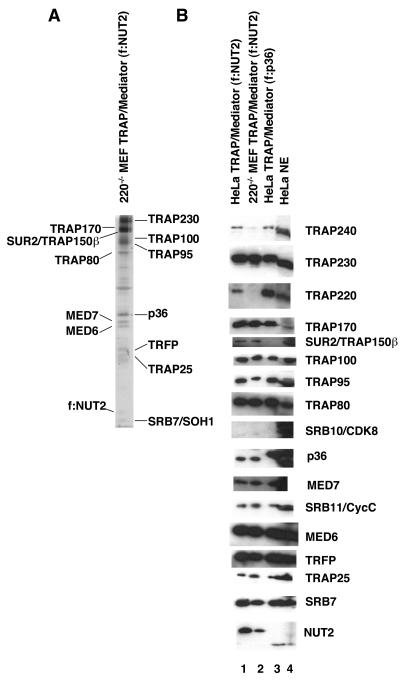
FIG. 1.
Composition of a residual TRAP/Mediator complex in TRAP220−/− MEFs. (A) Purified complex isolated from TRAP220−/− MEFs that had been stably transfected with a construct expressing FLAG-tagged NUT2 (f:NUT2). Nuclear extracts from the cells were chromatographed over P11, and a pool of 0.5 and 0.85 M KCl eluates was subjected to M2 agarose affinity chromatography. Eluates were analyzed by SDS-10.5% PAGE and stained with silver. Easily identifiable bands are marked. (B) Comparative immunoblot analysis of f:NUT2-derived complexes from HeLa cells (lane 1) and TRAP220−/− MEFs (lane 2). Total HeLa cell nuclear extract (NE) (lane 4) and an affinity-purified TRAP/Mediator preparation (lane 3) obtained via FLAG tagging of the p36 subunit (S. Malik and R. G. Roeder, unpublished results) were also included as controls. Following SDS-PAGE and transfer to polyvinylidene difluoride membranes, the complexes were probed with antibodies against the indicated TRAP/Mediator subunit. Note the altered mobility of the NUT2 polypeptide in the case of complexes (lanes 1 and 2) obtained by epitope tagging this subunit.
Comparison of the subunit composition of the complex thus obtained from Trap220−/− cells with an equivalent complex from HeLa cells revealed that, except for the absence of TRAP220 in the former, the two complexes were very similar, at least with respect to 15 of the 17 subunits that were tested (Fig. 1B, compare lanes 1 and 2). Although the TRAP220 subunit appears to be underrepresented in the NUT2-derived preparation from HeLa cells relative to a p36-derived complex (compare lanes 1 and 3), a clear contrast (in TRAP220 content) between the wild-type (lanes 1 and 3) and Trap220−/− (lane 2) complexes was seen. Furthermore, subunits from all three major modules of the TRAP/Mediator complex were represented in the immunoblot analysis. Thus, essentially all of the core subunits (including NUT2, SRB7, TRAP25, TRFP, MED6, MED7, p36, TRAP80, and TRAP170) are present in the Trap220−/− complex in the same proportions as in the complex from wild-type HeLa cells. Among members of the presumptive SRB10-SRB11-TRAP240-TRAP230 module, TRAP230 is present in both complexes (lanes 1 and 2). Consistent with previous results (25), the SRB11/cyclin C subunit was detectable in complexes from both control and Trap220−/− cells, while SRB10/CDK8 was not. We also note that the level of TRAP240 is significantly lower in the Trap220−/− complex relative to that in the wild-type complex from HeLa cells. Whether this is due to an inability of TRAP240 to associate with the complex in the absence of TRAP220 (indicative of a degree of modularity), a reduced expression of TRAP240 in MEFs or, more trivially, significant differences in immunological cross-reactivity between human and mouse proteins is being investigated.
These results indicate that knockout of the TRAP220 gene leaves behind a residual complex that selectively lacks TRAP220 and, at most, a few other subunits. The results also are consistent with a relatively peripheral location of TRAP220 within the TRAP/Mediator complex.
Functional analysis of the Trap220−/− TRAP/Mediator complex.
To characterize the function of the residual TRAP/Mediator complex purified from Trap220−/− MEFs, we assessed the complex's ability to interact with and support activation by VP16 and TR. To evaluate physical interactions with these activators, we incubated wild-type and Trap220−/− TRAP/Mediator preparations with appropriate GST fusions (Fig. 2A), washed the beads to remove unbound material, and probed the bound proteins for TRAP/Mediator subunits by immunoblotting. Both preparations displayed significant specific binding of TRAP/Mediator (monitored by antibodies to TRAP80 and TRFP subunits) to GST-VP16 (Fig. 2A, lanes 3 and 8) relative to control GST (lanes 2 and 7). In contrast, whereas wild-type TRAP/Mediator efficiently bound to liganded TR (compare lanes 9 and 10), the Trap220−/− preparation was significantly impaired in this interaction (compare lanes 4 and 5).
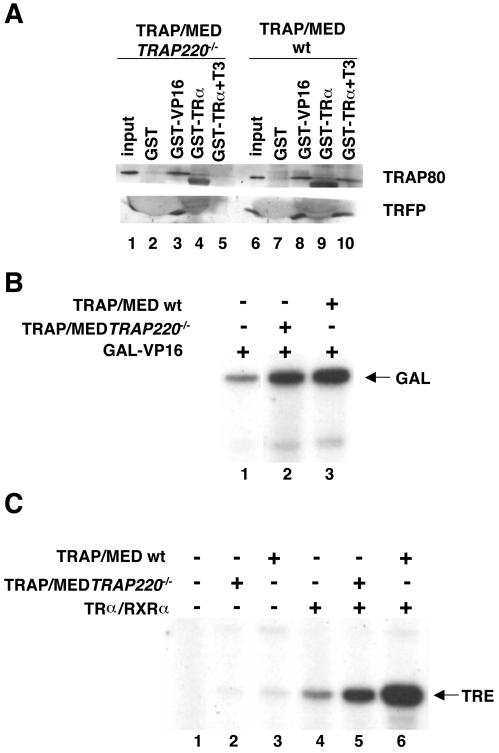
FIG. 2.
Functional analysis of the residual TRAP/Mediator complex from TRAP220−/− MEFs. (A) Wild-type (wt) TRAP/Mediator complex (f:NUT2, HeLa) and a complex obtained from TRAP220−/− MEFs were incubated with GST (lanes 2 and 7), GST-VP16 (lanes 3 and 8), or GST-TRα (lanes 4, 5, 9, and 10). In the case of interactions with GST-TRα, incubations were done in either the absence (lanes 4 and 9) or the presence (lanes 5 and 10) of T3. After being washed, the bound material was eluted from the beads and analyzed by SDS-PAGE and immunoblotting with the indicated TRAP/Mediator antibodies. (B) In vitro transcription reaction mixtures were reconstituted from highly purified transcription factors. All reaction mixtures contained 50 ng of GAL-VP16. TRAP/Mediator was added as indicated: lane 2, TRAP/MED Trap220−/− (f:NUT2); lane 3, TRAP/MED wt (f:NUT2). (C) In vitro transcription reactions were carried out as described for panel B, except that baculovirus-expressed TRα (10 ng) and RXRα (20 ng) were added to lanes 4 to 6. No exogenous ligand was added.
We next checked the two TRAP/Mediator complexes by in vitro transcription assays (Fig. 2B and C). As previously described (15, 26), the assay mixtures were reconstituted from near-homogeneous preparations of all factors: TFIIA, TFIIB, TFIIE, TFIIF, and PC4 were of recombinant origins; TFIID was derived from a cell line expressing FLAG-tagged TATA box binding protein; and RNA Pol II and TFIIH were isolated from HeLa cells following several rounds of conventional chromatography. In the absence of TRAP/Mediator, this system, as set up, supports only low to moderate levels of activator-dependent transcription (Fig. 2B, lane 1; Fig. 2C, compare lanes 1 and 4). Addition of normal (wild-type) TRAP/Mediator substantially stimulated GAL4-VP16-dependent transcription from the cognate template (Fig. 2B, compare lanes 1 and 3) as did the addition of the Trap220−/− complex (compare lanes 1 and 2), in agreement with the above data showing wild-type levels of interaction with VP16. However, in the case of TR-dependent transcription (Fig. 2C), consistent with the binding data, the level of activated transcription observed with the Trap220−/− TRAP/Mediator complex was significantly lower than that seen with the wild-type complex (compare lanes 5 and 6). Nonetheless (see below), the TRAP220 complex did show some coactivator activity for TR (compare lanes 4 and 5).
Collectively, these results indicate that, even though the residual complex from Trap220−/− cells is selectively impaired for TR interaction and function, it suffices for the function of transcriptional activators (VP16) that do not target TRAP220.
The N terminus of TRAP220 is sufficient for TRAP220 incorporation into the TRAP complex.
To identify regions in TRAP220 that are necessary for incorporation of the polypeptide into the complete complex, we constructed vectors expressing epitope-tagged TRAP220 and various derivatives and used these to generate corresponding stable cell lines. Given variations in the strength of the association of TRAP220 with the TRAP/Mediator complex and/or the possibility that TRAP220-containing complexes constitute only a subpopulation of the total TRAP/Mediator pool (25) (Fig. 1B, compare lanes 1 and 3), this approach also was expected to yield complexes with full occupancy of TRAP220. In addition to the full-length TRAP220, we generated three truncated derivatives (Fig. 3A). The N-terminal AB fragment spans amino acid residues 1 to 670 and, importantly, contains both NR boxes that previously were implicated in nuclear receptor binding (NR1 and NR2) (17, 52). The C-terminal CD fragment, which partially overlaps the AB fragment, spans residues 624 to 1581 and contains only the second NR box, NR2. The ED fragment, which was derived from the CD fragment, spans residues 1076 to 1581 and lacks both of the NR boxes.
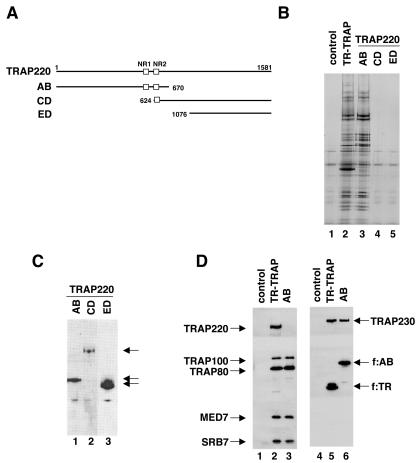
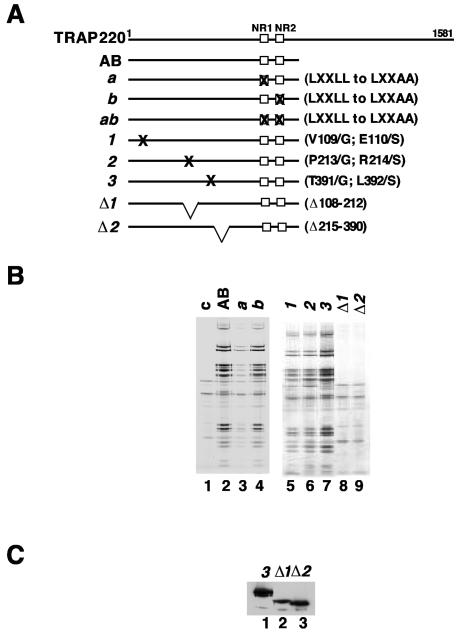
FIG. 3.
Identification of TRAP220 regions that are sufficient for interaction with the TRAP/Mediator complex. (A) Schematic representation of TRAP220 truncations that were FLAG tagged for generation of stable cell lines. The two LXXLL-motif-containing boxes (NR1 and NR2) are represented by open squares. (B) Silver staining of TRAP/Mediator complexes isolated from cell lines expressing f:TR (lane 2) or the f:TRAP220 mutants (lane 3, AB; lane 4, CD; lane 5, ED). A control M2 eluate from normal HeLa cell nuclear extract (lane 1) was included to allow identification of background (nonspecific) bands. Note that, in most experiments, TRAP220 stains negatively with silver. Therefore, the band in the AB complex that migrates around 220 kDa is likely a heterologous polypeptide. (C) Immunoblot analysis (using anti-FLAG antibodies) of purified FLAG-tagged TRAP220 mutants (and putative complexes). The FLAG-tagged polypeptide is identified for each mutant. (D) Immunoblot analysis (using antibodies against selected TRAP/Mediator subunits, as indicated) of the purified TRAP220 AB mutant. The TR-TRAP complex was used as a reference.
Upon stable transfection into HeLa cells and subsequent purification of tagged proteins from derived extracts by M2 agarose affinity chromatography, we failed to isolate either the full-length TRAP220 peptide or any associated complexes (data not shown). Although the actual basis of this observation is uncertain, we attribute this to the masking by the C terminus of epitopes necessary for affinity purification, since the tagged polypeptide could be detected in corresponding cell extracts. Nonetheless, the truncated AB, CD, and ED fragments were all expressed and amenable to efficient isolation via their epitope tags, as detected by immunoblotting with the anti-FLAG antibody (Fig. 3C). Further analysis by SDS-PAGE and silver staining revealed that only the AB fragment copurified with additional (specific) polypeptides (Fig. 3B, lanes 3 to 5). Comparison of the polypeptides contained in the AB-derived preparation with those in the TR-TRAP complex indicated near identity and, thus, an ability of this region of TRAP220 to associate with the complex (compare lanes 2 and 3). Immunoblotting with antibodies to selected TRAP/Mediator subunits (TRAP230, TRAP100, TRAP80, MED6, and SRB7) further confirmed that the AB fragment associates with the TRAP/Mediator complex (Fig. 3D). Note that the ca. 80-kDa AB fragment fully substituted for the endogenous (full-length) TRAP220, which was not detected in the AB complex (Fig. 3D, compare lanes 2 and 3), indicating that TRAP220 likely exists in a monomeric form in the complex. We conclude that an N-terminal region of TRAP220 (up to residue 670) is sufficient for proper physical association with the core TRAP/Mediator complex.
Functional analysis of the AB complex.
We next asked if the complex that contains only the N-terminal region of TRAP220 (hereafter referred to as the AB complex) could recapitulate TR-mediated transcriptional activation as effectively as the TRAP/Mediator complex that contains the full-length TRAP220. We first assessed whether the AB complex still retained its ability to interact with TR in a ligand-dependent manner. GST-TRα (and control GST) were immobilized on beads and incubated with affinity-purified AB complex in either the absence or the presence of the TR cognate ligand (T3). After the unbound material was washed, the proteins retained on the beads were eluted and analyzed by SDS-PAGE and silver staining (Fig. 4A). In the presence of T3, but not in its absence, the AB complex displayed efficient interaction with GST-TRα (Fig. 4A, compare lanes 1 and 4 and lanes 3 and 4) with almost half of the input (lane 5) being specifically retained on the beads. This is essentially equivalent to what is seen with a native complex containing full-length TRAP220 and indicates that, despite the absence of more than half of its carboxyl region, the residual TRAP220 fragment can be incorporated into the TRAP/Mediator complex and can mediate ligand-dependent interactions with TR (likely via the NR boxes) (see below).
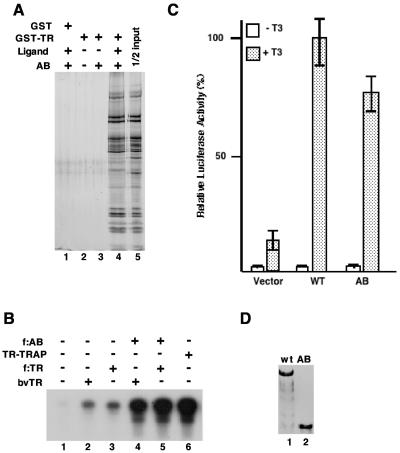
FIG. 4.
The N-terminal AB domain of TRAP220 is sufficient both for interaction with TR and for coactivator function. (A) In the GST pull-down assay, the AB complex (50% of the input is shown in lane 5) was incubated with GST (lane 1) or GST-TRα (lanes 2 to 4) either with no ligand (lanes 2 and 3) or in the presence of 10−7 M T3 (lanes 1 and 4). The retained material was eluted and, following SDS-PAGE, was visualized by silver staining. (B) In vitro transcription assay mixtures were reconstituted from highly purified factors (15), and transcription of a template bearing TR cognate sites (TRE) was monitored. Purified f:AB complex was added to reaction mixtures (lanes 4 and 5). These reaction mixtures also contained TR either from HeLa cells (lanes 3 and 5) or obtained via a baculovirus (bv) expression system (lanes 2 and 4). The TR-TRAP complex (lane 6) was used as a reference. RXRα, but not exogenous ligand, was added to all reaction mixtures. A control template (ML200) that does not contain TR-responsive elements was also included in the reaction mixtures to monitor effects on basal transcription. Although transcription from this template is not apparent at the autoradiography exposure shown, longer exposure revealed no effect of the AB complex on basal transcription. (C) Transient transfection assays were performed with TRAP220−/− MEF cells. Plasmid constructs containing either the full-length TRAP220 (wild type [WT]) or the AB derivative were transfected into the cells in either the presence or absence of T3, as indicated. The empty vector was used as a control. Following transfection, the normalized luciferase activity of the resulting extracts was measured. (D) Immunoblotting to monitor expression of the protein. After TRAP220 and mutant AB proteins were transiently expressed in TRAP220−/− MEF cells, the expressed proteins in nuclei were extracted and probed with anti-HA antibodies.
To determine if the AB complex actually supports TR-dependent transcription, we utilized both an in vitro transcription assay and a transfection-based approach. In the in vitro assay (Fig. 4B), we reconstituted the reaction mixture with highly purified components (15) and compared the ability of the AB complex to mediate activation by TR (and RXRα) relative to that of the previously described TR-TRAP complex (8). Since the latter complex contains TR (through which it is normally purified), thereby precluding a direct comparison, we tested whether the incubation of the AB complex with an amount of TR that is equivalent to that contained in a reference TR-TRAP complex leads to comparable stimulation of transcription from a TR-responsive template. TR from two sources (expressed in baculovirus or in HeLa cells) was used for this experiment. Inclusion of either preparation, in the presence of a saturating amount of RXRα, yielded low levels of transcription (Fig. 4B, compare lane 1 to lanes 2 and 3) relative to that observed with the TR-TRAP complex (lane 6). However, upon addition of the AB complex to the reaction mixtures, TR-dependent transcription was stimulated to levels comparable to that observed with the TR-TRAP complex (compare lanes 4 and 5 to lane 6). This indicates that the AB complex (and the truncated TRAP220 contained therein) is functionally competent in vitro. No effect of the AB complex on TR-independent (basal) transcription, monitored by a template that lacked TR-responsive elements, was seen under the conditions of the assay (data not shown).
To evaluate the activity of the AB fragment in vivo, we compared the effects of full-length TRAP220 to those of the AB fragment on ligand-dependent TR function in transfected Trap220−/− MEFs (Fig. 4C). As in earlier studies (19), TR activity (with T3) on a transfected reporter containing thyroid response element (TRE) sites was severely compromised, albeit not abolished (see below), in these cells. However, coexpression (with TR) of ectopic wild-type TRAP220 greatly (eightfold) increased the stimulation of the ligand-dependent activity of TR. Similarly, coexpression of the AB derivative of TRAP220 also led to a further (circa sixfold) enhancement of liganded TR activity. Parallel control experiments further established that full-length TRAP220 and AB were expressed at comparable levels in these assays (Fig. 4D). These results thus corroborate the conclusions from the in vitro analysis indicating essentially full functionality of the AB complex, at least with respect to TR activity.
Fine mapping of TRAP220 sequences required for incorporation into the TRAP complex or for interaction with TR.
To further delineate the exact sequences required for TRAP220 function, we introduced several mutations (outlined in Fig. 5A) into the AB fragment and asked if they affected the ability of TRAP220 to enter the TRAP/Mediator complex, the ability to interact with TR, or both. Mutant a contained a disruption of the first LXXLL motif (converted to LXXAA), mutant b contained a similar disruption of the second LXXLL motif, and mutant ab contained a joint disruption of both LXXLL motifs. Mutants 1 (V109G, E110S), 2 (P213G, R214S), and 3 (T391G, L392S) contained double point mutations in regions that are highly conserved between human TRAP220 and its Drosophila ortholog (34). Two additional mutants (Δ1 [Δ108-212] and Δ2 [Δ215-390]) were generated by deletions in the conserved regions. Stable HeLa cell lines expressing the corresponding FLAG-tagged TRAP220 AB mutants were generated, and M2 agarose affinity chromatography was employed to purify the mutant polypeptides and any associated complexes from the derived nuclear extracts. Analysis of purified protein preparations by SDS-PAGE and silver staining (Fig. 5B) showed that, except for the deletion mutants Δ1 and Δ2 (which were expressed at levels comparable to those of the other derivatives [Fig. 5C]), all mutants, like the parental AB fragment, were capable of interacting with the TRAP/Mediator complex whose constituent polypeptides were readily discernible. These experiments thus indicate that a region spanning amino acid residues 108 to 390 in TRAP220 is essential for the incorporation of this subunit into the TRAP/Mediator complex, whereas the LXXLL motifs important for nuclear receptor interactions and certain other phylogenetically conserved residues are not.
FIG. 5.
Mutational analysis of the TRAP220 AB region. (A) Schematic representation of mutations introduced in the TRAP220 AB domain. The resulting constructs were FLAG tagged for generation of stable cell lines. (B) Silver staining of purified proteins and putative TRAP/Mediator complexes isolated from cell lines expressing various FLAG-tagged AB derivatives shown in panel A. Lane 1 contains a negative control (M2 agarose eluate from a control HeLa nuclear extract). Note that Δ1 and Δ2 polypeptides stain negatively with silver. (C) Immunoblotting with anti-FLAG antibodies to ascertain expression of selected AB mutants.
Functional analysis of TRAP/Mediator complexes containing mutant NR boxes.
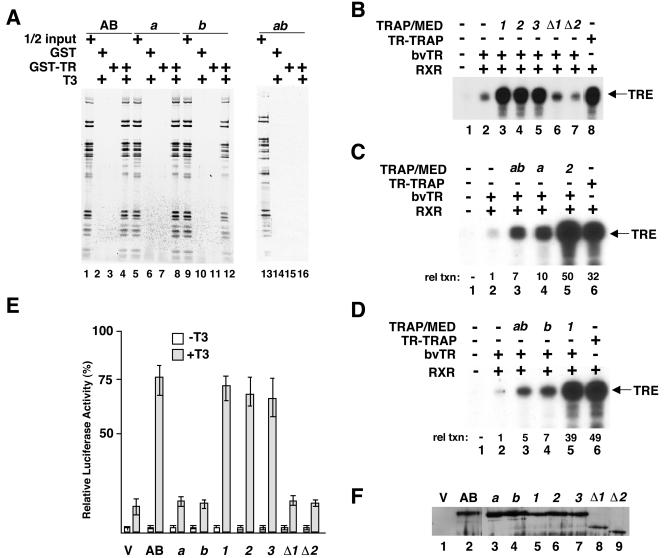
The ability of some variants of the TRAP220 AB fragment (especially a, b, and ab) to be incorporated into the TRAP/Mediator complex is significant from a practical standpoint, because it allows biochemical analysis of the role of the mutated LXXLL motifs in the natural context of the complex. Therefore, essentially as described for the experiment illustrated in Fig. 4, we tested the effects of mutations in the NR boxes on the interactions of TRAP/Mediator complexes carrying these mutations with GST-TRα (Fig. 6A). Complexes derived from either mutant a (compare lane 8 to lanes 6 and 7) or mutant b (compare lane 12 to lanes 10 and 11) bound to TRα (in a ligand-dependent fashion) relatively efficiently compared to the parental AB complex (lane 4). A slightly weaker interaction of the b complex (compare lanes 8 and 12) is consistent with the earlier demonstration that an NR2-containing polypeptide interacts more strongly with TR than an NR1-containing polypeptide (36). However, the ab complex, in which both NR boxes were mutated, did not interact with liganded TRα (compare lane 16 to lanes 14 and 15) at detectable levels.
FIG. 6.
Role of NR boxes in TRAP220 coactivator function. (A) In the GST pull-down assay, the AB (lanes 1 to 4), a (lanes 5 to 8), b (lanes 9 to 12), and ab (lanes 13 to 16) complexes were incubated with GST (lanes 2, 6, 10, and 14) or with GST-TRα (lanes 3, 4, 7, 8, 11, 12, 15, and 16) either with no ligand (lanes 3, 7, 11, and 15) or in the presence of 10−7 M T3 (lanes 2, 4, 6, 8, 10, 12, 14, and 16). Half of the input is shown for each complex tested (lanes 1, 5, 9, and 13). The retained material was eluted and, following SDS-PAGE, was visualized by silver staining. (B to D) In vitro transcription assay mixtures were reconstituted from highly purified factors (16, 22), and transcription of a template bearing TR cognate sites (TRE) was monitored. Purified complexes (derived from AB or mutants a, b, 1, 2, 3, Δ1, Δ2, and ab) were added to the reaction mixtures as indicated. These reaction mixtures also contained TR that was purified via a baculovirus (bv) expression system. bv-expressed RXRα was also included as indicated. The TR-TRAP complex was used as a reference for the experiment shown in panel B; for panels C and D, mutant complexes 2 and 1 also served as references. All reaction mixtures contained RXRα; no ligand was added. Reaction mixtures also contained ML200 template to monitor TR-independent effects, but, at the exposure shown, transcription from this template was not detected. For experiments illustrated in panels C and D, the RNA transcripts were quantitated by phosphorimaging. Transcription levels (rel txn) relative to those obtained in the absence of TRAP/Mediator (lane 2) are indicated. Experiments similar to those shown here were repeated several times. Although the precise magnitude of the effects varied, the general trends as reflected in the data shown were consistently observed. The fold stimulation (and other numbers mentioned in the text) are based on quantitation of the data shown. (E) Transient transfection assays were performed with TRAP220−/− MEF cells. Plasmid constructs containing either the parental TRAP220 AB fragment or the indicated AB derivative were transfected into the cells in either the presence or absence of T3. The empty vector was used as a control. Following transfection, the normalized luciferase activity of the resulting extracts was measured. (F) Immunoblotting to monitor expression of the TRAP220 proteins. After TRAP220 and mutant AB proteins were transiently expressed in TRAP220−/− MEF cells, the expressed nuclear proteins were extracted and probed with anti-FLAG antibodies.
In vitro transcription experiments were carried out to determine if the TRAP/Mediator complexes containing TRAP220 AB mutants were capable of supporting activation by TRα and RXRα in assay mixtures reconstituted from highly purified components (Fig. 6B to D). As described for the experiment illustrated in Fig. 4B, TRα and RXRα displayed very little transcriptional activity from the cognate template in the absence of added TRAP/Mediator coactivator (Fig. 6B, compare lanes 1 and 2). Supplementation with TRAP/Mediator complexes containing TRAP220 mutants 1, 2, and 3 (lanes 3 to 5) resulted in activated transcriptional levels equivalent to that of the reference complex (TR-TRAP in this analysis [lane 8]) but had no effect on basal transcription (data not shown). Thus, these complexes display essentially wild-type levels of coactivator activity. In contrast, the TRAP220 mutants Δ1 and Δ2 (Fig. 6B, lanes 6 and 7), which are impaired in their ability to interact with the TRAP/Mediator complex (see above), showed no significant coactivator activity.
TRAP/Mediator complexes containing NR box mutations (complexes a, b, and ab) (Fig. 5A) were also analyzed in similar in vitro functional assays (Fig. 6C and D). As described above, we tested their ability to support TRα (and RXRα) activity in a reconstituted transcription system that carries out only low-level activator-dependent transcription in the absence of TRAP/Mediator complex (Fig. 6C, lane 2, and D, lane 2). Supplementation with either mutant a complex (Fig. 6C, compare lanes 2 and 4) or mutant b complex (Fig. 6D, compare lanes 2 and 4) stimulated this activity by 10- or 7-fold, respectively. While significant (see Discussion), these levels represented only about 17 to 20% of the activity of complexes 1 (Fig. 6D, compare lanes 4 and 5) and 2 (Fig. 6C, compare lanes 4 and 5), which, as described above, contain silent mutations in the TRAP220 AB fragment. Furthermore, relevant to the issue of selective NR box usage, we reproducibly found that the activity of the mutant b complex was lower (by about 25%) (Fig. 6C and D) (data not shown) than that of the mutant a complex, consistent with its relatively diminished TRα binding potential (see above).
When the mutant ab complex was analyzed in this assay (Fig. 6C and D), it too resulted in significant enhancement (circa five- to sevenfold) of transcription over that seen with TRα and RXRα alone (Fig. 6C and D, compare lanes 2 and 3). However, the ab complex exhibited the lowest in vitro coactivator level of all the complexes (other than the non-Mediator-interacting mutant complexes Δ1 and Δ2) (Fig. 6B), consistent with the failure of the ab complex to interact with TR. Indeed, the coactivator activity of this complex was noticeably lower (by about 30%) (Fig. 6D, compare lanes 3 and 4) than that of the mutant b complex and markedly lower (by 86 to 88%) than those of mutant 1 (Fig. 6D, compare lanes 3 and 5) and 2 complexes (Fig. 6C, compare lanes 3 and 5).
To investigate the effects of the TRAP220 AB mutations in vivo, Trap220−/− MEFs were cotransfected with vectors expressing TR and mutant AB proteins and a luciferase reporter with TRE sites. Transcription was measured in the presence and absence of ligand (Fig. 6E). Of note, and as observed previously (19) (Fig. 4C), there was significant T3-dependent (TR) activity in the TRAP220 null cells. This is also consistent with the residual in vitro TR coactivator function of the ab complex, which, effectively, could have a completely nonfunctional TRAP220. However, in contrast to what was observed in the in vitro analysis and despite expression levels comparable to that of the parental AB fragment (Fig. 6F), mutants a and b were unable to stimulate TR activity beyond this background level. Mutants Δ1 and Δ2, which did not bind the TRAP/Mediator complex (Fig. 5B), were similarly defective. In contrast, mutants 1, 2, and 3, like the reference mutant, AB, showed a five- to sixfold increase in stimulation of TR activity.
Altogether, the analyses of TRAP/Mediator complexes containing TRAP220 NR boxes (Fig. 6) reveal that although single mutants in either NR1 or NR2 are unaffected with respect to binding to TR, their coactivation potential is greatly diminished. Furthermore, when both NR boxes are mutated, both binding to TR and coactivator activity are compromised.
DISCUSSION
The main conclusions of this paper are that (i) Trap220−/− cells contain a residual TRAP/Mediator complex that is largely intact, thus accounting for the function of the complex in routine transcriptional events and, hence, the viability of these cells in culture; (ii) TRAP220 interacts with the TRAP/Mediator complex through its N-terminal domain; and (iii) the TRAP220 NR boxes that mediate ligand-dependent interactions with nuclear receptors are conditionally required for coactivator function. The implications of these findings for TRAP220-dependent TRAP/Mediator function in vitro and in vivo are discussed below.
TRAP220 and the modular organization of the TRAP/Mediator complex.
An implication of our finding that Trap220−/− MEFs retain a form of the TRAP/Mediator complex that is substantially unaltered compared to that found in wild-type cells is that association of TRAP220 with the complex is tangential. This contrasts with what would be expected for a subunit that is integral to the overall structural organization of the complex. For example, in the case of TFIID, the TAFII250 subunit is thought to form a scaffold onto which the other TFIID subunits assemble (6), and its elimination in cells would be predicted to significantly disrupt the complex. Together with the observation that certain TRAP/Mediator preparations contain apparently substoichiometric levels of TRAP220 (25), this property of TRAP220 may have further implications for mechanisms by which TRAP220 function within the context of the TRAP/Mediator complex is effected (see below).
A recent bioinformatic study has suggested that TRAP220 is the metazoan ortholog of the MED1 subunit of the yeast SRB/Mediator complex (4). Underscoring this similarity, med1 disruption in yeast cells, in a manner reminiscent of the effects of TRAP220 elimination, displays a conditional phenotype that resembles that of the dispensable SRB10 gene (1). The yeast data are thus consistent with the present conclusion that the residual TRAP/Mediator complex in Trap220−/− cells is essentially intact and retains many of the functions of the wild-type complex. However, whereas our results would seem to suggest that TRAP220 is not an integral component of the human TRAP/Mediator core, biochemical studies of the yeast complex place MED1 in one of the core subcomplexes (21). Nonetheless, it is important to note that the identified homologies between TRAP220 and MED1 are rather limited and that evolutionary divergence could yet account for acquisition of novel (metazoan-specific) properties.
Multiple functional domains in TRAP220.
Based on our structural and functional analyses, TRAP220 appears to contain several domains. Given that the AB fragment (amino acid residues 1 to 670) is sufficient for interaction both with the residual TRAP/Mediator complex (as defined in this study) and with TR, the key domains that are responsible for the coactivator function, per se, reside toward the N terminus. These include an extended region (minimally encompassing amino acid residues 108 to 390) that interacts with other TRAP/Mediator subunits. In addition, a distinct central region (amino acid residues 600 to 670), which includes the two NR boxes, is critical for the strong ligand-dependent interactions with nuclear receptors (see below). Curiously, neither the in vitro (interaction with core TRAP/Mediator complex, interaction with receptors, and TR-dependent transcriptional activation) nor the in vivo (transfection) assays that we used in this study uncovered a role for the large C-terminal half of the TRAP220 molecule. Preliminary results indicate that the C terminus (1,076 to 1,581 amino acid residues) is heavily phosphorylated (31; C.-X. Yuan and R. G. Roeder, unpublished results), but its role remains unknown.
Potential role of NR boxes in TRAP220 function.
Three-dimensional structural analyses of nuclear receptors bound to NR-box-containing coactivator fragments have revealed that their signature LXXLL motifs and flanking residues closely pack into a groove created by the helices that comprise the receptor's AF2 domain (7, 33, 40). These studies have provided a physical basis for the critical role of NR boxes in nucleating specific coactivator assemblies (49). Our present functional analyses, aimed at understanding the role of the TRAP220 NR boxes in TR activity within the context of the intact TRAP/Mediator complex, have revealed that there may be several distinct underlying modalities. On the one hand, the results of our functional analyses (both in vivo transient transfection and in vitro transcription) showed that individual mutations in the NR1 and NR2 boxes of TRAP220 dramatically affect the ability of TRAP/Mediator to function as a coactivator for TR (Fig. 6). In contrast, the corresponding in vitro binding studies indicated that TRAP/Mediator complexes bearing these mutations bind essentially normally to liganded TR. However, in the case of the TRAP/Mediator complex (ab) that contains mutations in both the NR1 and NR2 complexes, the binding and functional data are consistent in that each type of assay showed compromised potential. However, like the complex derived from Trap220−/− MEFs (Fig. 2C), the complex with mutations in both NR boxes still shows significant in vitro coactivator activity for TR (relative to the activity observed in the complete absence of TRAP/Mediator). This level of activity is qualitatively similar to that of complexes containing TRAP220 with mutations in either NR1 or NR2 (mutants a and b). Moreover, the Trap220−/− MEFs also show significant residual TR activity that likely depends upon the residual TRAP complex (19) (Fig. 4C and 6E). Thus, our in vivo and in vitro data collectively reveal two aspects of Mediator-dependent TR function: (i) an inherent capability for low-level TRAP220-independent TR coactivator function of the Mediator and (ii) a potentially conditional nature of the requirement for the TRAP220 NR boxes. With regard to the latter point, the apparent inconsistency in the binding and functional data, if taken at face value, further implies that binding of TR to TRAP220 (via the NR boxes) is not sufficient to effect high levels of activated transcription. Indeed, as discussed below, our results suggest additional roles for NR boxes besides providing a structural anchor for TR.
In the present study, interactions between TR and mutant TRAP220-containing Mediator complexes were assessed by conventional non-steady-state GST pull-down assays. It is likely that this type of assay only scores strong aggregate protein-protein interactions. Yet in the complex environment of the cell, and likely under the conditions prevailing in our in vitro assay system, other factors may affect the final equilibrium. Thus, parameters that could impact NR box usage in both our in vivo transfection and in vitro assays include involvement of RXRα, the obligatory heterodimerization partner of TRα. It has become clear that RXR's role extends beyond dimerization with TR for DNA binding. Studies with diverse RXR dimerization partners have established that RXR can play an active role even when it is not activated by its cognate ligand (9-cis-retinoic acid) (2, 9, 39). These studies have invoked a “phantom ligand” effect to account for the assumption by RXR of an active conformation that enables it to interact with coactivators. In the specific case of the TR-RXR heterodimer and its interaction with a TRAP220 polypeptide, Ren et al. (36) showed preferential ligand-dependent association of in vitro translated NR2- and NR1-containing fragments (outside their normal complex context) with TRα and RXRα, respectively. In addition, efficient interaction between TRAP220 and the DNA-bound RXR-TR heterodimer was found to require an optimal spacing between the two NR boxes and, importantly, the intact AF2 domains of both TR and RXR. Thus, it is possible that in the context of a higher-order complex (DNA-TRα-RXRα-TRAP/Mediator), wherein TRAP220 is bound to both TR and RXR, the otherwise subtle effects of individual NR box mutations become more pronounced. This could be the case if, for example, critical NR box residues are needed not just for binding to TRα but for establishment of essential RXRα-specific contacts via other hitherto poorly characterized mechanisms, as discussed above. Indeed, in the study by Ren et al. (36), effects of individual NR box mutations on recruitment of a TRAP220 polypeptide fragment were clearly evident in the context of a DNA-bound TRα-RXRα complex.
In the context of the cell, constraints imposed by the natural chromatin template and the consequent requirement for additional cofactors could also be expected to have dominant effects. Based on our present understanding, the TR-dependent activation pathway is thought to be a multistep process (13, 28, 37). Beginning with unliganded TR that is bound to its cognate site within repressed chromatin, one expects the receptor to be engaged with an appropriate corepressor (e.g., N-CoR). Activation (in response to T3) triggers recruitment of chromatin-modifying and other related coactivators (e.g., ACTR), an exchange process that ends with the TRAP/Mediator complex and other components of the Pol II PIC being recruited to initiate transcription. Therefore, it may be that at some point (likely via TRα and RXRα interactions with the NR boxes of TRAP220), an intermediate consisting of TRα, RXRα, TRAP220, and one or more chromatin coactivators forms transiently (47). This would be in accord not only with a conditional requirement for the NR boxes but also with the present indications of a variable and peripheral association of TRAP220 with the TRAP/Mediator complex. Furthermore, this would be analogous to a recent proposal that describes how a ternary complex of TRβ, the coactivator ACTR, and the corepressor N-CoR could represent an intermediate along the activation pathway (24).
According to this scheme, additional constraints (such as prior association of TR with other cofactors) would impose more strict requirements for stronger NR-box-dependent interactions. Thus, under some conditions (e.g., those prevailing in relatively simplified in vitro systems), it may be sufficient to rely on the low-affinity interactions afforded by any compensatory interactions. By contrast, in the more physiological conditions of the transient transfection assay, a requirement for active displacement of prebound TR cofactors (corepressors and coactivators) may call for additional high-affinity interactions mediated by the individual NR boxes, which alone might help overcome a certain critical threshold. Therefore, our future studies directed at understanding the role of TRAP220 in nuclear receptor function are projected to utilize more-natural chromatin templates and additional cofactors in in vitro experiments.
Acknowledgments
We thank M. Ito and Y. Tao for antibodies and members of our laboratory for many fruitful discussions.
This work was supported by grants from the National Institutes of Health to R.G.R. and institutional funding for the laboratory.
REFERENCES
- 1.Balciunas, D., C. Galman, H. Ronne, and S. Bjorklund. 1999. The Med1 subunit of the yeast mediator complex is involved in both transcriptional activation and repression. Proc. Natl. Acad. Sci. USA 96:376-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bettoun, D. J., T. P. Burris, K. A. Houck, D. W. Buck II, K. R. Stayrook, B. Khalifa, J. Lu, W. W. Chin, and S. Nagpal. 2003. Retinoid X receptor is a nonsilent major contributor to vitamin D receptor-mediated transcriptional activation. Mol. Endocrinol. 17:2320-2328. [DOI] [PubMed] [Google Scholar]
- 3.Borggrefe, T., R. Davis, H. Erdjument-Bromage, P. Tempst, and R. D. Kornberg. 2002. A complex of the Srb8, -9, -10, and -11 transcriptional regulatory proteins from yeast. J. Biol. Chem. 277:44202-44207. [DOI] [PubMed] [Google Scholar]
- 4.Boube, M., L. Joulia, D. L. Cribbs, and H. M. Bourbon. 2002. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110:143-151. [DOI] [PubMed] [Google Scholar]
- 5.Carlson, M. 1997. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu. Rev. Cell Dev. Biol. 13:1-23. [DOI] [PubMed] [Google Scholar]
- 6.Chen, J. L., L. D. Attardi, C. P. Verrijzer, K. Yokomori, and R. Tjian. 1994. Assembly of recombinant TFIID reveals differential coactivator requirements for distinct transcriptional activators. Cell 79:93-105. [DOI] [PubMed] [Google Scholar]
- 7.Darimont, B. D., R. L. Wagner, J. W. Apriletti, M. R. Stallcup, P. J. Kushner, J. D. Baxter, R. J. Fletterick, and K. R. Yamamoto. 1998. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 12:3343-3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fondell, J. D., H. Ge, and R. G. Roeder. 1996. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl. Acad. Sci. USA 93:8329-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forman, B. M., K. Umesono, J. Chen, and R. M. Evans. 1995. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell 81:541-550. [DOI] [PubMed] [Google Scholar]
- 10.Freedman, L. P. 1999. Increasing the complexity of coactivation in nuclear receptor signaling. Cell 97:5-8. [DOI] [PubMed] [Google Scholar]
- 11.Ge, H., E. Martinez, C. M. Chiang, and R. G. Roeder. 1996. Activator-dependent transcription by mammalian RNA polymerase II: in vitro reconstitution with general transcription factors and cofactors. Methods Enzymol. 274:57-71. [DOI] [PubMed] [Google Scholar]
- 12.Ge, K., M. Guermah, C. X. Yuan, M. Ito, A. E. Wallberg, B. M. Spiegelman, and R. G. Roeder. 2002. Transcription coactivator TRAP220 is required for PPAR gamma 2-stimulated adipogenesis. Nature 417:563-567. [DOI] [PubMed] [Google Scholar]
- 13.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 14.Gu, W., S. Malik, M. Ito, C. X. Yuan, J. D. Fondell, X. Zhang, E. Martinez, J. Qin, and R. G. Roeder. 1999. A novel human SRB/MED-containing cofactor complex, SMCC, involved in transcription regulation. Mol. Cell 3:97-108. [DOI] [PubMed] [Google Scholar]
- 15.Guermah, M., S. Malik, and R. G. Roeder. 1998. Involvement of TFIID and USA components in transcriptional activation of the human immunodeficiency virus promoter by NF-κB and Sp1. Mol. Cell. Biol. 18:3234-3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guermah, M., Y. Tao, and R. G. Roeder. 2001. Positive and negative TAFII functions that suggest a dynamic TFIID structure and elicit synergy with TRAPs in activator-induced transcription. Mol. Cell. Biol. 21:6882-6894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heery, D. M., E. Kalkhoven, S. Hoare, and M. G. Parker. 1997. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 387:733-736. [DOI] [PubMed] [Google Scholar]
- 18.Ito, M., H. J. Okano, R. B. Darnell, and R. G. Roeder. 2002. The TRAP100 component of the TRAP/Mediator complex is essential in broad transcriptional events and development. EMBO J. 21:3464-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito, M., C. X. Yuan, H. J. Okano, R. B. Darnell, and R. G. Roeder. 2000. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol. Cell 5:683-693. [DOI] [PubMed] [Google Scholar]
- 20.Kaiser, K., and M. Meisterernst. 1996. The human general cofactors. Trends Biochem. Sci. 21:342-345. [PubMed] [Google Scholar]
- 21.Kang, J. S., S. H. Kim, M. S. Hwang, S. J. Han, Y. C. Lee, and Y. J. Kim. 2001. The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 276:42003-42010. [DOI] [PubMed] [Google Scholar]
- 22.Kang, Y. K., M. Guermah, C. X. Yuan, and R. G. Roeder. 2002. The TRAP/Mediator coactivator complex interacts directly with estrogen receptors alpha and beta through the TRAP220 subunit and directly enhances estrogen receptor function in vitro. Proc. Natl. Acad. Sci. USA 99:2642-2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee, T. I., and R. A. Young. 2000. Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34:77-137. [DOI] [PubMed] [Google Scholar]
- 24.Li, X., E. A. Kimbrel, D. J. Kenan, and D. P. McDonnell. 2002. Direct interactions between corepressors and coactivators permit the integration of nuclear receptor-mediated repression and activation. Mol. Endocrinol. 16:1482-1491. [DOI] [PubMed] [Google Scholar]
- 25.Malik, S., W. Gu, W. Wu, J. Qin, and R. G. Roeder. 2000. The USA-derived transcriptional coactivator PC2 is a submodule of TRAP/SMCC and acts synergistically with other PCs. Mol. Cell 5:753-760. [DOI] [PubMed] [Google Scholar]
- 26.Malik, S., M. Guermah, and R. G. Roeder. 1998. A dynamic model for PC4 coactivator function in RNA polymerase II transcription. Proc. Natl. Acad. Sci. USA 95:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malik, S., and R. G. Roeder. 2003. Isolation and functional characterization of the TRAP/Mediator complex. Methods Enzymol. 364:257-284. [DOI] [PubMed] [Google Scholar]
- 28.Malik, S., and R. G. Roeder. 2000. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 25:277-283. [DOI] [PubMed] [Google Scholar]
- 29.Malik, S., A. E. Wallberg, Y. K. Kang, and R. G. Roeder. 2002. TRAP/SMCC/Mediator-dependent transcriptional activation from DNA and chromatin templates by orphan nuclear receptor hepatocyte nuclear factor 4. Mol. Cell. Biol. 22:5626-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, et al. 1995. The nuclear receptor superfamily: the second decade. Cell 83:835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Misra, P., E. D. Owuor, W. Li, S. Yu, C. Qi, K. Meyer, Y. J. Zhu, M. S. Rao, A. N. Kong, and J. K. Reddy. 2002. Phosphorylation of transcriptional coactivator peroxisome proliferator-activated receptor (PPAR)-binding protein (PBP). Stimulation of transcriptional regulation by mitogen-activated protein kinase. J. Biol. Chem. 277:48745-48754. [DOI] [PubMed] [Google Scholar]
- 32.Myers, L. C., and R. D. Kornberg. 2000. Mediator of transcriptional regulation. Annu. Rev. Biochem. 69:729-749. [DOI] [PubMed] [Google Scholar]
- 33.Nolte, R. T., G. B. Wisely, S. Westin, J. E. Cobb, M. H. Lambert, R. Kurokawa, M. G. Rosenfeld, T. M. Willson, C. K. Glass, and M. V. Milburn. 1998. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature 395:137-143. [DOI] [PubMed] [Google Scholar]
- 34.Park, J. M., B. S. Gim, J. M. Kim, J. H. Yoon, H.-S. Kim, J.-G. Kang, and Y.-J. Kim. 2001. Drosophila Mediator complex is broadly utilized by diverse gene-specific transcription factors at different types of core promoters. Mol. Cell. Biol. 21:2312-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rachez, C., Z. Suldan, J. Ward, C. P. Chang, D. Burakov, H. Erdjument-Bromage, P. Tempst, and L. P. Freedman. 1998. A novel protein complex that interacts with the vitamin D3 receptor in a ligand-dependent manner and enhances VDR transactivation in a cell-free system. Genes Dev. 12:1787-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren, Y., E. Behre, Z. Ren, J. Zhang, Q. Wang, and J. D. Fondell. 2000. Specific structural motifs determine TRAP220 interactions with nuclear hormone receptors. Mol. Cell. Biol. 20:5433-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roeder, R. G. 1998. Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harbor Symp. Quant. Biol. 63:201-218. [DOI] [PubMed] [Google Scholar]
- 38.Roeder, R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 21:327-335. [PubMed] [Google Scholar]
- 39.Schulman, I. G., C. Li, J. W. Schwabe, and R. M. Evans. 1997. The phantom ligand effect: allosteric control of transcription by the retinoid X receptor. Genes Dev. 11:299-308. [DOI] [PubMed] [Google Scholar]
- 40.Shiau, A. K., D. Barstad, P. M. Loria, L. Cheng, P. J. Kushner, D. A. Agard, and G. L. Greene. 1998. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell 95:927-937. [DOI] [PubMed] [Google Scholar]
- 41.Stallcup, M. R. 2001. Role of protein methylation in chromatin remodeling and transcriptional regulation. Oncogene 20:3014-3020. [DOI] [PubMed] [Google Scholar]
- 42.Stevens, J. L., G. T. Cantin, G. Wang, A. Shevchenko, and A. J. Berk. 2002. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science 296:755-758. [DOI] [PubMed] [Google Scholar]
- 43.Taatjes, D. J., A. M. Naar, F. Andel III, E. Nogales, and R. Tjian. 2002. Structure, function, and activator-induced conformations of the CRSP coactivator. Science 295:1058-1062. [DOI] [PubMed] [Google Scholar]
- 44.Treuter, E., L. Johansson, J. S. Thomsen, A. Warnmark, J. Leers, M. Pelto-Huikko, M. Sjoberg, A. P. Wright, G. Spyrou, and J. A. Gustafsson. 1999. Competition between thyroid hormone receptor-associated protein (TRAP) 220 and transcriptional intermediary factor (TIF) 2 for binding to nuclear receptors. Implications for the recruitment of TRAP and p160 coactivator complexes. J. Biol. Chem. 274:6667-6677. [DOI] [PubMed] [Google Scholar]
- 45.Tsai, M. J., and B. W. O'Malley. 1994. Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Annu. Rev. Biochem. 63:451-486. [DOI] [PubMed] [Google Scholar]
- 46.Verrijzer, C. P., and R. Tjian. 1996. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem. Sci. 21:338-342. [PubMed] [Google Scholar]
- 47.Wallberg, A. E., S. Yamamura, S. Malik, B. M. Spiegelman, and R. G. Roeder. 2003. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol. Cell 12:1137-1149. [DOI] [PubMed] [Google Scholar]
- 48.Warnmark, A., T. Almlof, J. Leers, J. A. Gustafsson, and E. Treuter. 2001. Differential recruitment of the mammalian mediator subunit TRAP220 by estrogen receptors ERalpha and ERbeta. J. Biol. Chem. 276:23397-23404. [DOI] [PubMed] [Google Scholar]
- 49.Warnmark, A., E. Treuter, A. P. Wright, and J. A. Gustafsson. 2003. Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol. Endocrinol. 17:1901-1909. [DOI] [PubMed] [Google Scholar]
- 50.Xu, L., C. K. Glass, and M. G. Rosenfeld. 1999. Coactivator and corepressor complexes in nuclear receptor function. Curr. Opin. Genet. Dev. 9:140-147. [DOI] [PubMed] [Google Scholar]
- 51.Yang, W., C. Rachez, and L. P. Freedman. 2000. Discrete roles for peroxisome proliferator-activated receptor gamma and retinoid X receptor in recruiting nuclear receptor coactivators. Mol. Cell. Biol. 20:8008-8017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan, C. X., M. Ito, J. D. Fondell, Z. Y. Fu, and R. G. Roeder. 1998. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl. Acad. Sci. USA 95:7939-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu, Y., C. Qi, S. Jain, M. S. Rao, and J. K. Reddy. 1997. Isolation and characterization of PBP, a protein that interacts with peroxisome proliferator-activated receptor. J. Biol. Chem. 272:25500-25506. [DOI] [PubMed] [Google Scholar]