Significance
The valley of Tehuacán in Mexico is an important center of early Mesoamerican agriculture. To characterize the genetic constitution of the earliest phase of maize cultivation, we reexamined San Marcos cave in Tehuacán and sequenced DNA from three newly discovered maize samples dating at a similar age of 5,000 y B.P. The genomes of these samples reveal unforeseen levels of genetic diversity as compared with modern maize, indicating that the effects of domestication were not yet complete. We find that their genetic constitution was similar and influenced by inbreeding, suggesting that the corresponding plants come from a reduced population of isolated and perhaps self-pollinated individuals.
Keywords: maize, paleogenomics, teosinte, domestication, Tehuacán
Abstract
Pioneering archaeological expeditions lead by Richard MacNeish in the 1960s identified the valley of Tehuacán as an important center of early Mesoamerican agriculture, providing by far the widest collection of ancient crop remains, including maize. In 2012, a new exploration of San Marcos cave (Tehuacán, Mexico) yielded nonmanipulated maize specimens dating at a similar age of 5,300–4,970 calibrated y B.P. On the basis of shotgun sequencing and genomic comparisons to Balsas teosinte and modern maize, we show herein that the earliest maize from San Marcos cave was a partial domesticate diverging from the landraces and containing ancestral allelic variants that are absent from extant maize populations. Whereas some domestication loci, such as teosinte branched1 (tb1) and brittle endosperm2 (bt2), had already lost most of the nucleotide variability present in Balsas teosinte, others, such as teosinte glume architecture1 (tga1) and sugary1 (su1), conserved partial levels of nucleotide variability that are absent from extant maize. Genetic comparisons among three temporally convergent samples revealed that they were homozygous and identical by descent across their genome. Our results indicate that the earliest maize from San Marcos was already inbred, opening the possibility for Tehuacán maize cultivation evolving from reduced founder populations of isolated and perhaps self-pollinated individuals.
Botanical, archaeological, and genetic evidence indicate that maize (Zea mays ssp. mays L.) was domesticated in Mexico from Balsas teosinte (Zea mays ssp. parviglumis) as its single wild ancestor. The term “teosinte” refers to all annual or perennial species of the genus Zea that do not include maize, and that currently spread from northern Mexico to southwestern Nicaragua (1, 2). Mexico has the largest diversity of native maize germplasm, with no fewer than 59 native landraces that maintain more nucleotide diversity and less genetic differentiation from their ancestor than other crop species (3, 4). Extensive molecular analysis indicated that maize arose in central Mexico through a single domestication event that occurred ∼9,000 y B.P. (5). These same studies resolved that the populations of Balsas teosinte that are most closely related to extant maize are currently located at the intersection of the states of Michoacán, Guerrero, and Estado de Mexico, suggesting that maize diverged from an ancestral teosinte population in the Balsas river drainage (5–8). Domestication resulted in a group of ancient landraces that subsequently spread throughout the continent, adapting to a wide diversity of human practices, environmental conditions, and ecological niches (8, 9). Because cross-pollination prevails as a reproductive habit, it is believed that maize diversified through continuous divergent selection, favoring heterozygosity and distinct local adaptation. Because of this large diversity, extant native populations in Mexico show a large phenotypic variation in quantitative traits, such as plant height, ear size, kernel row number, or flowering time (10).
Pioneering archaeological expeditions lead by Richard MacNeish identified the valley of Tehuacán as an important center of early Mesoamerican agriculture (11, 12). After extensively exploring five caves (Coxcatlan, Purron, El Riego, Tecorral, and San Marcos), the MacNeish expedition uncovered more than 24,100 specimens that were identified as maize. In particular, the San Marcos cave yielded a total of 1,248 maize specimens in a well-defined stratigraphic sequence covering an evolutionary period of ∼6,500 y. Direct accelerator mass spectrometry (AMS) confirmed that the earliest maize found in Tehuacán was from San Marcos cave, with specimens dating 5,640–5,000 calibrated y B.P. (13). Although phytolith and starch grain evidence indicate that maize was present in the Balsas river valley by 8,700 y B.P. (14), the most ancient Mexican maize specimens reported to date are two inflorescence fragments found in the Guila Naquitz cave (Oaxaca) and averaging 6,235 y B.P. (15, 16). Contrary to specimens from Guila Naquitz (16), the earliest specimens of San Marcos are 27 remarkably uniform and tunicated cobs measuring 19–25 mm in length (17). A morphometric reexamination of the earliest specimens from San Marcos concluded that the cobs exhibited morphological traits indistinguishable from those found in some of the extant landraces, suggesting that the corresponding plants were already fully domesticated (17).
Limited analysis has been performed to investigate the process of maize domestication from a paleogenomic perspective. DNA extraction from repository specimens found at Ocampo caves (Tamaulipas, Mexico) showed that artificially selected contemporary alleles for teosinte branched1 (tb1), prolamin box binding factor (pbf), and sugary1 (su1)—involved in plant architecture, storage protein synthesis, and starch production, respectively—were present in cultivated maize by 4,400–4,300 y B.P. (18). More recently, nuclear DNA sequencing of hybridization-capture targets spanning 33 genes in 32 ancient maize samples from highly diverse geographic locations, and dating from 5141 ± 29 to 710 ± 50 14C y B.P., determined that maize was brought to the American Southwest from the central highlands, with subsequent gene flow from the Pacific coastal corridor (19). Although the analysis included three samples from Tehuacán, the genetic constitution and diversity of the earliest maize has not been elucidated and compared with extant landraces or Balsas teosinte. In particular, it remains unclear if the earliest maize from Tehuacán was partially or fully domesticated, and if the degree of genetic diversity found in the corresponding ancient populations could be similar to the degree of genetic diversity found in extant landraces.
In 2012 we initiated a new series of excavations in Tehuacán caves with the purpose of uncovering organic remains corresponding to ancient Mesoamerican crops. Our reexamination of San Marcos cave yielded several macrospecimens of maize dating 5,300–1,950 calibrated y B.P. Using whole-genome shotgun sequencing, we characterized the genome of three specimens dating 5,300–4,970 y B.P. and corresponding to the earliest cultural phase of Tehuacán. To reveal the population context in which initial maize domestication took place, we compared their genomic information to the genome of Balsas teosinte and extant maize. We also explored the level of genetic diversity that prevailed in loci that were artificially selected during domestication, and determined the degree of genetic similarity that prevailed among these ancient samples of similar age. Our results provide evidence of an unforeseen evolutionary context in which the initial phase of maize cultivation in Tehuacán included partially domesticated inbred individuals that prevailed in specific regions of the valley close to 5,000 y B.P.
Results and Discussion
New Excavation and Sampling in San Marcos Cave.
The 2012 expedition to San Marcos cave is illustrated in Fig. 1 A and B, where we recovered nine well-preserved macrospecimens of maize (Fig. 1 C and D). All except one (SM4, a carbonized cob) were morphologically analyzed and sampled for AMS dating (SI Appendix, Table S1). The most ancient specimen (SM10) was dated 4,240 ± 30 14C y B.P. (5,300–5,040 2σ calibrated age y B.P. at 95% confidence). Three other specimens found in distinct quadrants (SM3, SM5, and SM9) were dated 4,220–4,180 14C y B.P. (5,300–4,970 2σ calibrated age years B.P. at 95% confidence). SM9 and SM10 were two cobs morphologically reminiscent to those found in Zone E during the MacNeish expedition (Fig. 1D); their soft and long spikelet glumes confirmed that the earliest maize found in San Marcos was tunicated. In contrast, SM3 was a well-preserved basal stalk and SM5 was an aerial leaf sheet containing part of the internode (Fig. 1C). Overall, these specimens were equivalent in age and state of preservation to those originally collected during the MacNeish expedition, and currently preserved in several private or public collections (11, 20).
Fig. 1.
New archeological excavations in San Marcos cave. (A) The caves of San Marcos (cave on the left) and Tecorral (cave on the right). (B) Archaeobotanical sampling in San Marcos cave conducted in February 2012. (C) Maize specimens SM3 dating 5,280–4,970 cal. y B.P. (Left) and SM5 dating 5,300–4,980 calibrated y B.P. (Right). (Scale bar, 1.5 cm.) (D) Maize specimens SM9 dating 5,280–4,970 cal. y B.P. (Left) and SM10 5,300–5,040 cal. y B.P. (Right). (Scale bar, 43 mm.)
Paleogenomic Characterization of Ancient Maize Samples.
To determine the genomic constitution and degree of genetic variability present in the 5,300–4,970 y B.P. maize of San Marcos, we extracted DNA from specimens SM3, SM5, SM9, and SM10 and conducted whole-genome shotgun sequencing. Whereas SM9 did not yield sufficient endogenous DNA, recovered DNA from SM3, SM5, and SM10 ranged between 775 and 15,334 pg/μL. Whole-genome shotgun sequencing of high-quality libraries under SOLiD and Illumina platforms generated close to 388.4 × 106 (SM3) and 234.2 × 106 (SM10) quality-filtered reads (SI Appendix, Table S2). Comparison with version 3 of the B73 maize reference genome (21, 22) resulted in 8,479,668 (SM3) and 35,590,282 (SM10) sequences mapping to either repetitive (54.8% for SM3 and 44.5% for SM10) or unique (45.2% for SM3 and 55.5% for SM10) genomic regions, for a total length of 0.31 Gb (SM3) and 1.26 Gb (SM10) of the nonrepetitive genome (SI Appendix, Table S2). Sequences contained signatures of DNA damage typical of postmortem degradation in ancient samples, including overhangs of single-stranded DNA, unusual rates of cytosine deamination, and fragmentation of purines. A total of 33–37% of all sites had signatures of molecular damage and were excluded (23, 24) (SI Appendix, Fig. S1). Final mapping shows that the distribution of SM3 and SM10 sequences spreads over all 10 maize chromosmes (SI Appendix, Table S3). Although coverage depth was variable among both samples, SM3 and SM10 yielded no less than 560,914 and 613,893 unique genomic sites spread across the genome, with a coverage depth of at least 10× (SI Appendix, Fig. S2 and Table S4). Overall, these results indicate that SM3 and SM10 provide an authentic paleogenomic representation of 5,300–4,970 y B.P. maize that can be compared with Balsas teosinte and extant maize to assess its genetic diversity and determine their evolutionary relationship.
Relationship Between Ancient Maize, Extant Landraces, and Balsas Teosinte.
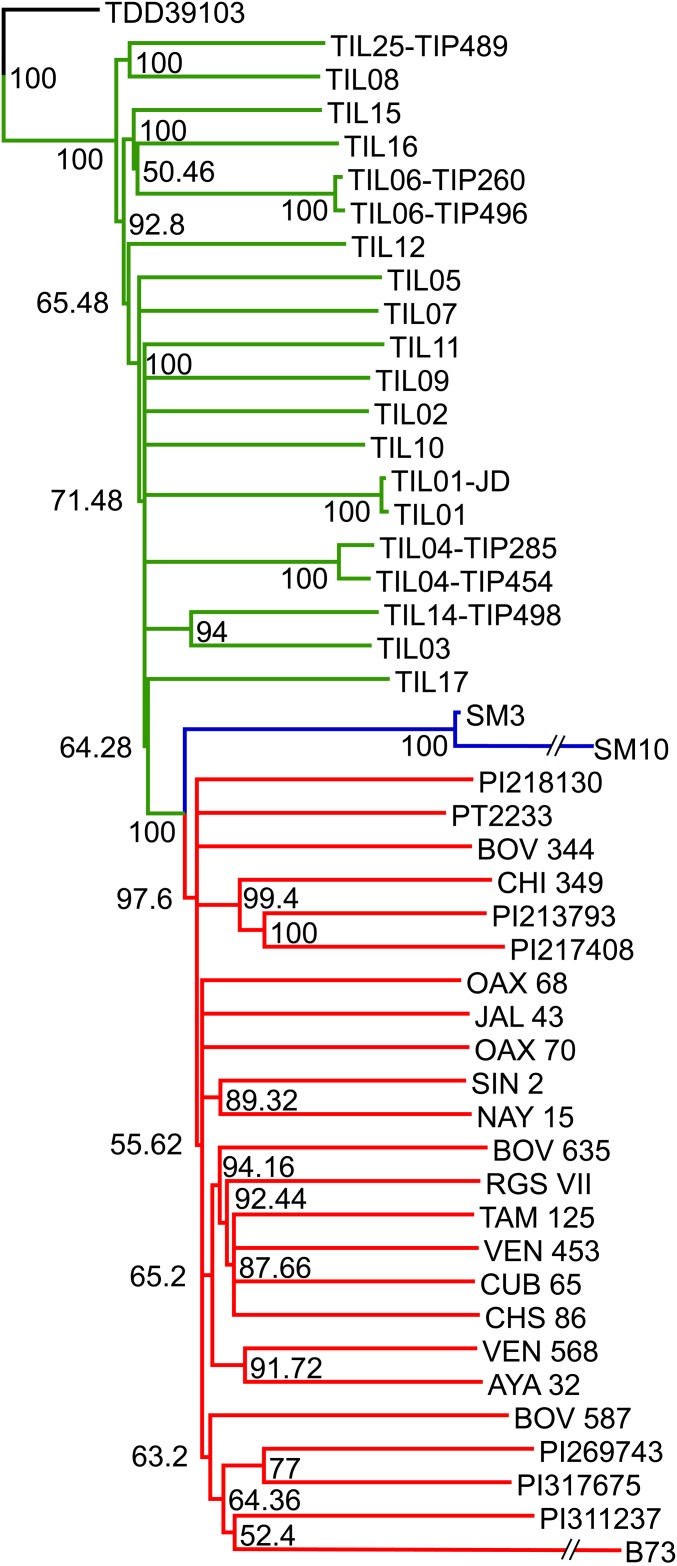
To understand the evolutionary relationship between 5,300 and 4,970 y B.P. maize, its wild ancestor, and extant landraces, we inferred a bootstrapped maximum-likelihood (ML) topology through patterns of population divergence applied to genome-wide polymorphisms common to SM3, SM10, and the HapMap3 dataset available for B73 as a reference genome, 22 maize landraces, 15 Balsas teosinte inbred lines, two accessions of Z. mays spp. mexicana (mexicana teosinte), and a single accession of Tripsacum dactyloides acting as an outgroup (SI Appendix, Figs. S3–S9 and Table S5). Using a previously reported pipeline (25–28), we obtained a total of 100,540 genome-wide SNPs. As illustrated in Fig. 2, the resulting tree shows all maize landraces and teosinte accessions separated in two distinct groups. The two ancient maize samples cluster together outside the diversity of all maize landraces and teosintes. The ancient maize from San Marcos cave diverged from the landraces in all 10,000 bootstrap samples (Fig. 2 and SI Appendix, Fig. S4), indicating that their evolutionary separation is far more likely than their inclusion in the landrace group. Previous studies have demonstrated widespread genomic introgression of mexicana teosinte in extant landraces from the Mexican central highlands but not in other landraces, such those included in the HapMap3 panel (29). To test the possibility that widespread introgression of mexicana teosinte into ancient maize from San Marcos could cause its genomic similarity to teosinte, we included in the ML topology, a previously sequenced central highland accession of the Palomero Toluqueño (PT2233) landrace (30). The tree presented in Fig. 2 placed PT2233 in the landrace group, suggesting that an eventual introgression of mexicana teosinte in ancient San Marcos maize is not the cause of its divergence from the landraces. The divergence between ancient San Marcos maize, teosinte, and the extant landraces is maintained when the analysis is based on single ancient maize samples tested across a significantly larger set of informative SNPs (201,450 in the case of SM3, and 892,033 in the case of SM10) (SI Appendix, Figs. S5 and S6). To determine if the topology could be biased by abundant SNPs represented at low depth coverage, causing a differential in branch length between SM3 and SM10, we generated a distinct tree that only considered the set of 13,079 heterozygous SNPs showing at least 10× coverage, common to both ancient samples and the HapMap3 dataset (SI Appendix, Fig. S7). This analysis produced the same average topology as in Fig. 2, but eliminated the difference in branch length between SM3 and SM10. We also generated an additional ML topology that included a collection of OAX70 SNPs obtained from the same HapMap3 sequence dataset, but independently called with our pipeline. The resulting tree respected the separation between Balsas teosinte and the extant landraces, and grouped both OAX70 datasets adjacently within the landrace group (SI Appendix, Fig. S8), indicating that the inclusion of a sequenced sample to our analysis does not cause a methodological bias resulting in the separation of ancient samples from the landraces and teosintes. Finally, a standard neighbor-joining dendogram based on pairwise p-distances also resulted in a topology similar to Fig. 2, separating the ancient maize samples from the landraces and teosintes (SI Appendix, Fig. S9).
Fig. 2.
Evolutionary relationships between ancient Tehuacán maize and its wild or cultivated relatives. ML tree from an alignment of 100,540 genome-wide SNPs covering nonrepetitive regions of the reference maize genome. SM3 and SM10 represent two maize samples dating 5,300–4,970 calibrated y B.P.; SNPs obtained from 77,960,582 mapped reads of the Palomero Toluqueño landrace (PT2233) were also included. The teosinte group is highlighted in green, the maize landrace group in red, and ancient maize samples from San Marcos in blue. The teosinte and landrace accessions follow previously reported nomenclatures (29); full details of bootstrap values are given in SI Appendix, Fig. S4.
These results indicate that the topology of Fig. 2 best reflects the evolutionary relationships between the ancient San Marcos maize and extant Zea populations, standing in contrast to those implying that the earliest maize from San Marcos was already fully domesticated (17). Although SM10 clearly exhibited the morphological phenotypes specific to a cob derived from a maize and not a teosinte female inflorescence, our evidence suggests that the genomic constitution of the earliest maize from San Marcos is intermediate to Balsas teosinte and maize landraces, maintaining some of the genetic diversity found in its wild ancestor but not in modern maize.
Genomic Evidence of Partial Domestication.
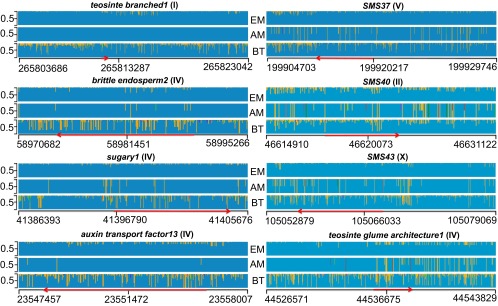
Because genome-wide patterns of diversity can reflect local demography rather than the effects of artificial selection, we conducted a detailed analysis of nucleotide variability in a selected group of loci previously found to be affected by domestication. The group included genomic regions spanning tb1, su1, teosinte glume architecture1 (tga1), brittle endosperm2 (bt2), auxin response factor13 (arf13), and three additional genes discovered during the genomic characterization of the Palomero Toluqueño landrace (SMS37, SMS40, and SMS43) (30). To this aim, we identified all SM10 nucleotide variants corresponding to the overall dataset of informative SNPs available in HapMap3 for extant maize and Balsas teosinte, and this within a genomic region spanning ∼20 kb across each of the selected genes (SI Appendix, Tables S6 and S7). As illustrated in Fig. 3, tb1, su1, bt2, and arf13 show strong loss of nucleotide variability across the genomic region spanning their coding and regulatory sequence in both SM10 and modern maize, suggesting that by 5,300–5,100 y B.P. these genes were already severely affected by domestication. In contrast, SM37, SMS40, SMS43, and tga1 show significantly higher levels of nucleotide variability in SM10 compared with modern maize (Fig. 3), either across the genomic region containing the corresponding gene, or in regions confined to the regulatory or coding sequence.
Fig. 3.
Genomic evidence of partial domestication in ancient Tehuacán maize. Admixture diagrams comparing SM10 nonconsecutive nucleotide variants to the corresponding SNP frequencies reported in modern maize or Balsas teosinte accessions, over 20-kb intervals spanning a selected gene affected by domestication. Blue and yellow correspond to predominant nucleotide variants in extant maize (EM) and their match in SM10 ancient maize (AM) and Balsas teosinte (BT); additional variants in Balsas teosinte or SM10 are depicted in variable colors. Chromosomal locations are indicated adjacent to the locus acronym, in parenthesis; the horizontal scale shows the chromosomal coordinates following the B73 reference genome, and the location and transcriptional orientation of the corresponding gene (red arrow).
In the case of tb1, SM10 contains the Tb1-M1 allele that occurs in close to 97% of modern maize, and this is the same for all 224 SNPs located within 10-kb upstream of its transcription initiation site, suggesting limited tillering and inflorescence phenotypic traits that facilitated harvesting (31, 32). In the case of bt2, a gene involved in starch biosynthesis and kernel composition (33, 34), loss of nucleotide variability prevails throughout a 20-kb region located either up- or downstream of the gene sequence. Reduction of nucleotide variability is less severe in the su1 locus than in the bt2 locus, suggesting that components of the starch biosynthetic pathway were artificially selected at different evolutionary rates. In the case of arf13, only 3 of 442 SNPs spanning the locus correspond to the least-represented SNP modern maize variant. In contrast, genomic regions spanning SMS37 (encoding a topoisomerase II-like protein), SMS40 (encoding a putative potassium channel protein), and SMS43 (encoding a methyl binding domain protein) show partial loss of nucleotide variability in the ancient San Marcos sample, compared with modern maize. For all three loci, a significant number of the SM10 nucleotide variants do not correspond to the prevailing haplotype of modern maize. Of particular interest is tga1, a SBP-domain transcriptional regulator that alters the development of the teosinte cupulate fruit case so that the kernel is exposed on the maize ear (35). Recent studies showed that a single fixed nucleotide difference—at position 18 of the ORF—is sufficient to transform tga1 into a transcriptional repressor (36), but we could not find ancient sequence aligning to tga1 at position 18 of the ORF. For SM10, we found 750 informative SNPs spanning ∼20 kb across the tga1 locus. Although the comparison of nucleotide diversity between Balsas teosinte and modern maize shows that the reduction of nucleotide diversity is significant across this 20-kb region (Fig. 3), in SM10 the region contains 6 of 750 nucleotide variants that are absent from modern maize, and 48 additional sites in which the SM10 SNP variant corresponds to the less-represented nucleotide modern maize variant. Three of six nucleotide variants not corresponding to modern maize map are within 10-kb upstream of the transcriptional initiation site, whereas most of the nucleotide variability found in SM10 is found downstream of the first exon. A comparison of the genetic diversity index θ, and of the frequency of segregating sites per individual at each locus, confirmed these tendencies by showing that in most cases θ and frequency of segregating sites per individual values in SM10 are intermediate to Balsas teosinte and extant maize (SI Appendix, Tables S7 and S8). In addition, we identified SM10 coverage of at least 100 nt in 451 of 462 regions of the maize genome previously identified as being under selection during domestication (37). In at least seven of these segments spread across six different chromosomes, genetic diversity in SM10 is higher than in extant maize at a level beyond the 1 σ value of θ, confirming a tendency toward incomplete domestication of the ancient sample (SI Appendix, Table S9). Overall, these results indicate that the genome of the earliest maize from San Marcos contained multiple loci in which the effect of the artificial selection imposed by domestication was not yet completed.
Genomic Evidence of Identity by Descent and Inbreeding.
Because SM3 and SM10 have a similar age and show a close association in the topology of Fig. 2, we hypothesize that the corresponding maize could have been genetically related. To assess and compare their genetic constitution, for each sample we identified the full set of single nucleotide sites originating from qualilty- and damage-filtered reads mapped to the B73 reference genome and having a minimal coverage of 10×, and compared the full set of single nucleotide sites common to SM3 and SM10 (1,076,063 sites spread in all 10 chromosomes) (SI Appendix, Fig. S10). The large majority of genomic sites common to SM3 and SM10 corresponds to identical homozygous or heterozygous single nucleotide variants (SNVs; 1,066,095 sites or 99.05%), with few sites sharing either one (9,382 SNVs or 0.87%) or no nucleotide variants (586 SNVs or 0.05%) (SI Appendix, Fig. S11 and Tables S10 and S11). In contrast, the sum of identical heterozygous and homozygous SNVs for a pair of randomly selected individuals belonging to an open-pollinated population of 4,500 plants of the Cacahuacintle (CCH) landrace represents 55.9% of all shared sites (SI Appendix, Fig. S11 and Table S10). On the basis of these results, SM3 and SM10 have a 99.1% probability of being identical by descent across their genome when all shared sites are included in the estimation, and a 90% probability if only polymorphic sites with respect to B73 are used.
To determine if this unusually high genetic similarity was fortuitously specific to SM3 and SM10, or if it reflected a trend affecting the earliest Tehuacán maize, we sequenced a DNA genomic library from specimen SM5, generating 1,171,216 Illumina sequences that mapped to unique regions of the maize reference genome. As in the case of the SM3 vs. SM10 comparison, the vast majority of common sites between SM5 and SM10 (97.4%), and between SM5 and SM3 (97.3%), correspond to identical homozygous or heterozygous SNVs (SI Appendix, Fig. S10 and Table S11), confirming the high degree of genetic similarity that prevails among these three samples. Finally, to confirm the tendency toward homozygosity shown by all three ancient genotypes, we performed a comparison of shared SNP identity between SM10 and a group of inbred or out-crossed accessions included in the HapMap3 (SI Appendix, Table S12). SM10 shows a frequency of heterozygous SNPs equivalent to Balsas teosinte inbred lines that have undergone four rounds of self-pollination; this frequency is 2.5-times smaller than the frequency of heterozygous sites found in RIMMA0438 (PI514809), an out-crossed landrace from Peru included in HapMap3. Because the HapMap3 panel tends to underestimate heterozygosity because it is designed for comparison of inbred genotypes, we compared SM10 to a CCH individual resulting from open-pollinated field conditions. The frequency of heterozygous polymorphic sites obtained by comparison with B73 is 6.1-times lower in SM10 than in a CCH landrace individual originating from an open-pollinated population of 4,500 plants.
Our overall results imply that the earliest maize of San Marcos cave was partially domesticated and belonged to a reduced population of individuals that could have originated by self-pollination, although mating within close relatives originating from a small isolated population could also result in similar genetic patterns. The high level of genetic similarity shared by SM3 and SM10 confirms that the corresponding individuals were close to contemporaneous, perhaps within the error range of 30 14C y. These results are in agreement with the homogenous morphology reported for the 27 earliest specimens of maize cobs found in San Marcos cave and corresponding to the early cultural horizon of the Tehuacán Valley (Coxcatlán phase). All previous studies described these specimens as being remarkably uniform in size (12, 17), but small and fragile compared with the cobs of the subsequent cultural phase (Abejas phase). Our results open a possibility for testing models in which early maize cultivation in the Tehuacán Valley evolved from an early phase dominated by small populations grown in isolation, to a subsequent phase in which these populations were intercrossed. Over the coming years, the comprehensive and systematic analysis of a larger set of paleogenomic datasets from samples of subsequent age will provide broader and more assertive insight into the evolutionary mechanisms by which teosinte was gradually transformed into maize.
Materials and Methods
Detailed descriptions of samples and methods are provided in SI Appendix.
Archaeological Excavation, Sampling, and Dating.
Sampling was performed following all necessary procedures to avoid human-related or cross-sample contamination; 10–20 mg of each specimen was dated by AMS using the service provided by Beta Analytic.
Sequencing of Ancient Samples.
Three SOLiD (for SM3 and SM10) and four indexed Illumina (SM3, SM5, and SM10) DNA libraries were built for subsequent shotgun sequencing. SOLiD libraries were sequenced at the Genomic Core Facility of Pennsylvania State University, and Illumina libraries were sequenced at Unidad de Genómica Avanzada, Laboratorio Nacional de Genómica para la Biodiversidad, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional or the Core Services at the University of California, Davis.
Read Processing, Mapping, and Genotyping.
Index sequences of 16 nucleotides were used to tag libraries described above. All libraries were filtered to remove adaptors and low-quality reads using Cutadapt (38) and keep reads longer than 50 bp with a quality above 10 Phred score. Filtered reads were mapped using the Burrows–Wheeler analysis (BWA) MEM algorithm with default conditions (39). Z. mays B73 RefGen_v3 (28) was used as the reference sequence after masking repetitive genomic regions with RepeatMasker (40).
Metagenomic Analysis and Postmortem Damage.
Cytosine deamination rates and fragmentation patterns were estimated using mapDamage2.1 (41). All sites behaving as molecular damage (CG→TA) were excluded (23, 24). A metagenomic filter was applied to discard reads that aligned to sequences in the GenBank National Center for Biotechnology Information database of all bacterial and fungal genomes using default mapping-quality parameters of BWA (39).
Evolutionary Analysis and SNP Genotype Comparisons.
Patterns of divergence were analyzed by generating ML trees using Treemix (42) and the intersection of SNPs passing quality filters for the ancient specimens and 44 selected individuals of the publically available database HapMap3 without imputation (43). For each tree, no fewer than 10,000 bootstrap pseudoreplicas were generated with a parallelized version of a public script (https://github.com/mgharvey/misc_python/blob/master/bin/TreeMix/treemix_tree_with_bootstraps.py), which uses the sumtree function in DendroPy (44) to obtain a consensus ML bootstrapped tree.
Nucleotide Variability at Domestication Loci.
The genomic coordinates of selected loci previously reported as affected by domestication were obtained from B73 RefGen_V3 from MaizeGDB (21, 22). All SNPs represented in HapMap3 (43) from more than 1,180 extant maize and 15 Balsas teosinte accessions were identified and compared with quality-mapped sequences obtained for SM10.
Estimation of Identity by Descent.
Identity by descent was calculated using plink V1.9 (45) using either all SNV common ancient samples (in pairwise comparisons) or all heterozygous SNPs shared between ancient maize samples and B73.
Supplementary Material
Acknowledgments
We thank Luis Delaye, Andrés Moreno, Angélica Cibrián, and two reviewers for their constructive suggestions; Qi Sun and David Vallejo-Díaz for kindly sharing raw sequence data of maize landraces; and Hilda Ramos-Aboites, Rigel Salinas-Gamboa, and Christian Martinez-Guerrero for providing technical support. M.V-E. and I.R.-A. are recipients of a graduate scholarship from Consejo Nacional de Ciencia y Tecnología (CONACyT). This research was supported by CONACyT Grant CB256826 and the Instituto Nacional de Antropología e Historia, through the Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional–Instituto Nacional de Antropología e Historia collaboration.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Sequence data generated for this study have been deposited in the European Nucleotide Archive (accession no. PRJEB16754).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1609701113/-/DCSupplemental.
References
- 1.Weatherwax P. The phylogeny of Zea mays. Am Midl Nat. 1935;1(16):1–71. [Google Scholar]
- 2.Sanchez J, et al. Distribución y Caracterizacion del Teocintle. Instituto Nacional de Investigaciones Forestales, Agricolas y Pecuarias; Guadalajara, Mexico: 1998. [Google Scholar]
- 3.Caicedo AL, et al. Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 2007;3(9):1745–1756. doi: 10.1371/journal.pgen.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lam HM, et al. Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet. 2010;42(12):1053–1059. doi: 10.1038/ng.715. [DOI] [PubMed] [Google Scholar]
- 5.Matsuoka Y, et al. A single domestication for maize shown by multilocus microsatellite genotyping. Proc Natl Acad Sci USA. 2002;99(9):6080–6084. doi: 10.1073/pnas.052125199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doebley J. The genetics of maize evolution. Annu Rev Genet. 2004;38(38):37–59. doi: 10.1146/annurev.genet.38.072902.092425. [DOI] [PubMed] [Google Scholar]
- 7.van Heerwaarden J, et al. Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc Natl Acad Sci USA. 2011;108(3):1088–1092. doi: 10.1073/pnas.1013011108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mercer K, Martínez-Vásquez Á, Perales HR. Asymmetrical local adaptation of maize landraces along an altitudinal gradient. Evol Appl. 2008;1(3):489–500. doi: 10.1111/j.1752-4571.2008.00038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corral J, et al. Climatic adaptation and ecological descriptors of 42 Mexican maize races. Crop Sci. 2008;4(48):1502–1512. [Google Scholar]
- 10.Pressoir G, Berthaud J. Population structure and strong divergent selection shape phenotypic diversification in maize landraces. Heredity (Edinb) 2004;92(2):95–101. doi: 10.1038/sj.hdy.6800388. [DOI] [PubMed] [Google Scholar]
- 11.Mangelsdorf PC, Macneish RS, Galinat WC. Domestication of corn. Science. 1964;143(3606):538–545. doi: 10.1126/science.143.3606.538. [DOI] [PubMed] [Google Scholar]
- 12.MacNeish R, Cook AG. 1972. Excavations in the San Marcos locality in the travertine slopes. The Prehistory of the Tehuacan Valley. Volume 5: Excavations and Reconnaissance, MacNeish R, et al. (Univ of Texas Press, Austin) pp 137–160.
- 13.Long A, Benz B, Donahue D, Jull A, Toolin L. First direct AMS dates on early maize from Tehuacán. Radiocarbon. 1989;1(210):1035–1040. [Google Scholar]
- 14.Piperno DR, Ranere AJ, Holst I, Iriarte J, Dickau R. Starch grain and phytolith evidence for early ninth millennium B.P. maize from the Central Balsas River Valley, Mexico. Proc Natl Acad Sci USA. 2009;106(13):5019–5024. doi: 10.1073/pnas.0812525106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piperno DR, Flannery KV. The earliest archaeological maize (Zea mays L.) from highland Mexico: New accelerator mass spectrometry dates and their implications. Proc Natl Acad Sci USA. 2001;98(4):2101–2103. doi: 10.1073/pnas.98.4.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benz BF. Archaeological evidence of teosinte domestication from Guilá Naquitz, Oaxaca. Proc Natl Acad Sci USA. 2001;98(4):2104–2106. doi: 10.1073/pnas.98.4.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benz B, Iltis H. Studies in archeological maize I: The “wild” maize from San Marcos cave reexamined. Am Antiq. 1990;55(3):500–511. [Google Scholar]
- 18.Jaenicke-Després V, et al. Early allelic selection in maize as revealed by ancient DNA. Science. 2003;302(5648):1206–1208. doi: 10.1126/science.1089056. [DOI] [PubMed] [Google Scholar]
- 19.da Fonseca RR, et al. The origin and evolution of maize in the Southwestern United States. Nat Plants. 2015;1(14003):14003. doi: 10.1038/nplants.2014.3. [DOI] [PubMed] [Google Scholar]
- 20.MacNeish R. 1967. A summary of the subsistence. The Prehistory of the Tehuacan Valley. Volume 1: Environment and Subsistence, eds Byers D (Univ of Texas Press, Austin), pp. 290–309.
- 21.Schnable PS, et al. The B73 maize genome: Complexity, diversity, and dynamics. Science. 2009;326(5956):1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 22.Andorf CM, et al. MaizeGDB update: New tools, data and interface for the maize model organism database. Nucleic Acids Res. 2016;44(D1):D1195–D1201. doi: 10.1093/nar/gkv1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofreiter M, Jaenicke V, Serre D, von Haeseler A, Pääbo S. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 2001;29(23):4793–4799. doi: 10.1093/nar/29.23.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilbert MT, et al. Distribution patterns of postmortem damage in human mitochondrial DNA. Am J Hum Genet. 2003;72(1):32–47. doi: 10.1086/345378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schubert M, et al. Characterization of ancient and modern genomes by SNP detection and phylogenomic and metagenomic analysis using PALEOMIX. Nat Protoc. 2014;9(5):1056–1082. doi: 10.1038/nprot.2014.063. [DOI] [PubMed] [Google Scholar]
- 26.Schubert M, et al. Prehistoric genomes reveal the genetic foundation and cost of horse domestication. Proc Natl Acad Sci USA. 2014;111(52):E5661–E5669. doi: 10.1073/pnas.1416991111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seguin-Orlando A, et al. Paleogenomics. Genomic structure in Europeans dating back at least 36,200 years. Science. 2014;346(6213):1113–1118. doi: 10.1126/science.aaa0114. [DOI] [PubMed] [Google Scholar]
- 28.Li H, et al. 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hufford MB, et al. The genomic signature of crop-wild introgression in maize. PLoS Genet. 2013;9(5):e1003477. doi: 10.1371/journal.pgen.1003477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vielle-Calzada J-Ph, et al. The Palomero genome suggests metal effects on domestication. Science. 2009;326(5956):1078. doi: 10.1126/science.1178437. [DOI] [PubMed] [Google Scholar]
- 31.Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386(6624):485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 32.Studer A, Zhao Q, Ross-Ibarra J, Doebley J. Identification of a functional transposon insertion in the maize domestication gene tb1. Nat Genet. 2011;43(11):1160–1163. doi: 10.1038/ng.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bae J, Giroux M, Hannah L. Cloning and characterization of the brittle-2 gene of maize. Maydica. 1990;35(4):317–322. [Google Scholar]
- 34.Comparot-Moss S, Denyer K. The evolution of the starch biosynthetic pathway in cereals and other grasses. J Exp Bot. 2009;60(9):2481–2492. doi: 10.1093/jxb/erp141. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, et al. The origin of the naked grains of maize. Nature. 2005;436(7051):714–719. doi: 10.1038/nature03863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Studer AJ, Zhao Q, Meeley R, Doebley JF. Evidence that the origin of naked kernels during maize domestication was caused by a single amino acid substitution in tga1. Genetics. 2015;200(3):965–974. doi: 10.1534/genetics.115.175752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hufford MB, et al. Comparative population genomics of maize domestication and improvement. Nat Genet. 2012;44(7):808–811. doi: 10.1038/ng.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing. EMBnet. 2011;1(17):10–12. [Google Scholar]
- 39.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smit A, Hubley R, Green P. 2010 RepeatMasker Open-3.0. Available at www.repeatmasker.org. Accessed November 10, 2014.
- 41.Jónsson H, Ginolhac A, Schubert M, Johnson PL, Orlando L. mapDamage2.0: Fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics. 2013;29(13):1682–1684. doi: 10.1093/bioinformatics/btt193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8(11):e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bukowski R, et al. Contruction of the third generation Zea mays haplotype map. bioRxiv. 2015 doi: 10.1101/026963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sukumaran J, Holder MT. DendroPy: A Python library for phylogenetic computing. Bioinformatics. 2010;26(12):1569–1571. doi: 10.1093/bioinformatics/btq228. [DOI] [PubMed] [Google Scholar]
- 45.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.